Abstract
A single species, Candida albicans, causes half of all invasive fungal infections in humans. Unlike other fungal pathogens, this organism switches between growth as budding yeast and as pseudohyphal and hyphal filaments in host organs and in vitro. Both cell types play a role in invasive disease: while hyphal and pseudohyphal filaments penetrate host cells and tissues, yeast cells are likely to facilitate dissemination through the bloodstream and establishment of distant foci of infection. Many regulators of the yeast-to-hypha switch have emerged from intensive investigations of this morphogenetic step, but the hypha-to-yeast switch remains poorly understood. Using a forward genetic approach, a novel putative regulator involved in the hypha-to-yeast switch was identified, the C. albicans pescadillo homolog, PES1. In eukaryotes from yeast to human, pescadillo homologs are involved in cell cycle control and ribosome biogenesis, and are essential. We find a pescadillo homolog to act in fungal morphogenesis, specifically in lateral yeast growth on filamentous cells. We also find essentiality of PES1 in C. albicans to be dependent on cell type, because hyphal cells, but not yeast cells, tolerate its loss. PES1 is therefore critical for completion of the C. albicans life cycle, in which the fungus switches between filamentous and yeast growth. Consistent with these in vitro findings, PES1 is required for C. albicans virulence in an in vivo insect model of infection.
Keywords: filament, virulence, essential gene, cell cycle
In humans, Candida albicans causes more invasive disease than any other fungal species. Mortality from C. albicans bloodstream infections is ≈49% (1). In addition to its role as a pathogen, C. albicans colonizes the mucous membranes of 30–60% of humans (2). How has C. albicans evolved as the most successful human fungal commensal and pathogen? This fungus's dramatic and frequent switch between growth as ovoid yeast and as filamentous pseudohyphae and hyphae has long been implicated in its success as a pathogen (3). Work from many laboratories has elucidated regulatory mechanisms for the switch from yeast to filamentous growth, and the role of hyphal growth in virulence (3). Little is known about the reverse switch, from filamentous to yeast growth, although mutants defective in the hypha-to-yeast switch show reduced virulence in each case in which this trait was examined (4–10).
The switch from yeast to hyphal growth is induced by specific conditions, such as low cell density, alkaline pH, high temperature, a poor carbon source, and serum. In yeast-inducing conditions, yeast do not produce hyphal cells. In contrast, hyphal and pseudohyphal filaments constitutively produce yeast cells on their subapical segments, while apical segments continue to produce filamentous cells. We call these cells lateral yeast (11). Lateral yeast growth presumably occurs in vivo and in vitro, because filamentous forms are practically never seen without yeast forms in deep organs of individuals with invasive candidiasis.
We propose that the filament-to-yeast switch is a critical part of the life cycle of C. albicans, because yeast cells fulfill specific roles during colonization and infection. Yeast possess suitable dimensions and physical properties to provide the fungus facile access to the host's bloodstream. While cultured endothelial cells from human umbilical cord veins phagocytose hyphae (12), other types of endothelial cells phagocytose yeast. Endocytosis of yeast by endothelial cells was observed in cells of the human blood brain barrier (13), endothelium of porcine whole blood vessel strips (14), and bovine aortic endothelial cells (15). In addition to their function in bloodborne dispersal during invasive disease, C. albicans yeast cells have been found to play distinct roles from hyphal cells in the interaction with the host immune system. Exposure of human monocytes to C. albicans yeast blocks their development into dendritic cells, a critical class of antigen-presenting cells (16). The ability to manipulate specific features of the host immune system by its different cell types is likely to contribute to the success of C. albicans in maintaining its commensal status and in invasive disease.
So far, two genes are known to affect lateral yeast growth: mutants in PDE2 have decreased, and those in CAP1 have increased lateral yeast growth (9, 17). Given the dearth of studies of the hypha-to-yeast switch, our goal was to identify novel regulators of this switch. This search identified a hyperfilamentous mutant with a transposon insertion in the promoter of the C. albicans pescadillo homolog, PES1.
Pescadillo was originally identified in a screen for genes required for development of zebrafish embryos (18). In organisms from yeast to human, the highly conserved homologs of this gene participate in cell cycle control, DNA replication, and ribosome biogenesis (19, 20). In all organisms studied to date, pescadillo homologs are essential (19, 20). However, the biochemical activity of pescadillo homologs in proliferation, cell cycle control, and ribosome biogenesis is unknown.
Here, we report that the C. albicans pescadillo homolog is required for normal growth of cells induced as yeast, and for normal production of lateral yeast on filaments. While a heterozygote retaining an intact allele of the C. albicans pescadillo homolog is viable (21), a previous large-scale study found this gene to be essential in C. albicans (22). In contrast, we find that C. albicans cells tolerate loss of the pescadillo homolog under hyphae-inducing, but not under yeast-inducing conditions. Delineating the pathways in which PES1 functions in the hypha-to-yeast switch will begin to elucidate this step of the C. albicans life cycle.
Results
Lateral Yeast Are Produced in Conditions That Induce Hyphal Tip Growth and Hyphal Branching.
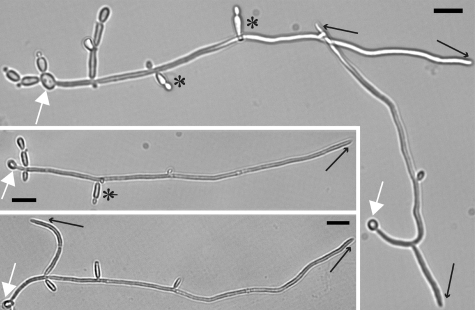
In all media tested, lateral yeast were produced from subapical cells of both pseudohyphal and hyphal filaments, whose apical cells were producing new filamentous cells. The same cultures contained filaments producing hyphal branches (Fig. 1 and data not shown). Because in liquid cultures, microenvironments of apical and subapical filamentous cells are expected to be similar, we conclude that lateral yeast growth occurs in hyphae-inducing conditions.
Fig. 1.
Lateral yeast are produced under conditions that induce hyphal tip growth and hyphal branching. Yeast cells of SC5314 (PES1/PES1) were inoculated into Spider medium, permitted to settle on a fibronectin-coated coverslip, and incubated at 37 °C for 6.5 h. Cells of the same culture on the same coverslip were photographed. White arrow, original yeast from which hyphal outgrowth occurred. Black arrow, hyphal branch or tip. Black star, lateral yeast. (Scale bar, 10 μm.)
A Hyperfilamentous Transposon Mutant in the C. albicans pescadillo Homolog, PES1.
We sought to identify novel regulators of lateral yeast growth. A previous forward genetic approach to filamentous growth successfully used haploinsufficiency phenotypes of C. albicans heterozygous mutants to identify regulators of this process (23). From a new heterozygous mariner transposon mutant collection, we isolated a hyperfilamentous mutant with a transposon insertion 47 nt upstream of the putative translational start site of ORF 19.4093. This ORF encodes a homolog of a protein conserved from yeast to human, originally called pescadillo when it was cloned in zebrafish (18), and YPH1 (yeast pescadillo homolog) or NOP7 in Saccharomyces cerevisiae (24, 25). We named this gene CaPES1 and abbreviate it as PES1 here. We created de novo mutations in PES1 in the prototrophic wild type, SC5314 (26), by deleting PES1 on one chromosome from the putative start to the putative stop codons, and replacing the second allele with a conditional allele as described below. The strains carrying targeted mutations in PES1 did not recapitulate the phenotype of the transposon mutant, which grew exclusively as pseudohyphal and hyphal filaments under all conditions tested. Instead, they showed specific defects in yeast growth when PES1 was depleted, as described below.
PES1 Is an Ortholog of the S. cerevisiae pescadillo Homolog YPH1.
The Pes1 predicted protein sequence is highly homologous to a large number of eukaryotic pescadillo orthologs, including S. cerevisiae YPH1. To examine whether the C. albicans pescadillo homolog can functionally substitute for loss of the essential gene YPH1, its ability to rescue growth of S. cerevisiae yph1 mutants was tested in two experiments. First, in a S. cerevisae yph1 deletion mutant in the Σ1278b genetic background, PES1 was able to substitute for YPH1 when expressed in either low- or high-copy number. Supporting information (SI) Fig. S1, published on the PNAS Web site, shows complementation of yph1Δ by PES1 in low-copy number. Second, expression of PES1 from a high-copy plasmid permitted growth of S. cerevisiae yph1Δ temperature-sensitive (ts) mutants in the W303 genetic background (19) at the restrictive temperature (not shown). C. albicans PES1 can complement loss of S. cerevisiae YPH1 in two genetic backgrounds, establishing that it is an ortholog of YPH1.
PES1 Is Essential in C. albicans in Yeast-Inducing Conditions.
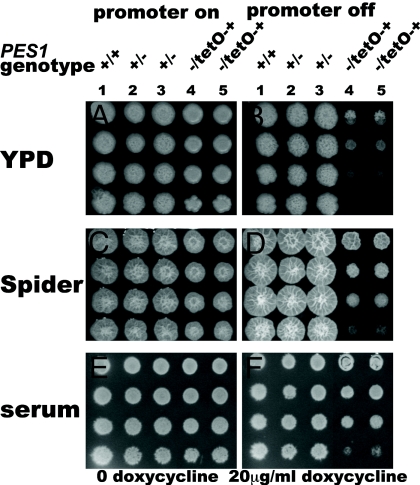
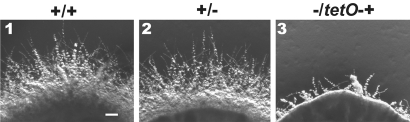
The effect of loss of Pes1 function was examined by constructing three conditional alleles of PES1. Given the essentiality of pescadillo homologs in other organisms, we did not attempt to construct a homozygous PES1 deletion strain. Two different repressible promoters were used to control expression of PES1 in heterozygotes that contain a deletion of one allele and a replacement of the native PES1 promoter of the remaining allele with the repressible MAL2 or tetO promoters (pMAL2 and tetO) (27, 28). Transcription from pMAL2 is repressed by glucose in the growth medium, partially repressed by mannitol, and induced by maltose. In our system, transcription from tetO is repressed by doxycycline or tetracycline and induced in the absence of these drugs. When PES1 transcription was repressed from either promoter on rich yeast-inducing medium, YPD, there was minimal growth of pes1Δ/pMAL2-PES1 or pes1Δ/tetO-PES1 strains (Fig. 2 and Fig. S2).
Fig. 2.
Hyphae-inducing conditions permit growth when expression of PES1 is repressed from tetO. Strains were grown overnight in YPD without doxycycline at 30 °C and diluted to an initial density of OD600 0.2. Fivefold dilutions were spotted onto YPD (A), YPD with 20 μg/ml doxycycline (B), Spider (C), Spider with 20 μg/ml doxycycline (D), 20% calf serum 2% glucose (E), and 20% calf serum 2% glucose with 20 μg/ml doxycycline (F). Plates were incubated at 37 °C for 4 days. (1) JKC915 (HIS1/his1::FRT-tetR PES1/PES1). (2) JKC956 (HIS1/his1::FRT-tetR PES1/pes1::FRT). (3) JKC962 (HIS1/his1::FRT-tetR PES1/pes1::FRT). (4) JKC1137 (HIS1/his1::FRT-tetR pes1::FRT/FRT-tetO-PES1). (5) JKC1143 (HIS1/his1::FRT-tetR pes1::FRT/FRT-tetO-PES1).
To test the effect of an abrupt perturbation of Pes1 function, we constructed heterozygotes in which the only allele of PES1 encodes a ts protein Pes1W416R (Fig. S3). A homologous amino acid substitution confers temperature sensitivity on S. cerevisiae mutants in YPH1 (25). Mutants with the genotype pes1Δ/PES1W416R had a growth defect on YPD at 24 °C and did not grow at 37 °C (Fig. S3). On rich yeast-inducing medium, functional Pes1 is required for growth.
Loss of Pes1 Is Not Lethal in Hyphae.
The effect of loss of Pes1 function on survival and growth of yeast or hyphal cells was examined in mutants containing three different conditional PES1 alleles. On yeast-inducing medium ts Pes1W416R mutants were not viable after 3 days at the restrictive temperature, 37 °C. In contrast, on hyphae-inducing media, these mutants were able to survive for 3 days at the restrictive temperature and then resume growth at the semipermissive temperature, 24 °C (Fig. S3). Like the Pes1W416R ts mutants, mutants expressing the repressible promoter alleles of PES1 showed a differential requirement for PES1 in yeast- vs. hyphae-inducing conditions. Cells in which PES1 expression was repressed from either pMAL2 or tetO grew under hyphae-inducing conditions (Fig. 2 and Fig. S2), indicating that in hyphal cells, loss of PES1 is not lethal.
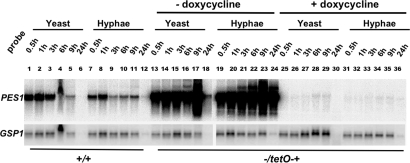
To confirm that PES1 message is depleted during promoter repression of pes1Δ/tetO-PES1 strains in both yeast- and hyphae-inducing conditions, levels of PES1 message were examined by Northern blot. Cells were grown under yeast- and hyphae-inducing conditions, in medium with and without doxycycline to repress transcription from tetO. pes1Δ/tetO-PES1 cells grown in medium without doxycycline strongly overexpressed PES1 compared to wild-type cells, irrespective of morphogenetic conditions. Conversely, PES1 expression was strongly repressed by doxycycline in both conditions, although a minimal amount of PES1 message was still detectable (Fig. 3). PES1 expression in these strains is therefore controlled effectively from the tetO promoter, independently of morphogenesis-inducing conditions.
Fig. 3.
PES1 is expressed in both yeast and hyphal cells, and control of PES1 expression from the tetO promoter is independent of yeast- or hyphae-inducing conditions. Strains were grown overnight in YPD at 30 °C, diluted to an initial density of OD600 0.2, and incubated in four conditions: YPD at 30 °C (samples 1-6 and 13-18), YPD with 10% calf serum at 37 °C (samples 7-12 and 19-24), YPD with 100 μg/ml doxycycline at 30 °C (samples 25-30), and YPD with 10% calf serum and 100 μg/ml doxycycline at 37 °C (samples 31-36). Cells were harvested at the indicated time points, and RNA preparations were blotted and probed with PES1. GSP1 was used as a loading control probe. (1-12) SC5314 (PES1/PES1). (13-36) JKC1137 (HIS1/his1::FRT-tetR pes1::FRT/FRT-tetO-PES1).
PES1 Is Expressed in Yeast and Hyphal Cells.
We examined whether PES1 is differentially expressed in yeast- vs. hyphae-inducing conditions. We used conditions previously described in large-scale microarrray analyses of the yeast-to-hypha switch (29). The wild type was found to express PES1 at comparable levels in both yeast- and hyphae-inducing conditions (Fig. 3).
Loss of PES1 Perturbs Lateral Yeast Growth.
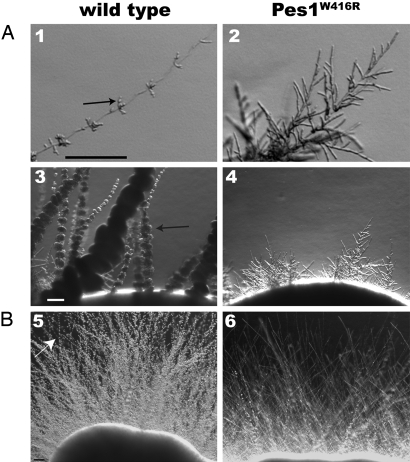
The effect of loss of Pes1 function on morphogenesis was examined. pes1Δ/pMAL2-PES1 strains growing on media containing mannitol, which partially represses PES1 expression, had considerably reduced lateral yeast growth (Fig. S4). Pes1W416R ts mutants were also defective in lateral yeast growth at the semipermissive temperature (Fig. 4). Loss of Pes1 function therefore results in defective lateral yeast growth.
Fig. 4.
Lateral yeast growth is defective during Pes1 inactivation. Wild-type and pes1Δ/PES1W416R strains were grown for 9 days at 24 °C on YP 2% galactose (A), and on RPMI1640, buffered to pH 7.5 with 165 mM Mops (B). (1, 3, and 5) SC5314 (PES1/PES1). (2 and 4) JKC1155 (pes1::FRT/FRT-PES1W416R). (6) JKC1160 (pes1::FRT/FRT-PES1W416R). (1 and 2) Tips of young filaments. (3–6) Filaments emanating from colony rim. Arrows, clusters of lateral yeast. (Scale bar, 100 μm.)
Loss of PES1 Does Not Lead to Constitutive Polarized Growth.
Formation of the ovoid shape of a budding yeast cell requires that the polarized growth direction of an emerging bud switches to isotropic expansion around the entire cell circumference as the growing bud prepares for mitosis (30). We tested whether defective lateral yeast growth of PES1 mutants is the result of an inability to appropriately switch to isotropic growth, i.e., whether these mutants remain trapped in a polarized growth mode. Terminal phenotypes of Pes1W416R ts mutants were examined in hyphae- and yeast-inducing conditions. Mutant yeast cells inoculated into hyphae-inducing conditions at the restrictive temperature produced germ tubes indistinguishable from those of wild type, which arrested as short hyphae (Fig. S5). When Pes1W416R ts yeast cells were inoculated into yeast-inducing conditions at the restrictive temperature, they arrested in the yeast form (Fig. S5).
The terminal phenotype of pes1Δ/pMAL2-PES1 yeast during promoter repression was followed for 3 days. Mother cells divided several times with each daughter receiving a nucleus, eventually forming clusters of attached large yeast. After 3 days of PES1 depletion, the nuclei appeared fragmented (Fig. S5). Thus, both abrupt inactivation of Pes1 and depletion of PES1 from a repressible promoter did not interfere with appropriate switching of yeast cells from polarized to isotropic growth. Mutants in PES1 are not trapped in a polarized growth state.
Quorum Sensing Is Not Perturbed by Pes1 Inactivation.
To test whether PES1 mutants' defect in lateral yeast growth is the result of an inability to respond to quorum-sensing molecules that stimulate a switch from hyphal to yeast growth and could be secreted by subapical filamentous cells, we examined the response of Pes1W416R ts mutants to two compounds that activate the C. albicans quorum-sensing response: farnesol and dodecanol (31). At the restrictive temperature for Pes1W416R ts cells, 37 °C, wild-type yeast switched to filamentous growth at a significant rate when diluted into rich medium without quorum-signaling compounds (29). Filamentous growth was suppressed by addition of dodecanol or farnesol to the medium, with no observable difference between Pes1W416R ts cells and the wild type (Fig. S5). Defective lateral yeast growth in PES1 mutants is not the result of an inability to respond to quorum-signaling molecules.
PES1 Overexpression Results in Decreased Filamentous Growth and Increased Lateral Yeast Growth.
To investigate whether PES1 overexpression has the opposite effect from PES1 depletion, pes1Δ/tetO-PES1 mutants were grown on medium without doxycycline. In these conditions, PES1 is strongly overexpressed (Fig. 3). On all solid media tested without doxycycline, these strains showed decreased hyphal growth, and the hyphae that were formed showed overabundant lateral yeast (Fig. 5). In liquid rich medium, strains overexpressing PES1 did not have a significantly altered growth rate compared to the wild type (Fig. S2). Overexpression of PES1 increases, while its depletion decreases lateral yeast growth.
Fig. 5.
PES1 overexpression leads to decreased hyphal growth and increased lateral yeast growth. Strains were streaked on Spider medium without doxycycline and incubated at 37 °C for 4 days. (1) JKC915 (HIS1/his1::FRT-tetR PES1/PES1). (2) JKC962 (HIS1/his1::FRT-tetR PES1/pes1::FRT). (3) JKC1143 (HIS1/his1::FRT-tetR pes1::FRT/FRT-tetO-PES1). (Scale bar, 100 μm.)
PES1 Is Required for Virulence in the Galleria mellonella Larva Model.
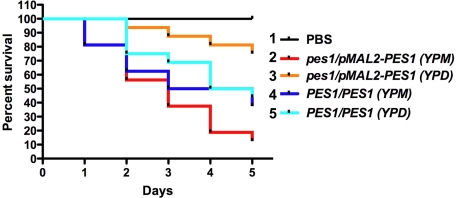
An insect model was used to examine the effect of Pes1 perturbation on C. albicans virulence (32). When injected at inocula between 5 × 105 and 106 cells, wild-type C. albicans SC5314 rapidly kill G. mellonella larvae held at 37 °C. pes1Δ/pMAL2-PES1 cells, preincubated in glucose to deplete PES1, were significantly reduced in virulence in this system (Fig. 6). Consistent results were obtained with the pes1Δ/tetO-PES1 and pes1Δ/PES1W416R mutants (data not shown). PES1 is therefore required for C. albicans virulence in this insect model.
Fig. 6.
PES1 depletion leads to loss of virulence in a Galleria mellonella model. C. albicans pes1Δ/pMAL2-PES1 mutant and wild-type strains were grown overnight at 30 °C in YPD to repress PES1 expression in the mutant, or in YP 2% maltose (YPM) to induce PES1 expression in the mutant, and resuspended in PBS to an inoculum of 106 cells. Sixteen larvae per condition were injected and kept at 37 °C. (1) Larvae injected with PBS only. (2) Larvae injected with JKC681 (pes1::FRT/FRT-pMAL2-PES1) pregrown in YPM. (3) Larvae injected with JKC681 (pes1::FRT/FRT-pMAL2-PES1) pregrown in YPD. (4) Larvae injected with SC5314 (PES1/PES1) pregrown in YPM. (5) Larvae injected with SC5314 (PES1/PES1) pregrown in YPD. The P value for difference in survival curves between larvae injected with JKC681 pregrown in YPD and with JKC681 pregrown in YPM is 0.0001 (Mantel-Cox test). The P value for difference in survival curves between larvae injected with SC5314 pregrown in YPD and with SC5314 pregrown in YPM is 0.547.
Discussion
In C. albicans, the pescadillo homolog PES1 is essential in yeast, whereas hyphal cells tolerate PES1 depletion (Fig. 2 and Fig. S2). The pes1 yeast growth defect has implications for host–pathogen interactions because C. albicans uses its yeast form during its commensal state and during dissemination and deep organ proliferation in invasive disease. Wild-type C. albicans constitutively produce lateral yeast on filaments in conditions that induce hyphal tip growth (Fig. 1), and this process is disturbed when functional Pes1 is lacking (Fig. 4 and Fig. S4). In an insect model, virulence of cells deficient in Pes1 was attenuated. A caveat here is that attenuation of pes1 mutant cells is likely to be potentiated by any growth defect. PES1-depleted cells have the least growth defect when they are exposed to a combination of strong hyphae-inducing signals (Fig. 2 and Fig. S2). Larvae were held at the hyphae-inducing temperature of 37 °C, but we do not know whether other hyphae-inducing signals prevail in the larva. Virulence attenuation during loss of PES1 is nevertheless consistent with a pathogenic role of Pes1-dependent yeast growth.
Pescadillo homologs of yeast, mouse, and human act in proliferation control and ribosome biogenesis (19, 20). It is not clear whether these distinct functions are based in distinct biochemical activities. PES1 is cotranscribed with genes involved in rRNA processing and ribosome biogenesis (33) (Candida Genome Database), suggesting that it may participate in these functions in C. albicans as well. One possibility is that growth arrest during PES1 depletion is caused by disruption of rRNA processing, leading to activation of a “nucleolar stress” cell cycle checkpoint (34), or to depletion of ribosomes and slowing of translation. If this is the case, tolerance of hyphae for PES1 depletion might indicate that hyphal cells need less ribosomes to grow than yeast cells. Alternatively, Pes1 may have separate roles in proliferation control and rRNA processing.
Cell cycle control in S. cerevisiae is linked to determination of cell shape (30). To form an ovoid yeast cell, new material is secreted toward the tip of a young bud from S phase until the G2/M transition, in a polarized or apical direction. At that point, growth is redirected to distribute new material isotropically around the entire cell circumference. Delay of mitosis, by prolonging the duration of polarized growth, leads to the elongated cells that form a pseudohyphal filament, and this effect is enhanced by mutations in cell cycle regulators required for progression through S and for the G2/M transition (30). The link between cell cycle control and morphogenesis is more complex in C. albicans because perturbation of many C. albicans cell cycle regulators, involved at all phases of the cell cycle, results in constitutive polarized growth (35). In C. albicans, the gene encoding G1 cyclin Cln3 is specifically required for growth in yeast-inducing conditions (36, 37), similarly to PES1. But unlike PES1 depletion, depletion of CLN3 in yeast-inducing conditions leads to growth arrest and then outgrowth of a hyphal or pseudohyphal filament (36, 37). In contrast, the terminal phenotype of pes1 mutants consists of growth arrest of yeast cells (Fig. S5). Consequently, inability to switch from polarized to isotropic growth cannot be the reason for the growth defect of PES1-depleted yeast cells. In S. cerevisiae, the pescadillo homolog Yph1 is required for entry into the cell cycle from G0, for progression from G1 to S, and for progression from G2 to M (25). It remains to be determined whether Pes1 is required in one or in multiple phases of the C. albicans cell cycle and how this activity differs between yeast and hyphal cells.
In S. cerevisiae, cell cycle entry and ribosome biogenesis are tightly coordinated at the transcriptional level in response to nutrient availability (38, 39). Du and Stillman found evidence that in S. cerevisiae, the pescadillo homolog Yph1 participates in nutritional signaling (25). Pes1 could participate in a signaling network that relays information about the cell's nutritional status to both the cell cycle machinery and the ribosomal biogenesis machinery simultaneously to coordinate these two processes. High levels of PES1 expression in fresh rich media, and decrease of PES1 levels over time (Fig. 3), may be consistent with this idea. The finding that overexpression of PES1 has the opposite morphogenetic effect as its depletion (Fig. 5) may support a regulatory role for Pes1. If Pes1 is linked to a nutritional signaling network, the cAMP-dependent protein kinase (PKA) pathway is a candidate for functional interactions with Pes1. The only previously described C. albicans mutants with altered lateral yeast growth, in PDE2 and CAP1, have abnormal intracellular cAMP levels (9, 17). In addition, a phosphatase linked to the PKA pathway, Yvh1, interacts physically with the pescadillo homolog in S. cerevisiae and in Plasmodium falciparum (40, 41). Signaling through the PKA pathway is required for ribosome biogenesis (38), consistent with a model whereby Pes1, in interaction with the PKA pathway, responds to availability of nutrients to promote rRNA processing and cell proliferation. Whether C. albicans hyphal cells require less PES1 because hyphal growth is less dependent on such a nutritional signal remains to be determined.
In summary, Pes1 is a putative regulator implicated both in the filament-to-yeast switch and in proliferation of yeast, two processes that are critical for the vegetative life cycle of this principal fungal pathogen of humans.
Materials and Methods
Strains and Culture Conditions.
The C. albicans and S. cerevisiae strains used in this study are described in Table S1. All C. albicans strains were generated in the SC5314 genetic background using the CaNAT1 selectable marker, as described in ref. 42. Each of the three conditional alleles of PES1 was generated from two independent heterozygotes. PES1 deletion and replacement of the PES1 promoter with pMAL2 and tetO, and introduction of the Pes1W416R ts-encoding mutation were confirmed by Southern blot. The genotype of S. cerevisiae strains was confirmed by PCR spanning the upstream and downstream homologous recombination junctions of transforming constructs. For routine growth of yeast strains, YPD (1% yeast extract/2% peptone/2% glucose) was used. Spider medium was made as in ref. 43. Solid serum medium was made with 10% or 20% calf serum (Gibco/Invitrogen cat. no.16170–078), water, and 2% agar; 2% glucose or maltose were added as indicated.
The heterozygous transposon mutant collection will be described elsewhere in detail. Briefly, a C. albicans genomic library, consisting of 4.9 × 104 independent clones (44), was mutagenized by transposon insertion in vitro. A hyperactive C9 mariner transposase was purified as a fusion with maltose binding protein, and transposition conditions were optimized according to considerations described in ref. 45. The transposon was adapted to contain the CaNAT1 marker, which confers resistance to the aminoglycoside nourseothricin on C. albicans and on Escherichia coli (42). A transposon-mutagenized genomic C. albicans plasmid library consisting of 1.26 × 105 clones was generated. In our system, each clone represents an independent insertion event because in vitro mutagenesis was followed by electroporation of products of transposition into E. coli and selection of E. coli transformants on nourseothricin (clonNAT, Werner BioAgents).
C. albicans wild-type strain SC5314 was transposon mutagenized by transformation with inserts excised from the in vitro mutagenized genomic library and selection on rich medium (YPD) containing nourseothricin. Mutagenesis occurred by integration of a fragment of genomic DNA carrying a transposon, into the C. albicans chromosome by homologous recombination, thereby integrating the transposon into the transformant's chromosome. A total of 17,000 mutants was arrayed in 96-well dishes; of these, 14,000 were viable in standard conditions. The PES1/pes1::tn transposon mutant was isolated as a hyperfilamentous clone directly from the transformation plate (YPD + nourseothricin).
Plasmid Construction.
Plasmids constructed for this study are described in SI Text on the PNAS Web site. Previously published plasmids used in this study are shown in Table S1, and primers are shown in Table S2, both of which are published as SI Text.
Microscopy of Lateral Yeast Growth in Liquid Medium.
Wild-type strain SC5314 was grown in YPD to saturation overnight at 24 °C, diluted to an OD600 of 0.005 in Spider medium, and inoculated into wells of 6 well plates containing fibronectin-coated coverslips. After 6.5 h incubation at 37 °C, cultures were fixed with formaldehyde and examined by brightfield microscopy.
Growth Assays.
For cell dilutions spotted onto solid media, cells were grown overnight in YPD at 24 °C for experiments using Pes1W416R-containing mutants, and at 30 °C for all other experiments. Cells were diluted to an OD600 of 0.2 in PBS and further diluted in fivefold steps in PBS. Cell suspensions were spotted onto appropriate media using a replicator with prongs calibrated to deliver 1.5 μl (V&P Scientific, VP407). For growth curves in liquid media, all strains were grown to saturation overnight in YPD with 0.01 μg/ml of doxycycline at 24 °C. They were washed once in the cognate growth medium (YPD, Spider, or 20% serum and 2% glucose in water) without or with 20 μg/ml of doxycycline and diluted to an OD600 of 0.05. One hundred microliters of each cell suspension was inoculated into wells of flat bottom 96-well dishes, and cultures were incubated at the cognate temperature without shaking in a Spectramax250 plate reader. OD600 readings were obtained every 30 min and imported into Excel at the end of the experiment. Means and standard deviations were calculated and graphed in Excel.
Northern Blot.
Cells of the wild-type strain SC5314, and a pes1/tetO-PES1 strain, JKC1137, were grown to saturation (OD600 >12) in YPD at 30 °C overnight. They were diluted to an initial density of OD600 0.2 into 12 ml medium per sample. The pes1/tetO-PES1 strain was incubated at 200 rpm in four conditions: YPD at 30 °C, YPD with 10% calf serum at 37 °C, YPD with 100 mg/ml doxycycline at 30 °C and YPD with 10% calf serum and 100 mg/ml doxycycline at 37 °C. The wild type was incubated in the first two conditions. Cells were harvested at six time points: 0.5, 1, 3, 6, 9, and 24 h. Total RNA was extracted using TRIzol (Invitrogen) according to the manufacturer's instructions. Total RNA was blotted to a nylon membrane (Hybond XL) and probed with the insert of pJK945, which was labeled with [32]ATP by random priming. As the loading control probe, a portion of the GSP1 ORF was amplified with primers 871 and 998. GSP1 encodes a small G protein whose expression level is not affected during morphogenesis (D. Kadosh, personal communication).
Quorum-Sensing Response Assay.
Strains were grown in YPD overnight at 24 °C, and diluted to an OD600 of 0.025 into 250 μl of each medium: YPD with 10% calf serum, YPD with 0.5% DMSO, and YPD with 200 μM dodecanol or farnesol, and incubated in 24-well dishes at 37 °C without shaking.
G. mellonella Killing Assay.
Injection of C. albicans in the G. mellonella killing assay was performed essentially as described (46). In brief, larvae in the final instar larval stage were obtained from Canadian Feeders. Inocula of C. albicans for the injections were prepared by growing 50 ml YPD cultures overnight at 30 °C. Cells were pelleted by centrifugation at 3,000 rpm for 10 min followed by three washes in PBS. Cells were resuspended in 1 ml of PBS and cell densities were determined by hemacytometer count and additionally confirmed by OD600 comparison for PES1-depleted cultures in which yeast cells formed aggregates of unseparated yeast. Final dilutions for injection of 106 cells were prepared in PBS. Sixteen larvae (330 ± 25 mg) were used per group and 5-μl injections were performed via the last left proleg. Larvae were incubated in Petri dishes at 37 °C in the dark and the number of dead larvae was scored daily. Kill curves were plotted and estimation of differences in survival by log-rank (Mantel-Cox) test was analyzed by the Kaplan-Meier method using GraphPad Prism statistical software.
Supplementary Material
Acknowledgments.
We thank Feng Zhi Shao, Gege Tan, Mrinalini Tavag, and Sabrina Hepburn for excellent technical assistance. Joong-Wook Park contributed to data shown in Fig. S2. Thanks to Angelika Amon for crucial logistical aid. The Harvard Digestive Diseases Imaging Core and Jessica Wagner are gratefully acknowledged. Thanks to Eric Rubin, Chris Sassetti, Joachim Morschhäuser, John Woolford, Todd Milne, Phil Hieter and Bill Fonzi for plasmids and strains. We are grateful to Simon Dove, Paula Watnick, Haoping Liu, Joyce Fingeroth, and Mike Lorenz for valuable comments on the manuscript. This work was funded by a Charles A. Janeway Child Health Research Center Award and by NIH R21 AI064715 to J.R.K. L.E.C. is supported by a Career Award in the Biomedical Sciences from the Burroughs Wellcome Fund, by a Canada Research Chair in Microbial Genomics and Infectious Disease, and by Canadian Institutes of Health Research Grant MOP-86452.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0809147105/DCSupplemental.
References
- 1.Wisplinghoff H, et al. Nosocomial bloodstream infections in US hospitals: analysis of 24,179 cases from a prospective nationwide surveillance study. Clin Infect Dis. 2004;39(3):309–317. doi: 10.1086/421946. [DOI] [PubMed] [Google Scholar]
- 2.Odds FC. London: Bailliere Tindall; 1988. Candida and Candidosis. [Google Scholar]
- 3.Kumamoto CA, Vinces MD. Contributions of hyphae and hypha-co-regulated genes to Candida albicans virulence. Cell Microbiol. 2005;7(11):1546–1554. doi: 10.1111/j.1462-5822.2005.00616.x. [DOI] [PubMed] [Google Scholar]
- 4.Braun BR, Kadosh D, Johnson AD. NRG1, a repressor of filamentous growth in C. albicans, is down-regulated during filament induction. EMBO J. 2001;20(17):4753–4761. doi: 10.1093/emboj/20.17.4753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Murad AM, et al. NRG1 represses yeast-hypha morphogenesis and hypha-specific gene expression in Candida albicans. EMBO J. 2001;20(17):4742–4752. doi: 10.1093/emboj/20.17.4742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kadosh D, Johnson AD. Rfg1, a protein related to the Saccharomyces cerevisiae hypoxic regulator Rox1, controls filamentous growth and virulence in Candida albicans. Mol Cell Biol. 2001;21(7):2496–2505. doi: 10.1128/MCB.21.7.2496-2505.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Laprade L, Boyartchuk VL, Dietrich WF, Winston F. Spt3 plays opposite roles in filamentous growth in Saccharomyces cerevisiae and Candida albicans and is required for C. albicans virulence. Genetics. 2002;161(2):509–519. doi: 10.1093/genetics/161.2.509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bensen ES, Filler SG, Berman J. A forkhead transcription factor is important for true hyphal as well as yeast morphogenesis in Candida albicans. Eukaryot Cell. 2002;1(5):787–798. doi: 10.1128/EC.1.5.787-798.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bahn YS, Staab J, Sundstrom P. Increased high-affinity phosphodiesterase PDE2 gene expression in germ tubes counteracts CAP1-dependent synthesis of cyclic AMP, limits hypha production and promotes virulence of Candida albicans. Mol Microbiol. 2003;50(2):391–409. doi: 10.1046/j.1365-2958.2003.03692.x. [DOI] [PubMed] [Google Scholar]
- 10.Li Y, Su C, Mao X, Cao F, Chen J. Roles of Candida albicans Sfl1 in hyphal development. Eukaryot Cell. 2007;6(11):2112–2121. doi: 10.1128/EC.00199-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Köhler JR. Mos10 (Vps60) is required for normal filament maturation in Saccharomyces cerevisiae. Mol Microbiol. 2003;49(5):1267–1285. doi: 10.1046/j.1365-2958.2003.03556.x. [DOI] [PubMed] [Google Scholar]
- 12.Phan QT, et al. Als3 is a Candida albicans invasin that binds to cadherins and induces endocytosis by host cells. PLoS Biol. 2007;5(3):e64. doi: 10.1371/journal.pbio.0050064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jong AY, Stins MF, Huang SH, Chen SH, Kim KS. Traversal of Candida albicans across human blood-brain barrier in vitro. Infect Immun. 2001;69(7):4536–4544. doi: 10.1128/IAI.69.7.4536-4544.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Klotz SA, Drutz DJ, Harrison JL, Huppert M. Adherence and penetration of vascular endothelium by Candida yeasts. Infect Immun. 1983;42(1):374–384. doi: 10.1128/iai.42.1.374-384.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zink S, Nass T, Rosen P, Ernst JF. Migration of the fungal pathogen Candida albicans across endothelial monolayers. Infect Immun. 1996;64(12):5085–5091. doi: 10.1128/iai.64.12.5085-5091.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Torosantucci A, et al. Candida albicans yeast and germ tube forms interfere differently with human monocyte differentiation into dendritic cells: a novel dimorphism-dependent mechanism to escape the host's immune response. Infect Immun. 2004;72(2):833–843. doi: 10.1128/IAI.72.2.833-843.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bahn YS, Sundstrom P. CAP1, an adenylate cyclase-associated protein gene, regulates bud-hypha transitions, filamentous growth, and cyclic AMP levels and is required for virulence of Candida albicans. J Bacteriol. 2001;183(10):3211–3223. doi: 10.1128/JB.183.10.3211-3223.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Allende ML, et al. Insertional mutagenesis in zebrafish identifies two novel genes, pescadillo and dead eye, essential for embryonic development. Genes Dev. 1996;10(24):3141–3155. doi: 10.1101/gad.10.24.3141. [DOI] [PubMed] [Google Scholar]
- 19.Kinoshita Y, et al. Pescadillo, a novel cell cycle regulatory protein abnormally expressed in malignant cells. The J Biol Chem. 2001;276(9):6656–6665. doi: 10.1074/jbc.M008536200. [DOI] [PubMed] [Google Scholar]
- 20.Lerch-Gaggl A, et al. Pescadillo is essential for nucleolar assembly, ribosome biogenesis, and mammalian cell proliferation. The J Biol Chem. 2002;277(47):45347–45355. doi: 10.1074/jbc.M208338200. [DOI] [PubMed] [Google Scholar]
- 21.Xu D, et al. Genome-wide fitness test and mechanism-of-action studies of inhibitory compounds in Candida albicans. PLoS Pathog. 2007;3(6):e92. doi: 10.1371/journal.ppat.0030092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Roemer T, et al. Large-scale essential gene identification in Candida albicans and applications to antifungal drug discovery. Mol Microbiol. 2003;50(1):167–181. doi: 10.1046/j.1365-2958.2003.03697.x. [DOI] [PubMed] [Google Scholar]
- 23.Uhl MA, Biery M, Craig N, Johnson AD. Haploinsufficiency-based large-scale forward genetic analysis of filamentous growth in the diploid human fungal pathogen C. albicans. EMBO J. 2003;22(11):2668–2678. doi: 10.1093/emboj/cdg256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Harnpicharnchai P, et al. Composition and functional characterization of yeast 66S ribosome assembly intermediates. Mol Cell. 2001;8(3):505–515. doi: 10.1016/s1097-2765(01)00344-6. [DOI] [PubMed] [Google Scholar]
- 25.Du YC, Stillman B. Yph1p, an ORC-interacting protein: potential links between cell proliferation control, DNA replication, and ribosome biogenesis. Cell. 2002;109(7):835–848. doi: 10.1016/s0092-8674(02)00773-0. [DOI] [PubMed] [Google Scholar]
- 26.Fonzi WA, Irwin MY. Isogenic strain construction and gene mapping in Candida albicans. Genetics. 1993;134(3):717–728. doi: 10.1093/genetics/134.3.717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Brown DH, Jr., Slobodkin IV, Kumamoto CA. Stable transformation and regulated expression of an inducible reporter construct in Candida albicans using restriction enzyme-mediated integration. Mol Gen Genet. 1996;251(1):75–80. doi: 10.1007/BF02174347. [DOI] [PubMed] [Google Scholar]
- 28.Gossen M, Bujard H. Tight control of gene expression in mammalian cells by tetracycline-responsive promoters. Proc Natl Acad Sci USA. 1992;89(12):5547–5551. doi: 10.1073/pnas.89.12.5547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kadosh D, Johnson AD. Induction of the Candida albicans filamentous growth program by relief of transcriptional repression: a genome-wide analysis. Mol Biol Cell. 2005;16(6):2903–2912. doi: 10.1091/mbc.E05-01-0073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rua D, Tobe BT, Kron SJ. Cell cycle control of yeast filamentous growth. Curr Opin Microbiol. 2001;4(6):720–727. doi: 10.1016/s1369-5274(01)00274-0. [DOI] [PubMed] [Google Scholar]
- 31.Hogan DA, Vik A, Kolter R. A Pseudomonas aeruginosa quorum-sensing molecule influences Candida albicans morphology. Mol Microbiol. 2004;54(5):1212–1223. doi: 10.1111/j.1365-2958.2004.04349.x. [DOI] [PubMed] [Google Scholar]
- 32.Cotter G, Doyle S, Kavanagh K. Development of an insect model for the in vivo pathogenicity testing of yeasts. FEMS Immunol Med Microbiol. 2000;27(2):163–169. doi: 10.1111/j.1574-695X.2000.tb01427.x. [DOI] [PubMed] [Google Scholar]
- 33.Ihmels J, Bergmann S, Berman J, Barkai N. Comparative gene expression analysis by differential clustering approach: application to the Candida albicans transcription program. PLoS Genet. 2005;1(3):e39. doi: 10.1371/journal.pgen.0010039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pestov DG, Strezoska Z, Lau LF. Evidence of p53-dependent cross-talk between ribosome biogenesis and the cell cycle: effects of nucleolar protein Bop1 on G(1)/S transition. Mol Cell Biol. 2001;21(13):4246–4255. doi: 10.1128/MCB.21.13.4246-4255.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Berman J. Morphogenesis and cell cycle progression in Candida albicans. Curr Opin Microbiol. 2006;9(6):595–601. doi: 10.1016/j.mib.2006.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bachewich C, Whiteway M. Cyclin Cln3p links G1 progression to hyphal and pseudohyphal development in Candida albicans. Eukaryot Cell. 2005;4(1):95–102. doi: 10.1128/EC.4.1.95-102.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chapa y Lazo B, Bates S, Sudbery P. The G1 cyclin Cln3 regulates morphogenesis in Candida albicans. Eukaryot Cell. 2005;4(1):90–94. doi: 10.1128/EC.4.1.90-94.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jorgensen P, et al. A dynamic transcriptional network communicates growth potential to ribosome synthesis and critical cell size. Genes Dev. 2004;18(20):2491–2505. doi: 10.1101/gad.1228804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bernstein KA, Bleichert F, Bean JM, Cross FR, Baserga SJ. Ribosome biogenesis is sensed at the Start cell cycle checkpoint. Mol Biol Cell. 2007;18(3):953–964. doi: 10.1091/mbc.E06-06-0512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sakumoto N, Yamashita H, Mukai Y, Kaneko Y, Harashima S. Dual-specificity protein phosphatase Yvh1p, which is required for vegetative growth and sporulation, interacts with yeast pescadillo homolog in Saccharomyces cerevisiae. Biochem Biophys Res Commun. 2001;289(2):608–615. doi: 10.1006/bbrc.2001.6021. [DOI] [PubMed] [Google Scholar]
- 41.Kumar R, et al. A zinc-binding dual-specificity YVH1 phosphatase in the malaria parasite, Plasmodium falciparum, and its interaction with the nuclear protein, pescadillo. Mol Biochem Parasitol. 2004;133(2):297–310. doi: 10.1016/j.molbiopara.2003.11.005. [DOI] [PubMed] [Google Scholar]
- 42.Shen J, Guo W, Köhler JR. CaNAT1, a heterologous dominant selectable marker for transformation of Candida albicans and other pathogenic Candida species. Infect Immun. 2005;73(2):1239–1242. doi: 10.1128/IAI.73.2.1239-1242.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Liu H, Köhler J, Fink GR. Suppression of hyphal formation in Candida albicans by mutation of a STE12 homolog. Science. 1994;266(5191):1723–1726. doi: 10.1126/science.7992058. [DOI] [PubMed] [Google Scholar]
- 44.Feng Q, Summers E, Guo B, Fink G. Ras signaling is required for serum-induced hyphal differentiation in Candida albicans. J Bacteriol. 1999;181(20):6339–6346. doi: 10.1128/jb.181.20.6339-6346.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lampe DJ, Grant TE, Robertson HM. Factors affecting transposition of the Himar1 mariner transposon in vitro. Genetics. 1998;149(1):179–187. doi: 10.1093/genetics/149.1.179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mylonakis E, et al. Galleria mellonella as a model system to study Cryptococcus neoformans pathogenesis. Infect Immun. 2005;73(7):3842–3850. doi: 10.1128/IAI.73.7.3842-3850.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.