Abstract
Rad51 protein, involved in homologous recombination, is overexpressed in a variety of tumors, and its expression is correlated with a poor prognosis. Here we propose to exploit the overexpression of Rad51 in cancer cells to design a Rad51 promoter-based anticancer therapy. On average, Rad51 mRNA and protein levels are increased in cancer cells four- and sixfold, respectively. Serendipitously, we discovered that when the Rad51 ORF is replaced with another ORF, the difference in promoter activity between normal and cancer cells increases to an average of 840-fold with a maximum difference of 12,500-fold. This dramatic difference in activity has high therapeutic potential. We demonstrate that the fusion of Rad51 promoter to diphtheria toxin A (DTA) gene kills a variety of cancer cell types, including breast cancer, fibrosarcoma, and cervical cancer cells, with minimal effect on normal breast epithelial cells and normal fibroblasts. Our results suggest that therapies based on the Rad51 promoter will be highly tumor specific and open new avenues for targeting a broad range of cancers.
Keywords: cancer, transcriptionally targeted therapy
The recombinase protein Rad51 is essential in repairing DNA double-strand breaks (DSBs) by homologous recombination (HR) (1). It facilitates the search for homology and joint heteroduplex formation with the sister chromatid (2, 3). Rad51 expression is tightly controlled in normal cells as inappropriate recombination can lead to genomic instability (4, 5). However, Rad51 is overexpressed in the majority of human tumor cells (6–8). The reasons for Rad51 overexpression in cancer cells are not entirely understood. It is not the result of gene duplication or protein stability, but is thought to occur at the level of transcriptional regulation in the promoter region (6). The tumor suppressor protein p53, which is frequently mutated in cancer, interacts with the Rad51 core promoter and Rad51 protein to inhibit both its expression and activity (9, 10); while the transcription factor STAT5 has been shown to stimulate the expression of Rad51 (11, 12). Overexpression of Rad51 leads to increases in genomic instability (3, 13) and resistance to DSB-inducing cancer therapies (14, 15). Elevated levels of Rad51 correlate with increased invasiveness of breast cancer (16) and can be used as an independent prognostic marker for mean survival time in patients with non-small cell lung cancer (17). The inhibition of Rad51 has been explored as a way to sensitize cancer cells to radiotherapy (18–20).
The goal of cancer treatment is to selectively eliminate malignant cells while leaving normal tissue intact. Transcriptionally targeted anticancer therapy employs an elegant approach to selectively destroy cancer cells by placing a reporter and/or cytotoxic gene/oncolytic virus under the transcriptional control of the cancer or tissue-specific promoters (reviewed in refs. 21–24). Examples of promoters that have been used in previous studies include the telomerase RNA subunit hTER and catalytic subunit hTERT (25–30), tyrosinase (31), prostate antigen (32), survivin (33), and midkine genes (34). Although the results from these studies are promising, most notably those using hTERT, limitations of these promoters are insufficient expression of therapeutic genes, leaky expression resulting in toxicity to normal cells, or narrow specificity to a particular tumor type (23, 35). It is beneficial to investigate other cancer-specific promoters for their use in such therapy with hopes of finding one with high efficacy and selectivity in a broad range of cancers.
Here we present evidence that the Rad51 promoter can be a powerful tool in transcriptionally targeted gene therapy. Rad51 protein is overexpressed by an average of 5-fold in cancer cells. Unexpectedly, when the Rad51 ORF is replaced with a reporter ORF, the difference in promoter activity between normal and cancer cells reaches up to 12,500-fold. This can be explained by negative posttranscriptional regulation of Rad51 expression, which is removed when the Rad51 ORF is replaced. The dramatic difference in Rad51 promoter activity between normal and cancer cells allows for the targeting of cancer cells with high efficacy and selectivity. By transfecting cancer cells with the bacteria diphtheria toxin A (DTA) gene, an inhibitor of protein synthesis (36), we were able to decrease cell number and inhibit de novo protein synthesis up to 100,000-fold in a variety of cancer cells while having minimal effect on noncancerous cells. These results open new avenues for the development of transcriptionally targeted therapies using Rad51.
Results
Rad51 Protein and mRNA Are Elevated in Cancer Cells.
We hypothesized that because Rad51 is overexpressed in the majority of cancer cells, the Rad51 promoter can be exploited for transcriptionally targeted cancer therapy. We first examined the endogenous levels of Rad51 protein and transcripts in a panel of human cancer and normal cell lines including: four breast cancer cell lines HCC-1954, MDA-MB-468, T47-D, and MCF7; cervical cancer cell line HeLa; fibrosarcoma line HT1080; transformed kidney cells GP2–293; three lines of normal fibroblasts HCA2, IMR-90, WI-38; and three normal human mammary epithelial cell lines HMEC1, HMEC2, and HMEC4.
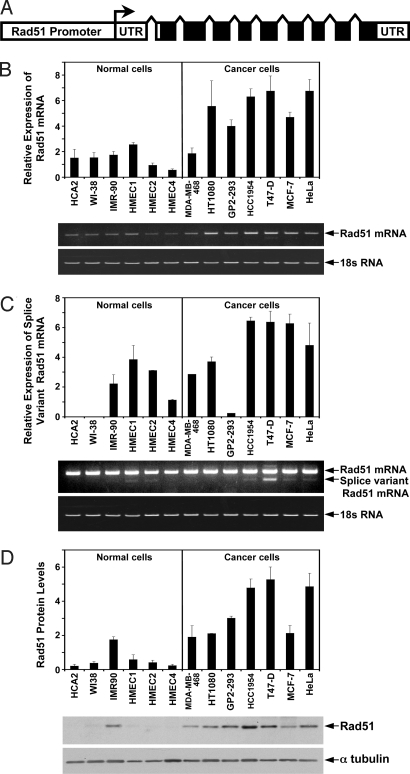
Rad51 transcript was examined using quantitative RT-PCR (Fig. 1 A and B) with primers to exons 1–3. The levels of Rad51 transcript were greater in cancer cells than in the normal cells (P = 0.001, t test). On average (by pooling the data for all of the noncancerous cells versus the cancerous cells) cancer cells had 3.5-fold increase in the transcript levels. The cell line with the strongest Rad51 expression was T47-D, which had a 12.2-fold increase when compared to HMEC4, which had the least amount.
Fig. 1.
Rad51 mRNA and protein levels are increased in cancer cells. (A) Diagram of the human Rad51 gene. Transcription start site is indicated by arrow. Coding exons are represented by solid black boxes. Upstream to the start of transcription is the Rad51 regulatory region. (B) Analysis of Rad51 transcript levels in normal and cancerous cells by quantitative RT-PCR. The top bands are RT-PCR products of Rad51 mRNA and the bottom bands are RT-PCR products of 18S subunit ribosomal RNA used as a reference. The histogram represents the relative intensity of the Rad51 band normalized to the 18S band. (C) Analysis of alternatively spliced Rad51 transcript levels by quantitative RT-PCR. Same PCR primers are used as in B but the number of the PCR cycles is increased and the gel is overexposed to visualize the less abundant alternative splice variant of Rad51. The top band is full length (not quantified because of saturation), and the band directly below it is the alternative slice variant. The histogram represents the relative intensity of the Rad51 splice variant band with the 18S band used as a reference. (D) Western blot analysis of Rad51 protein. Protein levels for each cell line were normalized using α-tubulin as a loading control and are displayed in the histogram above the gels. All of the experiments were repeated three times and error bars are SD.
An alternatively spliced form of Rad51 is thought to have a higher translation efficiency than the main transcript (37). This form is also associated with an increased cancer risk in BRCA2 carriers (38). Therefore, we also compared the levels of the alternatively spliced transcript in normal and cancer cells using quantitative RT-PCR (Fig. 1C). The cell lines HCA2, WI-38, and GP2–293 did not have detectable alternative transcripts; while HCC1954, T47-D, and MCF-7 had the highest levels (Fig. 1C). On average, cancer cells showed a 2.5-fold increase in the alternatively spliced Rad51 transcript, and this difference was statistically significant (P = 0.037, t test).
We next analyzed the Rad51 protein levels in the 13 cell lines by Western blot (Fig. 1D). The analysis shows that Rad51 is more abundant in cancer cells when compared to normal cells (P = 0.001, t test). Rad51 protein levels were the greatest in T47-D cells and the lowest in HCA2, resulting in a 25-fold differential. On average, cancerous cells displayed a 5.7-fold increase in the level of Rad51 protein.
Rad51 Promoter Activity is Dramatically Increased in Cancer Cells.
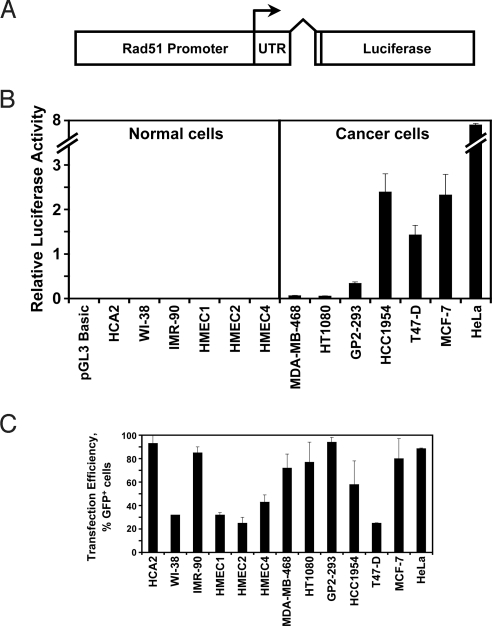
To test whether the differential expression of Rad51 can be used for anticancer therapy we cloned the putative Rad51 regulatory region including 2,931 nucleotides upstream to 3,601 nucleotides downstream from the start of transcription (37, 39) (Fig. 1A) from total DNA isolated from normal human cells. We then cloned the GFP ORF under the control of the Rad51 promoter. The resulting construct, pRad51-GFP contains the 2,931 bp of upstream regulatory sequences, the first noncoding exon of the Rad51, and the first 12 aa of the Rad51 ORF. pRad51-GFP, was transfected into HCA2, HT1080, and GP2–293 cells, and GFP expression was analyzed by flow cytometry. The two cancer cell lines, HT1080 and GP2–293 showed a large number of GFP+ cells (67% in GP2–293 and 34% in HT1080). Surprisingly, no GFP+ cells were detectable in the normal human fibroblasts HCA2. This result suggested that the difference in Rad51-GFP expression between the two cancer cell lines and the normal cells was much more dramatic than the difference in endogenous Rad51 levels.
Because the expression of Rad51-GFP was virtually undetectable in normal cells, we replaced GFP with firefly luciferase (Fig. 2A), a more sensitive reporter. The resulting construct, pRad51-Luc, was transfected into the panel of 13 cancer and normal cell lines and 72 h posttransfection cell extracts were tested for luciferase activity (Fig. 2B). To normalize for differences in transfection efficiency, cells were transfected with pEGFP-N1 vector and the number of cells with detectable GFP fluorescence was scored by flow cytometry (Fig. 2C). The ratio between luciferase activity and the number of GFP+ cells was used as a measure of Rad51-Luc expression. All of the cancer cell lines displayed dramatically elevated Rad51 promoter activity (Fig. 2B and supporting information (SI) Table S1). There was up to a 12,500-fold difference in luciferase activity between the lowest activity cell line (HCA2) and the highest (HeLa). On average, cancer cells displayed a >840-fold Rad51 promoter activity than the normal cells. This difference in promoter activity is striking and is much greater than the difference observed in the endogenous protein and transcript levels. We conclude that the constructs containing Rad51 promoter in which Rad51 ORF is replaced with a reporter or a cytotoxic gene hold a great promise for transcriptional gene therapy.
Fig. 2.
Rad51 promoter fused to luciferase gene shows dramatic difference in promoter activity between normal and cancer cells. (A) Diagram of the pRad51-Luc construct with the firefly luciferase gene under control of Rad51 promoter. Transcription start site is indicated by arrow. (B) Luciferase assays measuring Rad51 promoter activity in 13 cell lines. Cells were transfected with 2 μg of pRad51-Luc and luciferase activity was analyzed in cell extracts 72 h posttransfection. Luciferase activity was normalized for the efficiency of transfection determined by transfection with GFP-expressing plasmid, shown in C. The values for luciferase activity for all of the cell lines are provided in Table S1. The experiments were repeated three times and error bars show SD. (C) Transfection efficiency in 13 cell lines. In parallel with pRad51 luciferase transfections shown in B, cells were transfected with 2 μg of the GFP-expressing plasmid pEGFP-N1 and analyzed by flow cytometry 72 h posttransfection. The parameters for FACS analysis were set so as to detect all cells with green fluorescence above the background. This ensures that all transfected cells are scored regardless of the differences in expression in different cell lines. Efficiency of transfection is expressed as the percentage of GFP+ cells. The experiments were repeated three times and error bars show SD.
Rad51 Promoter Fused to Diphtheria Toxin A Selectively Kills Cancer Cells with Minimal Effect on Normal Cells.
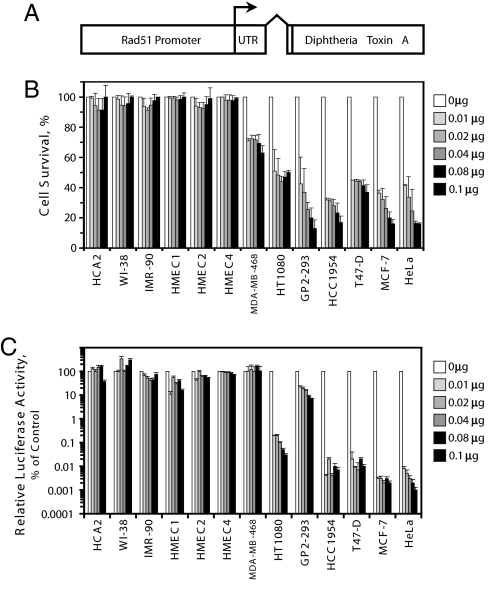
To test whether the Rad51 promoter fused to a cytotoxic gene will selectively kill cancer cells, we cloned the diphtheria toxin A ORF under the control of the Rad51 promoter (Fig. 3A). We then examined the effect of the Rad51-DTA fusion on cancer and normal cells. We used two approaches to measure the toxicity to the cells: decline in cell counts and inhibition of protein synthesis. The panel of 13 cell lines was transfected with increasing amounts of pRad51-DTA plasmid and/or the promoterless pGL3 plasmid. Complementing amounts of pGL3 were added so as to keep the amount of plasmid DNA equal in each transfection. Cells were allowed to express the transgene for 72 h, attached cells were harvested, and then counted using a Beckman Coulter cell counter. Transfection efficiency was determined by transfecting the cells with a GFP vector in the absence of DTA. DTA inhibits protein synthesis and triggers apoptosis and detachment of cells (40), although the attached fraction may contain some cells at early stages of apoptosis. The killing effect (Fig. 3B) at each dose of the pRad51-DTA was expressed as a percentage of attached cells transfected with Rad51-DTA construct relative to the transfection with the control plasmid pGL3 (see Materials and Methods for the details). pRad51-DTA did not cause a decline in cell counts in any of the normal cell lines. However, all cancer cell lines displayed 30–80% reduction in cell survival. The observed killing effect is likely to be an underestimate, as some early apoptotic cells are counted as attached cells.
Fig. 3.
The construct containing Rad51 promoter fused to diphtheria toxin A (DTA) gene selectively kills cancer cells. (A) Diagram of the pRad51-DTA construct. Transcription start site is indicated by arrow. (B) Decline in cell counts after transfection with pRad51-DTA. Cells were cotransfected with the indicated amounts of pRad51-DTA and promoterless plasmid pGL3 to bring the total amount of DNA in each transfection to 0.1 μg. Attached cells were harvested 72 h posttransfection and counted using a Beckman Coulter counter. For each cell line, the cell counts obtained after transfection with pRad51-DTA were divided by cell counts in the control transfections with 0 μg of pRad51-DTA and normalized for the efficiency of transfection. The experiments were repeated three times and error bars show SD. (C) Inhibition of protein synthesis by pRad51-DTA. In this experiment inhibition of SV40-luciferase expression is used as a model of reduction in de novo protein synthesis by DTA. Cells were transfected with pRad51-DTA and pGL3 as described above along with 1 μg of a plasmid encoding firefly luciferase under SV40 promoter. Cells were harvested 72 h posttransfection and subjected to luciferase assay. For each cell line, luciferase activity after transfection with pRad51-DTA was divided by luciferase activity in control transfections with 0 μg of pRad51-DTA. The experiments were repeated three times and error bars show SD.
To measure inhibition of protein synthesis, we cotransfected a pRad51-DTA and control plasmid, as described above, with the firefly luciferase gene under the control of the SV40 promoter/enhancer element. Cells were harvested 72 h posttransfection and luciferase activity was measured in the protein extracts (Fig. 3C). Reduction in luciferase activity was used as a measure of the inhibition of protein synthesis. Protein synthesis in the six normal cell lines either did not change or decreased at most 10-fold. One of the cancer cell lines, MDA-MB-468, did not show a change, but the other six cancer cell lines had reductions in protein synthesis that ranged from 10- up to 100,000-fold. Various amounts of transfected pRad51-DTA had similar toxicities, which is consistent with the fact that very low levels of DTA are sufficient to kill the cell (36). In summary, the Rad51-DTA construct displayed moderate to very strong toxicity to six out of seven cancer cell lines that were tested, and had minimal toxicity to normal cells. These results demonstrate the feasibility of using the Rad51 promoter for targeted anticancer therapy.
Discussion
Our study identifies the Rad51 gene promoter as a promising cancer-specific promoter for transcriptionally targeted therapy. Therapies based on the expression of suicide genes driven by cancer-specific promoters offer an exciting possibility of selectively eliminating cancer cells with no toxicity to normal tissue. This approach has been attempted with several promoters, most notably with the hTERT (human telomerase) promoter (reviewed in refs. 22, 23). These approaches, however, are slow to transition into clinical trials because of several challenges that need to be overcome, most important being safety and efficacy. Safety issues involve the inability for current promoters to be both absolutely selective for cancer cells and not significantly active in normal cells. Efficacy issues arise from the weak activity of the cancer-specific promoters.
Our results suggest that the Rad51 promoter offers superior strength and selectivity. The activity of the hTERT promoter was shown to be on average 10-fold higher in cancer cells than in the normal cells (27, 30, 41, 42), while with the Rad51 promoter we observe up to 12,500-fold increase in promoter activity. In a study that used an hTERT-DTA fusion for selective killing of cancer cells (25), hTERT-DTA decreased protein synthesis up to 68%, while with Rad51-DTA we observed up to 100,000-fold decrease in protein synthesis using similar amount of DTA-expressing construct. Furthermore, while hTERT is overexpressed in the majority of cancer cells, up to 15% of cancer cells do not express it and rely on alternative, recombination-based mechanisms to extend their telomeres, termed alternative lengthening of telomeres (ALT) (43). Thus, a therapy based on the hTERT promoter would not be effective in these cells. On the basis of the comparison of our results with hTERT studies, Rad51 promoter-driven therapeutic constructs seem very promising. These constructs may also be successfully used to treat ALT tumors, which use recombination rather than telomerase for telomere maintenance.
Overexpression of Rad51 in tumors has been described before (6–8), however, no attempts have been made to exploit it for transcriptionally targeted therapy, likely because the difference in endogenous protein expression between normal and cancer cells is relatively modest for this purpose. Serendipitously, we found that when the Rad51 promoter is fused to another ORF an unprecedented difference in promoter activity between normal and cancer cells is achieved. This can be explained by complex posttranscriptional regulation of Rad51 expression. Rad51 is a key protein in homologous recombination and its activity is tightly controlled in mammalian cells, because unrestrained recombination can wreak havoc in the repetitive human genome. Although the promoter is greatly activated in cancer cells the translation of the message may still be highly inhibited. By replacing the Rad51 message we remove the posttranscriptional regulation and reveal the dramatic difference in promoter activity. Another possibility could be that a critical inhibitory regulatory element has been (fortuitously) omitted in selecting the segment of the Rad51 gene used to create the promoter fusion.
We did not detect any GFP+ cells in the normal cell lines when we used Rad51-GFP construct. However, with the luciferase reporter we observed a very low level of activity in normal cells (Table S1). Because luciferase assay measures activity in cell extract rather than in individual cells, we cannot distinguish whether only a few normal cells express luciferase or all cells express it at a very low level. The former possibility seems more likely, considering extreme toxicity of diphtheria toxin (36). However, the latter scenario is also possible, because a more recent report has suggested that at least 10 molecules of DTA (rather than 1) are required to kill the cell (44).
The next step in developing the Rad51-based therapy is to test Rad51 promoter driven constructs in animal models in vivo. This can be accomplished through packaging the constructs in a viral vector system. Studies using the hTERT promoter have used both replication-defective (45) and replication-competent (46, 47) adenovirus as means for successful in vivo gene delivery. Pseudotyped lentiviral vectors targeted to prostate cells containing a prostate-specific promoter to control gene-of-interest expression have also been used (48). Similarly, the Rad51 promoter can be used to specifically express a cytoxic gene and/or a reporter gene in a replication-deficient virus by the substitution of the adenoviral E1A genes with our Rad51 construct. Alternatively, the Rad51 promoter can be used to control the expression of the E1A genes in what is known as a conditionally replicating adenovirus, thus allowing the virus to selectively kill cancer cells. The conditionally replicating system is likely to further improve the specificity of the construct and decrease the toxicity to normal cells.
In conclusion, we have demonstrated that fusion constructs containing the Rad51 promoter are highly active in cancer cells and repressed in normal cells. We performed the proof-of-principle experiments to show that Rad51 promoter fused to a cytotoxic gene kills a variety of cancer cells with high selectivity and efficacy. These results open new avenues for developing transcriptionally targeted therapies based on Rad51 promoter.
Materials and Methods
Cell Culture.
All cell lines were grown in monolayer on treated polystyrene cell culture dishes (Corning) at 37 °C in 3% O2, 5% CO2, and 97% relative humidity in HERA Cell 240 incubators. Human normal fibroblasts HCA2, IMR-90, and WI-38 used in this study were immortalized by constitutive expression of hTERT from integrated pBABE-Puro retrovirus. Immortalized human foreskin fibroblast line HCA2 and immortalized embryonic lung fibroblast IMR-90 and WI-38 were maintained in MEM (ATCC) supplemented with 15% FBS; FBS, (Gibco), and 1× Pen/Strep (Gibco). Normal human mammary epithelial cells HMEC1, HMEC2, and HMEC4 (Clonetics) were maintained in MEBM (Lonza) and supplemented with MEGM SingleQuots (Lonza), which contains BPE, hEGF, insulin, hydrocortisone, and GA-1000. Human fibrosarcoma cell line HT1080 (ATCC), human embryonic kidney line GP2–293 (Clontech), and human cervical carcinoma line HeLa (ATCC) were maintained in DMEM (Gibco) supplemented with 10% FBS (Gibco), 1× Pen/Strep (Gibco), and 1× nonessential amino acids (Gibco). Breast epithelial carcinoma line MDA-MB-468 (ATCC) was maintained in Leibovitz L-15 (ATCC) supplemented with 10% FBS and 1× Pen/Strep. Breast epithelial carcinoma line HCC-1954 (ATCC) was maintained in RPMI-1640 (ATCC) and supplemented with 10% FBS and 1× Pen/Strep. Breast epithelial carcinoma T47-D (ATCC) was maintained in RPMI-1640 supplemented with 10% FBS, 1× Pen/Strep, and 0.01 mg/mL bovine insulin (Sigma I 4011). Breast epithelial carcinoma line MCF7 (ATCC) was maintained in MEM and supplemented with 10% FBS, 1× Pen/Strep, and 0.01 mg/mL bovine insulin.
Cloning of the Human Rad51 Promoter Region and Construction of pRad51-GFP, pRad51-Luc, and pRad51-DTA Plasmids.
The 6,532-bp Rad51 regulatory region was cloned in two steps. In the first step, the region from 2,931 bp upstream to 230 bp downstream from the start of transcription was PCR amplified using the GC-rich PCR kit (Roche) with the primers 5′-AACATTAATGCACAGCAGGTGAGCAGCTAGCAAGCAAGC-3′ and 5′-CGCACCGGTGCCATTACTCGGTCCGCAGCGCTCCTCTCTCCAGC-3′, and subcloned into the pEGFP-N1 plasmid (Clontech) to replace the original CMV promoter by digesting both the PCR product and plasmid with the restriction enzymes AseI + AgeI resulting in pRad51(1/2) plasmid. In the second step, primers 5′-TCTGTAAACTCGCGCAGGATCAAGCTCTCG-3′ and 5′-TCCACCGGTGTATCTGCATTTGCTTCAAGCTGCATCTGC-3′ were used to PCR amplify 164 bp upstream to 3,601 bp downstream from the Rad51 transcription start site. An internal EcoRI site located 23 bp upstream of the start of transcription and the oligo-introduced AgeI sites were used to digest both the PCR product and pRad51(1/2) plasmid, followed by ligating the Rad51 gene fragment from 2,931 bp upstream to 24 bp upstream to the start of transcription with the fragment containing 23 bp upstream to 3,601 bp downstream of the start of transcription. This two-step method reconstitutes the wild-type full-length 6,532 bp Rad51 regulatory region, containing 2,931 bp upstream to 3,601 bp downstream from the start of transcription. The regulatory region includes the start of transcription, the first exon (noncoding), the first intron, and the first 40 bp of the second exon (coding), with the GFP gene ligated in frame after the 40-bp second exon fragment. Final pRad51-GFP plasmid was tested by restriction enzyme digestion and sequencing.
To transfer the full 6,532-bp Rad51 promoter region to the promoterless pGL3-Basic (Promega), which contains the gene for firefly luciferase, the restriction enzyme sites AgeI and AseI had to be introduced into pGL3-Basic polylinker by site-directed mutagenesis (Stratagene) with the following primers: 5′-CCGGAAGCTTACCGGTCGCCACCATGGAAGACGCC-3′ and 5′-GCCAAGCTTAATTAATTCGCAGATCTCGAGCC-3′, resulting in pGL3-Basic(Age/Ase) vector. The full-length Rad51 promoter region was then cut out of the pRad51-GFP plasmid by the restriction enzymes AseI and AgeI and cloned into the same sites in pGL3-Basic to create pRad51-Luc, with the translational start of the firefly luciferase gene in frame with the first 12 amino acids of the Rad51 coding region and under the Rad51 promoter.
To construct pRad51-DTA, which contains the Rad51 promoter controlling bacterial diphtheria toxin A gene, GFP was excised from pRad51-GFP with the restriction enzymes AgeI and NotI and replaced with the gene encoding DTA. The DTA gene was obtained by PCR amplifying the DTA coding sequence from plasmid pROSA26KPN (from P. Soriano, Fred Hutchinson Cancer Research Center, Seattle, WA) with the following primers to introduce an AgeI site at the 5′ end and a NotI site at the 3′ end: 5′TTAGCGGCCGCTTAGAGCTTTAAATCTCTGTAGGTAG-3′ and 5′-CCTACCGGTCGCCACCATGGATCCTGATGATGTTG-3′.
Western Blots.
Exponentially growing cells were harvested and counted on a Beckman Coulter Z2 particle counter. Cells were resuspended in PBS pH 7.4 (Gibco) with Complete Protease Inhibitor Mixture (Roche) and lysed by mixing with Laemmli sample buffer (BioRad) containing 5% 2-mercaptoethanol (J. T.Baker), followed by boiling for 10 min with vortexing every 5 min. Protein concentration was determined by DC protein assay (BioRad). Protein extracts (25 μg) from each cell line were separated on a 10% SDS/PAGE, blotted onto a nitrocellulose membrane (BioRad) and blocked in TBS-T with 1.25% dried milk (wt/vol). Membranes were probed with mouse monoclonal primary antibodies against human Rad51 (NeoMarkers) overnight or α-tubulin (Abcam) for 2 h and probed with HRP conjugated goat anti-mouse secondary antibodies (BioRad) for 2 h. The images were analyzed using ImageQuantTL (Amersham).
Quantitative RT-PCR.
Exponentially growing cells where harvested and counted on a Beckman Coulter Z2 particle counter. mRNA was extracted using the RNeasy Mini kit and QIAshredder (Qiagen) and concentrations were determined by A260 nM spectrophotometry on a SmartSpec Plus (BioRad). Titan one-tube RT-PCR (Roche) kit was used to amplify the 5′ of Rad51 mRNA using 0.4 μg of total mRNA and primers 5′CCAGAGACCGAGCCCTAAGGAGAGTGCG-3′ and 5′-TGGCATTTATGCCACACTGCTCTAACCGTG-3′. The following PCR program was used to quantify the main transcript: (1) heat 0.4 μg of RNA sample in 16 μL ddH2O at 85 °C for 3 min; (2) add enzyme/primer/dNTP mix; (3) 50 °C for 30 min; (4) 94 °C for 2 min; (5) 10 cycles of 94 °C for 1 min, 60 °C for 1 min, 68 °C for 1 min; (6) 15 cycles of 94 °C for 1 min, 60 °C for 1 min, 68 °C for 1 min + 5 sec/cycle; and (7) 68 °C for 7 min. A similar PCR program was run to examine the smaller alternative splice variant at the 5′ end, with an additional 5 cycles at step 6. Control PCR was performed at the same conditions as experimental reaction with primers for 18S ribosomal subunit from QuantumRNA 18S internal standards kit (Ambion) at 3:7 18S primer:competimer mix. PCR products were run on a 1.5% agarose gel and analyzed by ImagequantTL (Amersham).
Rad51 Promoter Activity and Luciferase Assays.
Two micrograms of pRad51-Luc or 2 μg of the pEGFP-N1 (Clontech) were transfected into 1 × 106 growing cells of each of the 13 cell lines by Amaxa Nucleofector II electroporation. The following Nucleofector programs and transfection solutions were used for each cell line: HCA2, program U-20 and solution NHDF; IMR-90, program X-001 and solution NHDF; WI-38, program V-001 and solution NHDF; HMEC1, HMEC2, and HMEC4, program Y-001 and solution HMEC; MDA-MB-468, program X-005 and solution V; HT1080, program L-005 and solution V; GP2–293, program A-023 and solution V; HCC1954, program A-023 and solution V; T47-D, program A-023 and solution V; MCF7, program P-020 and solution V; and HeLa, program I-013 and solution V. Cells transfected with pEGFP-N1 were harvested 72 h posttransfection and analyzed by FACS analysis to determine the percentage of cells with detectable GFP. Cells transfected with pRad51-Luc were harvested and counted 72 h posttransfection and lysed using passive lysis buffer (Promega) at a ratio of 200 μL/1 × 106 cells and then 20 μL of this extract was used in the luciferase assay (Promega) using a GloMax20/20 Luminometer (Promega).
Analysis of the Effect of Rad51 Promoter Driven DTA on the Cells: Cell Counts and Luciferase Assay.
Cells were split, and 24 h later 1 × 105 cells of each cell line were cotransfected with 0, 0.01, 0.02, 0.04, 0.08, or 0.1 μg of pRad51-DTA supplemented with the control pGL3 basic plasmid to bring the amount of DNA to 0.1 μg in each transfection, along with 1 μg of pGL3-control plasmid containing firefly luciferase under the SV40 promoter using a Fugene 6 transfection reagent (Roche). Cells were harvested 72 h posttransfection and counted by a Z2 particle counter (Beckman Coulter), and protein extracts were obtained by lysing cells with passive lysis buffer (Promega) at a ratio of 50 μL/50,000 cells. Twenty microliters of the extract were used for each luciferase assay.
To measure cell survival after pRad51-DTA transfection (Fig. 3B) it is essential to calculate the survival of transfected cells, because the total cell count obtained 3 days after transfection with pRad51-DTA includes nontransfected cells that continue to proliferate, while cells that are killed by DTA do not proliferate. To calculate the percent survival of transfected cells (ST) we used the formula:
where TSE is the number of transfected cells (cells that received the plasmid) that survived after transfection with pRad51-DTA, and TSC is the number of transfected cells that survived after control transfection with the GFP vector. TSE and TSC are calculated as:
where H is the total number of cells harvested 3 days after transfection, k is the growth rate of nontransfected cells, calculated as the number of cells harvested 3 days after the control (GFP) transfection divided by the number of cells plated. N is the number of cells that did not receive the plasmid, calculated as the total number of cells used for transfection multiplied by transfection efficiency.
The experiment measuring the decline in luciferase activity after pRad51-DTA transfection relative to the control transfection (Fig. 3C) did not require a correction for transfection efficiency. This is because only the transfected cells were expressing luciferase.
Acknowledgments.
We thank Michael Bozzella and Zhiyong Mao for critically reading the manuscript. This work was supported by grants from National Institutes of Health (NIH), Ellison Medical Foundation, and American Federation for Aging Research to V.G., Ellison Medical Foundation grant to A.S., and NIH Ruth L. Kirschstein National Research Service Award Fellowship to C.H.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0807990106/DCSupplemental.
References
- 1.Baumann P, West SC. Role of the human RAD51 protein in homologous recombination and double-stranded-break repair. Trends Biochem Sci. 1998;23:247–251. doi: 10.1016/s0968-0004(98)01232-8. [DOI] [PubMed] [Google Scholar]
- 2.Baumann P, Benson FE, West SC. Human Rad51 protein promotes ATP-dependent homologous pairing and strand transfer reactions in vitro. Cell. 1996;87:757–766. doi: 10.1016/s0092-8674(00)81394-x. [DOI] [PubMed] [Google Scholar]
- 3.Arnaudeau C, et al. RAD51 supports spontaneous non-homologous recombination in mammalian cells, but not the corresponding process induced by topoisomerase inhibitors. Nucleic Acids Res. 2001;29:662–667. doi: 10.1093/nar/29.3.662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Richardson C. RAD51, genomic stability, and tumorigenesis. Cancer Lett. 2005;218:127–139. doi: 10.1016/j.canlet.2004.08.009. [DOI] [PubMed] [Google Scholar]
- 5.Thacker J. The RAD51 gene family, genetic instability and cancer. Cancer Lett. 2005;219:125–135. doi: 10.1016/j.canlet.2004.08.018. [DOI] [PubMed] [Google Scholar]
- 6.Raderschall E, et al. Elevated levels of Rad51 recombination protein in tumor cells. Cancer Res. 2002;62:219–225. [PubMed] [Google Scholar]
- 7.Maacke H, et al. DNA repair and recombination factor Rad51 is over-expressed in human pancreatic adenocarcinoma. Oncogene. 2000;19:2791–2795. doi: 10.1038/sj.onc.1203578. [DOI] [PubMed] [Google Scholar]
- 8.Xia SJ, Shammas MA, Shmookler Reis RJ. Elevated recombination in immortal human cells is mediated by HsRAD51 recombinase. Mol Cell Biol. 1997;17:7151–7158. doi: 10.1128/mcb.17.12.7151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Arias-Lopez C, et al. p53 modulates homologous recombination by transcriptional regulation of the RAD51 gene. EMBO Rep. 2006;7:219–224. doi: 10.1038/sj.embor.7400587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Linke SP, et al. p53 interacts with hRAD51 and hRAD54, and directly modulates homologous recombination. Cancer Res. 2003;63:2596–2605. [PubMed] [Google Scholar]
- 11.Slupianek A, et al. Fusion tyrosine kinases induce drug resistance by stimulation of homology-dependent recombination repair, prolongation of G(2)/M phase, and protection from apoptosis. Mol Cell Biol. 2002;22:4189–4201. doi: 10.1128/MCB.22.12.4189-4201.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Slupianek A, et al. BCR/ABL regulates mammalian RecA homologs, resulting in drug resistance. Mol Cell. 2001;8:795–806. doi: 10.1016/s1097-2765(01)00357-4. [DOI] [PubMed] [Google Scholar]
- 13.Richardson C, Stark JM, Ommundsen M, Jasin M. Rad51 overexpression promotes alternative double-strand break repair pathways and genome instability. Oncogene. 2004;23:546–553. doi: 10.1038/sj.onc.1207098. [DOI] [PubMed] [Google Scholar]
- 14.Hannay JA, et al. Rad51 overexpression contributes to chemoresistance in human soft tissue sarcoma cells: a role for p53/activator protein 2 transcriptional regulation. Mol Cancer Ther. 2007;6:1650–1660. doi: 10.1158/1535-7163.MCT-06-0636. [DOI] [PubMed] [Google Scholar]
- 15.Collis SJ, et al. Ribozyme minigene-mediated RAD51 down-regulation increases radiosensitivity of human prostate cancer cells. Nucleic Acids Res. 2001;29:1534–1538. doi: 10.1093/nar/29.7.1534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Maacke H, et al. Over-expression of wild-type Rad51 correlates with histological grading of invasive ductal breast cancer. Int J Cancer. 2000;88:907–913. doi: 10.1002/1097-0215(20001215)88:6<907::aid-ijc11>3.0.co;2-4. [DOI] [PubMed] [Google Scholar]
- 17.Qiao GB, et al. High-level expression of Rad51 is an independent prognostic marker of survival in non-small-cell lung cancer patients. Br J Cancer. 2005;93:137–143. doi: 10.1038/sj.bjc.6602665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Russell JS, et al. Gleevec-mediated inhibition of Rad51 expression and enhancement of tumor cell radiosensitivity. Cancer Res. 2003;63:7377–7383. [PubMed] [Google Scholar]
- 19.Ohnishi T, et al. In vitro and in vivo potentiation of radiosensitivity of malignant gliomas by antisense inhibition of the RAD51 gene. Biochem Biophys Res Commun. 1998;245:319–324. doi: 10.1006/bbrc.1998.8440. [DOI] [PubMed] [Google Scholar]
- 20.Collis SJ, Deweese TL, Jeggo PA, Parker AR. The life and death of DNA-PK. Oncogene. 2004;24:949–961. doi: 10.1038/sj.onc.1208332. [DOI] [PubMed] [Google Scholar]
- 21.Hardcastle J, Kurozumi K, Chiocca EA, Kaur B. Oncolytic viruses driven by tumor-specific promoters. Curr Cancer Drug Targets. 2007;7:181–189. doi: 10.2174/156800907780058880. [DOI] [PubMed] [Google Scholar]
- 22.Wu L, Johnson M, Sato M. Transcriptionally targeted gene therapy to detect and treat cancer. Trends Mol Med. 2003;9:421–429. doi: 10.1016/j.molmed.2003.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lo HW, Day CP, Hung MC. Cancer-specific gene therapy. Adv Genet. 2005;54:235–255. doi: 10.1016/S0065-2660(05)54010-0. [DOI] [PubMed] [Google Scholar]
- 24.Fujiwara T, Urata Y, Tanaka N. Telomerase-specific oncolytic virotherapy for human cancer with the hTERT promoter. Curr Cancer Drug Targets. 2007;7:191–201. doi: 10.2174/156800907780058835. [DOI] [PubMed] [Google Scholar]
- 25.Abdul-Ghani R, et al. Use of transcriptional regulatory sequences of telomerase (hTER and hTERT) for selective killing of cancer cells. Mol Ther. 2000;2:539–544. doi: 10.1006/mthe.2000.0196. [DOI] [PubMed] [Google Scholar]
- 26.Koga S, et al. A novel telomerase-specific gene therapy: gene transfer of caspase-8 utilizing the human telomerase catalytic subunit gene promoter. Hum Gene Ther. 2000;11:1397–1406. doi: 10.1089/10430340050057477. [DOI] [PubMed] [Google Scholar]
- 27.Komata T, et al. Treatment of malignant glioma cells with the transfer of constitutively active caspase-6 using the human telomerase catalytic subunit (human telomerase reverse transcriptase) gene promoter. Cancer Res. 2001;61:5796–5802. [PubMed] [Google Scholar]
- 28.Kirch HC, et al. Tumor-specific activation of hTERT-derived promoters by tumor suppressive E1A-mutants involves recruitment of p300/CBP/HAT and suppression of HDAC-1 and defines a combined tumor targeting and suppression system. Oncogene. 2002;21:7991–8000. doi: 10.1038/sj.onc.1205965. [DOI] [PubMed] [Google Scholar]
- 29.Majumdar AS, et al. The telomerase reverse transcriptase promoter drives efficacious tumor suicide gene therapy while preventing hepatotoxicity encountered with constitutive promoters. Gene Ther. 2001;8:568–578. doi: 10.1038/sj.gt.3301421. [DOI] [PubMed] [Google Scholar]
- 30.Gu J, et al. Tumor-specific transgene expression from the human telomerase reverse transcriptase promoter enables targeting of the therapeutic effects of the Bax gene to cancers. Cancer Res. 2000;60:5359–5364. [PubMed] [Google Scholar]
- 31.Nettelbeck DM, et al. Novel oncolytic adenoviruses targeted to melanoma: specific viral replication and cytolysis by expression of E1A mutants from the tyrosinase enhancer/promoter. Cancer Res. 2002;62:4663–4670. [PubMed] [Google Scholar]
- 32.Latham JP, Searle PF, Mautner V, James ND. Prostate-specific antigen promoter/enhancer driven gene therapy for prostate cancer: construction and testing of a tissue-specific adenovirus vector. Cancer Res. 2000;60:334–341. [PubMed] [Google Scholar]
- 33.Chen JS, et al. Cancer-specific activation of the survivin promoter and its potential use in gene therapy. Cancer Gene Ther. 2004;11:740–747. doi: 10.1038/sj.cgt.7700752. [DOI] [PubMed] [Google Scholar]
- 34.Yu L, et al. Midkine promoter-driven suicide gene expression and -mediated adenovirus replication produced cytotoxic effects to immortalised and tumour cells. Eur J Cancer. 2004;40:1787–1794. doi: 10.1016/j.ejca.2004.04.014. [DOI] [PubMed] [Google Scholar]
- 35.Bilsland AE, Fletcher-Monaghan A, Keith WN. Properties of a telomerase-specific Cre/Lox switch for transcriptionally targeted cancer gene therapy. Neoplasia. 2005;7:1020–1029. doi: 10.1593/neo.05385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yamaizumi M, Mekada E, Uchida T, Okada Y. One molecule of diphtheria toxin fragment A introduced into a cell can kill the cell. Cell. 1978;15:245–250. doi: 10.1016/0092-8674(78)90099-5. [DOI] [PubMed] [Google Scholar]
- 37.Hasselbach L, et al. Characterisation of the promoter region of the human DNA-repair gene Rad51. Eur J Gynaecol Oncol. 2005;26:589–598. [PubMed] [Google Scholar]
- 38.Antoniou AC, et al. RAD51 135G–>C modifies breast cancer risk among BRCA2 mutation carriers: results from a combined analysis of 19 studies. Am J Hum Genet. 2007;81:1186–1200. doi: 10.1086/522611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schmutte C, et al. Characterization of the human Rad51 genomic locus and examination of tumors with 15q14–15 loss of heterozygosity (LOH) Cancer Res. 1999;59:4564–4569. [PubMed] [Google Scholar]
- 40.Chang MP, et al. Internucleosomal DNA cleavage precedes diphtheria toxin-induced cytolysis. Evidence that cell lysis is not a simple consequence of translation inhibition. J Biol Chem. 1989;264:15261–15267. [PubMed] [Google Scholar]
- 41.Abdul-Ghani R, et al. Use of transcriptional regulatory sequences of telomerase (hTER and hTERT) for selective killing of cancer cells. Mol Ther. 2000;2:539–544. doi: 10.1006/mthe.2000.0196. [DOI] [PubMed] [Google Scholar]
- 42.Takakura M, et al. Cloning of human telomerase catalytic subunit (hTERT) gene promoter and identification of proximal core promoter sequences essential for transcriptional activation in immortalized and cancer cells. Cancer Res. 1999;59:551–557. [PubMed] [Google Scholar]
- 43.Bryan TM, et al. Evidence for an alternative mechanism for maintaining telomere length in human tumors and tumor-derived cell lines. Nat Med. 1997;3:1271–1274. doi: 10.1038/nm1197-1271. [DOI] [PubMed] [Google Scholar]
- 44.Sarntinoranont M, Iadarola MJ, Lonser RR, Morrison PF. Direct interstitial infusion of NK1-targeted neurotoxin into the spinal cord: a computational model. Am J Physiol Regul Integr Comp Physiol. 2003;285:R243–254. doi: 10.1152/ajpregu.00472.2002. [DOI] [PubMed] [Google Scholar]
- 45.Gu J, Fang B. Telomerase promoter-driven cancer gene therapy. Cancer Biol Ther. 2003;2:S64–70. [PubMed] [Google Scholar]
- 46.Kishimoto H, et al. In vivo imaging of lymph node metastasis with telomerase-specific replication-selective adenovirus. Nat Med. 2006;12:1213–1219. doi: 10.1038/nm1404. [DOI] [PubMed] [Google Scholar]
- 47.Wirth T, et al. A telomerase-dependent conditionally replicating adenovirus for selective treatment of cancer. Cancer Res. 2003;63:3181–3188. [PubMed] [Google Scholar]
- 48.Pariente N, et al. A novel dual-targeted lentiviral vector leads to specific transduction of prostate cancer bone metastases in vivo after systemic administration. Mol Ther. 2007;15:1973–1981. doi: 10.1038/sj.mt.6300271. [DOI] [PubMed] [Google Scholar]