Abstract
Nature uses a limited set of metabolites to perform all of the biochemical reactions. To increase the metabolic capabilities of biological systems, we have expanded the natural metabolic network, using a nonnatural metabolic engineering approach. The branched-chain amino acid pathways are extended to produce abiotic longer chain keto acids and alcohols by engineering the chain elongation activity of 2-isopropylmalate synthase and altering the substrate specificity of downstream enzymes through rational protein design. When introduced into Escherichia coli, this nonnatural biosynthetic pathway produces various long-chain alcohols with carbon number ranging from 5 to 8. In particular, we demonstrate the feasibility of this approach by optimizing the biosynthesis of the 6-carbon alcohol, (S)-3-methyl-1-pentanol. This work demonstrates an approach to build artificial metabolism beyond the natural metabolic network. Nonnatural metabolites such as long chain alcohols are now included in the metabolite family of living systems.
Keywords: metabolic engineering, protein engineering, chain elongation, long chain alcohols
Nature uses a limited set of metabolites such as organic acids, amino acids, nucleotides, lipids and sugars as building blocks for biosynthesis. These chemicals support the biological functions of all organisms. So far, construction of artificial biological systems (1–5) is limited by the existing metabolic capabilities. By supplying living cells with chemically synthesized nonnatural amino acids (6, 7) and sugars (8) as new building blocks, it is possible to introduce novel physical and chemical properties into biological entities. These efforts raise an interesting question: Can we rewire metabolism in a bottom-up fashion to produce nonnatural metabolites from simple carbon source? If so, such engineered artificial metabolism should be able to expand the chemical repertoire that living systems can use and produce. To begin to address this question, we developed a strategy to produce 7-(C7) to 9-carbon (C9) 2-keto acids, which can lead to useful nonnatural alcohols (C6–C8).
Aliphatic alcohols with carbon chain length >5 (C > 5) are attractive biofuel targets because they have higher energy density, and lower water solubility [1-pentanol 23 g/L, 1-hexanol 6.2 g/L, 1-heptanol 1.2 g/L (9)] that could facilitate postproduction purification from culture medium through an aqueous/organic 2-phase separation process. The only well-characterized mechanism for aliphatic alcohol production is through the Ehrlich pathway (10), which converts branched-chain amino acids into alcohols. The carbon number (up to 5) of the alcohols derived from this type of pathway is limited by the carbon number in the branched chain amino acid pathways (11). To overcome this limitation, existing metabolic networks need to be expanded. This is a daunting task because a metabolic pathway usually involves the collective function of multiple enzymes, which have to be engineered by rational design (12) or directed evolution (13, 14) to perform nonnatural activities.
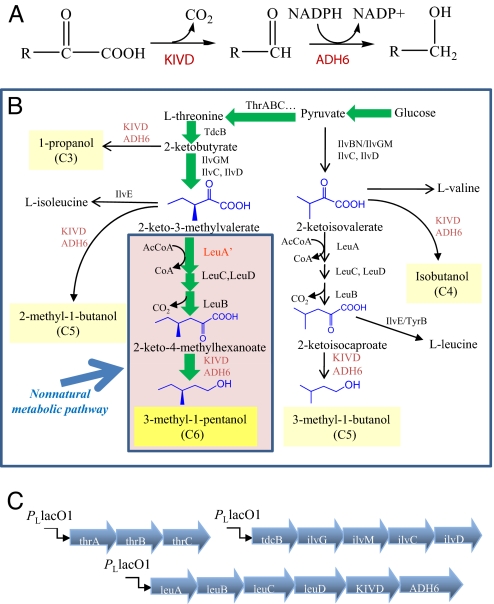
Acetyl-CoA is a common chemical unit for carbon chain elongation, such as reactions in tricarboxylic acid cycle, glyoxylate cycle, mevalonate pathway, and leucine biosynthesis (15). To explore the possibility of using acetyl-CoA related chemistry to produce C6 alcohol, we have engineered a nonnatural metabolic pathway (Fig. 1B, shaded region) into E. coli. First, we used the existing metabolic capability of E. coli to synthesize (S)-2-keto-3-methylvalerate, the 2-keto acid precursor of amino acid l-isoleucine. The chemical structure of 2-keto-3-methylvalerate is very similar to 2-ketoisovalerate (the 2-keto acid precursor of amino acid l-valine), containing only one more methyl group on the side chain. Because 2-ketoisovalerate is converted to 2-ketoisocaproate through a 3-step chain elongation cycle by 2-isopropylmalate synthase (LeuA), isopropylmalate isomerase complex (LeuC, LeuD) and 3-isopropylmalate dehydrogenase (LeuB), we reasoned that LeuA, LeuB, LeuC and LeuD may be promiscuous enough to allow 2-keto-3-methylvalerate go through the same elongation cycle and produce a novel compound 2-keto-4-methylhexanoate. Analogous to the Ehrlich pathway for production of fusel alcohols (Fig. 1A), we speculated that 2-keto-4-methylhexanoate could be converted to the corresponding aldehyde and then to a 6-carbon alcohol, (S)-3-methyl-1-pentanol, by the broad-substrate-range 2-ketoisovalerate decarboxylase (KIVD) from Lactococcus lactis (16) and alcohol dehydrogenase VI (ADH6) from Saccharomyces cerevisiae (17).
Fig. 1.
Pathway design. (A) Conversion of 2-keto acids to alcohols by a broad-substrate range 2-keto-acid decarboxylase (KIVD) and an alcohol dehydrogenase (ADH6). (B) Schematic representation of the biosynthetic pathway of 3-methyl-1-pentanol. The engineered nonnatural metabolic pathway is shaded in lavender. Similar to 2-ketoisovalerate, 2-keto-3-methylvalerate can have one more carbon added to its side chain by the leucine biosynthesis enzymes. (C) Synthetic operons for gene expression. Overexpression of ThrABC, TdcB and IlvGMCD drives the carbon flux toward 2-keto-3-methylvalerate.
Results and Discussion
Construction of a Nonnatural Metabolic Pathway for Biosynthesis of (S)-3-Methyl-1-Pentanol.
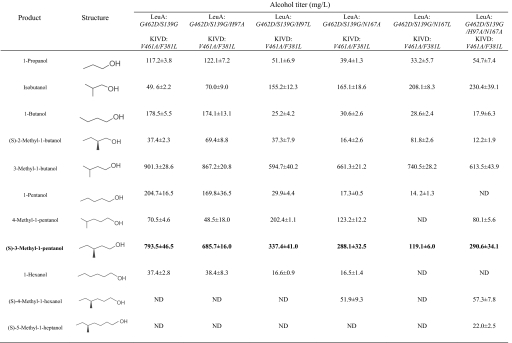
We constructed 3 synthetic operons (Fig. 1C) under the control of the PLlacO1 promoter: The first operon is composed of 3 genes on a low copy plasmid in the transcriptional order thrA-thrB-thrC; the second operon is composed of 5 genes on a medium copy plasmid in the transcriptional order tdcB-ilvG-ilvM-ilvC-ilvD, and the third operon is composed of 6 genes on a high copy plasmid in the transcriptional order leuA-leuB-leuC-leuD-KIVD-ADH6 (and a control operon without leuABCD). Except for KIVD, ADH6 and ThrA (G433R mutant (18) insensitive to threonine feedback inhibition), all other genes encode wild-type E. coli enzymes. As a result of overexpressing these 14 genes in a modified threonine-hyperproduction strain (ATCC98082, ΔilvE, ΔtyrB), 6.5 mg/L of (S)-3-methyl-1-pentanol was produced from 20 g/L glucose (Table 1, column 4), whereas a leucine-feedback insensitive G462D mutant (19) LeuA produced 40.8 mg/L of C6 alcohol (Table 1, column 5). In contrast, without overexpression of LeuABCD, no C6 alcohol production was detected (Table 1, column 3).
Table 1.
Production profile of alcohols from the designed pathway, with different KIVD mutants
Note that the V461A/F381L mutant gives the highest titer of 3-methyl-1-pentanol. E. coli cultures were grown in M9 medium with 20 g/L glucose plus 0.1 mM IPTG at 30 °C for 40 h. These products were identified by GC-MS and quantified by GC-FID. ND, not detectable.
Structure-Based Redesign of KIVD.
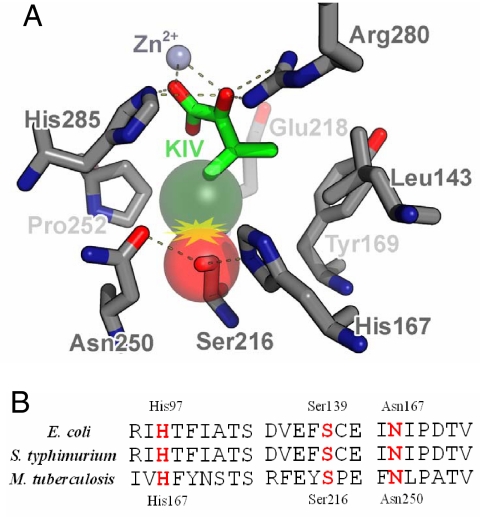
Because KIVD and ADH6 are promiscuous enzymes, they can also convert other intracellular 2-keto acids into alcohols (Fig. 1B, Table 1). To reduce the formation of byproducts and drive the carbon flux toward the target C6 alcohol, we examined the effect of engineering KIVD with higher selectivity toward 2-keto-4-methylhexanoate. The protein sequence alignment shows that KIVD has 40% and 31% sequence identities with Enterobacter cloacae indolepyruvate decarboxylase IPDC (20) and Zymomonas mobilis pyruvate decarboxylase ZmPDC (21) respectively. A homology model for the substrate-binding region of KIVD and IPDC was built based on the crystal structures of ZmPDC (PDB: 1ZPD). As can be seen from Fig. 2, 4 residues, Ser-286, Phe-381, Val-461, and Met 538, in combination with cofactor thiamine diphosphate (ThDP), delineate the keto-acid binding pocket of KIVD. Noticeably, the corresponding residues of ZmPDC have bulkier side chains, Tyr-290, Trp-392, Ile-472, and Trp-551; and those of IPDC have smaller ones, Thr-290, Ala-387, Val-467 and Leu-542. These differences can explain the substrate spectrum of these 2-keto acid decarboxylases and suggests that substitution of related amino acids might be able to change substrate specificity. A ZmPDC I472A variant was shown to be more active on longer-chain keto acids other than pyruvate (22). The corresponding residue of KIVD, V461, was thus mutated to alanine. Compared with the wild-type KIVD, the V461A mutant produced 3 times more 3-methyl-1-pentanol (Table 1, column 6). Further optimization was performed by mutating either F381 or M538 to smaller hydrophobic side chains such as leucine or alanine (Table 1). The F381L/V461A mutant was the best variant obtained and produced 384.3 mg/L of 3-methyl-1-pentanol.
Fig. 2.
Stereoview of active site of Z. mobilis pyruvate decarboxylase ZmPDC (green) and the corresponding homology model of Enterobacter cloacae indolepyruvate decarboxylase IPDC (cyan) and KIVD (purple), using ZmPDC as the template. The multiple sequence alignment was performed with ClustalW. Residues Y290, W392, and W551 of ZmPDC restrict the size of the binding pocket and prevent activating substrates larger than pyruvate. Residues F381, V461, and M538 of KIVD were mutated to smaller hydrophobic residues such as alanine and leucine to allow the enzyme accept substrates larger than 2-ketoisovalerate.
Fig. 4.
Structures of biosynthesized alcohols. (A) Nonnatural alcohols produced and their corresponding metabolic pathways. (B) 3-methyl-1-pentanol is S-isomer as confirmed by chiral GC analysis after MTBSTFA (N-Methyl-N-[tert-butyldimethyl-silyl]trifluoroacetimide) derivatization.
Both wild-type and F381L/V461A KIVD were added an N-terminal 6xHis-tag, overexpressed and purified through Ni-NTA columns. The kinetic parameters for activation of 2-ketoisovalerate (cognate substrate) and 2-keto-4-methylhexanoate (nonnatural substrate) were determined using a coupled enzymatic assay (22). Compared with the wild-type KIVD, for the smaller substrate, 2-ketoisovalerate, F381L/V461A KIVD has a significantly lower kcat (2.7 s−1 versus 38.3 s−1) and higher Km (7.7 mM versus 2.2 mM); for 2-keto-4-methylhexanoate, F381L/V461A KIVD has a comparable kcat (3.0 s−1 versus 10.8 s−1) and a slightly higher Km (0.22 mM versus 0.14 mM). Thus, the specificity constant kcat/Km of F381L/V461A KIVD toward 2-keto-4-methylhexanoate is 40-fold higher than that toward 2-ketoisovalerate. In comparison, the specificity constant kcat/Km of wild-type KIVD toward 2-keto-4-methylhexanoate is only 4-fold higher than that toward 2-ketoisovalerate (Table 2). Such a change in KIVD specificity distinguishably affects the distribution profile of alcohol products (more long-chain alcohols and less short- chain alcohols).
Table 2.
Kinetic parameters of wild type and mutant KIVD
Enlarging the Binding Pocket of LeuA.
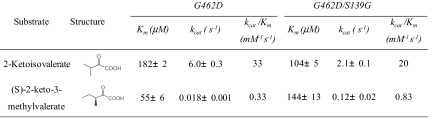
Besides KIVD, the other key enzyme determining the carbon flux toward 3-methyl-1-pentanol production is LeuA. LeuA catalyzes the condensation of acetyl-CoA with 2-keto-3-methylvalerate, which is the first step of the expanded metabolic pathway (Fig. 1B). LeuA also competes with KIVD for substrate 2-keto-3-methylvalerate, and thus reduces the formation of side product 2-methyl-1-butanol. Engineering KIVD with higher activity toward 2-keto-3-methylvalerate should help increase 3-methyl-1-pentanol production. As inferred from the crystal structure of Mycobacterium tuberculosis LeuA (23), residues His-167, Ser-216 and Asn-250 are within a radius of 4 Å of the γ-methyl group of bound 2-ketoisovalerate (Fig. 3A). Nonnatural substrate (S)-2-keto-3-methylvalerate contains one more methyl group that would cause steric hinderance with Ser-216, which could be relieved by mutating serine to the smallest amino acid glycine. Multiple protein sequence alignment shows that E. coli LeuA shares 92% and only 21% sequence identity with Salmonella typhimurium LeuA and M. tuberculosis LeuA respectively. Fortunately, the binding pocket is well conserved and the corresponding residues of E. coli LeuA are His-97, Ser-139 and Asn-167 (Fig. 3B). The G462D/S139G mutant LeuA was cloned and produced 793.5 mg/L 3-methyl-1-pentanol (Table 3, column 3), twice the amount by G462D LeuA.
Fig. 3.
Residues in the active site of LeuA. (A) Binding pocket of Mycobacterium tuberculosis LeuA (PDB: 1SR9) complexed with its natural substrate 2-ketoisovalerate (KIV, green). (S)-2-keto-3-methylvalerate has one more methyl group (dark green sphere) that would cause steric conflict with Ser 216 (red sphere), His-167, and Asn-250. (B) Multiple sequence alignment of M. tuberculosis, E. coli, and Salmonella typhimurium LeuA. The binding pocket is conserved, and the corresponding residues of E. coli LeuA are His-97, Ser-139, and Asn-167. These residues were subjected to site-specific mutagenesis.
Table 3.
Alcohol production with different LeuA mutants
Note that the G462D/S139G mutant gives the highest titer of 3-methyl-1-pentanol). E. coli cultures were grown in M9 medium with 20 g/L glucose plus 0.1 mM IPTG at 30 °C for 40 h. These products were identified by GC-MS and quantified by GC-FID. ND, not detectable.
Enzymatic assay indicates that G462D LeuA has an extremely low kcat (0.018 s−1) for (S)-2-keto-3-methylvalerate, which is 333-fold less than that for 2-ketoisovalerate (6.0 s−1). Because G462D LeuA has a comparable Km for both substrates (55 μM versus 182 μM), the low kcat may be why a previous report showed that 2-keto-3-methylvalerate is a strong inhibitor of LeuA (24). However, the S139G mutation increases the kcat 7-fold for (S)-2-keto-3-methylvalerate to 0.12 s−1 (Table 4).
Table 4.
Kinetic parameters of wild type and mutant LeuA
Additional mutations were then performed on His-97 and Asn-167. Even though better mutant was not found for production of 3-methyl-1-pentanol, interestingly, the G462D/S139G/N167A triple mutant produced 51.9 mg/L 4-methyl-1-hexanol (C7), and the G462D/S139G/H97A/N167A quadruple mutant produced 57.3 mg/L 4-methyl-1-hexanol (C7) and 22.0 mg/L 5-methyl-1-heptanol (C8).
Biosynthesis of a Repertoire of Nonnatural Alcohols.
Because the engineered LeuA has larger binding pockets, the chain elongation activities may continue several more rounds by LeuA on the 2-keto acids produced from the LeuABCD or other pathways (Fig. 4A). For example, 2-ketobutyrate can be converted to 2-ketovalerate, then to 2-ketocaproate, and finally to 2-ketoheptanoate by LeuABCD. In parallel, 2-keto-3-methylvalerate can be converted to 2-keto-4-methylhexanoate, then to 2-keto-5-methylhepatanoate and to 2-keto-6-methyloctanoate. All these keto acids are substrates of F381L/V461A KIVD. Upon decarboxylation, the corresponding aldehydes are reduced to the corresponding alcohols by ADH6. Indeed, we observed accumulation of 5 other nonnatural alcohols: 1-pentanol, 1-hexanol, 4-methyl-1-pentanol, 4-methyl-1-hexanol and 5-methyl-1-heptanol (Tables 1 and 3). The anteiso-methyl-branched alcohols are all derived from the same chiral precursor, (S)-2-keto-3-methylvalerate. The S-configuration of the stereogenic center in these alcohols remains unchanged during biosynthesis as confirmed by chiral GC analysis of (S)-2-methyl-1-butanol [see supporting information (SI)] and (S)-3-methyl-1-pentanol (Fig. 4B). These enantiomerically pure alcohols may be useful chiral synthons for chemical synthesis (25).
Conclusions
In this work, we have shown that by combining protein engineering and metabolic engineering approaches, it is possible to expand the intermediary metabolism of E. coli to produce various C5 to C8 alcohols that are not readily produced by microorganisms. Because of their specific physical and chemical properties, these long chain alcohols could be good candidates as biofuels or renewable chemical reagents. For practical applications, further metabolic engineering (26) and enzyme engineering (27) will be needed to increase the production yield and rate of these compounds. Because the 2-keto acid precursors of alcohols can be converted to amino acids by aminotransferases, we hereby also provide a biosynthetic way, instead of traditional organic synthesis, to expand the repertoire of nonnatural amino acids that have recently found broad applications (6, 28).
Materials and Methods
Vector Construction.
All cloning procedures (see SI for cloning scheme) were carried out in the E. coli strain XL10-gold (Stratagene). Oligos were synthesized by Operon Biotechnologies (see SI for sequence details). PCRs were performed with KOD polymerase (Novagen).
Fermentation Procedure.
The aminotransferase genes, ilvE and tyrB, of a threonine-hyperproduction E. coli strain ATCC98082 were inactivated by P1 transduction (29). This modified strain was transformed with pZS_thrO, pZAlac_tdcBilvGMCD and pZE_LeuABCDKA6 for alcohol production. Overnight cultures incubated in LB medium were diluted 100-fold into 5 mL of M9 medium supplemented with 1× trace metal mix A5 (11), 0.5% yeast extract and 2% glucose in 125-ml conical flasks. Antibiotics were added appropriately (ampicillin 100 mg/L, spectinomycin 25 mg/L, kanamycin 25 mg/L). Cells were grown to an optical density at 600 nm of ≈1.0 at 37 °C, followed by adding 0.1 mM isopropyl-b-d-thiogalactoside (IPTG). Cultures were then transferred to a 30 °C shaker (250 rpm) and incubated for 40 h.
GC-MS Analysis.
The GC-MS system is composed of model 6890N network GC system (Agilent Technologies), a model 7883B injector and autosampler (Agilent Technologies) and a model 5973 network mass selective detector (Agilent Technologies). Samples were separated through a DB-5ms capillary column (30 m, 0.25-mm internal diameter, 0.25-μm film thickness; Agilent Technologies) with helium (1 mL·min−1) as the carrier gas. Alcohols extracted by 200 μL of toluene from 1 mL of fermentation medium were directly injected for mass analysis.
GC–FID Analysis.
Alcohol compounds were quantified by a gas chromatograph equipped with flame ionization detector. The system is composed of a model 5890A gas chromatograph (Hewlett Packard) and a model 7673A automatic injector, sampler and controller (Hewlett Packard). Samples were separated through A DB-FFAP capillary column (30 m, 0.32-mm internal diameter, 0.25-μm film thickness; Agilent Technologies). GC oven temperature was initially placed at 40 °C for 2 min, increased with a gradient of 5 °C min−1 until 45 °C, and held for 4 min. Then it was increased with a gradient 15 °C min−1 until 230 °C and held for 4 min. Helium was used as the carrier gas. The temperature of injector and detector was set at 225 °C. Alcohol standards were purchased from either Sigma–Aldrich or TCI America.
For chiral GC analysis, samples were separated through a HP-CHIRAL 20ß column (30 m, 0.32-mm internal diameter, 0.25-μm film thickness; Agilent Technologies). The racemic mixture of 3-methyl-1-pentanol could not be directly resolved. However, after reaction with N-Methyl-N-[tert-butyldimethyl-silyl]trifluoroacetimide (Pierce), the conjugated product could be resolved into 2 peaks. GC oven temperature was initially placed at 50 °C for 4 min, increased with a gradient of 10 °C min−1 until 90 °C, and held for 2 min. And then it was increased with a gradient 2 °C min−1 until 130 °C and held for 2 min. Finally the temperature was increased with a gradient 35 °C min−1 until 235 °C and held for 2 min. Helium was used as the carrier gas. The temperature of injector and detector was set at 225 °C.
Protein Expression and Purification.
Both gene fragments encoding wild-type and F381L/V461A KIVD were amplified from plasmid pZE_LeuABCDKA6, using primers hiskivd_tevfwd and hiskivd_bamrev. After digestion with BamHI, the gene fragments were inserted into expression plasmid pQE9 (Qiagen) to yield pQE_hiskivd_wt and pQE_hiskivd_FL. The ADH6 gene fragment was amplified from yeast genomic DNA, using primers hisadh_tevfwd and hisadh_bamrev, digested with BamHI and inserted into pQE9 to generate pQE_hisadh6. Similarly, genes encoding G462D and G462D/S139G LeuA were amplified from plasmid pZE_LeuABCDKA6, using primers hisleua_tevfwd and hisleua_bamrev. After digestion with BamHI, the PCR products were ligated into pQE9 to create pQE_hisleua_GD and pQE_hisleua_GS. The resulting expression plasmids pQE_hiskivd_wt, pQE_hiskivd_FL, pQE_hisadh6, pQE_hisleua_GD and pQE_hisleua_GS were transformed into E. coli strain BL21(DE3) harboring pREP4 (Qiagen). Cells were inoculated from an overnight preculture at 1/100 dilution and grown in 200 mL of 2XYT rich medium containing 50 mg/L ampicillin and 25 mg/L kanamycin. At an OD600 of 0.6, recombinant proteins were expressed by induction of the cell cultures with 0.1 mM IPTG, followed by incubation at 30 °C overnight. Cell pellets were lysed by sonication in a buffer containing 250 mM NaCl, 2 mM DTT, 5 mM imidazole and 50 mM Tris pH 9.0. By applying a stepwise gradient of imidazole (up to 250 mM), enzymes were purified from crude cell lysates through Ni-NTA column chromatography. The fractions of highest purity were pooled and buffer-exchanged using Amicon Ultra centrifugal filters (Millipore). Storage buffer 1 [50 μM Tris buffer (pH 8.0), 1 mM MgSO4, 20% glycerol] was used for LeuA and ADH6, and storage buffer 2 [50 μM Tris buffer (pH 8.0), 1 mM MgSO4, 0.2 mM ThDP, 20% glycerol] was used for KIVD. The concentrated protein solutions were aliquoted (100 μL) into PCR tubes and flash frozen at −80 °C for long term storage.
Enzymatic Assay of KIVD.
Substrate 2-ketoisovalerate (KIV) was purchased from Sigma–Aldrich, and (S)-2-keto-4-methylhexanoate (KHV) was custom synthesized by AsisChem Inc. Protein concentration was determined by measuring UV absorbance at 280 nm. The decarboxylation activity of KIVD was measured at 30 °C, using a coupled enzymatic assay method. Excess ADH6 was used to reduce aldehyde into alcohol, and concomitantly, cofactor NADPH was oxidized to NADP+. The assay mixture contained 0.2 mM NADPH, 0.1 μM ADH6 and 0.1–20 mM 2-keto acids in assay buffer (50 mM potassium phosphate buffer, pH 6.8, 1 mM MgSO4, 0.5 mM ThDP) with a total volume of 0.2 mL. The reactions were started by adding 2 μL of KIVD (final concentrations: for KIV, 20 nM wild-type KIVD, 200 nM F381L/V461A KIVD; for KHV, 50 nM both), and the consumption of NADPH was monitored at 340 nm (extinction coefficient, 6.22 mM−1·cm−1). Kinetic parameters (kcat and Km) were determined by fitting initial velocity data to the Michaelis–Menten equation, using Origin software.
Measurement of LeuA activity.
The assay mixture contained 100 mM KCl, 2 mM MgCl2, 1 mM acetyl-CoA and 100 mM Tris (pH 8.0) with a total volume of 100 μL. G462D or G462D/S139G LeuA (each 100 nM) was reacted with 2-ketoisovalerate in a concentration range from 25 μM to 1 mM for 10 min at 30 °C, while 4 μM G462D or 1.5 μM G462D/S139G LeuA was reacted with (S)-2-keto-3-methylvalerate in a concentration range from 50 μM to 2 mM for 30 min at 30 °C. The reactions were stopped by adding 0.3 mL of ethanol. Then 0.2 mL of a fresh 1 mM solution of 5,5′-Dithio-Bis (2 Nitrobenzoic Acid) in 100 mM Tris buffer (pH 8.0) was added, and the yellow color product was measured at 412 nm. The values obtained were corrected for unspecific hydrolysis by subtracting the absorbance of controlled samples without addition of 2-keto acids. A molar extinction coefficient of 13,600 M−1·cm−1 was used in the final calculations.
Supplementary Material
Acknowledgments.
This work was supported in part by the University of California, Los Angeles Department of Energy Institute for Genomics and Proteomics.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0807157106/DCSupplemental.
References
- 1.Elowitz MB, Leibler S. A synthetic oscillatory network of transcriptional regulators. Nature. 2000;403:335–338. doi: 10.1038/35002125. [DOI] [PubMed] [Google Scholar]
- 2.Gardner TS, Cantor CR, Collins JJ. Construction of a genetic toggle switch in Escherichia coli. Nature. 2000;403:339–342. doi: 10.1038/35002131. [DOI] [PubMed] [Google Scholar]
- 3.Basu S, Gerchman Y, Collins CH, Arnold FH, Weiss R. A synthetic multicellular system for programmed pattern formation. Nature. 2005;434:1130–1134. doi: 10.1038/nature03461. [DOI] [PubMed] [Google Scholar]
- 4.Fung E, et al. A synthetic gene-metabolic oscillator. Nature. 2005;435:118–122. doi: 10.1038/nature03508. [DOI] [PubMed] [Google Scholar]
- 5.Gibson DG, et al. Complete chemical synthesis, assembly, and cloning of a Mycoplasma genitalium genome. Science. 2008;319:1215. doi: 10.1126/science.1151721. [DOI] [PubMed] [Google Scholar]
- 6.Wang L, Brock A, Herberich B, Schultz PG. Expanding the genetic code of Escherichia coli. Science. 2001;292:498–500. doi: 10.1126/science.1060077. [DOI] [PubMed] [Google Scholar]
- 7.Yoo TH, Link AJ, Tirrell DA. Evolution of a fluorinated green fluorescent protein. Proc Natl Acad Sci USA. 2007;104:13887–13890. doi: 10.1073/pnas.0701904104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Prescher JA, Dube DH, Bertozzi CR. Chemical remodelling of cell surfaces in living animals. Nature. 2004;430:873–877. doi: 10.1038/nature02791. [DOI] [PubMed] [Google Scholar]
- 9.Yalkowsky SH, He Y. CRC Handbook of Aqueous Solubility Data. Boca Raton, FL: CRC; 2003. [Google Scholar]
- 10.Ehrlich F. Über die Bedingungen der Fuselölbildung und über ihren Zusammenhang mit dem Eiweissaufbau der Hefe. Berichte der Deutschen Chemischen Gesellschaft. 1907;40:1027–1047. [Google Scholar]
- 11.Atsumi S, Hanai T, Liao JC. Non-fermentative pathways for synthesis of branched-chain higher alcohols as biofuels. Nature. 2008;451:86–89. doi: 10.1038/nature06450. [DOI] [PubMed] [Google Scholar]
- 12.Handel TM, Williams SA, DeGrado WF. Metal ion-dependent modulation of the dynamics of a designed protein. Science. 1993;261:879–885. doi: 10.1126/science.8346440. [DOI] [PubMed] [Google Scholar]
- 13.Atwell S, Ultsch M, De Vos AM, Wells JA. Structural plasticity in a remodeled protein–protein interface. Science. 1997;278:1125. doi: 10.1126/science.278.5340.1125. [DOI] [PubMed] [Google Scholar]
- 14.Arnold FH. Combinatorial and computational challenges for biocatalyst design. Nature. 2001;409:253–257. doi: 10.1038/35051731. [DOI] [PubMed] [Google Scholar]
- 15.Berg JM, Tymoczko JL, Stryer L. Biochemistry. 5th Ed. New York: W.H. Freeman & Co; 2002. [Google Scholar]
- 16.de la Plaza M, Fernandez de Palencia P, Pelaez C, Requena T. Biochemical and molecular characterization of alpha-ketoisovalerate decarboxylase, an enzyme involved in the formation of aldehydes from amino acids by Lactococcus lactis. FEMS Microbiol Lett. 2004;238:367–374. doi: 10.1016/j.femsle.2004.07.057. [DOI] [PubMed] [Google Scholar]
- 17.Larroy C, Rosario Fernandez M, Gonzalez E, Pares X, Biosca JA. Properties and functional significance of Saccharomyces cerevisiae ADHVI. Chem Biol Interact. 2003;143–144:229–238. doi: 10.1016/s0009-2797(02)00166-7. [DOI] [PubMed] [Google Scholar]
- 18.Shiio I, Nakamori S, Sano K. US Patent 3. 1971;580:810. [Google Scholar]
- 19.Gusyatiner MM, Lunts MG, Kozlov YI, Ivanovskaya LV, Voroshilova EB. US Patent 6. 2002;403:342. [Google Scholar]
- 20.Schutz A, et al. Crystal structure of thiamindiphosphate-dependent indolepyruvate decarboxylase from Enterobacter cloacae, an enzyme involved in the biosynthesis of the plant hormone indole-3-acetic acid. Eur J Biochem. 2003;270:2312–2321. doi: 10.1046/j.1432-1033.2003.03601.x. [DOI] [PubMed] [Google Scholar]
- 21.Dobritzsch D, Konig S, Schneider G, Lu G. High resolution crystal structure of pyruvate decarboxylase from Zymomonas mobilis. Implications for substrate activation in pyruvate decarboxylases. J Biol Chem. 1998;273:20196–20204. doi: 10.1074/jbc.273.32.20196. [DOI] [PubMed] [Google Scholar]
- 22.Siegert P, et al. Exchanging the substrate specificities of pyruvate decarboxylase from Zymomonas mobilis and benzoylformate decarboxylase from Pseudomonas putida. Protein Eng Des Sel. 2005;18:345–357. doi: 10.1093/protein/gzi035. [DOI] [PubMed] [Google Scholar]
- 23.Koon N, Squire CJ, Baker EN. Crystal structure of LeuA from Mycobacterium tuberculosis, a key enzyme in leucine biosynthesis. Proc Natl Acad Sci USA. 2004;101:8295. doi: 10.1073/pnas.0400820101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wiegel J, Schlegel HG. a-Isopropylmalate synthase from Alcaligenes eutrophus H 16. Arch Microbio. 1977;112:239–246. doi: 10.1007/BF00413087. [DOI] [PubMed] [Google Scholar]
- 25.Negishi E, Tan Z, Liang B, Novak T. An efficient and general route to reduced polypropionates via Zr-catalyzed asymmetric CC bond formation. Proc Natl Acad Sci USA. 2004;101:5782–5787. doi: 10.1073/pnas.0307514101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Causey TB, Zhou S, Shanmugam KT, Ingram LO. Engineering the metabolism of Escherichia coli W3110 for the conversion of sugar to redox-neutral and oxidized products: Homoacetate production. Proc Natl Acad Sci USA. 2003;100:825–832. doi: 10.1073/pnas.0337684100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bloom JD, et al. Evolving strategies for enzyme engineering. Curr Opin Struct Biol. 2005;15:447–452. doi: 10.1016/j.sbi.2005.06.004. [DOI] [PubMed] [Google Scholar]
- 28.Dieterich DC, Link AJ, Graumann J, Tirrell DA, Schuman EM. Selective identification of newly synthesized proteins in mammalian cells using bioorthogonal noncanonical amino acid tagging (BONCAT) Proc Natl Acad Sci USA. 2006;103:9482–9487. doi: 10.1073/pnas.0601637103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Baba T, et al. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: The Keio collection. Mol Syst Biol. 2006;2 doi: 10.1038/msb4100050. 10.1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.