Abstract
Type I IFNs were discovered as the primary antiviral cytokines and are now known to serve critical functions in host defense against bacterial pathogens. Accordingly, established mediators of IFN antiviral activity may mediate previously unrecognized antibacterial functions. RNase-L is the terminal component of an RNA decay pathway that is an important mediator of IFN-induced antiviral activity. Here, we identify a role for RNase-L in the host antibacterial response. RNase-L−/− mice exhibited a dramatic increase in mortality after challenge with Bacillus anthracis and Escherichia coli; this increased susceptibility was due to a compromised immune response resulting in increased bacterial load. Investigation of the mechanisms of RNase-L antibacterial activity indicated that RNase-L is required for the optimal induction of proinflammatory cytokines that play essential roles in host defense from bacterial pathogens. RNase-L also regulated the expression of the endolysosomal protease, cathepsin-E, and endosome-associated activities, that function to eliminate internalized bacteria and may contribute to RNase-L antimicrobial action. Our results reveal a unique role for RNase-L in the antibacterial response that is mediated through multiple mechanisms. As a regulator of fundamental components of the innate immune response, RNase-L represents a viable therapeutic target to augment host defense against diverse microbial pathogens.
Keywords: cathepsin-E; interferon; 2′, 5′-oligoadenylate; cytokine; endosome
Type 1 IFNs were discovered based on their antiviral activity; however, their role in the innate immune response to nonviral pathogens has recently gained recognition (1, 2). The elucidation of Toll-Like Receptor (TLR) and non-TLR signaling pathways, which function to detect microbial infection and activate expression of host innate immune genes, revealed that the induction of type 1 IFNs is a central component of the genetic response to both viral and bacterial pathogens (3). Viral and bacterial nucleic acids and LPS from Gram-negative bacteria activate an overlapping signal transduction pathway that converges on IFN-regulatory factor-3 (IRF3), a transcription factor that is required for IFNβ induction (4). Importantly, the induction of IFN by bacteria is required for the successful resolution of infections by a diverse profile of bacteria, demonstrating its functional role in host defense from bacterial challenge (5–7). For example, we recently reported a critical role for IFNβ in the IL-1β-dependent killing of Bacillus anthracis (BA) spores (8). Thus, a current challenge is to determine the mechanisms by which IFNs exert their antibacterial activity. The broader role for type 1 IFNs in the innate immune response to viral and bacterial pathogens suggested that common downstream effectors are involved. Specifically, established mediators of IFN antiviral action may serve previously unrecognized roles in antibacterial immunity.
RNase-L is an established mediator of IFN antiviral and antiproliferative activities that functions as the terminal component of an RNA decay pathway (9). Enzymatic activation of RNase-L requires the binding of its activator, 2′,5′-linked oligo-adenylate (2–5A, pppA(2′p5′A)n n ≥ 2), which is produced by a family of IFN-regulated 2′,5′-oligoadenylate synthetases (OAS). Activated RNase-L cleaves single-stranded viral and cellular RNAs; however, the precise molecular events linking RNase-L-mediated degradation of specific RNAs to its biologic activities remain to be determined. For example, although the degradation of viral RNAs by RNase-L is clearly an important component of its antiviral activity (10), RNase-L also mediates antimicrobial activities, such as apoptosis, that involve the modulation of host gene expression (11). Importantly, microarray analyses determined that a finite number of cellular mRNAs are regulated in an RNase-L-dependent manner, demonstrating its capacity to selectively regulate host transcripts (12) (see Table 1). Taken together, these findings suggest that RNase-L mediates its biologic activities, in part, through the regulation of specific host mRNAs. Information on the identities of these transcripts in the context of an innate immune response, and the contribution of this regulation to defense against microbial pathogens in vivo, is essential to determine the mechanisms by which RNase-L functions in host defense.
Table 1.
Microarray of B. anthracis infected C57Bl/6 and RNase-L−/− macrophages
| Gene title | Symbol | KO unt vs. WT unt | KO 8 h vs. WT 8 h | KO 8 h vs. KO unt | WT 8 h vs. WT unt | P value | KO vs. WT fold induction at 8 hpi |
|---|---|---|---|---|---|---|---|
| 1. Cathepsin E | Ctse | 18.64 | 19.56 | 1.13 | 1.07 | 0.02 | 1.05 |
| 2. Chemokine (C-X-C motif) receptor 4 | Cxcr4 | 3.48 | 4.41 | −1.78 | −2.25 | 0.03 | 1.27 |
| 3. Ral guanine nucleotide dissociation stimulator,-like 1 | Rgl1 | −2.5 | −2.4 | −1.2 | −1.3 | 0.02 | 1.06 |
| 4. IFN-induced protein with tetratricopeptide repeats 2 | Ifit2 | 1.96 | −1.12 | 145 | 317.4 | <0.001 | −2.19 |
| 5. Tumor necrosis factor | Tnf | 1.59 | −1.21 | 10.2 | 19.7 | <0.001 | −1.93 |
| 6. IL-1 beta | Il1b | 1.69 | −1.35 | 9.25 | 21.11 | <0.001 | −2.28 |
| 7. Chemokine (C-X-C motif) ligand 1 | Cxcl1 | 1.57 | −1.25 | 5.21 | 10.2 | <0.001 | −1.96 |
| 8. CD86 antigen | Cd86 | 1.17 | −1.62 | 1.04 | 1.99 | <0.001 | −1.91 |
| 9. Chemokine (C-C motif) ligand 7 | Ccl7 | 1.68 | −1.16 | 2.39 | 4.66 | 0.009 | −1.95 |
| 10. CD55 antigen | Cd55 | 1.58 | −1.24 | −1.64 | 1.2 | 0.033 | −1.96 |
Microarray analysis was performed on untreated (unt) RNase-L−/− (KO) and WT macrophages and after BA infection for 8 h. Values in columns 3–6 are the N-fold change in the average of 3 log2 signals between KO and WT samples from the indicated conditions; positive and negative values indicate increased and decreased expression respectively in KO as compared with WT macrophages. P values were calculated by ANOVA and corrected for multiple comparisons to control ″false discovery rate″; P values shown for rows 1–3 are for the untreated KO vs. WT comparison, and P values for rows 4–10 are for the N-fold induction comparison (column 8). The data were filtered to identify mRNAs for which the fold change between KO and WT samples in identical conditions was +/− 2.0 (rows 1–3, columns 3 and 4). The data were further filtered to identify mRNAs for which the N-fold change in gene induction (i.e., expression at 8 hpi vs. untreated for WT as compared with 8 hpi vs. untreated for KO) was +/− 1.9; mRNAs that met this criterion and encode proteins with established immune functions are shown in column 8, rows 4–10; other mRNAs that exhibited differential induction are listed in Table S1.
In light of work from our group and others indicating a critical role for type 1 IFNs in host defense from bacterial challenge, and the established role of RNase-L as a mediator of IFN action, we investigated whether RNase-L functioned in antibacterial activity against the Gram-positive and -negative bacteria, BA and Escherichia coli. Remarkably, RNase-L−/− mice exhibited a dramatic increase in mortality after infection with both bacteria that reflected a compromised immune response and increased bacterial load. Induction of the proinflammatory cytokines, IL-1β, TNFα, and IFNβ, which are essential for defense from BA (8) and E. coli (13), was diminished after bacterial infection of RNase-L−/− macrophages suggesting that the antibacterial action of RNase-L is mediated, in part, through the regulation of cytokine induction. RNase-L−/− mice also displayed a delay in endotoxin-induced lethality, providing further evidence for the physiologic relevance of the impaired cytokine induction phenotype. In addition, we identified an unexpected impact of RNase-L on the expression of the mRNA encoding the endolysosomal protease, cathepsin-E (CatE) and on associated endosomal activities including the elimination of phagocytic vacuoles and bactericidal killing. Lysosomal targeting via endosomal pathways is a fundamental mechanism for the elimination of microbial pathogens; accordingly, the RNase-L-dependent modulation of this activity represents a mechanism by which RNase-L may elicit its antimicrobial activity. Taken together, our studies identify a previously uncharacterized role for RNase-L in antibacterial immunity that is mediated, in part, through its regulation of cytokine induction and endolysosomal functions, two critical components of the host antibacterial response.
Results and Discussion
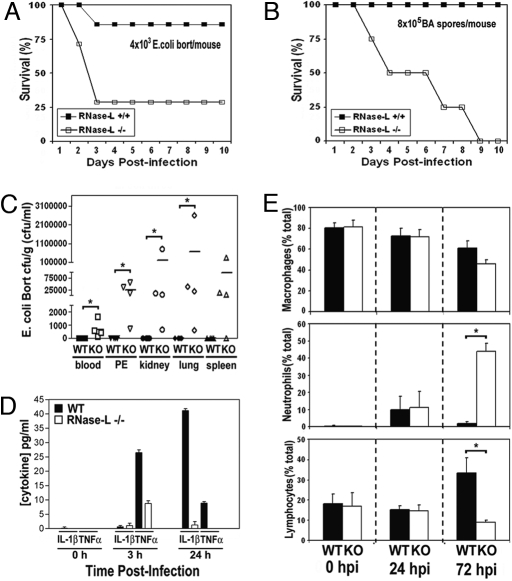
IFN is a critical component of host defense from BA (8) and E. coli (6); therefore, we investigated the role of RNase-L, a key effector of IFN action, in antibacterial immunity against these pathogens. RNase-L−/− and WT C57BL/6 mice were injected i.p. with Gram-positive BA Stern 34F2 spores, or Gram-negative E. coli O18:K1:H7 Bort strain and monitored for signs of infection and survival. RNase-L−/− mice exhibited a dramatic increase in mortality after infection with either of the two bacteria, as compared with WT mice (Fig. 1 A and B). Whereas the WT mice did not succumb to BA infection during the course of the experiment, 50% of the BA-infected RNase-L−/− mice died by 4 days, and 100% died by day 9. RNase-L−/− mice exhibited a similarly enhanced susceptibility to E. coli challenge. These findings identify an essential and previously unrecognized role for RNase-L in the host immune response to bacterial pathogens.
Fig. 1.
The immune response to bacterial challenge is compromised in RNase-L−/− mice. (A and B) RNase-L−/− and WT mice (7 mice per group) were challenged with E. coli O18:K1:H7 Bort or BA at the indicated doses, and Kaplan–Meier survival analyses are shown (34). (C) Mice were inoculated with 2.5 × 103 cfu E. coli, and bacterial titers in organs (cfu/g), and peritoneal exudates (PE) and blood (cfu/mL) were determined at 72 hpi. Lines indicate mean values, and * denotes P < 0.05. (D) Plasma IL-1β and TNFα levels were measured by ELISA at the indicated times after E. coli infection; each value is the average for 3 mice; error bars are standard deviation (s.d.). (E) Macrophages, neutrophils, and lymphocytes in peritoneal fluid at the indicated hours after E. coli infection are expressed as the average of the percentage of a given cell type in ≥400 total cells counted per mouse from 4 independent mice +/− s.d. *, P < 0.05.
To identify differences between RNase-L−/− and WT mice in their immune responses to E. coli and BA infections, the bacterial load, induction of proinflammatory cytokines, and immune cell profile of peritoneal infiltrates were measured after infection. The bacterial burden in liver, lung, kidney, spleen, blood, and peritoneal fluid was small at early time points after E. coli infection and did not dramatically differ between RNase-L−/− and WT mice; however, by 72 h post infection (hpi), RNase-L−/− mice were unable to control the infection as indicated by elevated bacterial loads (Fig. 1C). The increased bacterial load corresponded with dramatically diminished circulating levels of plasma IL-1β and TNFα (Fig. 1D). Similarly, BA was increased in tissues from RNase-L−/− mice and the expression of IL-1β and TNFα were decreased (Fig. S1A and Fig. 2A). Quantification of peritoneal immune cell infiltrates revealed further striking differences between RNase-L−/− and WT mice. In the absence of infection, there were no differences in the numbers of macrophages, neutrophils, or lymphocytes; however, the impaired capacity of RNase-L−/− mice to clear the infection corresponded with a dramatic recruitment of neutrophils, and a reduced influx of lymphocytes as compared with WT mice at 72 hpi (Fig. 1E). An enhanced recruitment of neutrophils was also observed after BA infection (Fig. S1B) but was not effective in resolving the infection, suggesting that the activity of host phagocytes may be impaired (see Fig. 3 D and E). Taken together, these findings indicate that multiple components of the host immune response are compromised in RNase-L−/− mice, and that the impaired capacity of RNase-L−/− mice to clear a bacterial infection, rather than the overproduction of host cytokines, is responsible for the increased mortality observed.
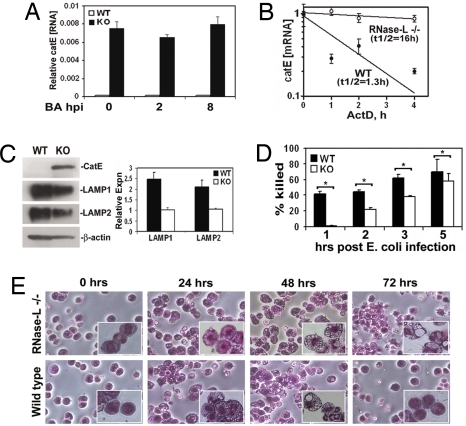
Fig. 3.
Elevated expression of CatE is associated with impaired phagosome clearance and bactericidal activity in RNase-L−/− macrophages. (A) Steady-state expression of CatE mRNA in macrophages +/− BA infection was determined by qRTPCR; values are normalized to constitutively expressed GAPDH mRNA. (B) Decay of CatE mRNA was quantified by qRTPCR after transcriptional arrest in WT and RNase-L−/− macrophages. (C) LAMP 1/2, CatE, and α-actin proteins in WT and RNase-L−/− macrophages were measured by Western blot. Right shows densitometric quantitation of LAMP1/2 signals normalized to α-actin; blot shown is representative of 3 independent experiments. (D) Peritoneal macrophages from WT and RNase-L−/− mice were infected with E. coli for the indicated times, and the percentage reduction in viable bacteria as compared with the 1 hour value is shown (percentage killing); macrophages from 4 separate mice were analyzed in 2 independent experiments; the mean percentage killing +/− standard error is shown. *, P < 0.05. (E) At the indicated times after infection with E. coli (2.5 × 103 cfu), cells in the peritoneal fluid were isolated and stained; representative fields are shown at ×200 and ×400 (Inset).
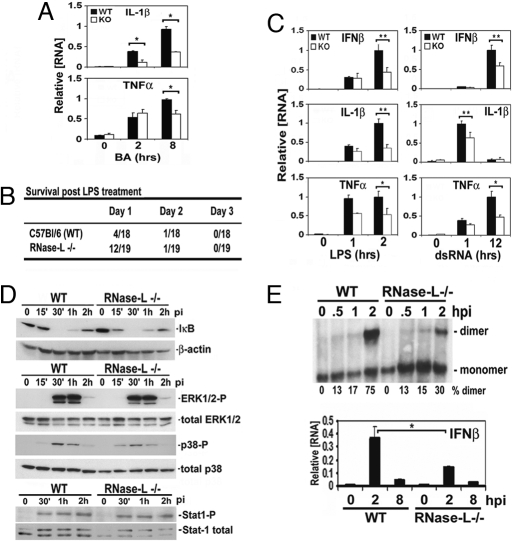
The diminished induction of cytokine expression and altered recruitment of immune cells after E. coli infection of RNase-L−/− mice suggested that the antibacterial activity of RNase-L may occur through the modulation of host gene expression. To identify host genes that are regulated in an RNase-L-dependent manner, we performed microarray analysis on WT and RNase-L−/− macrophages after BA infection. Macrophages were chosen for this study, because they represent key components of the innate immune response to the BA spore and K1-encapsulated E. coli models used here (7, 14). The induction of host antimicrobial genes is an important component of the innate immune response to bacterial infection; therefore, we sought to identify RNase-L-dependent changes in gene induction that may contribute to its antibacterial activity. The microarray data were filtered to detect mRNAs for which the induction differed by +/−1.9-fold (Tables 1, rows 4–10, and Table S1). This cutoff was chosen based on the differential expression of TNFα that was validated for WT and RNase-L−/− macrophages (Figs. 1D and 2A). Seventeen transcripts met this criterion, 7 of which encoded proteins with established immune functions. The BA-stimulated expression of all 7 transcripts was reduced in RNase-L−/− as compared with WT macrophages and may contribute to the compromised immune response in RNase-L−/− mice. In agreement with the reduced levels of plasma IL-1β, and TNFα protein after E. coli challenge of RNase-L−/− mice (Fig. 1D), the induction of these mRNAs was diminished in BA-infected RNase-L−/− macrophages (Fig. 2A). In addition, basal- and bacteria-induced chemokine or chemokine receptor expression was altered in RNase-L−/− macrophages (Table 1), consistent with the altered recruitment of immune cells observed in RNase-L−/− mice (Figs. 1E and S1B). Thus, cytokine and chemokine induction was altered in an RNase-L-dependent manner after infection with both E. coli and BA, suggesting that this regulation is a fundamental component of the host response to diverse microbial stimuli. Consistent with this view, cytokine induction by the TLR agonists LPS, dsRNA, and CpG was impaired in RNase-L−/− as compared with WT macrophages at 1 or more of the time points tested (Figs. 2C and S2). Importantly, the diminished induction of proinflammatory cytokines in RNase-L−/− mice corresponded with a marked delay in endotoxin-induced lethality, demonstrating the physiologic relevance of this phenotype (Fig. 2B). Thus, the compromised cytokine response in RNase-L−/− mice is associated with increased susceptibility to infection with a live bacterium (E. coli) but is protective from the cytokine storm that contributes to the pathologic activity of LPS.
Fig. 2.
Bacteria- and TLR agonist-induced signaling and gene expression are diminished in RNase-L−/− macrophages. (A) qRTPCR analysis of IL-1β and TNFα mRNA expression after BA infection for the indicated times. (B) LPS-induced mortality is delayed in RNase-L−/− after i.p. injection with 35 mg/kg LPS; P < 0.05. (C) cytokine expression after treatment of WT and RNase-L−/− macrophages with the TLR3 and 4 agonists, dsRNA (25 μg/mL) and LPS (100 ng/mL), respectively, was measured by qRTPCR. (D and E). ERK1/2 and Stat1 phosphorylation, the degradation of IκB, and dimerization of IRF-3 protein were measured after E. coli infection. Experiments were conducted in triplicate; IRF-3 dimers were quantified by densitometry. Induction of IFNβ mRNA (E Lower) was measured by qRTPCR after E. coli infection. For all bar graphs, error bars are s.d. *, P < 0.05; **, P < 0.001.
To determine whether the diminished cytokine induction in RNase-L−/− macrophages reflected a defect in signal transduction, the status of signaling proteins that are activated in response to E. coli infection was measured in RNase-L−/− and WT macrophages (3). Specifically, phosphorylation of Stat1, p38, and ERK, and degradation of IκB were comparable in RNase-L−/− and WT macrophages (Fig. 2D). In contrast, the dimerization of IRF-3, an essential transcription factor for the induction of proinflammatory cytokines by microbial stimuli (4), was reduced by 2-fold after E. coli infection of RNase-L−/− macrophages (Fig. 2E). Consistent with this impaired activation of IRF-3, the induction of IFNβ, a primary target of IRF-3 signaling, was diminished in RNase-L−/− macrophages (Fig. 2E Lower). Thus, RNase-L is required for the optimal activation of IRF-3, and the diminished induction of IRF-3 target genes may account, in part, for the increased susceptibility of RNase-L−/− mice to bacterial challenge.
In addition to its impact on the induction of immune response genes, the RNase-L-dependent regulation of basal gene expression may contribute to its antibacterial activity. Indeed, our microarray analysis of >34,000 unique mRNAs identified just 3 mRNAs encoding proteins of known function that exhibited differential expression in RNase-L−/− as compared with WT macrophages when filtered for a +/− 2-fold change (P ≤ 0.05; rows 1–3 in Table 1; also see Table S2). Among these transcripts, the mRNA encoding CatE stood out as the transcript that exhibited the most dramatic (≈20-fold) RNase-L-dependent regulation. CatE mRNA was increased in RNase-L−/− macrophages suggesting that it is a direct target of RNase-L regulation. Accordingly, we focused on examining the RNase-L-dependent regulation of CatE, and its potential role in antibacterial activity. The microarray data were validated for CatE mRNA and protein and confirmed that its up-regulation in RNase-L−/− macrophages was independent of bacterial infection (Fig. 3 A and C). The increase in steady-state CatE mRNA corresponded with a dramatic 12-fold increase in the CatE mRNA half-life in RNase-L−/− as compared with WT macrophages (Fig. 3B). In contrast, the half-lives of other cellular mRNAs encoding unstable (TNFα, IL-1β, c-fos) and stable (TLR3) mRNAs did not markedly differ (Table S3). Thus, CatE mRNA is targeted for degradation in an RNase-L-dependent manner, suggesting that it represents a host RNase-L substrate.
CatE is an aspartic proteinase that mediates multiple immune functions as a component of the endolysosomal pathway (15–17); accordingly, CatE-mediated functions may contribute to the antibacterial activity of RNase-L. The lysosome-associated membrane proteins LAMP 1 and 2 (LAMP 1/2) are increased in CatE−/− mice, indicating these proteins are regulated by CatE (15). LAMP1/2 are required for the terminal step in phagosome maturation in which late endosomes fuse with lysosomes to eliminate phagocytosed microbial cargo (18, 19). As expected for CatE-regulated proteins, the expression of LAMP 1/2 was decreased in RNase-L−/− macrophages that overexpress endogenous CatE (Fig. 3C); therefore, phagosome clearance may be impaired in these cells. Consistent with this idea, macrophages in peritoneal exudates of E. coli-infected RNase-L−/− mice accumulated phagocytic vacuoles and retained an activated morphology at postinfection times when WT macrophages had eliminated these vacuoles and returned to a quiescent state (see the 72 hpi in Figs. 3E and S1C for BA infection). In addition, CatE colocalized with BA in infected macrophages, suggesting it may impact antibacterial activity by modulating the endocytic trafficking of internalized spores (Fig. S3). Phagosome maturation is essential for macrophage antimicrobial functions; therefore, the impaired clearance of phagocytic vacuoles in RNase-L−/− macrophages may result in reduced bactericidal activity. Indeed, E. coli and BA spore killing was reduced in RNase-L−/− as compared with WT macrophages at early times postinfection (Figs. 3D and S1D). At later times after infection, bactericidal activity was comparable in RNase-L−/− and WT macrophages; however, this activity was without apparent effect on in vivo outcome, suggesting the early reduction in bacterial clearance had a profound impact on resolution of the infection in mice. Thus, in our murine infection model, multiple components, including bactericidal activity, cytokine action, and immune cell regulators, contribute to the antibacterial activity of RNase-L.
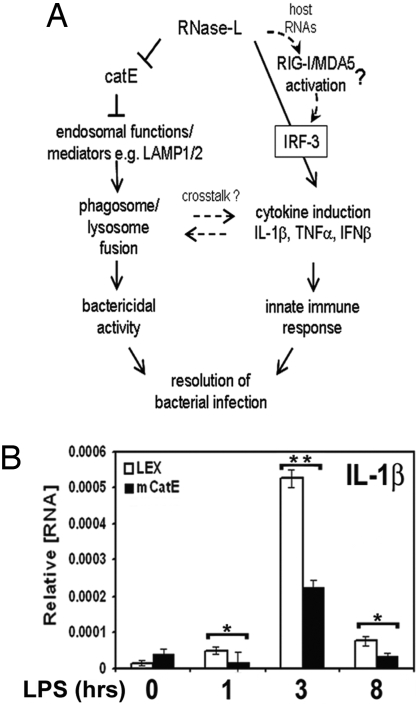
Our findings identify an essential role for RNase-L in antibacterial immunity and support a model in which this activity involves at least two mechanisms (Fig. 4A). First, RNase-L is required for the optimal induction of proinflammatory cytokines. Second, RNase-L regulates CatE, and associated endolysosomal functions, that in turn are required for the elimination of phagocytosed bacteria. Consistent with the role of RNase-L as an upstream regulator of IFN β and IL-1β as a mechanism of its antibacterial activity, we recently reported that IFNβ is essential for the IL-1β-dependent killing of BA spores (8), and the roles of these cytokines in defense from E. coli are well established (3, 6). The reciprocal regulation of immune signaling/cytokine induction and endolysosomal pathways was recently demonstrated, providing a link between these key components of the innate immune response (20–22). In light of this cross-talk, and to examine the potential role of CatE in RNase-L-regulated cytokine induction, we determined whether CatE overexpression alone, independent of other RNase-L-mediated changes in gene expression, could mimic any components of the RNase-L−/− phenotype. Remarkably, similar to RNase-L−/− macrophages, induction of IL-1β was diminished after LPS treatment of CatE-transduced, as compared with vector control, RAW264.7 macrophages, thus linking CatE regulation by RNase-L to its impact on cytokine induction (Fig. 4B). However, ectopic expression of CatE did not alter LPS induction of TNFα, suggesting that RNase-L-mediated activities independent of CatE regulation are required (data not shown). An alternative mechanism that may contribute to the compromised cytokine induction in RNase-L−/− macrophages involves a recently reported role for RNase-L in the activation of the cytosolic nucleic acid sensors, RIG-I and MDA5 (23). Specifically, RNase-L activation resulted in the generation of small host RNAs that activate RIG-I/MDA5 signaling to augment IFN-β induction by viruses (24). In the context of bacterial challenge, we demonstrated that IRF-3 activation and the induction of IL-1β, IFNβ, and TNFα, key downstream effectors of RIG-I/MDA5 signaling (23, 25), were impaired in RNase-L−/− macrophages (Fig. 4). Thus, the RNase-L-dependent regulation of bacteria-induced cytokine expression may also occur through the production of RIG-I/MDA5-activating RNAs (Fig. 4A). Furthermore, the RNase-L-dependent regulation of IFNβ (Fig. 2 and ref. 24) and the IFN-mediated regulation of RNase-L (26) may comprise a positive feedback loop to amplify the IFN response. Finally, RNase-L may impact the host immune response through additional mechanisms, as suggested by the RNase-L-dependent regulation of other immune mediators (e.g., chemokines); the specific roles of these mediators in RNase-L action remain to be determined.
Fig. 4.
RNase-L-dependent regulation of CatE and cytokine induction as mechanisms of its antibacterial action. (A) A model depicting a role for RNase-L in the host antibacterial response through modulation of cytokine induction and lysosome targeting pathways. (B) RAW264.7 macrophages were stably transfected with CatE or vector control to mimic the elevated expression of endogenous CatE in RNase-L−/− macrophages, and IL1β induction was measured by qRTPCR after LPS stimulation; expression of transduced CatE is shown in Fig. S4.
All established biological activities of RNase-L require its activation by 2–5A, the synthesis of which is mediated by dsRNA-dependent OAS enzymes. dsRNA is produced in the course of most viral infections; however, the source of dsRNA in virus-independent activities of RNase-L remains to be determined. We did not observe rRNA cleavage, an indicator of RNase-L activation, after bacterial challenge (Fig. S5), suggesting that low levels of 2–5A, possibly produced in a localized manner (27), are sufficient for RNase-L activity. The dsRNA required to produce 2–5A may be derived from pathogen or host RNAs; indeed, certain host mRNAs contain secondary structure that is capable of activating OAS (28). In addition, 2–5A linkages have been reported in bacteria (29) and RNase-L expression in bacteria results in cell RNA degradation and antiproliferative activity (30), suggesting that bacteria may produce endogenous activators of RNase-L. The validation of specific RNase-L substrates will provide a sensitive readout of its activity and a more physiologic system in which to dissect the source(s) of RNase-L activators in cells.
Taken together, our findings indicate that RNase-L mediates fundamental components of the host innate immune response; therefore, it is likely that RNase-L functions in the defense against diverse microbial pathogens. The targeted modulation of RNase-L expression or activity may thus provide an approach to augment antimicrobial therapies.
Materials and Methods
For additional information on Materials and methods, please see SI Materials and Methods.
Bacterial Infections and LPS Treatment.
All experiments used 6- to 8-week-old RNase-L−/− mice on a C57BL/6 background (31) and WT C57BL/6 mice. Mice were housed in the University of Maryland, Baltimore, animal facility according to IACUC-approved protocols. For survival studies, mice (7 of each genotype) were injected i.p. with E. coli O18:K1:H7 Bort strain or BA Sterne 34F2 strain at the indicated doses, and monitored twice daily. In the LPS study, mice were injected i.p. with 35 mg/kg LPS (E. coli O111:B4; Sigma) in 3 independent experiments of 4, 5, and 10 mice per group, respectively. For analyses of in vivo infections, mice (4 per group) were i.p. injected with indicated doses of E. coli or BA spores. Both male and female mice were used for the in vivo analyses, and no sex-specific differences in the responses to infection or TLR agonists were detected. Mice were killed at the indicated times postinfection, and blood, peritoneal exudate, and organs (liver, lung, spleen, and kidney) were harvested for cytokine, bacterial load, and cell population analysis.
Measurement of Cytokines, Bacterial Load, and Immune Infiltrates.
Cytokines were analyzed by ELISA from blood serum (University of Maryland, Baltimore, cytokine core) and bacterial counts were obtained by culturing dilutions of blood, peritoneal exudate, and organ lysates on LB agar plates. Peritoneal exudates were adhered to slides and stained by using the Diff-Quick Stain Set (Dade Behring).
Preparation, Culture, and Treatment of Peritoneal Macrophages.
Mice were i.p. injected with 3 mL of 3% thioglycollate (Rennel). 3–4 days after injection, elicited macrophages were harvested by peritoneal lavage with PBS. Macrophages were maintained in RPMI containing 10% FBS, 1× antibiotic/antimycotic, and plasmocin (Invivogen). Sources for TLR agonists are as follows: LPS (E. coli O111:B4; Sigma), dsRNA (polyriboinosinic:polyribocytidylic acid; GE Healthcare), and CpG DNA (TCCATGACGTTCCT GACGTT, Coley Pharmaceutical Group).
Bactericidal Assay.
The assay for BA was conducted as described (7) and modified for E. coli as follows: 106 macrophages were adhered to culture dishes for 18 h then infected with 4 × 107 cfu of bacteria for 30 min. Gentamycin was then added for 30 min to kill residual bacteria, and cells were lysed immediately (time 0) and at the indicated times, and viable bacteria were quantified by culturing on LB agar plates. Bactericidal activity was calculated as the percentage reduction in viable bacteria as compared with the zero time point (percentage killing).
Microarray Analysis.
Thioglycollate-elicited macrophages (2 × 107) pooled from 2 mice per sample (three independent samples/time point) were infected with BA spores at MOI of 1 and harvested after 0 or 8 h. Total RNA was prepared by using TRIzol (Invitrogen) and further purified with Qiagen RNAeasy kit; microarray analysis used the Affymetrix Mouse Genome 430 2.0 array. Microarray data analysis was performed by using the Array Data Analysis and Management System (ADAMS) that uses publicly available tools for analysis of the data. Briefly, raw probe intensities were normalized and summarized by the gcRMA algorithm (32) using the R statistical environment (www.Rproject.org). Genes that were called “absent” in all samples by the Affymetrix MAS5 algorithm were removed from further analysis. Significance analysis was done by using ANOVA and linear modeling of microarrays by using the package Limma. P values obtained were corrected for multiple comparisons by using Benjamini and Hochberg false discovery-rate test. Additionally, absolute ratios (N-folds) were calculated for each comparison. The complete microarray dataset can be viewed at http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?token=pfyvxkyccmsxqxg&acc=GSE13530.
Analysis of mRNA Expression and Stability.
Total cellular RNA was isolated by using TRIzol reagent (Invitrogen) as directed. To analyze the mRNA half-life, macrophages were treated with 5 μg/ml actinomycin D (Sigma), and total RNA was harvested at the time points indicated. Specific mRNAs were quantified by reverse transcription/real time PCR (qRTPCR). For qRTPCR analysis, 2 μg of DNase1-treated total RNA was used for reverse transcription to synthesize first strand cDNA by using SuperScriptaseII RT (Invitrogen). Aliquots of the first-strand cDNA were used in qRTPCRs by using SYBR green as directed by the manufacturer (Superarray). For all primer sets, reactions were first conducted by using a temperature gradient to optimize annealing conditions, and the absence of primer-dimers was confirmed by melting condition analysis. RT reactions carried out in the absence of RT served as a negative control, and to detect PCR products generated from contaminating genomic DNA. All PCR primers are shown in Table S4. Each analysis represents at least 2 independent experiments, and within an experiment, all reactions were performed in triplicate, and the Ct values were converted to RNA concentration by using a standard curve. mRNA values from each time point were normalized to the constitutively expressed GAPDH mRNA. Values were graphed on semilog axes, and first-order turnover rates were calculated by nonlinear regression of the percentage of mRNA remaining as a function of time after actinomycin-D treatment.
Measurement of Protein Expression.
Cell lysates were prepared and analyzed by Western blot as described (20 μg of protein per lane) (33). The phosphor- or total p38 MAPK, ERK, JNK, IRF3, and IκB antibodies (Cell Signaling) were used at a 1:1,000 dilution. Anti-murine CatE (R&D Systems) antibody was used at 2 μg/ml, and the LAMP 1/2 antibodies (Developmental Studies Hybridoma Bank, University of Iowa) were used at 1:50, and 1:20 dilutions, respectively. Signals were quantified by using the Quantity One software from BioRad.
Lentiviral Transduction.
Murine RAW264.7 cells were transduced with mCatE, and stable transductants were selected according to the manufacturer's protocol (Open Biosystems). Briefly, HIV-1-based lentiviruses packaging the pLEX-mCatE, or pLEX vector were generated by using a TLA-HEK293T-based transfection system. All vectors were purchased from Open Biosystems. TransLentiviral Packaging Mix (28.5 μg) and 9 μg of pLEX vector were transfected into a 100-mm plate of TLA-HEK293T cells by using Arrest-In transfection reagent (Open Biosystems). Cells were incubated in 12 mL of DMEM containing 10% FBS, 100 units/mL of penicillin, and 100 μg/mL of streptomycin. After 48 h, virus-containing medium was collected and replaced with fresh medium. The collected medium was stored at +4 °C. Seventy-two hours after transfection, virus-containing medium was collected again and combined with the previous harvest. The cell medium was centrifuged at 1,000 × g for 20 min at +4 °C and then filtered through a 0.45-μm filter to remove cell debris. The lentiviral supernatant was then concentrated through ultracentrifugation at 95,000 × g for 90 min at +4 °C. The resulting pellet was resuspended in RPMI, aliquoted, and stored at −80 °C.
For transduction, RAW264.7 cells were infected with lentivirus at a MOI of 1 in 1 mL of serum-free RPMI. After at least 4 h of infection, 9 mL of serum-containing RPMI was added; 48 h after transduction, 1 μg/mL of puromycin was added to select for stable transductants.
Statistical Analysis.
Statistical significance for E. coli and BA counts isolated from organs, blood, or peritoneal exudate (PE) was calculated by using the Wilcoxon rank sum test by the University of Maryland Marlene and Stewart Greenebaum Cancer Center Biostatistics Core. P values for the bactericidal and gene expression analyses were determined by the Student's t test or the χ2 test (LPS toxicity).
Supplementary Material
Acknowledgments.
We thank Simeon Goldblum, Martin Flajnik, and Subhendu Basu (University of Maryland, Baltimore) for discussions of the manuscript and Clive Evans (Virginia Bioinformatics Institute) for RNA sample analysis and assistance with microarray analysis. We further acknowledge Dr. Robert Silverman (Cleveland Clinic Foundation) for providing the RNase-L−/− mice and Drs. Jian Ying Wang and Juren He (Baltimore Veterans Affairs Medical Center) for technical assistance with microscopy. Generation and analysis of Gene Expression data was funded through National Institutes of Health Cooperative Agreement number 1 U54 Al057168-01 to A. Cross and O. Crasta via subcontract from the University of Maryland, Baltimore, M. Levine, PI.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0807265105/DCSupplemental.
References
- 1.Stetson DB, Medzhitov R. Type I interferons in host defense. Immunity. 2006;25:373–381. doi: 10.1016/j.immuni.2006.08.007. [DOI] [PubMed] [Google Scholar]
- 2.Decker T, Muller M, Stockinger S. The yin and yang of type I interferon activity in bacterial infection. Nat Rev. 2005;5:675–687. doi: 10.1038/nri1684. [DOI] [PubMed] [Google Scholar]
- 3.Uematsu S, Akira S. Toll-like receptors and innate immunity. J Mol Med. 2006;84:712–725. doi: 10.1007/s00109-006-0084-y. [DOI] [PubMed] [Google Scholar]
- 4.Hiscott J. Triggering the innate antiviral response through IRF-3 activation. J Biol Chem. 2007;282:15325–15329. doi: 10.1074/jbc.R700002200. [DOI] [PubMed] [Google Scholar]
- 5.Karaghiosoff M, et al. Central role for type I interferons and Tyk2 in lipopolysaccharide-induced endotoxin shock. Nat Immunol. 2003;4:471–477. doi: 10.1038/ni910. [DOI] [PubMed] [Google Scholar]
- 6.Mancuso G, et al. Type I IFN signaling is crucial for host resistance against different species of pathogenic bacteria. J Immunol. 2007;178:3126–3133. doi: 10.4049/jimmunol.178.5.3126. [DOI] [PubMed] [Google Scholar]
- 7.Kang TJ, et al. Murine macrophages kill the vegetative form of Bacillus anthracis. Infection Immun. 2005;73:7495–7501. doi: 10.1128/IAI.73.11.7495-7501.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kang TJ, et al. Bacillus anthracis spores and lethal toxin induce IL-1beta via functionally distinct signaling pathways. Eur J Immunol. 2008;38:1574–1584. doi: 10.1002/eji.200838141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Silverman RH. A scientific journey through the 2–5A/RNase L system. Cytokine Growth Factor Rev. 2007;18:381–388. doi: 10.1016/j.cytogfr.2007.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li XL, Blackford JA, Hassel BA. RNase L mediates the antiviral effect of interferon through a selective reduction in viral RNA during encephalomyocarditis virus infection. J Virol. 1998;72:2752–2759. doi: 10.1128/jvi.72.4.2752-2759.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Castelli JC, et al. A study of the interferon antiviral mechanism: Apoptosis activation by the 2–5A system. J Exp Med. 1997;186:967–972. doi: 10.1084/jem.186.6.967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Malathi K, et al. A transcriptional signaling pathway in the IFN system mediated by 2′-5′-oligoadenylate activation of RNase L. Proc Natl Acad Sci USA. 2005;102:14533–14538. doi: 10.1073/pnas.0507551102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cross AS, et al. Pretreatment with recombinant murine tumor necrosis factor alpha/cachectin and murine interleukin 1 alpha protects mice from lethal bacterial infection. J Exp Med. 1989;169:2021–2027. doi: 10.1084/jem.169.6.2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cross A, et al. The importance of a lipopolysaccharide-initiated, cytokine-mediated host defense mechanism in mice against extraintestinally invasive Escherichia coli. J Clin Invest. 1995;96:676–686. doi: 10.1172/JCI118110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yanagawa M., et al. Cathepsin E deficiency induces a novel form of lysosomal storage disorder showing the accumulation of lysosomal membrane sialoglycoproteins and the elevation of lysosomal pH in macrophages. J Biol Chem. 2007;282:1851–1862. doi: 10.1074/jbc.M604143200. [DOI] [PubMed] [Google Scholar]
- 16.Chain BM, et al. The expression and function of cathepsin E in dendritic cells. J Immunol. 2005;174:1791–1800. doi: 10.4049/jimmunol.174.4.1791. [DOI] [PubMed] [Google Scholar]
- 17.Zaidi N, Kalbacher H. Cathepsin E: A mini review. Biochem Biophys Res Commun. 2008;367:517–522. doi: 10.1016/j.bbrc.2007.12.163. [DOI] [PubMed] [Google Scholar]
- 18.Eskelinen EL. Roles of LAMP-1 and LAMP-2 in lysosome biogenesis and autophagy. Mol Aspects Med. 2006;27:495–502. doi: 10.1016/j.mam.2006.08.005. [DOI] [PubMed] [Google Scholar]
- 19.Huynh KK, et al. LAMP proteins are required for fusion of lysosomes with phagosomes. EMBO J. 2007;26:313–324. doi: 10.1038/sj.emboj.7601511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Blander JM, Medzhitov R. Regulation of phagosome maturation by signals from toll-like receptors. Science. 2004;304:1014–1018. doi: 10.1126/science.1096158. [DOI] [PubMed] [Google Scholar]
- 21.Husebye H, et al. Endocytic pathways regulate Toll-like receptor 4 signaling and link innate and adaptive immunity. EMBO J. 2006;25:683–692. doi: 10.1038/sj.emboj.7600991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sanjuan MA, et al. Toll-like receptor signalling in macrophages links the autophagy pathway to phagocytosis. Nature. 2007;450:1253–1257. doi: 10.1038/nature06421. [DOI] [PubMed] [Google Scholar]
- 23.Yoneyama M, Fujita T. Function of RIG-I-like receptors in antiviral innate immunity. J Biol Chem. 2007;282:15315–15318. doi: 10.1074/jbc.R700007200. [DOI] [PubMed] [Google Scholar]
- 24.Malathi K., Dong B, Gale M, Jr, Silverman RH. Small self-RNA generated by RNase L amplifies antiviral innate immunity. Nature. 2007;448:816–819. doi: 10.1038/nature06042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gitlin L, et al. Essential role of mda-5 in type I IFN responses to polyriboinosinic:polyribocytidylic acid and encephalomyocarditis picornavirus. Proc Natl Acad Sci USA. 2006;103:8459–8464. doi: 10.1073/pnas.0603082103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhou A, Molinaro RJ, Malathi K, Silverman RH. Mapping of the human RNASEL promoter and expression in cancer and normal cells. J Interferon Cytokine Res. 2005;25:595–603. doi: 10.1089/jir.2005.25.595. [DOI] [PubMed] [Google Scholar]
- 27.Nilsen TW, Baglioni C. Mechanism for discrimination between viral and host mRNA in interferon-treated cells. Proc Natl Acad Sci USA. 1979;76:2600–2604. doi: 10.1073/pnas.76.6.2600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Molinaro RJ, et al. Selection and cloning of poly(rC)-binding protein 2 and Raf kinase inhibitor protein RNA activators of 2′,5′-oligoadenylate synthetase from prostate cancer cells. Nucleic Acids Res. 2006;34:6684–6695. doi: 10.1093/nar/gkl968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Trujillo MA, et al. The occurrence of 2′-5′ oligoadenylates in Escherichia coli. Eur J Biochem. 1987;169:167–173. doi: 10.1111/j.1432-1033.1987.tb13594.x. [DOI] [PubMed] [Google Scholar]
- 30.Pandey M, Rath PC. Expression of interferon-inducible recombinant human RNase L causes RNA degradation and inhibition of cell growth in Escherichia coli. Biochem Biophys Res Commun. 2004;317:586–597. doi: 10.1016/j.bbrc.2004.03.083. [DOI] [PubMed] [Google Scholar]
- 31.Zhou A, et al. Interferon action and apoptosis are defective in mice devoid of 2′,5′-oligoadenylate-dependent RNase L. EMBO J. 1997;16:6355–6363. doi: 10.1093/emboj/16.21.6355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wu Z, Irizarry R, Gentleman R, Martinez M, Spencer F. A model-based background adjustment for oligonucleotide expression arrays. J Am Stat Assoc. 2004;99:909–917. [Google Scholar]
- 33.Li XL, Andersen JB, Ezelle HJ, Wilson GM, Hassel BA. Post-transcriptional regulation of RNase-L expression is mediated by the 3′-untranslated region of its mRNA. J Biol Chem. 2007;282:7950–7960. doi: 10.1074/jbc.M607939200. [DOI] [PubMed] [Google Scholar]
- 34.Kaplan EL, Meier P. Nonparametric estimation from incomplete observations. J Am Stat Assoc. 1958;53:457–481. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.