Abstract
Mutations in oncogenes and tumor suppressor genes are responsible for tumorigenesis and represent favored therapeutic targets in oncology. We exploited homologous recombination to knock-in individual cancer mutations in the genome of nontransformed human cells. Sequential introduction of multiple mutations was also achieved, demonstrating the potential of this strategy to construct tumor progression models. Knock-in cells displayed allele-specific activation of signaling pathways and mutation-specific phenotypes different from those obtainable by ectopic oncogene expression. Profiling of a library of pharmacological agents on the mutated cells showed striking sensitivity or resistance phenotypes to pathway-targeted drugs, often matching those of tumor cells carrying equivalent cancer mutations. Thus, knock-in of single or multiple cancer alleles provides a pharmacogenomic platform for the rational design of targeted therapies.
Keywords: cancer mutation, oncogene addiction, pharmacogenomic, targeted therapies, tumor progression model
The construction of model systems that accurately recapitulate the genetic alterations present in human cancer is a prerequisite to understand the cellular properties imparted by the mutated alleles and to identify genotype and tumor-specific pharmacological responses. In this regard, mammalian cell lines have been widely used as model systems to functionally characterize cancer alleles carrying point mutations and to develop and validate anticancer drugs. These models typically involve the ectopic expression (by means of plasmid transfection or viral infection) of mutated cDNAs in human or mouse cells (1). Although these approaches have yielded remarkable results, they are typically hampered by at least two caveats. First, the expression is achieved by transient or stable transfection of cDNAs, often resulting in over-expression of the target allele at levels that do not recapitulate what occurs in human cancers. Second, the expression of the mutated cDNA is achieved under the control of nonendogenous viral promoters. As a result, the mutated alleles cannot be appropriately (endogenously) modulated in the target cells. While such systems in which mutated oncogenes are ectopically expressed under exogenous promoters have been instrumental in dissecting their oncogenic properties, they have also led to controversial results. For example, studies focused on oncogene-mediated transformation and senescence have generated conflicting data depending on whether the cancer alleles were ectopically expressed or permanently introduced in the genome of mouse or human cells (2–5). To address the limitation of current models, we have used targeted homologous recombination to introduce (knock-in, KI) a panel of cancer alleles in human somatic cells. Specifically, we focused on EGFR, KRAS, BRAF, and PIK3CA mutated alleles that are found in multiple cancer types. Mutant cells have then been used to study the biochemical and transforming potential of common cancer alleles and to identify genotype-specific pharmacological profiles.
Results
KI of Mutated BRAF, EGFR, KRAS and PIK3CA Alleles in the Genome of Human Cells.
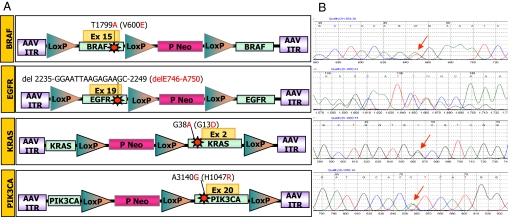
We used adeno-associated-viral (AAV) mediated homologous recombination to introduce somatic mutations commonly found in tumors in human somatic cells. Specifically, we focused on the following alleles: EGFR (delE746-A750), KRAS (G13D), BRAF (V600E), and PIK3CA (H1047R) that are found in multiple cancer types (Fig. 1A). These include among others, lung (EGFR and KRAS), colorectal (KRAS, BRAF, PIK3CA), breast (PIK3CA), pancreatic (KRAS), and prostate (KRAS, BRAF) carcinomas and melanoma (BRAF) (http://www.sanger.ac.uk/genetics/CGP/cosmic/). As recipient cells, we used three nontransformed epithelial cell lines of breast (MCF10A, hTERT-HME1) and retinal (hTERT RPE-1) origin. These cells display a number of features rendering them appealing for genetic and biological manipulation. They can be propagated indefinitely in vitro, but are not able to grow in anchorage-independent conditions or to form tumors when injected subcutaneously into nude mice, which makes them a suitable model to study oncogene-mediated transformation (6, 7). Furthermore, they have been previously used to assess a number of cellular phenotypes, including growth factor-dependent proliferation, motility, and invasive growth (7–10). A common strategy was used to generate the recombinant the AAV vectors required to knock-in each of the four cancer alleles (see Fig. 1A). In brief, the homologous recombination cassette was cloned within the AAV inverted terminal repeats and consisted of two ≈1-kb sequences (“homology arms”), one of which contained the specific mutation. A selectable marker was placed between the homology arms flanked by two LoxP sites, to allow Cre recombinase-mediated excision of the Neo cassette from the genome of the targeted cells (see Fig. 1A) and the possibility of recycling the resistance marker for the sequential introduction of multiple alleles in the same cell. After infection with rAAV and G418 selection, clones with locus-specific integration of the targeted alleles were identified through a PCR screening approach (see Methods for details). Positive clones were expanded and genomic DNA (gDNA) and RNA were extracted to sequence the targeted region to independently confirm the presence and the expression of the specific mutations (Fig. 1B). To account for clonal variability, multiple independent cell lines carrying each of the mutations were generated and analyzed at the biochemical, biological, and pharmacological levels.
Fig. 1.
Targeted knock-in (KI) of cancer mutations in human somatic cells. (A) Structure of AAV targeting constructs. AAV vectors carrying oncogenic alleles, either in the 5′ (BRAF, EGFR) or the 3′ arm (KRAS and PIK3CA), were used to introduce the indicated mutations in human cells by homologous recombination. ITR, inverted terminal repeat; Neo, geneticin-resistance gene; P, SV40 promoter; triangles, loxP sites. The nucleotide and amino acid changes are indicated. (B) The expression of the introduced genetic alterations in the targeted cells was determined by RT-PCR and sequencing of the BRAF, EGFR, KRAS, and PIK3CA transcripts. Red arrows indicate the introduced genetic alterations.
Biochemical Analysis of Mutated Alleles in Human Cells.
The cancer alleles that were knocked-in in human cells have been previously described to display distinct biochemical and biological properties (10–14). Indeed, we found that introduction of oncogenic mutations in the EGFR, KRAS, BRAF and PIK3CA genes in hTERT-HME1 breast cells resulted in activation of the corresponding proteins and triggered specific signaling pathways (Fig. S1). As expected, EGFR knock-in cells showed striking constitutive (ligand-independent) phosphorylation of EGFR (see Fig. S1A). Increased levels of total EGFR protein were also detected; these are likely because of the stabilization of the receptor and reduced degradation imparted by the E746-A750 deletion, as previously shown in lung cancer cells carrying the same allele (15–17). Interestingly, KRAS, BRAF and PIK3CA mutated cells also displayed allele-specific biochemical features. These included, respectively, PI3K-mediated AKT phosphorylation (see Fig. S1B), constitutive activation of the KRAS protein as measured by a GTP loading assay (see Fig. S1C), and BRAF-initiated activation of the MAPK kinase signaling pathway (see Fig. S1D). Similar results were obtained in multiple independent hTERT-HME1 clones of each genotype and in the MCF10A and hTERT RPE-1 KI cells carrying the same alleles (data not shown).
Transforming Potential of Cancer Alleles Ectopically Expressed or KI Human Somatic Cells.
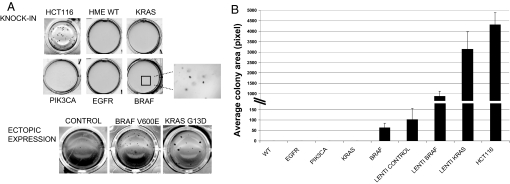
The in vitro measurable property that more closely correlates with the tumorigenic potential of cancer cells is their ability to grow in anchorage-independent fashion. Accordingly, ectopic expression of the cDNAs corresponding to the four cancer alleles had been previously shown to promote transformation of epithelial cells, such as those used in the present study (9, 10, 13, 18, 19). We evaluated the oncogenic properties of all KI cells by a conventional colony-formation assay in soft agar. The corresponding WT cells and the colon cancer cell line HCT 116 were used as negative and positive controls, respectively. We found that EGFR, KRAS, and PIK3CA KI hTERT-HME1 cells were unable to grow in soft agar, while BRAF mutated cells gave rise to few small colonies (Fig. 2A). Quantitative assessment of the number of colonies is provided in Fig. 2B. Similarly, no anchorage-independent growth was observed in either MCF10A or hTERT RPE-1 cells carrying cancer mutations (data not shown). Of note, the BRAF mutated cells were not tumorigenic when injected in immunocompromised mice (data not shown). These data are in contrast with previous results obtained by over-expression of the corresponding alleles in a number of human cellular models. We therefore decided to directly compare the KI versus the ectopic expression methodology. To achieve this goal, we engineered hTERT-HME1 cells to express the KRAS and BRAF mutated cDNAs under the control of viral promoters. The results were unequivocal in that hTERT-HME1 cells ectopically expressing any of the corresponding mutated cDNAs readily formed colonies (see Fig. 2). In particular, a remarkable difference in the number and size of colonies was observed.
Fig. 2.
Transforming potential of cells carrying oncogenic alleles. (A) An anchorage-independent growth assay was performed on hTERT-HME1 cells carrying the indicated genotypes. HCT 116 colorectal cancer cells were used as positive control. The same assay was performed on cells infected with lentiviral vectors expressing the G13D KRAS or V600E BRAF mutations. A lentiviral vector encoding for luciferase was used as a negative control. Representative photographs were taken after 3 weeks. (B) The area occupied by colonies was analyzed with BD Pathway HT bioimager and counted with BD AttoVision 1.5 software. Columns indicate mean area of four fields and error bars represent SD.
Genotype-Specific Clustering of KI Cells by Pharmarray Analysis.
We reasoned that our KI cell system could offer an opportunity to explore the pharmacogenomic properties of cancer alleles. To assess this possibility, we prepared a custom library of biologically-active drugs (Table S1) which comprised: (i) commonly used chemotherapeutic agents (e.g., 5-fluorouracil, cisplatin); (ii) recently developed tyrosine kinase inhibitors (e.g., dasatinib); (iii) drugs approved by the Food and Drug Administration (FDA) for a clinical indication other than cancer, but that were previously shown to have an antiproliferative effect in vitro (e.g., simvastatin); (iv) drugs currently undergoing oncology clinical trials (e.g., everolimus, triciribine); (v) a small collection of natural bioactive compounds (e.g., apigenin, deguelin); and (vi) a number of pathway specific pharmacological tools that were added to the library as controls (e.g., LY294002, PD98059). Parental and KI cells were seeded in complete growth medium and cell number was estimated by determining cellular ATP content. Under these conditions, no significant differences were observed in the proliferative potential of the KI cells as compared to their normal WT counterpart (Fig. S2). Each compound was then preliminarily tested on WT cells to determine the concentration referred to as the highest no-observed effect level (NOEL), the IC50, and the IC90 values. The effects of the drugs on cell viability were measured by the ATP bioluminescent assay.
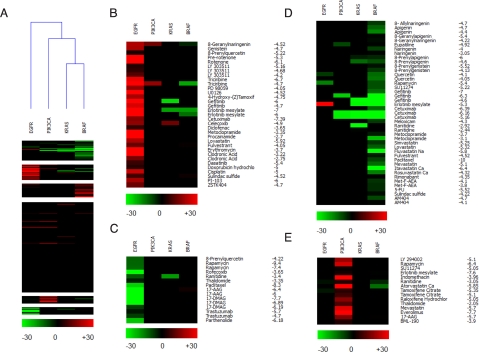
After the initial analysis, all KI clones and parental cells were assayed, testing at least three concentrations of each drug (range shown in Table S1) and using a minimum of two clones for each different genotype. The differential activity (ΔKI values, expressed as a percentage of cell-growth inhibition) between KI and parental cells was calculated for each compound at a given concentration. The results showed negligible variability among clones carrying the same mutation; therefore, the data obtained from multiple clones for each genotype were averaged. Data analysis details are described in SI Methods, and the full set of averaged data of pharmacological responses at each tested drug concentration is provided in Table S2. Normalized data were further analyzed using data clustering algorithms to better visualize the mutation-specific pharmacological phenotypes in isogenic cell pairs. For this purpose, we adopted a new software application that we had previously developed for microarray data clustering and visualization (20). Unclustered ΔKI values are depicted in Fig. S3, while analyzed data (that we define as pharmarray) are shown in Fig. 3 for the hTERT-HME1 cell model. Red-colored boxes indicate drugs that, at the indicated concentrations, preferentially inhibited the growth of mutated cells, while green boxes show compounds to which KI cells were more resistant than their WT counterpart does. Black boxes indicate no significant differences in response between KI and parental cells. The vast majority of drugs did not show selectivity toward any specific genotype, as shown by the predominant black columns. However, the approach successfully identified a set of clusters that were cell- and genotype-specific (see Fig. 3). When an unsupervised hierarchical clustering analysis of the pharmaco genomic data were performed using the pharmarray approach, a clear segregation of the KI cells was readily obtained (see Fig. 3A). Specifically, we discovered that the pharmarray analysis generated genotype-specific trees reflecting the signaling pathways in which the corresponding oncogenic mutations are known to act. These included on one side the cells carrying KRAS and BRAF mutations, on the other side the PIK3CA and the EGFR clones (see Fig. 3A). We then used a K-means algorithm to identify individual resistant and sensitive genotype-specific clusters (see Fig. 3 B–E). A distinct set of compounds that clustered according to their ability to selectively inhibit EGFR mutated cells was evident. These included gefitinib and erlotinib, and other less specific but already known EGFR inhibitors, such as genistein (21) (see Fig. 3B). Additional resistant and sensitive genotype-specific clusters were identified by the pharmarray approach (see Fig. 3 C–E), including a prominent red-inhibitory group of drugs affecting preferentially the PIK3CA mutated genotype (see Fig. 3E). This cluster included LY294002, indomethacin, rapamycin, and everolimus.
Fig. 3.
Pharmarray analysis of hTERT-HME1 cells carrying the indicated alleles. (A) Heatmap of the pharmacogenomic data (Pharmarray). Each column represents the average of multiple isogenic clones of the indicated genotype. Each row displays the results of differential response to drugs of the KI compared to WT cells. A color code is attributed at each drug concentration tested (log [M]). Drugs that, at the indicated concentrations, preferentially inhibit the growth of mutated cells are highlighted by the red color, while green indicates compounds to which KI cells are more resistant than the WT counterpart. For this reason, the same compound, depending upon its concentration, may segregate in a red (sensitive) or green (resistant) cluster. Black boxes indicate doses at which no significant differences in response between KI and parental cells were observed. Overall clustering of all of the compounds by K-means and of all of the genotypes by hierarchical clustering use an average cosine correlation coefficient. (B–E) Individual clusters composed of drugs with similar genotype-specific activity: (B) EGFR sensitive; (C) EGFR resistant; (D) KRAS/BRAF resistant; (E) PIK3CA-sensitive cluster.
Knock-in Cells Display Drug Responses Resembling Those of Tumors Carrying Equivalent Mutations.
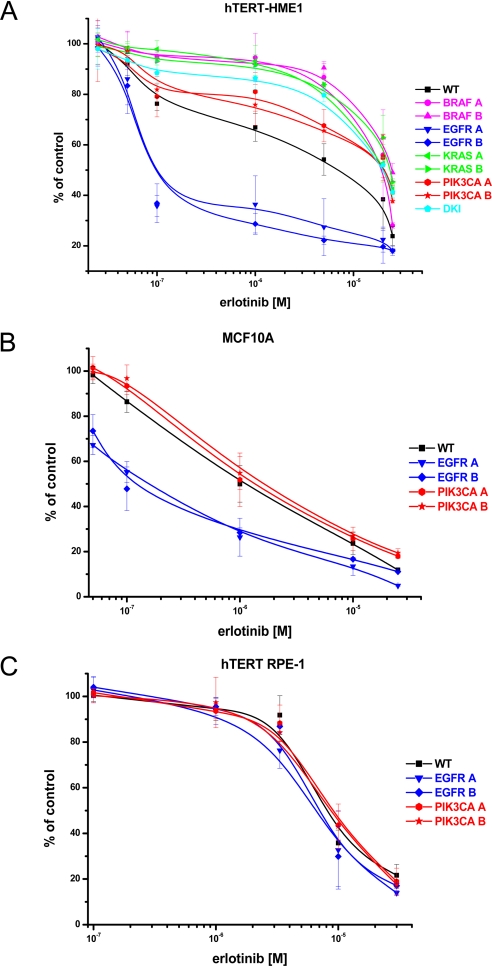
We assessed whether the response to targeted drugs of the KI cells may recapitulate that of naturally occurring cancer cells carrying equivalent cancer mutations. As a test case, we choose the EGFR KI cells, because it is well known that EGFR kinase inhibitors are most effective on cancer cells carrying a mutated EGFR gene (22, 23). Accordingly, we performed a detailed analysis of the effect of erlotinib on EGFR KI in multiple cellular backgrounds. We found that this drug preferentially inhibited the growth of hTERT-HME1 and MCF10A KI with the EGFR delE746-A750 allele (Figs. 4 A and B). Strikingly, the IC50 values of erlotinib in EGFR mutant cells (0.16 ± 0.06 μM, MCF10A, and 0.25 ± 0.14 μM, hTERT-HME1), were over 10-fold less than those of the corresponding WT cells. Gefitinib showed a similar selectivity pattern (Fig. S4A). Unexpectedly, no selectivity toward EGFR inhibitors was observed in the third cell line (hTERT RPE-1) carrying the EGFR delE746-A750 allele (Fig. 4C and Fig. S4B). We and others have previously shown that constitutive activation of the RAS/RAF pathway (for example by oncogenic KRAS mutations) can impair the response to drugs targeting EGFR (24, 25). We therefore hypothesized that a previously unreported activating alteration of the RAS/RAF pathway could be responsible for such lack of effect of erlotinib and gefitinib in hTERT RPE-1 cells. Indeed, mutational analysis of KRAS coding sequence in this line revealed that both the parental and KI cells carried a six-base pair insertion in exon 2 of this gene (Fig. S5A). Similar molecular alterations had been previously found in animal and human tumors (26, 27). Biochemical analysis demonstrated that this insertion strongly activates KRAS by permanently switching the corresponding mutated protein into the GTP-bound active state (Fig. S5B). Despite the presence of an activating KRAS mutation, we found that hTERT RPE-1 are not transformed (data not shown), thus further confirming our finding on the lack of transforming potential of endogenously expressed mutant KRAS alleles. Our results suggest that hTERT RPE-1 cells have acquired a KRAS gain-of-function mutation either during the immortalization procedure or during their continuous growth in culture. It is also possible (albeit unlikely) that the tissue of the individual from which the hTERT RPE-1 cells were established was already carrying the corresponding mutated KRAS allele.
Fig. 4.
Effect of the EGFR tyrosine kinase inhibitor erlotinib on KI cells. The effect of erlotinib treatment on cellular proliferation was assessed for hTERT-HME1 (A), MCF10A (B), and hTERT RPE-1 (C) isogenic clones carrying the indicated mutations. Cell viability was estimated by determining ATP content in three replicate wells. Results are normalized to growth of cells treated with DMSO and are represented as mean ± SD of at least three independent experiments.
Next, we investigated the mechanism responsible for the pronounced effect of erlotinib on the viability of EGFR KI clones in the hTERT-HME1 background. Flow cytometric analysis showed that 48-hours' treatment with erlotinib at 0.1 to 0.5 μM caused an accumulation of EGFR KI cells in the G1/G0 phase of the cell cycle, thereby decreasing the proportion of cells in the S and G2/M-phases (Table 1). At the same time-point, no significant apoptosis was observed. After more prolonged exposure (7 days) to 0.1–1 μM erlotinib (a range within clinically achievable concentrations), a sustained inhibition of cell proliferation was again observed only in hTERT-HME1 EGFR KI clones, while WT cells were only marginally affected (Fig. S6). Notably, on day 7 a modest but significant fraction of apoptotic cells was observed in EGFR KI cells, but not in parental cells (see Figs. S6 and S7). These findings suggest that oncogenic addiction is recapitulated in EGFR KI cells, as their proliferation and survival is clearly dependent upon EGFR kinase activity. Overall, these results are well in accordance with literature data on tumor cells carrying EGFR mutations (22, 28, 29).
Table 1.
Effects of erlotinib on cell cycle in KI cells
| % Cells in phase |
P | % Cells in phase |
P | |||||
|---|---|---|---|---|---|---|---|---|
| Vehicle |
Erlotinib 0.1 μM |
Erlotinib 0.5 μM |
||||||
| Mean | SD | Mean | SD | Mean | SD | |||
| hTERT-HME1 WT | ||||||||
| G1 | 71.6 | 6.6 | 77.4 | 1.0 | 0.208 | 77.0 | 1.2 | 0.227 |
| G2/M | 14.2 | 2.2 | 11.1 | 1.3 | 0.047 | 11.4 | 1.3 | 0.022 |
| S | 14.7 | 3.9 | 11.5 | 2.1 | 0.309 | 11.6 | 2.3 | 0.349 |
| G2 + S | 28.9 | 5.9 | 22.6 | 1.0 | 0.152 | 23.0 | 1.2 | 0.167 |
| Sub-G1 | 1.0 | 0.3 | 0.3 | 0.1 | 0.013 | 0.3 | 0.2 | 0.061 |
| KI EGFR delE746-A750 | ||||||||
| G1 | 70.3 | 3.0 | 87.3 | 2.2 | 0.006 | 85.3 | 1.5 | 0.007 |
| G2/M | 16.4 | 2.6 | 9.2 | 1.6 | 0.038 | 10.7 | 1.2 | 0.046 |
| S | 13.3 | 2.6 | 3.4 | 0.6 | 0.004 | 4.0 | 0.8 | 0.010 |
| G2 + S | 29.7 | 3.0 | 12.7 | 2.2 | 0.006 | 14.7 | 1.5 | 0.007 |
| Sub-G1 | 1.6 | 1.9 | 3.2 | 2.5 | 0.307 | 3.7 | 3.7 | 0.517 |
WT and EGFR KI cells were incubated for 48 h with the indicated concentrations of erlotinib, and the effect on cell cycle was assessed by FACS analysis. Erlotinib induced a significant arrest of EGFR KI clones in the G0/G1-phase of the cell cycle, while minimally affecting parental cells. Means of at least four independent experiments are shown. Significance by paired t test was taken at P < 0.01.
Sequential Introduction of Mutations to Model Drug Resistance.
It has been recently reported that, albeit rarely, EGFR and PIK3CA mutations can coexist in human tumors (30), and that activation of the PI3K/AKT signaling pathway can circumvent the effect of EGFR tyrosine kinase inhibitors. To verify whether we could recapitulate this phenomenon in our cellular model as well, we generated double KI clones (DKI) carrying both the PIK3CA (H1047R) and EGFR (delE746-A750) mutations. Identification of DKI mutant cells was achieved as described for the single KI approach. hTERT-HME1 DKI clones carrying both EGFR and PIK3CA mutations were then treated with gefitinib and erlotinib. Strikingly, the occurrence of both mutations abrogated the sensitization seen with the EGFR KI alone (see Fig. 4A and Fig. S4A). These results are in concordance with recent findings in brain tumors cells that carry similar pathway lesions (EGFR and PTEN alterations) and are resistant to anti-EGFR therapies (31).
Discussion
Until now, strategies to study cancer mutations in human cells have mainly involved ectopic expression of the corresponding mutated cDNA under the control of nonendogenous, constitutively active promoters These approaches do not accurately recapitulate the occurrence of cancer mutations in human tumors (32). To overcome these limitations, we used targeted homologous recombination to introduce cancer alleles in the genome of human cells by stable modification of the corresponding genomic locus. As a result, the heterozygously mutated genes are expressed under their endogenous promoters. Using this approach, we generated isogenic cell lines carrying mutations frequently found in human tumors, including KRAS G13D (33), BRAF V600E (34), EGFR delE746-A750 (28), and PIK3CA H1047R (11). Several studies have shown that single cancer alleles, when ectopically expressed, can transform human cells (9, 10, 13, 18, 19). In contrast, we found that the introduction of cancer alleles in the genome of immortalized human cells of epithelial origin was generally not sufficient to confer transforming properties. We propose that the sequential addition of multiple mutations by direct modification of the corresponding genomic loci could eventually lead to the transformation of human epithelial cells, but further work is needed to test this hypothesis. This technology could also be used to discriminate passengers from drivers' cancer mutations, and to assess the role of the germline variations (SNPs) that have been found to predispose to cancer (35). The latter frequently occurs in noncoding regions and consequently cDNA transfection approaches are often inapplicable to assess the functional role of SNPs. Understanding how common oncogenic alleles affect resistance and sensitivity to targeted drugs is key to defining individualized cancer therapies. To address this issue, we evaluated the response of the KI cells to a panel of over 90 compounds, including established (FDA approved) drugs and recently developed kinase inhibitors. This approach led to the identification of a number of drug-genotype interactions, including the striking response of EGFR mutant cells toward the EGFR kinase inhibitors erlotinib and gefitinib. These results indicate that our strategy can successfully identify validated pharmacogenomic interactions and suggest that it can be exploited to identify new genotype-targeted drugs. In conclusion, a number of general considerations can be drawn from our results. KI of cancer mutations generates isogenic cellular models carrying the same lesions observed in human tumors. Mutant cells show striking drug sensitivity phenotypes, when treated with targeted inhibitors, resembling the response and resistance mechanisms occurring in human tumors. Most excitingly, profiling of bioactive drugs on KI cells carrying single or multiple mutations can be rapidly performed to identify drug-genotype correlations, thus allowing the rational design of clinical trials based on the genetic milieu of individual tumors.
Methods
Cells and Cell Culture Reagents.
Parental and genetically modified cells were obtained and cultured as described in SI Methods.
Chemicals and Drugs.
Chemicals and drugs were purchased from several different commercial suppliers as indicated in Table S1. All compounds were reconstituted in the appropriate solvents and stored in aliquots at the temperature recommended by the manufacturers.
Plasmids and Viral Vectors.
All of the KI-targeting vectors were constructed using a modified pBluescript plasmid, which we named pSA-5A, containing a Neo resistance gene driven by a SV40 promoter; two loxP sites flank this G418 resistance cassette. The list of primers used to amplify the homology arms is available in Table S3. All experimental procedures for targeting vector construction, AAV production, cell infection, and screening for recombinants have already been described elsewhere (36). The list of primers used for screening is available from the authors upon request. The lentiviral vector expressing BRAF V600E was a kind gift of of Dr María S. Soengas (Ann Arbor, MI). The procedure to obtain the lentivirus expressing the KRAS G13D mutation has been described elsewhere (2).
Biochemical, Cellular, and Pharmacology (Pharmarray) Analysis.
Detailed descriptions of biochemical, cell based, and pharmacological (pharmarray) analysis are available in SI Methods.
Supplementary Material
Acknowledgments.
We thank Dr. Chris Torrance, Dr. Luca Cardone, Dr. Fonnet Bleeker for critically reading the manuscript, and Tommaso Renzulli for technical assistance with the BD Pathway HT bioimager. We thank all of the staff at the Institute for Cancer Research and Treatment Pharmacy, and in particular Dr. Franca Goffredo for supplying several drugs. Work in the laboratories of the authors is supported by the Italian Association for Cancer Research (A.B.and E.M.), Italian Ministry of Health (A.B.), Regione Piemonte (A.B.), Italian Ministry of University and Research (A.B. and E.M.), CRT Progetto Alfieri (A.B.), FP-7 EU Marie Curie Program (A.B.), and European Union Framework Program 6 (FP6), Migrating Cancer Stem Cells Contract 037297 (to A.B.).
Footnotes
Conflict of interest statement: A.B. is a consultant for Horizon Discovery (Cambridge UK) to which some of the cell lines described in this article have been licensed through the University of Turin.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0808757105/DCSupplemental.
References
- 1.Hahn WC, et al. Creation of human tumour cells with defined genetic elements. Nature. 1999;400:464–468. doi: 10.1038/22780. [DOI] [PubMed] [Google Scholar]
- 2.Arena S, et al. Knock-in of oncogenic kras does not transform mouse somatic cells but triggers a transcriptional response that classifies human cancers. Cancer Res. 2007;67:8468–8476. doi: 10.1158/0008-5472.CAN-07-1126. [DOI] [PubMed] [Google Scholar]
- 3.Guerra C, et al. Tumor induction by an endogenous K-ras oncogene is highly dependent on cellular context. Cancer Cell. 2003;4:111–120. doi: 10.1016/s1535-6108(03)00191-0. [DOI] [PubMed] [Google Scholar]
- 4.Konishi H, et al. Knock-in of mutant K-ras in nontumorigenic human epithelial cells as a new model for studying K-ras mediated transformation. Cancer Res. 2007;67:8460–8467. doi: 10.1158/0008-5472.CAN-07-0108. [DOI] [PubMed] [Google Scholar]
- 5.Tuveson DA, et al. Endogenous oncogenic K-ras(G12D) stimulates proliferation and widespread neoplastic and developmental defects. Cancer Cell. 2004;5:375–387. doi: 10.1016/s1535-6108(04)00085-6. [DOI] [PubMed] [Google Scholar]
- 6.Jiang XR, et al. Telomerase expression in human somatic cells does not induce changes associated with a transformed phenotype. Nat Genet. 1999;21:111–114. doi: 10.1038/5056. [DOI] [PubMed] [Google Scholar]
- 7.Shay JW, Wright WE, Brasiskyte D, Van der Haegen BA. E6 of human papillomavirus type 16 can overcome the M1 stage of immortalization in human mammary epithelial cells but not in human fibroblasts. Oncogene. 1993;8:1407–1413. [PubMed] [Google Scholar]
- 8.Debnath J, Muthuswamy SK, Brugge JS. Morphogenesis and oncogenesis of MCF-10A mammary epithelial acini grown in three-dimensional basement membrane cultures. Methods. 2003;30:256–268. doi: 10.1016/s1046-2023(03)00032-x. [DOI] [PubMed] [Google Scholar]
- 9.Isakoff SJ, et al. Breast cancer-associated PIK3CA mutations are oncogenic in mammary epithelial cells. Cancer Res. 2005;65:10992–11000. doi: 10.1158/0008-5472.CAN-05-2612. [DOI] [PubMed] [Google Scholar]
- 10.Brummer T, et al. Functional analysis of the regulatory requirements of B-Raf and the B-Raf(V600E) oncoprotein. Oncogene. 2006;25:6262–6276. doi: 10.1038/sj.onc.1209640. [DOI] [PubMed] [Google Scholar]
- 11.Samuels Y, et al. Mutant PIK3CA promotes cell growth and invasion of human cancer cells. Cancer Cell. 2005;7:561–573. doi: 10.1016/j.ccr.2005.05.014. [DOI] [PubMed] [Google Scholar]
- 12.Pao W, Miller VA. Epidermal growth factor receptor mutations, small-molecule kinase inhibitors, and non-small-cell lung cancer: current knowledge and future directions. J Clin Oncol. 2005;23:2556–2568. doi: 10.1200/JCO.2005.07.799. [DOI] [PubMed] [Google Scholar]
- 13.Campbell PM, et al. K-Ras promotes growth transformation and invasion of immortalized human pancreatic cells by Raf and phosphatidylinositol 3-kinase signaling. Cancer Res. 2007;67:2098–2106. doi: 10.1158/0008-5472.CAN-06-3752. [DOI] [PubMed] [Google Scholar]
- 14.Sato M, et al. Multiple oncogenic changes (K-RAS(V12), p53 knockdown, mutant EGFRs, p16 bypass, telomerase) are not sufficient to confer a full malignant phenotype on human bronchial epithelial cells. Cancer Res. 2006;66:2116–2128. doi: 10.1158/0008-5472.CAN-05-2521. [DOI] [PubMed] [Google Scholar]
- 15.Furukawa M, et al. Gefitinib-sensitive EGFR lacking residues 746–750 exhibits hypophosphorylation at tyrosine residue 1045, hypoubiquitination, and impaired endocytosis. DNA Cell Biol. 2007;26:178–185. doi: 10.1089/dna.2006.0573. [DOI] [PubMed] [Google Scholar]
- 16.Okabe T, et al. Differential constitutive activation of the epidermal growth factor receptor in non-small cell lung cancer cells bearing EGFR gene mutation and amplification. Cancer Res. 2007;67:2046–2053. doi: 10.1158/0008-5472.CAN-06-3339. [DOI] [PubMed] [Google Scholar]
- 17.Shtiegman K, et al. Defective ubiquitinylation of EGFR mutants of lung cancer confers prolonged signaling. Oncogene. 2007;26:6968–6978. doi: 10.1038/sj.onc.1210503. [DOI] [PubMed] [Google Scholar]
- 18.Vikis H, et al. EGFR-T790M is a rare lung cancer susceptibility allele with enhanced kinase activity. Cancer Res. 2007;67:4665–4670. doi: 10.1158/0008-5472.CAN-07-0217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ince TA, et al. Transformation of different human breast epithelial cell types leads to distinct tumor phenotypes. Cancer Cell. 2007;12:160–170. doi: 10.1016/j.ccr.2007.06.013. [DOI] [PubMed] [Google Scholar]
- 20.Fu L, Medico E. FLAME, a novel fuzzy clustering method for the analysis of DNA microarray data. BMC Bioinformatics. 2007;8:3. doi: 10.1186/1471-2105-8-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Akiyama T, et al. Genistein, a specific inhibitor of tyrosine-specific protein kinases. J Biol Chem. 1987;262:5592–5595. [PubMed] [Google Scholar]
- 22.Sordella R, Bell DW, Haber DA, Settleman J. Gefitinib-sensitizing EGFR mutations in lung cancer activate anti-apoptotic pathways. Science. 2004;305:1163–1167. doi: 10.1126/science.1101637. [DOI] [PubMed] [Google Scholar]
- 23.Carey KD, et al. Kinetic analysis of epidermal growth factor receptor somatic mutant proteins shows increased sensitivity to the epidermal growth factor receptor tyrosine kinase inhibitor, erlotinib. Cancer Res. 2006;66:8163–8171. doi: 10.1158/0008-5472.CAN-06-0453. [DOI] [PubMed] [Google Scholar]
- 24.Pao W, et al. KRAS mutations and primary resistance of lung adenocarcinomas to gefitinib or erlotinib. PLoS Med. 2005;2:e17. doi: 10.1371/journal.pmed.0020017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Benvenuti S, et al. Oncogenic activation of the RAS/RAF signaling pathway impairs the response of metastatic colorectal cancers to anti-epidermal growth factor receptor antibody therapies. Cancer Res. 2007;67:2643–2648. doi: 10.1158/0008-5472.CAN-06-4158. [DOI] [PubMed] [Google Scholar]
- 26.Higinbotham KG, Rice JM, Buzard GS, Perantoni AO. Activation of the K-ras gene by insertion mutations in chemically induced rat renal mesenchymal tumors. Oncogene. 1994;9:2455–2459. [PubMed] [Google Scholar]
- 27.Bollag G, et al. Biochemical characterization of a novel KRAS insertion mutation from a human leukemia. J Biol Chem. 1996;271:32491–32494. doi: 10.1074/jbc.271.51.32491. [DOI] [PubMed] [Google Scholar]
- 28.Lynch TJ, et al. Activating mutations in the epidermal growth factor receptor underlying responsiveness of non-small-cell lung cancer to gefitinib. N Engl J Med. 2004;350:2129–2139. doi: 10.1056/NEJMoa040938. [DOI] [PubMed] [Google Scholar]
- 29.Paez JG, et al. EGFR mutations in lung cancer: correlation with clinical response to gefitinib therapy. Science. 2004;304:1497–1500. doi: 10.1126/science.1099314. [DOI] [PubMed] [Google Scholar]
- 30.Yamamoto H, et al. PIK3CA Mutations and copy number gains in human lung cancers. Cancer Res. 2008;68:6913–6921. doi: 10.1158/0008-5472.CAN-07-5084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mellinghoff IK, Cloughesy TF, Mischel PS. PTEN-mediated resistance to epidermal growth factor receptor kinase inhibitors. Clin Cancer Res. 2007;13:378–381. doi: 10.1158/1078-0432.CCR-06-1992. [DOI] [PubMed] [Google Scholar]
- 32.Hua VY, Wang WK, Duesberg PH. Dominant transformation by mutated human ras genes in vitro requires more than 100 times higher expression than is observed in cancers. Proc Natl Acad Sci USA. 1997;94:9614–9619. doi: 10.1073/pnas.94.18.9614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pollock CB, Shirasawa S, Sasazuki T, Kolch W, Dhillon AS. Oncogenic K-RAS is required to maintain changes in cytoskeletal organization, adhesion, and motility in colon cancer cells. Cancer Res. 2005;65:1244–1250. doi: 10.1158/0008-5472.CAN-04-1911. [DOI] [PubMed] [Google Scholar]
- 34.Davies H, et al. Mutations of the BRAF gene in human cancer. Nature. 2002;417:949–954. doi: 10.1038/nature00766. [DOI] [PubMed] [Google Scholar]
- 35.Hemminki K, Forsti A, Lorenzo Bermejo J. Etiologic impact of known cancer susceptibility genes. Mutat Res. 2008;658(1–2):42–54. doi: 10.1016/j.mrrev.2007.07.001. [DOI] [PubMed] [Google Scholar]
- 36.Arena S, Pisacane A, Mazzone M, Comoglio PM, Bardelli A. Genetic targeting of the kinase activity of the Met receptor in cancer cells. Proc Natl Acad Sci USA. 2007;104:11412–11417. doi: 10.1073/pnas.0703205104. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.