Abstract
Phenylene ethynylenes comprise a prototypical class of synthetic antimicrobial compounds that mimic antimicrobial peptides produced by eukaryotes and have broad-spectrum antimicrobial activity. We show unambiguously that bacterial membrane permeation by these antimicrobials depends on the presence of negative intrinsic curvature lipids, such as phosphatidylethanolamine (PE) lipids, found in high concentrations within bacterial membranes. Plate-killing assays indicate that a PE-knockout mutant strain of Escherichia coli drastically out-survives the wild type against the membrane-active phenylene ethynylene antimicrobials, whereas the opposite is true when challenged with traditional metabolic antibiotics. That the PE deletion is a lethal mutation in normative environments suggests that resistant bacterial strains do not evolve because a lethal mutation is required to gain immunity. PE lipids allow efficient generation of negative curvature required for the circumferential barrel of an induced membrane pore; an inverted hexagonal HII phase, which consists of arrays of water channels, is induced by a small number of antimicrobial molecules. The estimated antimicrobial occupation in these water channels is nonlinear and jumps from ≈1 to 3 per 4 nm of induced water channel length as the global antimicrobial concentration is increased. By comparing to exactly solvable 1D spin models for magnetic systems, we quantify the cooperativity of these antimicrobials.
Keywords: antibiotic resistant bacteria, host defense peptides, innate immunity, protein-membrane interactions
The recent emergence of antibiotic-resistant bacteria is a worldwide public health problem (1). Antimicrobial peptides (AMPs) from innate immunity are known to have broad spectrum and selective activity against pathogens (2–7). Despite their diversity in sequence, secondary structures, and source, most of the more than 800 different AMPs that have been identified are amphiphilic and cationic (2, 3). It is thought that electrostatic interactions facilitate association of the peptide with the anionic bacterial membrane (2, 8). Moreover, such peptides are often implicated in pore formation in the bacterial membrane. Although the amphiphilicity is important in pore formation, the exact molecular mechanism by which membrane pores are formed is still not clear. Over the last decade, synthetic molecules that mimic these features have been designed and investigated, including stereoisomers of natural AMPs (9), α-peptides (10–12), β-peptides (13–16), peptoids (17), aromatic oligomers (18, 19), and synthetic polymers (20–24), such as phenylene ethynylene. The precise molecular mechanism of activity for most of these compounds is also unknown, as is the reason why it is difficult for bacteria to evolve immunity to them.
In this paper, we dissect the membrane activity of a prototypical synthetic antimicrobial (25, 26) from the bacterium level to the molecular self-assembly level. A number of biophysical differences exist between the membranes of bacteria and eukaryotes (2, 27). We begin by showing unambiguously that negative intrinsic curvature PE lipids, which are found in high concentrations in Gram-negative bacterial membranes, play a critical role in the activity of specifically active phenylene ethynylene antimicrobials. Plate-killing assays indicate that a PE-knockout mutant strain of Escherichia coli, AD93, drastically out-survives its WT parent strain W3899 against the membrane-active phenylene ethynylene antimicrobials, whereas the opposite is true when both strains are challenged with the traditional metabolic antibiotic tobramycin. This observed trend is consistent with the composition-dependent permeation behavior of phenylene ethynylene when using giant unilamellar vesicles (GUV) as the model bacterial cell membrane. Interestingly, this trend is different from those observed for natural antimicrobial peptides (28), which may suggest a qualitatively distinct class of antimicrobial behavior. We also show why PE lipids are important for the mechanism. PE lipids facilitate formation of negative-curvature circumferential barrels of transmembrane pores. Synchrotron x-ray scattering indicates that membranes rich in PE lipids can be induced to form an inverted hexagonal HII phase, which consists of arrays of water channels, by a surprisingly small number of antimicrobial molecules. In the induced HII phase, the antimicrobial occupation is nonlinear and jumps from ≈1 to 3 per 4 nm of water channel length as the global antimicrobial concentration is increased. By comparing to the exactly solvable 1-D spin models, the 3-Potts model and the Ising model, we quantify the cooperativity of antimicrobial occupancy, and find that the system is well described by a two-level system of singly and triply occupied states separated by a free energy difference E1–3 of ≈4.7 kT.
Results and Discussion
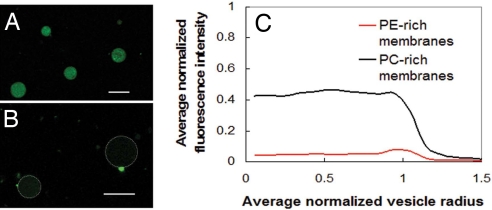
Variations of dye leakage assays on lipid vesicles are commonly performed to study the activity of membrane active antimicrobials. To illustrate both the potential and the limitation of this approach, we performed composition-dependent dye leakage experiments using GUVs. Phenylene ethynylene-treated giant unilamellar vesicles (GUVs) with PC-rich membranes (DOPG/DOPC = 20/80) (Fig. 1A) are compared with corresponding GUVs with PE-rich membranes with similar lipid compositions to typical E. coli membranes (DOPG/DOPE = 20/80) (Fig. 1B), both of which initially encapsulated fluorescently labeled (Alexa480) dextran. After treatment with phenylene ethynylene, the Alexa480 fluorescence intensity inside the PE-rich GUVs is drastically less than that inside the PC-rich GUVs, quantified by average circularly integrated fluorescence intensities in Fig. 1C. These results indicate that phenylene ethynylene induces more leakage from PE-rich GUVs, and suggest that the existence of PE lipids in the target membrane promotes phenylene ethynylene-induced membrane permeation. It can be seen, however, that there is significant nonspecific leakage. (The normalized intensity of untreated GUVs is set to one.) Moreover, bacteria are not vesicles and have, for example, more complex lipid distributions as well as lipopolysaccharides, both of which can modify results.
Fig. 1.
The limitation of dye leakage experiments. (A) Alexa480 fluorescence intensity from tagged dextran encapsulated in antimicrobial-treated DOPG/DOPC = 20/80 GUVs. (B) Alexa480 fluorescence intensity from tagged dextran encapsulated in antimicrobial-treated DOPG/DOPE = 20/80 GUVs is extremely weak because of leakage. Dotted white lines indicate the outline of vesicle membranes. (Scale bars, 10 μm, in A and B.) These dye leakage results and others suggest that a trend of increasing leakage with increasing PE lipid content in membranes after treated with the phenylene ethynylene antimicrobials. (C) Confocal microscopy results show circularly integrated intensities of fluorescently tagged dextran inside antimicrobial treated GUVs for PE-rich membranes composed of DOPG/DOPE = 20/80 and for PC-rich membranes composed of DOPG/DOPC = 20/80. Data from 10 GUVs are averaged into the PE-rich trace and 17 into the PC-rich trace. Fluorescence intensity is normalized so that untreated vesicles have fluorescence intensity of 1.0, so clearly there is significant nonspecific leakage, which complicates interpretation.
To evaluate directly the importance of PE lipids for antimicrobial activity, we performed bacteria plate-killing assays by using two strains of E. coli bacteria, a WT W3899 strain with a typical concentration of ≈78% PE lipids in its membrane, and a mutant ‘PE knockout’ strain (AD93) with essentially no PE in its membrane. The mutant strain AD93 is accomplished by constructing an inactivated allele of the gene encoding phosphatidylserine synthase, an enzyme which catalyzes the committing step to the synthesis of PE. Analysis of phospholipid composition confirms that AD93 has no measureable PE in its cell membrane (29, 30).
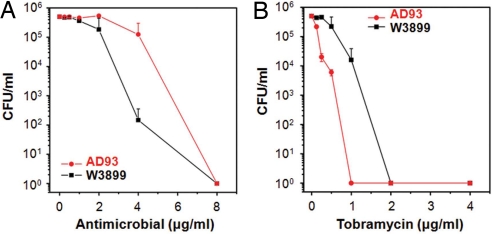
The results of comparative plate-killing assays in the presence of the phenylene ethynylene antimicrobial are shown in Fig. 2A. Because PE deletion is usually a lethal mutation to bacteria, growth of the PE-deficient mutant AD93 requires a supplement of certain divalent ions in the LB media (29). As a result, all comparative killing assays with phenylene ethynylene on both the mutant and WT strains are performed at the same Mg2+ concentration in sterile Hepes buffer solution supplemented with 400 mM sucrose to simulate natural molecular crowding effects in realistic environments, which osmotically stabilizes both strains of bacteria. PE-deficient AD93 cells survive at a significantly higher rate than the WT W3899 strain (≈3 orders of magnitude at 4 μg/ml). In an additional set of control experiments, we challenge both strains with tobramycin, a conventional metabolic antibiotic that targets the bacterial ribosomal machinery rather than the membrane. The mutant strain AD93 now survives at a drastically lower rate than its WT parent strain W3899 (≈4 orders of magnitude at 1 μg/ml), as shown in Fig. 2B (see Fig. S1 for additional information). These comparative plate-killing results using PE knockout mutants are consistent with our recent in vitro x-ray study (26), in which we show that the specificity of phenylene ethynylene antimicrobial activity is related to the PE content of the target membrane. The three basic toxicological profiles of the antimicrobials, nonactive against both bacteria and eukaryotic cells, specifically active against bacteria but not eukaryotic cells, and nonspecifically active against both bacteria and eukaryotic cells, correspond to decreasing minimal PE threshold values for inducing the inverted hexagonal phase. Moreover, that PE deletion is lethal in normative environments may help explain the unexpected absence of bacterial strains resistant to this class of antimicrobials despite repeated exposure (23): A lethal mutation is required in order for bacteria to gain immunity.
Fig. 2.
PE-deficient E. coli mutant out-survives the WT against the membrane-active antimicrobial. (A) Microbicidal assays show that, in the presence of the membrane-active phenylene ethynylene-based antimicrobial, the survival of the fragile PE-deficient mutant strain AD93 is much greater than that of its WT parent strain W3899 (≈3 orders of magnitude at 4 μg/ml) in sterile buffer solution (10 mM Hepes, 60 mM NaCl, 100 μM MgCl2, and 400 mM sucrose, pH 7.5). (B) When being exposed to tobramycin, a conventional antibiotic that targets the ribosome rather than the bacterial membrane, the WT parent strain (W3899) survived at a drastically higher rate than the mutant strain (AD93) (≈4 orders of magnitude at 1 μg/ml) in sterile buffer solution (10 mM Hepes, 60 mM NaCl, and 200 μM MgCl2, pH 7.5). Data points are reported as mean ± standard deviation.
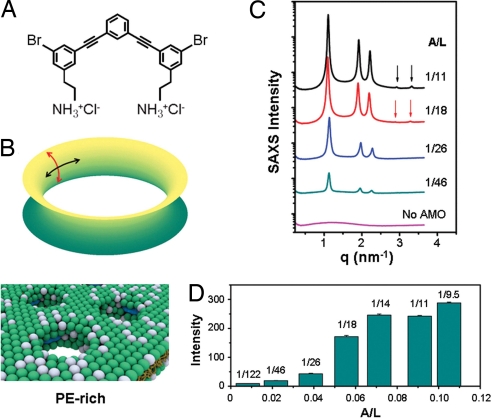
What is the role of the present prototypical phenylene ethynylene antimicrobial in the membrane permeation mechanism? In the case of natural innate immunity AMPs, it is currently thought that the peptides adopt two distinct states when membrane bound, one with induced pores and one lacking induced pores depending on the molar ratio of peptide-to-lipid. Because naturally occurring AMPs are protein-based and have electron densities that are close to those of lipids, we do not have experimental access to a more detailed picture, nor do we know how these self-assembled pore structures change as the AMP concentration is increased toward the MIC. Synthetic antimicrobials do not have this limitation. Phenylene ethynylene antimicrobials (Fig. 3A) have two high-electron-density bromines per molecule, so we can monitor the structural evolution of the induced water channels in PE-rich membranes by using high-resolution synchrotron small angle x-ray scattering (SAXS).
Fig. 3.
The structure of induced HII phase changes with antimicrobial concentration. (A) Structure of phenylene ethynylene-based antimicrobial. (B Top) Formation of a transmembrane pore requires both negative and positive curvatures (black and red arrows, respectively). (Bottom) Schematic representation of how this PE-dependent structural tendency may be realized on a membrane, in which the antimicrobials recruit PE lipids to achieve the negative circumferential curvature necessary for pore formation. The white and green spheres represent headgroups of zero intrinsic curvature (e.g., DOPG, DOPC) and negative intrinsic curvature lipids (e.g., DOPE) respectively. The antimicrobials are represented by blue spherocylinders. (C) Synchrotron SAXS data show that the inverted hexagonal phase induced by antimicrobial in DOPG/DOPE = 20/80 membranes evolves structurally as antimicrobial to lipid molar ratio (A/L) varies from 1/122 to 1/9.5. (D) The integrated diffraction intensities increase in a sigmoidal manner and saturate at A/L ∼ 1/14.
The specifically active phenylene ethynylene antimicrobial induces negative curvature by reorganizing lipids into an inverted hexagonal (HII) phase. Negative curvature is one of the structural ingredients of saddle-splay curvature (negative curvature along one principal direction and positive in the other, Fig. 3B), which is topologically required for pore formation. In fact, negative curvature can be seen in the circumferential barrel of an induced pore (Fig. 3B). SAXS data (Fig. 3C) show the self-assembled structure of the antimicrobial-membrane system (DOPG/DOPE = 20/80) as the antimicrobial/lipid molar ratio A/L is varied from 1/122, where the HII phase first forms, to 1/9.5, the saturation structure. At A/L = 1/122, the q positions of observed diffraction peaks have a relative ratio of 1:√3:2, and indicate the existence of a hexagonal structure with a lattice parameter a = 4π/(q10√3) = 6.43 nm. As A/L increases, the relative intensities of diffraction peaks change, and two extra diffraction peaks that index to a hexagonal structure appear (1:√3:2:√7:3), which together indicate that a significant structural change has occurred. Further, as A/L is increased, the integrated diffraction intensities increase in a nonlinear manner (Fig. 3D), and saturate at A/L ≈1/14. We use this saturation value of antimicrobial/lipid stoichiometry to estimate an upper limit of ≈3.5 antimicrobials per 4 nm of length along the water channels in the HII phase. (4 nm is the thickness of a typical membrane.)
It is interesting to assess the role of negative curvature lipids such as PE in the antimicrobial-induced formation of the HII phase. The first question is whether PE is targeted in the sense that the antimicrobial specifically binds with PE directly, and the presence of that bound complex ‘triggers’ the formation of a porous phase. This is likely not the case because a finite threshold of PE is necessary for the HII phase (26). However, the negative intrinsic curvature of PE reduces the free energy cost required to form the HII phase. It is known that cosurfactants can lower the bending modulus of a lipid membrane and provide an alternate route to high-curvature self-assembled phases, such as the HII phase, in membranes with low intrinsic curvature, such as those with little or no PE (31). We performed experiments by using hexanol-softened DOPG/DOPC/DOPE membranes, and found that the presence of small amounts of PE is insufficient for the induced generation of curvature, in agreement with earlier results (Table 1). For example, at DOPG/DOPE/DOPC = 20/00/80, the addition of hexanol does not result in HII type antimicrobial-lipid complexes. However, at PE concentrations below that required for the formation of HII phase, the addition of hexanol resulted in a transition from unilamellar vesicles to a precursor Im3m phase, a porous ‘plumber's nightmare’ phase that is rich in negative Gaussian curvature, which is topologically necessary for pore formation (32). This shows that these antimicrobials generate curvature in a manner that depends on the local lipid composition, and that the presence of PE alone is not a sufficient condition for curvature generation. Having demonstrated the above, the next question is whether PE is a necessary condition for antimicrobial-induced curvature generation. We find that it is possible to generate the HII phase even if PE were replaced with other negative intrinsic curvature lipids such as cardiolipin in the presence of divalent ions (see SI Text and unpublished results). This is consistent with the experimental observation that the PE-knockout mutants can still be killed because their membrane compositions contain cardiolipin. However, the importance of PE is highlighted by the observation that cardiolipin is much less efficient in generating the necessary negative curvature, and the antimicrobial/lipid molar ratio required to induce the HII phase with the same diffraction intensity is increased by more than a factor of ≈20× (see Fig. S2 for additional information).
Table 1.
Structure of lipid-antimicrobial complexes with hexanol
| Lipid Composition (DOPG/DOPE/DOPC) | Lipid Only | Hexanol:Lipid 3:1 |
|---|---|---|
| 20/80/00 | HII | HII |
| 20/64/16 | HII | HII |
| 20/56/24 | Unilamellar Vesicle | Im3m |
| 20/48/32 | Unilamellar Vesicle | Im3m |
| 20/20/60 | Unilamellar Vesicle | Unilamellar Vesicle |
| 20/00/80 | Unilamellar Vesicle | Unilamellar Vesicle |
Hexanol decreases the membrane bending modulus and can be used to drive phase transitions into nonlamellar phases without changing intrinsic curvature. The addition of hexanol does not result in an HII phase without PE. However, at finite PE concentrations, addition of hexanol can produce a precursor Im3m phase with negative Gaussian curvature, which is topologically required for pore formation. All samples have A/L = 1/14.
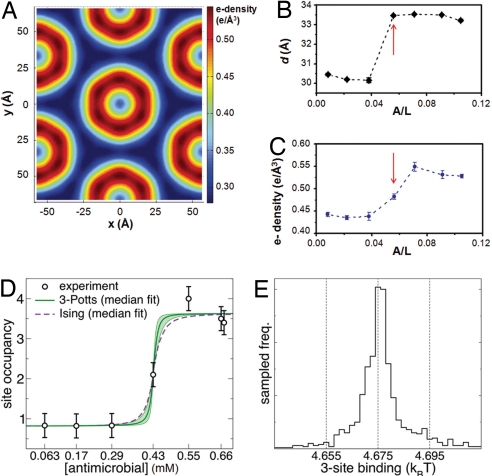
To quantify how efficiently phenylene ethynylene antimicrobials generate the necessary negative curvature, we examined the structural evolution of the observed hexagonal phase as a function of A/L. The electron density ρ(x,y) of the 2-D hexagonal unit cell has been reconstructed from the x-ray data (x, y are mutually perpendicular axes in the plane perpendicular to the channel axis). Our choices for the phases are (+,−,−) and (+,−,−,−,+) for the induced structures below with 3 peaks and 5 peaks, respectively, which are consistent with previous assigned phases for lipid-only phases (33). Fig. 4A shows a reconstructed electron density for a unit cell at A/L = 1/14. The dark blue regions have the lowest electron density (≈0.29 e/Å3) and correspond to hydrocarbon chains of the lipids, whereas the light blue circular central regions have intermediate electron density (≈0.33 e/Å3) and correspond to water within the hydrophilic channels. The circular “rims” in dark red (≈0.55 e/Å3) surrounding these water cores have the highest electron density, higher than those of typical phospholipid head groups (≈0.41 e/Å3), suggesting that the high-electron-density bromine atoms of the antimicrobials stay close to the amphiphilic interface; this is consistent with recent experiments on phenylene ethynylene-lipid interactions at an air-water interface (34).
Fig. 4.
Cooperativity of antimicrobial occupation is described by a Potts model. (A) A typical electron density profile of the 2D hexagonal unit cell confirms the formation of an inverted hexagonal structure. Here, we show the 2D plot of the reconstructed electron density profile at A/L = 1/14. The light blue regions have low electron density (≈0.29 e/Å3) and correspond to hydrocarbon chains of the lipids, whereas the “rims” in dark red have the highest electron density (≈0.55 e/Å3) and correspond to the regions near phospholipid head groups. The rim centers in lighter blue have intermediate electron density (≈0.33 e/Å3) and correspond to water within hydrophilic channels. (B and C) The diameter of water channels (B) and the electron density at the hydrophilic surface of lipid membranes (C) change abruptly as A/L increases (red arrows). (D and E) By analogy with exactly solvable 1D magnetic systems, we deduce thermodynamic parameters governing antimicrobial cooperativity in generating the necessary negative membrane curvature, using the antimicrobial occupancy data in the induced HII phase. We estimated the antimicrobial occupancy based on the relative enhancement in electron density around the hydrophilic membrane surface. Using Bayesian sampling of model fits, we show the range of predictions obtained from 1D 3-Potts and the simpler Ising model fits (D). Both 3-Potts model and Ising model give excellent fits with identical values for E1–3. We find that the antimicrobial occupation as a function of system-wide antimicrobial concentration is highly nonlinear, but is well described by a simple two-level system of singly and triply occupied states separated by a well defined free energy difference E1–3 of ≈ 4.68 kT, which quantifies the cooperativity in these antimicrobials. (E) The posterior distribution with normal likelihood from the experimental data are sampled by using Monte Carlo for 105 steps, and an ensemble of 103 3-Potts model parameter choices are taken. The median predicted site occupancy with upper and lower quartiles show the range of model predictions. The E1–3 binding free energy has the narrowest distribution with a full width of 0.04 kT (a measure of the statistical uncertainty of the fit, independent of the experimental error), whereas the other model parameters have variability of at least 1 kT. The histogram of E1–3 binding parameters from the Bayesian sampling shows this narrow distribution.
Based on the reconstructed electron density profile, we can estimate the size of the water core, d, by measuring the diameter of the “rim” with highest electron density (Fig. 4B). From these measurements, we can see that the water core diameter d changes abruptly: For A/L <1/18, d stays almost constant at 30.3 ± 0.2 Å. At A/L = 1/18, d abruptly increases to ≈33.5 Å and stays nearly constant at 33.4 ± 0.2 Å for larger A/L. Another way to consider these changes in the water channel diameter is membrane thinning induced by increases in A/L, which is reminiscent of the behavior of multilamellar stacks of lipids with antimicrobial peptides (35–37). In contrast with natural antimicrobial peptides, where membrane thinning may generate pores, the most pronounced membrane thinning induced by these synthetic antimicrobials occur at A/L ratios much higher than those required to form HII water channels. This membrane thinning correlates with changes in the highest electron density (ρhead/antimicrobial) at the ‘rim’ around the water channel near the phospholipid head groups. Fig. 4C shows the average value of the high electron density rims ρhead/antimicrobial of the 2D hexagonal unit cell at different A/L. When A/L <1/18, ρhead/antimicrobial stays approximately constant at ≈0.44 e/Å3; when A/L >1/18, ρhead/antimicrobial increases to ≈0.54 ± 0.01 e/Å3 and saturates. Based on the extent of enhancement in ρhead/antimicrobial and the electron density of the Br-rich antimicrobial, we estimate a saturation value of ≈3.4 ± 1.3 antimicrobials per 4 nm of water channel. This is in good agreement with our previous independent, stoichiometry-based estimate above. This corresponds to one antimicrobial per ≈1,227 Å2 (≈20 lipid molecules). Given that the effective surface area of a curved transmembrane pore is larger than that for a schematic cylinder, the number of antimicrobials in an actual transmembrane pore is expected to be larger (≈7.8 ± 3 antimicrobials per pore if the surface coverage density were constant. In this estimate, we approximate the edge of an induced pore by using a rim with constant radius and assume a constant antimicrobial surface density. The precise value of the number of antimicrobials will depend on how the head group area per lipid changes in this geometric organization).
We use our antimicrobial occupancy data on the induced HII phase to deduce thermodynamic parameters governing antimicrobial cooperativity in generating the necessary negative membrane curvature, by analogy to exactly solvable 1D spin systems. Electron density reconstructions indicate that each 4-nm segment of an HII channel is occupied by 1, 2, or 3 antimicrobials. The antimicrobials interact with each other on a 4-nm segment, but also with their neighboring segments because it is part of an HII channel. To understand trends in the behavior of antimicrobial occupancy within an HII channel, we first use a general 1D 3-Potts model (a 1D chain of sites 4 nm apart, each with three possible states) with a maximum of five adjustable parameters. The measured data indicate that sites essentially change from singly to triply occupied (Fig. 4D). This suggests that we can describe the results by using a simpler Ising model with only two states (and described by two adjustable parameters). Each model contains a single parameter for the difference in free energy of triply and singly occupied states, E1–3. In the Ising model, E1–3 = kT·log (cmid/c0), where cmid is the antimicrobial concentration at the transition between single and triple occupation, and c0 = 46.1 mM is the concentration at which one antimicrobial is expected per site without any interactions. Fig. 4D shows the result of Bayesian sampling of model fits, and the range of predictions from different 3-Potts and Ising model fits. An initial set of 3-Potts model parameters are fit by using a maximum likelihood estimate with a normal distribution; these parameters are used to define a uniform prior distribution within 5 kT. Both models give excellent fits with essentially identical values for E1–3 near 4.68 kT which justifies the simpler Ising approximation. In fact, the posterior distribution estimates a narrow range of good fit values for this parameter (Fig. 4E). It is interesting to note that the present model implicates two different types of cooperativity, intrasite and intersite. The former describes antimicrobial interactions within a site, and is usually described by the Hill analysis used to quantify cooperativity in biophysical systems (25), and the latter describes interactions between such sites, which likely involves strong coupling to the membrane elasticity. Both are necessary for a satisfactory fit to the data (See SI Text and Fig. S3).
These results show that phenylene ethynylene antimicrobials are extremely efficient in generating negative curvature in PE-rich membranes; only 1 antimicrobial per ≈20 lipid molecules are needed to initiate formation of an HII phase of water channels. We find that the antimicrobial occupation as a function of increasing global antimicrobial concentration is highly nonlinear, but is well described by a surprisingly simple two-level system of singly and triply occupied states separated by a well defined free energy difference E1–3 of ≈4.7 kT, which quantifies the extent of cooperativity in these membrane-active antimicrobials for the first time. These occupancy changes coincide with reorganization of the PE-rich membrane, and results in observed membrane thinning and resultant increases in the water channel diameter.
Materials and Methods
Phenylene Ethynylene Synthesis.
The phenylene ethynylene molecule, referred to as antimicrobial oligomer 2 (AMO 2) (26), was synthesized by coupling tert-butyl 3,5-dibromophenethylcarbamate with 1,3-diethynylbenzene by using Sonogoshira coupling in a 4 to 1 stoichiometry. The compound was then purified to homogeneity by silica gel chromatography to yield a white solid (yield 45%). 1H NMR (CDCl3): δ 7.68 (t, 1H, phenyl H), 7.56 (t, 2H, phenyl H), 7.51 (t, 1H, phenyl H), 7.48 (d, 1H, phenyl H), 7.37 (d, 1H, phenyl H), 7.34 (m, 2H, phenyl H), 7.31 (m, 2H, phenyl H), 4.58 (s, 2H, 2NH), 3.38 (m, 4H, 2CH2), 2.79 (t, 4H, 2CH2), 1.46 (s, 18H, 6CH3) ppm. 13C NMR (CDCl3): δ 156.1, 141.7, 135.1, 132.8, 132.5, 132.0, 131.1, 129.0, 125.4, 123.6, 122.7, 90.0, 88.9, 79.9, 41.8, 36.1, 31.3, 28.7 ppm. m/z [M++Na]: 745.3 [M+-2Boc + 2Na]: 567.2. This Boc-protected compound was treated with 4 M HCl-dioxane to quantitatively yield the ammonium salt. 1H NMR (DMSO-D6): δ 7.92 (s, 4H, 2NH2), 7.77 (t, 1H, phenyl H), 7.71 (t, 2H, phenyl H), 7.65 (t, 1H, phenyl H), 7.62 (m, 2H, phenyl H), 7.58 (d, 2H, phenyl H), 7.52 (m, 2H, phenyl H), 3.11 (m, 4H, 2CH2), 2.92 (t, 4H, 2CH2) ppm. 13C NMR (DMSO-D6): δ 138.7, 134.6, 133.0, 131.8, 131.6, 128.6, 125.3, 122.9, 122.5, 89.8, 87.9, 60.3, 58.2, 48.0, 41.6, 40.4, 32.9 ppm.
Bacteria Plate-Killing Assay.
The PE-deficient strain AD93 was constructed from the WT E. coli strain W3899 (29). Both AD93 and W3899 were grown in LB medium (10 g of Bacto-tryptone, 5 g of Bacto-yeast, and 5 g of NaCl per liter) supplemented with 20 mM MgCl2 at pH 7.5. Cells were first grown to stationary phase at 37°C. A 25-μl culture was diluted with fresh media by one hundredfold and regrown at 37°C to midlog growth phase (OD600 ≈ 0.5–0.7). The cells were harvested and washed twice with a sterile buffer solution (10 mM Hepes, 60 mM NaCl, 400 mM sucrose, and 100 μM MgCl2, pH 7.5) via centrifugation at 10,000 rpm for 5 min, and within 15 min were adjusted with sterile buffer solution to ≈5 × 106 CFU/ml before being inoculated into wells of a polystyrene 96-well microplate.
Serial 2-fold dilutions of phenylene ethynylene antimicrobial stock solutions in DMSO were first made with buffer solution in Eppendorf centrifuge cups. We inoculated 20 μl of adjusted bacteria suspensions into each zero-dilution well in preset microplates to achieve ≈5 × 105 CFU/ml in each well (200 μl), and incubated the plates at 37°C for 3 h. Tenfold serial dilutions were subsequently made with sterile Hepes buffer solution. Each dilution (100 μl) was plated onto LB agar plates that were incubated at ≈35°C overnight to yield visible colonies.
Each plate-killing assay trial was carried out in triplicate and the reported results are the averages of two independent trials. Controls included broth-only and buffer-only samples to test the sterility and provide blank values for the assay readings, as well as untreated bacterial suspension samples to indicate the final inoculum size. A set of controls containing DMSO at 2% (vol. conc.) indicated that DMSO does not affect bacterial survival. Tobramycin was prepared in a sterile buffer (10 mM Hepes and 60 mM NaCl, pH 7.5) and stored in aliquots at −20°C.
Sample Preparation and Experiments Using Confocal Microscopy.
GUVs were prepared using the swelling method. DOPG {1,2-dioleoyl-sn-glycero-3-[phospho-rac-(1-glycerol)] (sodium salt)}, DOPC (1,2-dioleoyl-sn-glycero-3-phosphocholine), and DOPE (1,2-dioleoyl-sn-glycero-3-phosphoethanolamine) were purchased from Avanti Polar Lipids and used as supplied without further purification. Specific mixtures of DOPG, DOPC, and DOPE in chloroform or chloroform/methanol were deposited (100 μl at 20 mg/ml) onto roughened, cleaned Teflon, dried under vacuum, and then hydrated with 5 ml of 100 mM sucrose solution followed by incubation for 2–3 days at 37°C (38, 39). Dextran (3 kDa) labeled with Alexa Fluor 480 (Invitrogen) was incorporated into the sucrose swelling solution at concentrations of 20 μM. After swelling, GUV suspensions were diluted by 40-fold into 200 mM glucose. For confocal viewing of composition-dependent dye release, phenylene ethynylene antimicrobial in DMSO solution was added to the vesicle suspension at two different global antimicrobial concentrations (28 mol% or 14 mol% of lipid used). A Leica SP2 laser scanning confocal microscope was used to observe GUVs. Similar results were observed at both examined antimicrobial concentrations.
Sample Preparation and Experiments Using Synchrotron Small Angle X-ray Scattering.
DOPG stock solutions in chloroform/methanol solution were mixed with DOPE stock solutions in chloroform at desired lipid ratio. The mixture was dried under N2, desiccated under vacuum for overnight, and rehydrated with Millipore water to a final lipid concentration of 20 mg/ml at 37°C. The resultant solution was sonicated to clarity and then extruded through a 0.2-μm Nucleopore filter. To create membranes with reduced bending modulus, the cosurfactant hexanol was added at a 1:3 hexanol:lipid molar ratio.
Antimicrobial stock solutions in DMSO/water = 1/11 (vol/vol) and lipid vesicle solutions were mixed at the desired phenylene ethynylene antimicrobial to lipid molar ratio (A/L) and sealed in quartz capillaries. SAXS experiments were performed at the Stanford Synchrotron Radiation Laboratory (BL4–2) and the Advanced Photon Source (BESSRCAT) as described (26). Fourier synthesis techniques for centrosymmetric systems were used to calculate the electron density profiles of unit cells (33, 40, 41). For samples using membranes containing hexanol, SAXS experiments were performed by using an in-house x-ray source as described (42).
For additional information see the SI Text.
Supplementary Material
Acknowledgments.
We thank Prof. Matt Parsek, Dr. Lori K. Sanders, and Dr. Wujing Xian for helpful discussions and Robert Coridan, Mike Lee, and Vivek N. Patel for technical assistance. We also thank Prof. William Dowhan for his invaluable help with the PE-deficient E. coli mutant. X-ray work was performed at the Stanford Synchrotron Radiation Lab, the Advanced Photon Source, and the Frederick Seitz Materials Research Lab. This work was supported in part by National Science Foundation Grants DMR08–04363 and CBET08–27293, the Center of Advanced Materials for Purification of Water with Systems Science and Technology Centers, and the Rensselaer Polytechnic Institute-University of Illinois at Urbana-Champaign Nanoscale Science and Engineering Center, National Institutes of Health Grants R01-Al-074866 and R01 AI15650, and Office of Naval Research Grant N00014–03-1–0503.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. I.S. is a guest editor invited by the Editorial Board.
This article contains supporting information online at www.pnas.org/cgi/content/full/0806456105/DCSupplemental.
References
- 1.Tomasz A. Multiple-antibiotic-resistant pathogenic bacteria – A report on the Rockefeller University workshop. N Engl J Med. 1994;330:1247–1251. doi: 10.1056/NEJM199404283301725. [DOI] [PubMed] [Google Scholar]
- 2.Zasloff M. Antimicrobial peptides of multicellular organisms. Nature. 2002;415:389–395. doi: 10.1038/415389a. [DOI] [PubMed] [Google Scholar]
- 3.Brogden KA. Antimicrobial peptides: Pore formers or metabolic inhibitors in bacteria? Nat Rev Microbiol. 2005;3:238–250. doi: 10.1038/nrmicro1098. [DOI] [PubMed] [Google Scholar]
- 4.Hancock REW, Lehrer R. Cationic peptides: A new source of antibiotics. Trends Biotechnol. 1998;16:82–88. doi: 10.1016/s0167-7799(97)01156-6. [DOI] [PubMed] [Google Scholar]
- 5.Hancock REW, Sahl H-G. Antimicrobial and host-defense peptides as new anti-infective therapeutic strategies. Nat Biotech. 2006;24:1551–1557. doi: 10.1038/nbt1267. [DOI] [PubMed] [Google Scholar]
- 6.Boman HG. Innate immunity and the normal microflora. Immunol Rev. 2000;173:5–16. doi: 10.1034/j.1600-065x.2000.917301.x. [DOI] [PubMed] [Google Scholar]
- 7.Shai Y. Mechanism of the binding, insertion, and destabilization of phospholipid bilayer membranes by α-helical antimicrobial and cell non-selective membrane-lytic peptides. Biochim Biophys Acta, Biomembr. 1999;1462:55–70. doi: 10.1016/s0005-2736(99)00200-x. [DOI] [PubMed] [Google Scholar]
- 8.Yeaman MR, Yount NY. Mechanisms of antimicrobial peptide action and resistance. Pharmacol Rev. 2003;55:27–56. doi: 10.1124/pr.55.1.2. [DOI] [PubMed] [Google Scholar]
- 9.Oren Z, Shai Y. Selective lysis of bacteria but not mammalian cells by diastereomers of melittin: Structure-function study. Biochemistry. 1997;36:1826–1835. doi: 10.1021/bi962507l. [DOI] [PubMed] [Google Scholar]
- 10.Chen YX, et al. Rational design of α-helical antimicrobial peptides with enhanced activities and specificity/therapeutic index. J Biol Chem. 2005;280:12316–12329. doi: 10.1074/jbc.M413406200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Won H-S, Jung S-J, Kim HE, Seo M-D, Lee B-J. Systematic peptide engineering and structural characterization to search for the shortest antimicrobial peptide analogue of Gaegurin 5. J Biol Chem. 2004;279:14784–14791. doi: 10.1074/jbc.M309822200. [DOI] [PubMed] [Google Scholar]
- 12.Fernandez-Lopez S, et al. Antibacterial agents based on the cyclic D,L-α-peptide architecture. Nature. 2001;412:452–455. doi: 10.1038/35086601. [DOI] [PubMed] [Google Scholar]
- 13.Hamuro Y, Schneider JP, DeGrado WF. De novo design of antibacterial β-peptides. J Am Chem Soc. 1999;121:12200–12201. [Google Scholar]
- 14.Porter EA, Wang X, Lee H-S, Weisblum B, Gellman SH. Non-haemolytic β-amino-acid oligomers. Nature. 2000;404:565. doi: 10.1038/35007145. [DOI] [PubMed] [Google Scholar]
- 15.Liu D, DeGrado WF. De novo design, synthesis, and characterization of antimicrobial β-peptides. J Am Chem Soc. 2001;123:7553–7559. doi: 10.1021/ja0107475. [DOI] [PubMed] [Google Scholar]
- 16.Porter EA, Weisblum B, Gellman SH. Mimicry of host-defense peptides by unnatural oligomers: Antimicrobial β-Peptides. J Am Chem Soc. 2002;124:7324–7330. doi: 10.1021/ja0260871. [DOI] [PubMed] [Google Scholar]
- 17.Patch JA, Barron AE. Helical peptoid mimics of magainin-2 amide. J Am Chem Soc. 2003;125:12092–12093. doi: 10.1021/ja037320d. [DOI] [PubMed] [Google Scholar]
- 18.Tew GN, et al. De novo design of biomimetic antimicrobial polymers. Proc Natl Acad Sci USA. 2002;99:5110–5114. doi: 10.1073/pnas.082046199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu D, et al. Nontoxic membrane-active antimicrobial arylamide oligomers. Angew Chem Int Ed. 2004;43:1158–1162. doi: 10.1002/anie.200352791. [DOI] [PubMed] [Google Scholar]
- 20.Arnt L, Nüsslein K, Tew GN. Nonhemolytic abiogenic polymers as antimicrobial peptide mimics. J Polym Sci A Polym Chem. 2004;42:3860–3864. [Google Scholar]
- 21.Kuroda K, DeGrado WF. Amphiphilic polymethacrylate derivatives as antimicrobial agents. J Am Chem Soc. 2005;127:4128–4129. doi: 10.1021/ja044205+. [DOI] [PubMed] [Google Scholar]
- 22.Ilker MF, Nüsslein K, Tew GN, Coughlin EB. Tuning the hemolytic and antibacterial activities of amphiphilic polynorbornene derivatives. J Am Chem Soc. 2004;126:15870–15875. doi: 10.1021/ja045664d. [DOI] [PubMed] [Google Scholar]
- 23.Tew GN, Clements D, Tang H, Arnt L, Scott RW. Antimicrobial activity of an abiotic host defense peptide mimic. Biochim Biophys Acta, Biomembr. 2006;1758:1387–1392. doi: 10.1016/j.bbamem.2006.03.001. [DOI] [PubMed] [Google Scholar]
- 24.Mowery BP, et al. Mimicry of antimicrobial host-defense peptides by random copolymers. J Am Chem Soc. 2007;129:15474–15476. doi: 10.1021/ja077288d. [DOI] [PubMed] [Google Scholar]
- 25.Som A, Tew GN. Influence of lipid composition on membrane activity of antimicrobial phenylene ethynylene oligomers. J Phys Chem B. 2008;112:3495–3502. doi: 10.1021/jp077487j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yang L, et al. Synthetic antimicrobial oligomers induce a composition-dependent topological transition in membranes. J Am Chem Soc. 2007;129:12141–12147. doi: 10.1021/ja072310o. [DOI] [PubMed] [Google Scholar]
- 27.Matsuzaki K. Why and how are peptide-lipid interactions utilized for self-defense? Magainins and tachyplesins as archetypes. Biochim Biophys Acta, Biomembr. 1999;1462:1–10. doi: 10.1016/s0005-2736(99)00197-2. [DOI] [PubMed] [Google Scholar]
- 28.Matsuzaki K, et al. Relationship of membrane curvature to the formation of pores by magainin 2. Biochemistry. 1998;37:11856–11863. doi: 10.1021/bi980539y. [DOI] [PubMed] [Google Scholar]
- 29.DeChavigny A, Heacock PN, Dowhan W. Sequence and inactivation of the pss gene of Escherichia coli. Phosphatidylethanolamine may not be essential for cell viability. J Biol Chem. 1991;266:5323–5332. [PubMed] [Google Scholar]
- 30.Rietveld AG, et al. Regulation of lipid polymorphism is essential for the viability of phosphatidylethanolamine-deficient Escherichia coli cells. J Biol Chem. 1994;269:28670–28675. [PubMed] [Google Scholar]
- 31.Koltover I, Salditt T, Rädler JO, Safinya CR. An inverted hexagonal phase of cationic liposome-DNA complexes related to DNA release and delivery. Science. 1998;281:78–81. doi: 10.1126/science.281.5373.78. [DOI] [PubMed] [Google Scholar]
- 32.Mishra A, Gordon VD, Yang L, Coridan R, Wong GCL. HIV TAT forms pores in membranes by inducing saddle-splay curvature: Potential role of bidentate hydrogen bonding. Angew Chem Int Ed. 2008;47:2986–2989. doi: 10.1002/anie.200704444. [DOI] [PubMed] [Google Scholar]
- 33.Turner DC, Gruner SM. X-ray diffraction reconstruction of the inverted hexagonal (HII) phase in lipid-water systems. Biochemistry. 1992;31:1340–1355. doi: 10.1021/bi00120a009. [DOI] [PubMed] [Google Scholar]
- 34.Ishitsuka Y, et al. Amphiphilic poly(phenyleneethynylene)s can mimic antimicrobial peptide membrane disordering effect by membrane insertion. J Am Chem Soc. 2006;128:13123–13129. doi: 10.1021/ja061186q. [DOI] [PubMed] [Google Scholar]
- 35.Lee M-T, Hung W-C, Chen F-Y, Huang HW. Many-body effect of antimicrobial peptides: On the correlation between lipid's spontaneous curvature and pore formation. Biophys J. 2005;89:4006–4016. doi: 10.1529/biophysj.105.068080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chen F-Y, Lee M-T, Huang HW. Evidence for membrane thinning effect as the mechanism for peptide-induced pore formation. Biophys J. 2003;84:3751–3758. doi: 10.1016/S0006-3495(03)75103-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Heller WT, et al. Membrane thinning effect of the β-sheet antimicrobial protegrin. Biochemistry. 2000;39:139–145. doi: 10.1021/bi991892m. [DOI] [PubMed] [Google Scholar]
- 38.Needham D, Evans E. Structure and mechanical properties of giant lipid (DMPC) vesicle bilayers from 20°C below to 10°C above the liquid crystal-crystalline phase transition at 24°C. Biochemistry. 1988;27:8261–8269. doi: 10.1021/bi00421a041. [DOI] [PubMed] [Google Scholar]
- 39.Tamba Y, Yamazaki M. Single giant unilamellar vesicle method reveals effect of antimicrobial peptide magainin 2 on membrane permeability. Biochemistry. 2005;44:15823–15833. doi: 10.1021/bi051684w. [DOI] [PubMed] [Google Scholar]
- 40.Harper PE, Mannock DA, Lewis RNAH, McElhaney RN, Gruner SM. X-ray diffraction structures of some phosphatidylethanolamine lamellar and inverted hexagonal phases. Biophys J. 2001;81:2693–2706. doi: 10.1016/S0006-3495(01)75912-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Henry NFM, Lonsdale K. International Tables for X-ray Crystallography. Birmingham, United Kingdom: Kynoch Press; 1969. [Google Scholar]
- 42.Yang L, et al. Self-assembled virus-membrane complexes. Nat Mater. 2004;3:615–619. doi: 10.1038/nmat1195. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.