Abstract
Mutations in the a disintegrin and metalloprotease with thrombospondin motifs (ADAMTS) family of secreted proteases cause diseases linked to ECM abnormalities. However, the mechanisms by which these enzymes modulate the ECM during development are mostly unexplored. The Caenorhabditis elegans MIG-17/ADAMTS protein is secreted from body wall muscle cells and localizes to the basement membrane (BM) of the developing gonad where it controls directional migration of gonadal leader cells. Here we show that specific amino acid changes in the ECM proteins fibulin-1C (FBL-1C) and type IV collagen (LET-2) result in bypass of the requirement for MIG-17 activity in gonadal leader cell migration in a nidogen (NID-1)-dependent and -independent manner, respectively. The MIG-17, FBL-1C and LET-2 activities are required for proper accumulation of NID-1 at the gonadal BM. However, mutant FBL-1C or LET-2 in the absence of MIG-17 promotes NID-1 localization. Furthermore, overexpression of NID-1 in mig-17 mutants substantially rescues leader cell migration defects. These results suggest that functional interactions among BM molecules are important for MIG-17 control of gonadal leader cell migration. We propose that FBL-1C and LET-2 act downstream of MIG-17-dependent proteolysis to recruit NID-1 and that LET-2 also activates a NID-1-independent pathway, thereby inducing the remodeling of the BM required for directional control of leader cell migration.
Keywords: ECM, fibulin-1, organogenesis, type IV collagen
The interaction between basement membranes (BMs) and migrating cells or the epithelial layer is a complicated but carefully controlled process that involves remodeling of the ECM. For example, the migration of border cells required for oogenesis and patterning of the early embryo in Drosophila melanogaster is accompanied by precise regulation of the synthesis and degradation of type IV collagen, laminin, and perlecan (1, 2). Fibronectin expression is required for branching morphogenesis of the submandibular salivary gland, lung, and kidney in mouse (3). However, the mechanisms of cell-ECM interactions and the roles of BMs in cell migration remain largely unknown.
Gonadogenesis in the nematode Caenorhabditis elegans serves as a simple model system for elucidating the function of BMs in organ morphogenesis. The development of hermaphrodite gonads is regulated by the migration of gonadal distal tip cells (DTCs), which promote directional elongation of the gonad arms during the larval stages to form the U-shaped gonads found in adult animals (4, 5). Two secreted a disintegrin and metalloprotease with thrombospondin motifs (ADAMTS) family metalloproteases, MIG-17 and GON-1, are involved in this process. GON-1 is required for DTC motility, whereas MIG-17 controls the direction of DTC movement but does not control DTC motility per se (6, 7). MIG-17 is expressed in the body wall muscles and is localized to the BM of the gonad surface, where it is required for DTC migration (7, 8). It has been proposed that MIG-17 and GON-1 remodel BMs via proteolysis (6, 7). Dominant mutations in fbl-1, which encodes fibulin-1 (FBL-1), an ECM protein, can bypass the requirement for MIG-17 activity in DTC migration (9). Furthermore, fbl-1 deletion suppresses the gonadal elongation defects of gon-1 null mutants (10). These observations suggest that mutation of FBL-1 or depletion of FBL-1 may give rise to alterations in BM ECM architecture that mimic the downstream events normally elicited by MIG-17 or GON-1 activities, respectively.
In this study, we analyzed a novel genetic locus, saf-2 [suppressor of a disintegrin and metalloprotease (ADAM) family defect], in which mutations act as dominant suppressors of DTC migration defects in mig-17 mutants. saf-2 was found to be equivalent to let-2, which was previously identified by lethal mutations and encodes the type IV collagen α2 chain (11). Two suppressor let-2 mutations were identified in the C-terminal region of LET-2, one in the noncollagenous (NC1) domain and the other in a Gly-X-Y repeat just upstream of the NC1 domain. let-2; fbl-1 double mutants exhibited severe gonadal defects that were not observed in either single mutant, revealing a synthetic interaction between these independently identified suppressor genes. Interestingly, fbl-1 mutants suppressed mig-17 in a nid-1-dependent manner, whereas let-2 mutants suppressed mig-17 in a nid-1-independent manner. Genetic analysis of protein localization to the gonadal BM revealed that MIG-17 is required for active accumulation of FBL-1C, which contributes to NID-1 localization. Our findings suggest that intermolecular interactions among FBL-1C, NID-1, and LET-2 are involved in MIG-17/ADAMTS-dependent regulation of gonadal leader cell migration.
Results
Mutations in the Type IV Collagen α2 Chain Suppress DTC Migration Defects in mig-17 Mutants.
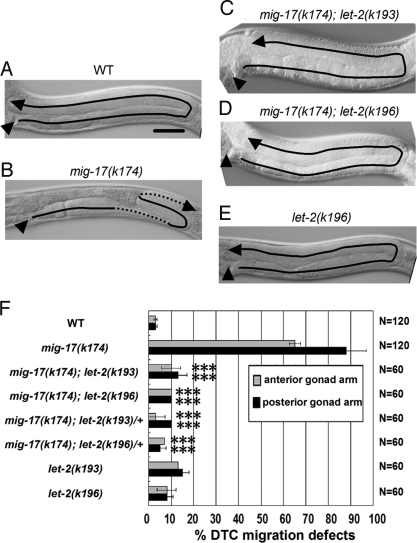
Mutations in mig-17 resulted in misshapen gonads due to misdirected migration of the gonadal DTCs (Fig. 1 A and B). To identify molecules that interact with MIG-17 to regulate DTC migration, we used ethylmethane sulfonate mutagenesis to isolate suppressors of mig-17(k174), a null allele containing a nonsense mutation. The suppressor mutations let-2(k193) and let-2(k196) were dominant and strongly suppressed the DTC migration defects in homozygotes and heterozygotes (Fig. 1 C, D, and F). mig-17(k174) single mutants show gonad morphogenesis defects, whereas mig-17; let-2(k193) and mig-17; let-2(k196) double mutants mostly had U-shaped gonad arms similar to those in WT animals, although the gonads were slightly thicker and shorter than in the WT (Fig. 1 C, D, and F). let-2 encodes LET-2A and LET-2B, two spliced isoforms of the α2 chain of type IV collagen (12). The k193 and k196 mutants contained amino acid substitutions in the NC1 domain and Gly-X-Y repeats of LET-2, respectively [supporting information (SI) Fig. S1]. The serine mutated in k193 is evolutionarily conserved between the C. elegans and human α2 chains, suggesting its functional importance in type IV collagen (Fig. S1A).
Fig. 1.
Gonadal phenotypes of the WT and mutants. Anterior is left and dorsal is up. Posterior gonads of WT (A), mig-17(k174) (B), mig-17(k174); let-2(k193) (C), mig-17(k174); let-2(k196) (D) and let-2(k196) (E) hermaphrodites are shown. The predicted migratory routes of DTCs are indicated by arrows. The stippled lines in (B) indicate that the corresponding regions of the gonad arm are out of focus. Arrowheads point to the vulvae. (Scale bar, 50 μm.) (F) Percentages of anterior and posterior gonad arms with abnormal DTC migration. Error bars represent the mean SD. Results for Fisher's exact test against mig-17(k174) are indicated: ***, P < 0.001; NS, not significant.
Because the conventional let-2 mutations are temperature-sensitive (ts) embryonic or larval lethal (12), we examined these phenotypes in the suppressor let-2 mutants. We found that k193 has weak nonconditional embryonic lethality and k196 has ts embryonic and larval lethality (Table S1). The suppressor let-2 mutants showed minimal DTC migration defects by themselves (Fig. 1 E and F). To analyze the nature of the mutant LET-2 proteins, we examined the effect of transgenic extrachromosomal arrays containing multiple let-2 genes on the suppression of DTC migration defects when introduced into mig-17(k174) mutants (Fig. S1B). Although WT let-2 arrays weakly suppressed mig-17 defects in the anterior gonads, mutant let-2 transgene arrays comprised of the k193 and k196 mutations strongly suppressed mig-17 defects (Fig. S1B). These results suggest that the suppressor let-2 mutations are gain-of-function mutations that bypass the MIG-17 requirement for control of DTC migration.
We also investigated the effects of known let-2 mutations on gonad morphogenesis in mig-17 mutants. Two ts lethal alleles, g25 and b246 (12), suppressed the DTC migration defects very weakly at 20 °C (Table S2). When animals were shifted from 20 °C to 22.5 °C at the L2 stage, b246 and g25 partially suppressed the posterior gonadal defects of mig-17. Very weak suppression was observed in L3 stage shifted animals (Table S3). Shifting to 25 °C resulted in severe gonadal defects, which precluded the analysis of mig-17 suppression. To examine whether reduction of type IV collagen in the BM affects the suppression, we depleted a single copy of emb-9/α1 chain of type IV collagen using the null allele emb-9(g23cg46) (13). We found that g23cg46/+ partially suppressed mig-17(k174) (Fig. S1C). These results suggest that a defect in or reduction of type IV collagen suppresses the DTC migration defect of mig-17 mutants.
Expression of Suppressor LET-2 Proteins.
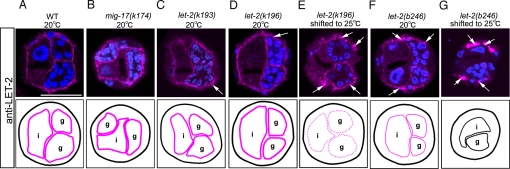
The LET-2 protein is secreted from the body wall muscle cells and localizes to the BM. We examined whether suppressor LET-2 mutant proteins localize to the BM. An Ab against LET-2 clearly stained cross-sections of gonadal and intestinal BMs of WT and mig-17 hermaphrodites (Fig. 2 A and B). The Ab similarly stained BMs of let-2(k193) and let-2(k196) mutants at 20 °C although weak accumulation of LET-2 proteins was detectable in the cytoplasm of the muscle cells (Fig. 2 C and D). BM localization was also observed in let-2(b246) mutants (13), but less LET-2 protein localized in let-2(b246) mutants than in the suppressor let-2 mutants grown at 20 °C (Fig. 2 C, D, and F) or 25 °C (Fig. 2 E and G). In contrast, accumulation of LET-2 proteins in the muscle cells in let-2(b246) mutants (13) was much greater than in the suppressor mutants (compare Fig. 2 C, D, and F with E and G). Because LET-2 is highly expressed in DTCs (14), we also examined LET-2 expression in DTCs in animals shifted to 25 °C. As in the case for the muscle cells, some accumulation of mutant LET-2(k193) and LET-2(k196) proteins was observed, although no LET-2 accumulation was detected in WT DTCs (Fig. S2).
Fig. 2.
LET-2 localization to the gonadal BM. Localization of LET-2 proteins in WT (A), mig-17 (B) and let-2 (C–G) animals. Cross-sections of L4 hermaphrodites were stained with anti-LET-2 (magenta) and DAPI (blue). Arrows indicate accumulation of LET-2 proteins in the muscle cells. Borders of the gonads and intestines are illustrated below the photos. Levels of LET-2 localization to the BM are indicated in magenta by thick lines (normal levels), thin lines (low levels), and dotted lines (very low levels). Black lines indicate undetectable levels of protein. g, gonad; i, intestine. (Scale bar, 25 μm.) The accumulation of LET-2(k196) in the muscle cells was higher at 25 °C than at 20 °C. The result of let-2(k193) shifted to 25 °C was similar to (C).
Genetic Interactions Between Suppressor Alleles of let-2 and fbl-1.
We previously showed that specific amino acid substitutions in the BM protein FBL-1C can suppress mig-17 mutations in a dominant gain-of-function manner (9)—a mode of suppression reminiscent of that observed in the let-2 suppressors. Therefore, we tested potential genetic interactions among let-2, fbl-1, and mig-17 mutants by constructing double and triple mutants. We used two fbl-1 alleles, k201 and k206. When combined with let-2 or let-2; mig-17 double mutants, they showed strong gonad morphogenesis defects that were not observed in either the fbl-1 or the let-2 single mutants. The gonad arms of the double and triple mutants were about 1.5- to 2-fold thicker than those of WT. In particular, the DTCs of the anterior gonad arms often ceased migration shortly after the first or second turn, which resulted in severe swelling of the proximal part of the gonad arms (Fig. S3).
We next examined whether fbl-1(k201) and let-2(k196) interacted genetically with mutations in other BM proteins. fbl-1(k201) and let-2(k196) enhanced the DTC migration defect of unc-52(e1421)/perlecan but not of cle-1(cg120)/typeXVIII collagen (Fig. S4). A weak allele of epi-1/lamininα, gm57 was also enhanced by let-2(k196) (Fig. S4). These results suggest that the gonadal BMs in let-2 and fbl-1 mutants are partially compromised and that LET-2 and FBL-1 act together in the gonadal BM to achieve proper morphogenesis of the gonad.
NID-1 Dependency of Suppression.
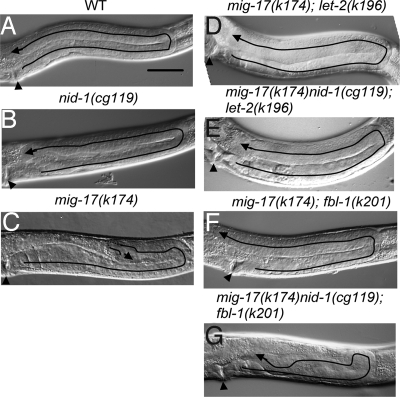
Mammalian nidogen is known to specifically bind to type IV collagen in vitro (15). We therefore examined whether suppression of mig-17 mutations by mutant LET-2 proteins depends on NID-1. C. elegans has a single nidogen gene and a null mutation in this gene, nid-1(cg119) (16), showed only weak DTC migration defects by itself (Fig. 3 A and B and Fig. S5A). nid-1(cg119) enhanced the DTC migration defect in mig-17(k174) mutants, suggesting that the functions of mig-17 and nid-1 are partially redundant with respect to control of DTC migration (Fig. S5A). nid-1(cg119) did not enhance the weak DTC migration defects of the let-2 mutants (Fig. S5B). The introduction of nid-1(cg119) into the mig-17(k174); let-2(k193) or mig-17(k174); let-2(k196) backgrounds did not influence the suppressive activity of the let-2 mutations, indicating that the suppression by mutant LET-2 proteins does not depend on NID-1 (Fig. 3 D and E and Fig. S5B).
Fig. 3.
NID-1-independent and -dependent suppression of mig-17. (A–G) Phenotypes of WT and mutant gonads. (Scale bar, 50 μm.) The predicted migratory routes of DTCs are depicted by lines, and the distal tips of the gonad arms are indicated by arrows. Arrowheads point to the vulvae.
Because human fibulin-1 has also been reported to physically interact with nidogen in vitro (17), we examined whether mig-17 suppression by the FBL-1C mutant proteins requires NID-1. We introduced nid-1(cg119) into the mig-17(k174); fbl-1(k201) or mig-17(k174); fbl-1(k206) backgrounds. Interestingly, we observed that suppression of DTC migration defects by the fbl-1 mutations was severely impaired in the absence of NID-1, in contrast to that observed for the let-2 mutations (Fig. 3 F and G and Fig. S5C). The triple mutants exhibited meandering DTC migration phenotypes similar to those observed in mig-17 (Fig. 3C). To examine whether the intact NID-1 is required for suppression of mig-17 by fbl-1, an in-frame deletion allele nid-1(cg118) lacking the G2 domain was introduced into the fbl-1(k201); mig-17(k174) or fbl-1(k206); mig-17(k174) backgrounds (16). The suppression was partially weakened in fbl-1(k206); mig-17(k174), and was not affected in fbl-1(k201); mig-17(k174) (Fig. S5C). These results indicate that the G2 domain of NID-1 is not essential for suppression of mig-17 by mutant FBL-1 proteins.
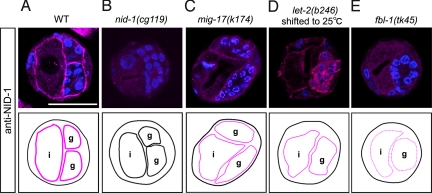
NID-1 Accumulation Is Increased in mig-17; fbl-1 and mig-17; let-2 Double Mutants.
To examine localization of NID-1, we generated a specific Ab that recognizes the C-terminal G3 domain of NID-1. Using immunohistochemistry, we demonstrated that this Ab stained the gonadal and intestinal BMs in WT but not in nid-1(cg119) animals (Fig. 4 A and B). The signal was fainter in mig-17(k174) and let-2(b246) and was very faint in fbl-1(tk45) (Fig. 4 C–E). These results suggest that NID-1 localization is strongly dependent on FBL-1C and partially dependent on MIG-17 and LET-2. We therefore examined whether NID-1 localization is affected by the fbl-1 and let-2 suppressors. Interestingly, NID-1 accumulation in the gonadal BM was increased in both mig-17(k174); fbl-1 and mig-17(k174); let-2 double mutants (Fig. 5 A–D). Because the G2 domain of nidogen binds type IV collagen in mammals (18, 19), we examined NID-1(cg118) localization in the mig-17(k174), mig-17(k174); let-2(k193) and mig-17(k174); let-2(k196) mutants. In contrast to the weak localization of NID-1(cg118) observed in the mig-17(k174) mutants, NID-1(cg118) localization was increased in both mig-17(k174); let-2(k193) and mig-17(k174); let-2(k196) mutants (Fig. S6), indicating that mutant LET-2 proteins do not require the G2 domain to recruit NID-1. Thus, the recruitment of NID-1 to the gonadal BM by LET-2 may be indirect. These results indicate that one of the molecular consequences of the suppressor let-2 and fbl-1 mutations is enhancement of the activity to localize NID-1 in the gonadal BMs which is weakened in the absence of MIG-17.
Fig. 4.
Genetic analysis of NID-1 localization. Localization of NID-1 in (A) WT, (B) nid-1(cg119), (C) mig-17(k174), (D) let-2(b246), and (E) fbl-1(tk45) animals. Cross-sections of L4 hermaphrodites were stained with anti-NID-1 (magenta) and DAPI (blue). The Ab recognizes an unknown antigen in the gonad of let-2(b246) from animals reared at 25 °C (D). Levels of NID-1 localization to the BM are indicated by magenta thick lines (normal levels), thin lines (low levels) and dotted lines (very low levels). g, gonad; i, intestine. (Scale bar, 25 μm.)
Fig. 5.
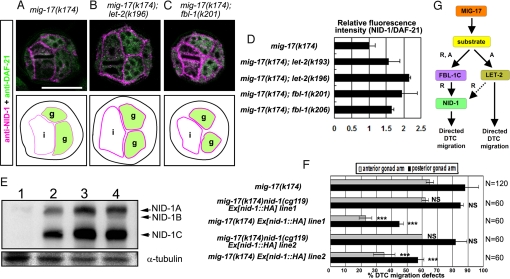
NID-1 overexpression suppresses mig-17. (A–C) Cross-sections of L4 hermaphrodites stained with anti-NID-1 (magenta) and anti-DAF-21 (HSP90) (green). DAF-21 was used as an internal standard for quantification of NID-1. (A) mig-17, (B) mig-17; let-2(k196), (C) mig-17; fbl-1(k201). Borders of gonads and intestines are illustrated below the photos. Levels of NID-1 localization to the BM are indicated by magenta thick lines (normal levels) and thin lines (low levels). g: gonads, i: intestines. (Scale bar, 25 μm.) (D) Fluorescence intensity of anti-NID-1 relative to that of anti-DAF-21. The relative fluorescence intensity for each sample was normalized to that of mig-17. See Fig. S6E legend for methods. (E) Extracts from nid-1(cg119) (lane 1), mig-17 (k174) (lane 2), mig-17 Ex[nid-1::HA] line1 (lane 3) or mig-17 Ex[nid-1::HA] line2 (lane 4) hermaphrodites were analyzed. nid-1 expresses three spliced isoforms, NID-1A, B, and C (arrows). Expression of NID-1 proteins are 2.5-fold (line 3) and 2.8-fold (line 4) higher in these transgenic animals compared to mig-17 animals (n = 3). (F) Quantification of DTC migration defects in mig-17 and mig-17 Ex[nid-1::HA] (a NID-1-HA overexpression strain). Results for Fisher's exact test against mig-17(k174) are indicated: ***, P < 0.001; NS, not significant. Error bars represent the mean SD. (G) Model for the protein cascade downstream of MIG-17. MIG-17-dependent proteolysis of an unknown substrate recruits and activates FBL-1C, which then recruits NID-1 to the BM to control DTC migration. MIG-17-dependent proteolysis also activates LET-2 to induce NID-1-dependent and -independent MIG-17 pathways. Dashed arrow, potentially indirect interaction. R, recruitment; A, activation.
These observations raise the possibility that reduction of NID-1 in the gonadal BM is the cause of misdirected DTC migration in mig-17 mutants. We therefore examined whether overexpression of NID-1 in mig-17 mutants suppresses the DTC migration defects. We used HA epitope-tagged NID-1, which can restore suppression when expressed in mig-17(k174) nid-1(g119); fbl-1, in this experiment. Surprisingly, the DTC migration in mig-17 animals was markedly ameliorated by NID-1-HA overexpression (Fig. 5 E and F). Thus, it is likely that NID-1 accumulation in the gonadal BM plays an important role in MIG-17-dependent control of DTC migration.
We previously showed that the gonadal localization of FBL-1C is reduced in mig-17 mutants (9). Therefore, it is possible that the decreased localization of NID-1 in mig-17 might reflect the reduction of FBL-1C. We examined whether overexpression of fbl-1C::3HA, which rescues the fbl-1(tk45) null mutant, can enhance NID-1 accumulation in the gonadal BM and rescue the DTC migration defects of mig-17 mutants. FBL-1C-3HA weakly localized to the BM (Fig. S7C). However, accumulation of NID-1 was not affected and was similar to that observed in mig-17 single mutants (Fig. S7 B and D). Consistent with these results, overexpression of FBL-1C-3HA did not rescue the DTC migration defects in mig-17 mutants (Fig. S7E).
Discussion
MIG-17 is an ADAMTS protease localized to the gonadal BM, where it is likely required for proper remodeling of the BM to promote gonad morphogenesis. Using C. elegans as a model system, we isolated two suppressor mutations of the gonadogenesis defects of mig-17 mutants and found that they occur in the C-terminal region of the α2 chain of type IV collagen, which is encoded by let-2. The suppressor let-2 mutations showed synergistic effects when combined with suppressor fbl-1 mutations. The suppression by fbl-1 was dependent on nid-1, whereas that by let-2 was not. Analysis of protein localization revealed that NID-1 localization to the gonadal BM was reduced in mig-17 mutants, but suppressor let-2 or fbl-1 mutations promoted NID-1 localization in mig-17 mutants.
In this study, we analyzed protein localization to the BM of gonad arms rather than to the DTCs because the latter analysis is technically difficult. However, we have previously shown that MIG-17 localizes throughout, and recruits FBL-1C to, the gonadal BM (8, 9). We also demonstrated that MIG-17 is activated in the BM of gonad arms specifically during directional migration of DTCs (20). These results are consistent with our observation that NID-1 localization to the BM of gonad arms correlates with MIG-17-dependent control of DTC migration, suggesting that MIG-17 recruitment of FBL-1C and NID-1 in the BM of gonad arms reflects its activity in DTC migration.
Partial Dysfunction of BMs in Suppressor Mutants.
The let-2(b246) animals showed strong accumulation of type IV collagen in muscle cells even at 20 °C and had weak suppressor activity, whereas the secretion of type IV collagens was mostly normal in suppressor let-2(k196) and let-2(k193) mutants and mig-17 mutant suppression was substantial at 20 °C, implying that the latter let-2 mutants exhibited strong suppression due to defects in the BM that are induced by the secreted mutant LET-2 proteins. The k193 mutation is in the C-terminal NC1 domain that is important for initiating the assembly of three α chains (two α1 and one α2), as well as for the covalent linkage of NC1 domains of two type IV collagen trimers (21). Because the k193 and k196 mutants are viable and fertile, the gonadal BMs are presumably not as seriously impaired as in the lethal let-2 mutants. Thus, it may be that the α chain trimer is partially malformed in k193 mutants. Alternatively, the type IV collagen network that is formed in k193 mutants could be partially disorganized due to aberrant formation of intermolecular bonds involving NC1 domains. Because the k196 mutation is an amino acid substitution that can perturb triple helix formation near the NC1 domain, it may also affect the function of the NC1 domain.
The dysfunction of BMs in the let-2 and fbl-1 suppressor mutants is supported by the fact that the let-2(k196) mutation genetically interacts with fbl-1 mutations k201 and k206 and that the let-2; fbl-1 double mutants exhibit a strong swollen gonad phenotype. The enhancement of DTC migration defects of unc-52 and epi-1 by let-2 and fbl-1 suppressors also support this idea.
NID-1-Dependent and -Independent Pathways Downstream of MIG-17.
Because the nid-1(cg119) null mutant has essentially WT U-shaped gonads, it appears that MIG-17 and NID-1 have redundant functions in DTC migration. However, the fact that NID-1 localization is reduced in mig-17 mutants and overexpression of NID-1 can markedly rescue the DTC migration defects suggests that MIG-17 also acts through NID-1. Thus, it is likely that both NID-1-dependent and -independent pathways are downstream of MIG-17 and that these two pathways are redundant. Although fbl-1 function in the WT background was essential for normal NID-1 localization, overexpression of FBL-1C in the mig-17 background did not enhance NID-1 localization. Therefore, MIG-17 activity is probably required not only for recruiting FBL-1C (9), but also for activating FBL-1C to recruit NID-1. Although it is possible that FBL-1C or LET-2 is the substrate of MIG-17, so far we have failed to detect such interactions. We speculate that the cleavage of an unknown substrate by MIG-17 activates FBL-1C by, for example, altering the conformation of FBL-1C, which then recruits NID-1 and causes the remodeling of the gonadal BM that is required for directional DTC migration (Fig. 5G).
We observed that accumulation of NID-1 at the BM increased in the suppressor fbl-1 and let-2 mutants in the absence of MIG-17. The specific amino acid substitutions in mutant FBL-1C or LET-2 might mimic MIG-17 function to increase the affinity of the BM for NID-1. Because the suppression by fbl-1 is nid-1-dependent, it is reasonable that mutant FBL-1 gained the function of NID-1 localization. However, enhancement of NID-1 localization in let-2 mutants is not expected because of the nid-1-independent suppression by let-2 mutants. Because abnormal DTC migration in mig-17 mutants can be substantially suppressed by NID-1 overexpression, it seems that the recovery of NID-1 gonadal BM accumulation in suppressor mutants is functionally important for suppression of DTC migration defects. It might be possible that both NID-1-dependent and -independent MIG-17 pathways are activated in suppressor let-2 mutants and that the latter pathway is still intact even in the nid-1; let-2 double mutants. The reduced localization of NID-1 at the gonadal BM in let-2(b246) mutants supports the idea that WT LET-2 is also required for efficient accumulation of NID-1. Furthermore, we showed that gonadal FBL-1C-Venus localization was not affected in let-2(b246) mutants grown at nonpermissive temperatures (see Fig. S7 F and G), indicating that LET-2 recruits NID-1 independently of FBL-1C. Although it is not required for localization of LET-2, MIG-17 may regulate the function of WT LET-2 in the BM to induce NID-1-dependent and -independent pathways for DTC migration control (Fig. 5G). For example, LET-2 may contain a cryptic guidance cue that is exposed by MIG-17-dependent proteolysis or by suppressor let-2 mutations. Cryptic fragments of type IV collagen having angiogenic and anti-angiogenic activities have been described in mammals (22–24).
Possible Functions of Nidogens in Cell Migration.
Recently, regulated expression of two α-integrins, INA-1 and PAT-2, was shown to be important for turning and cessation of movement of DTCs: INA-1 is required for continuous migration, whereas PAT-2 is required for pathfinding during migration (25). Because integrins are major receptors for ECM proteins, the meandering migration of DTCs in mig-17 mutants may be due to misrecognition of the BM by the integrin receptors. In mammals, integrins expressed in neutrophils bind the Arg-Gly-Asp (RGD) sequence in the rod domain of nidogen and activate chemotaxis (26). Integrins containing the β1 subunit have been shown to be required for nidogen-dependent migration of Schwann cells (27). Therefore, nidogen plays an important role in the interaction between integrins and the BM.
In DTC pathfinding, dorsal migration is guided by the membrane-bound receptors UNC-5 and UNC-40, which respond to the UNC-6/netrin gradient (28). Because we previously showed that mig-17(k174) enhances a weak allele of unc-6 (7), the DTCs in mig-17 mutants may not be sufficiently sensitive to the UNC-6 signal. Interestingly, NID-1 has been reported to interact with UNC-40 in the guidance of commissural axons in C. elegans; NID-1 appears to negatively regulate UNC-40 when the dorsally migrating axon of the SDQR neuron reorients anteriorly (29). Reduction of NID-1 in the gonadal BM may also reduce the guidance function of UNC-40 expressed in DTCs.
Interactions Among Fibulin-1, Nidogen and Type IV Collagen May Be Evolutionarily Conserved.
Type IV collagen is a major component of the BM and is associated with other ECM molecules such as nidogen, laminin, and heparan sulfate proteoglycans (30–32). Mammalian fibulin-1 binds to nidogen-1, laminin-1, and some proteoglycans (15, 17, 33). Mouse fibulin-1 also interacts weakly with type IV collagen (34). Therefore, LET-2 and FBL-1 may bind directly or indirectly through other ECM molecules to form a supramolecular network in the BM to support proper gonad morphogenesis. Our findings that both mutant FBL-1C and LET-2 can enhance NID-1 BM accumulation and that the suppression of DTC migration defects of mig-17 by mutant FBL-1C depends on NID-1 implies functional interactions among these four molecules.
There are two nidogen genes in mammals (35). nidogen-1 and nidogen-2 double-KO mice and fibulin-1 KO mice exhibit similar delayed embryonic lung development with thickened parenchymal septa and improperly expanded saculi (35, 36). These results suggest that fibulin-1 and nidogens also functionally interact in mammals.
Finally, our finding that in C. elegans MIG-17 interacts with fibulin-1, nidogen, and type IV collagen, all major components of the BM, suggests the possibility that a similar functional interaction between ADAMTS and these ECM molecules may also occur in mammals during organogenesis.
Materials and Methods
Strains and Genetic Analysis.
Culture, handling and ethylmethane sulfonate mutagenesis of C. elegans were performed using standard methods (37). Worms were grown at 20 °C unless otherwise noted. The following mutations and genetic balancers were used in this work: cle-1(cg120), emb-9(g23cg46), epi-1(gm57), fbl-1(k201, k206 and tk45), let-2(b246, g25, k193 and k196), mig-17(k174), nid-1(cg118 and cg119), unc-7(e139), unc-42(e270), unc-52(e1421), unc-119(e2498), nT1[qIs51] and hT2[qIs48].
Microscopy.
DTC migration was analyzed as described (38). The strains with let-2 suppressor mutations often showed somewhat shorter phase III migration than WT animals. This phenotype was not scored as a defect here. The localization of LET-2, FBL-1C, and NID-1 was analyzed using a confocal laser scanning microscope (LSM5, Zeiss) equipped with a C-apochromat 63× (water immersion; NA 1.2) lens and controlled by PASCAL version 3.2 SP2 software.
Constructs and Germline Transformation.
Construction of the nid-1::HA and let-2 plasmids and methods for germline transformation are described in the SI Materials and Methods.
Preparation of Antisera and Ab Staining.
The nid-1 cDNA clone yk1531C10 was obtained from Yuji Kohara at the National Institute of Genetics, Mishima, Japan. Part of the G3 domain (residues 1281–1414 for NID-1A) (17) of NID-1 tagged with histidines was cloned into pET-28a(+) (Novagen). The tagged protein was isolated from E. coli. Rabbit antiserum against NID-1 was purified on a column fixed with the histidine-tagged NID-1. L3 to L4 animals were fixed with 4% paraformaldehyde in PBS for 12 h on ice. Frozen sections were prepared as described (39). After blocking the sections with 0.1% (wt/vol) Triton X-100 and 3% BSA in PBS, they were immunostained by incubation with anti-LET-2 (NW68, 1:200) (14), mouse anti-GFP IgG (3E6, 1:200; Molecular Probes), mouse monoclonal anti-DAF-21(HSP90) (608F, 1:10) (40), anti-NID-1 (5 μg/ml) or rat anti-HA IgG (3F10, 2 μg/ml; Roche) for 12 h at 4 °C followed by incubation with the appropriate Alexa Fluor-conjugated secondary Ab (1:500; Molecular Probes) for 1 h at room temperature and then stained with DAPI (2 μg/ml; Wako) for 10 min at room temperature. Anti-NID-1 was preabsorbed with the acetone powder of nid-1(cg119) null mutants.
Western Blotting.
Mixed stage worm extracts (20 μg) were immunoblotted with rabbit anti-NID-1 (2 μg/ml) or mouse monoclonal anti-α-tubulin (12G10, 1:1,000, J. Frankel and M. Nelson, provided by Developmental Studies Hybridoma Bank at the University of Iowa).
Supplementary Material
Acknowledgments.
We thank Jim Kramer for type IV collagen Abs, Yasunori Yamaguchi for the DAF-21 mAb, Alan Coulson for cosmid clones, Yuji Kohara for cDNA clones, Andy Fire for GFP fusion vectors, and Theresa Stiernagle for strains. We also thank Mitsue Sano and Asami Sumitani for technical assistance, and Kunihiro Matsumoto, Naoki Hisamoto, and Shinji Ihara for critical reading of the manuscript. Some nematode strains used in this work were provided by the Caenorhabditis Genetics Center, which is funded by the NIH National Center for Research Resources. This work was supported by grants from the Japanese Ministry of Education, Culture, Sports, Science and Technology.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0804055106/DCSupplemental.
References
- 1.Montell DJ. Border-cell migration: the race is on. Nat Rev Mol Cell Biol. 2003;4:13–24. doi: 10.1038/nrm1006. [DOI] [PubMed] [Google Scholar]
- 2.Medioni C, Noselli S. Dynamics of the basement membrane in invasive epithelial clusters in Drosophila. Development. 2005;132:3069–3077. doi: 10.1242/dev.01886. [DOI] [PubMed] [Google Scholar]
- 3.Sakai T, Larsen M, Yamada KM. Fibronectin requirement in branching morphogenesis. Nature. 2003;423:876–881. doi: 10.1038/nature01712. [DOI] [PubMed] [Google Scholar]
- 4.Kimble JE, White JG. On the control of germ cell development in Caenorhabditis elegans. Dev Biol. 1981;81:208–219. doi: 10.1016/0012-1606(81)90284-0. [DOI] [PubMed] [Google Scholar]
- 5.Hedgecock EM, Culotti JC, Hall DH, Stern BD. Genetics of cell and axon migrations in Caenorhabditis elegans. Development. 1987;100:365–382. doi: 10.1242/dev.100.3.365. [DOI] [PubMed] [Google Scholar]
- 6.Blelloch R, Kimble J. Control of organ shape by a secreted metalloprotease in the nematode Caenorhabditis elegans. Nature. 1999;399:586–590. doi: 10.1038/21196. [DOI] [PubMed] [Google Scholar]
- 7.Nishiwaki K, Hisamoto N, Matsumoto K. A metalloprotease disintegrin that controls cell migration in Caenorhabditis elegans. Science. 2000;288:2205–2208. doi: 10.1126/science.288.5474.2205. [DOI] [PubMed] [Google Scholar]
- 8.Ihara S, Nishiwaki K. Prodomain-dependent tissue targeting of an ADAMTS protease controls cell migration in Caenorhabditis elegans. EMBO J. 2007;26:2607–2620. doi: 10.1038/sj.emboj.7601718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kubota Y, Kuroki R, Nishiwaki K. A fibulin-1 homolog interacts with an ADAM protease that controls cell migration in C. elegans. Curr Biol. 2004;14:2011–2018. doi: 10.1016/j.cub.2004.10.047. [DOI] [PubMed] [Google Scholar]
- 10.Hesselson D, Newman C, Kim KW, Kimble J. GON-1 and fibulin have antagonistic roles in control of organ shape. Curr Biol. 2004;14:2005–2010. doi: 10.1016/j.cub.2004.11.006. [DOI] [PubMed] [Google Scholar]
- 11.Sibley MH, Johnson JJ, Mello CC, Kramer JM. Genetic identification, sequence, and alternative splicing of the Caenorhabditis elegans α2(IV) collagen gene. J Cell Biol. 1993;123:255–264. doi: 10.1083/jcb.123.1.255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sibley MH, Graham P.L, von Mende N, Kramer JM. Mutations in the α2(IV) basement membrane collagen gene of Caenorhabditis elegans produce phenotypes of differing severities. EMBO J. 1994;13:3278–3285. doi: 10.1002/j.1460-2075.1994.tb06629.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gupta MC, Graham PL, Kramer JM. Characterization of α1(IV) collagen mutations in Caenorhabditis elegans and the effects of α1 and α2(IV) mutations on type IV collagen distribution. J Cell Biol. 1997;137:1185–1196. doi: 10.1083/jcb.137.5.1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Graham PL, et al. Type IV collagen is detectable in most, but not all, basement membranes of Caenorhabditis elegans and assembles on tissues that do not express it. J Cell Biol. 1997;137:1171–1183. doi: 10.1083/jcb.137.5.1171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ries A, et al. Recombinant domains of mouse nidogen-1 and their binding to basement membrane proteins and monoclonal antibodies. Eur J Biochem. 2001;268:5119–5128. doi: 10.1046/j.0014-2956.2001.02437.x. [DOI] [PubMed] [Google Scholar]
- 16.Kang SH, Kramer JM. Nidogen is nonessential and not required for normal type IV collagen localization in Caenorhabditis elegans. Mol Biol Cell. 2000;11:3911–3923. doi: 10.1091/mbc.11.11.3911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sasaki T, et al. Structural characterization of two variants of fibulin-1 that differ in nidogen affinity. J Mol Biol. 1995;245:241–250. doi: 10.1006/jmbi.1994.0020. [DOI] [PubMed] [Google Scholar]
- 18.Aumailley M, et al. Nidogen mediates the formation of ternary complexes of basement membrane components. Kidney Int. 1993;43:7–12. doi: 10.1038/ki.1993.3. [DOI] [PubMed] [Google Scholar]
- 19.Reinhardt D, et al. Mapping of nidogen binding sites for collagen type IV, heparan sulfate proteoglycan, and zinc. J Biol Chem. 1993;268:10881–10887. [PubMed] [Google Scholar]
- 20.Ihara S, Nishiwaki K. Stage specific activation of MIG-17/ADAMTS controls cell migration in. Caenorhabditis elegans FEBS J. 2008;275:4296–4305. doi: 10.1111/j.1742-4658.2008.06573.x. [DOI] [PubMed] [Google Scholar]
- 21.Yurchenco PD, Schittny JC. Molecular architecture of basement membranes. FASEB J. 1990;4:1577–1590. doi: 10.1096/fasebj.4.6.2180767. [DOI] [PubMed] [Google Scholar]
- 22.Colorado PC, et al. Anti-angiogenic cues from vascular basement membrane collagen. Cancer Res. 2000;60:2520–2526. [PubMed] [Google Scholar]
- 23.Xu J, et al. Proteolytic exposure of a cryptic site within collagen type IV is required for angiogenesis and tumor growth in vivo. J Cell Biol. 2001;154:1069–1079. doi: 10.1083/jcb.200103111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ortega N, Werb Z. New functional roles for non-collagenous domains of basement membrane collagens. J Cell Sci. 2002;115:4201–4214. doi: 10.1242/jcs.00106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Meighan CM, Schwarzbauer JE. Control of C. elegans hermaphrodite gonad size and shape by vab-3/Pax6-mediated regulation of integrin receptors. Genes Dev. 2007;21:1615–1620. doi: 10.1101/gad.1534807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gresham HD, et al. Domain-specific interactions between entactin and neutrophil integrins. J Biol Chem. 1996;271:30587–30594. doi: 10.1074/jbc.271.48.30587. [DOI] [PubMed] [Google Scholar]
- 27.Lee HK, et al. Nidogen is a prosurvival and promigratory factor for adult Schwann cells. J Neurochem. 2007;102:686–698. doi: 10.1111/j.1471-4159.2007.04580.x. [DOI] [PubMed] [Google Scholar]
- 28.Hedgecock EM, Norris CR. Netrins evoke mixed reactions in motile cells. Trends Genet. 1997;13:251–253. doi: 10.1016/s0168-9525(97)01177-3. [DOI] [PubMed] [Google Scholar]
- 29.Kim S, Wadsworth WG. Positioning of longitudinal nerves in C. elegans by nidogen. Science. 2000;288:150–154. doi: 10.1126/science.288.5463.150. [DOI] [PubMed] [Google Scholar]
- 30.Fujiwara S, et al. Structure and interactions of heparan sulfate proteoglycans from a mouse tumor basement membrane. Eur J Biochem. 1984;143:145–157. doi: 10.1111/j.1432-1033.1984.tb08353.x. [DOI] [PubMed] [Google Scholar]
- 31.Charonis AS, Tsilibary EC, Yurchenco PD, Furthmayr H. Binding of laminin to type IV collagen: a morphological study. J Cell Biol. 1985;100:1848–1853. doi: 10.1083/jcb.100.6.1848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Aumailley M, Wiedemann H, Mann K, Timp R. Binding of nidogen and the laminin-nidogen complex to basement membrane collagen type IV. Eur J Biochem. 1989;184:241–248. doi: 10.1111/j.1432-1033.1989.tb15013.x. [DOI] [PubMed] [Google Scholar]
- 33.Aspberg A, et al. Fibulin-1 is a ligand for the C-type lectin domains of aggrecan and versican. J Biol Chem. 1999;274:20444–20449. doi: 10.1074/jbc.274.29.20444. [DOI] [PubMed] [Google Scholar]
- 34.Pan TC, et al. Sequence of extracellular mouse protein BM-90/fibulin and its calcium-dependent binding to other basement-membrane ligands. Eur J Biochem. 1993;215:733–740. doi: 10.1111/j.1432-1033.1993.tb18086.x. [DOI] [PubMed] [Google Scholar]
- 35.Bader BL, et al. Compound genetic ablation of nidogen 1 and 2 causes basement membrane defects and perinatal lethality in mice. Mol Cell Biol. 2005;25:6846–6856. doi: 10.1128/MCB.25.15.6846-6856.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kostka GR, et al. Perinatal lethality and endothelial cell abnormalities in several vessel compartments of fibulin-1-deficient mice. Mol Cell Biol. 2001;21:7025–7034. doi: 10.1128/MCB.21.20.7025-7034.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Brenner S. The genetics of Caenorhabditis elegans. Genetics. 1974;77:71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nishiwaki K. Mutations affecting symmetrical migration of distal tip cells in Caenorhabditis elegans. Genetics. 1999;152:985–997. doi: 10.1093/genetics/152.3.985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kubota Y, et al. The conserved oligomeric Golgi complex acts in organ morphogenesis via glycosylation of an ADAM protease in C. elegans. Development. 2006;133:263–273. doi: 10.1242/dev.02195. [DOI] [PubMed] [Google Scholar]
- 40.Inoue T, et al. Caenorhabditis elegans DAF-21 (HSP90) is characteristically and predominantly expressed in germline cells: spatial and temporal analysis. Dev Growth Differ. 2003;45:369–376. doi: 10.1046/j.1440-169x.2003.00706.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.