Abstract
On their entry into the thymus, developing lymphocyte progenitors depend on signaling from the pre-T cell receptor (pre-TCR), which orchestrates differentiation, cell proliferation, and survival. The exact mechanism of pre-TCR-mediated suppression of T cell death remains unclear and controversial. Here, we identify Bim and Bid, 2 members of the BH3-only group of the BCL2 family, as important regulators of pre-T cell death. Both factors are highly expressed in proapoptotic thymocytes and their expression is suppressed on signaling through the pre-TCR. Their expression is directly regulated by the transcription factors FoxO3a and p53. Bid expression and p53 activity are related to the ongoing rearrangement of the TCR loci and induced DNA damage responses. Bim expression and FoxO3a nuclear translocation are directly controlled by the pre-TCR by means of its downstream kinase Akt/PKB. Interestingly, deletion of either gene on a pre-TCR−/− background rescues survival, but fails to induce further progenitor differentiation uncoupling the 2 processes.
Keywords: BCL2 family, cell death, pre-TCR signaling, T cell development
T cell development is a strictly regulated process initiated by the entry of hematopoietic progenitors into the thymus (1). During this developmental progression, thymocytes are “screened” for their ability to successfully rearrange T cell receptor (TCR) loci and express a functional TCR on the surface. The first checkpoint during T cell development is controlled by the expression of the pre-TCR. Pre-TCR signaling is important for cell proliferation, differentiation, and suppression of cell death. One of the main functions of the receptor is the suppression of cell death (2, 3), because the majority of pre-TCR deficient thymocytes fail to differentiate to the CD4+8+ (DP) stage and display an apoptotic phenotype. However, little is known about the molecular mechanisms that either enforce or suppress apoptosis in early thymocytes.
Initially, it was suggested that the death adaptor Fas-associated death domain (FADD) is an essential regulator of pre-T cell death, becasue deletion of FADD activity induced the differentiation of pre-TCR-deficient DN cells to the DP stage (4). Also, several studies proposed that the p53 pathway is an important regulator of death of pre-TCR deficient pre-T cells. These reports have shown that introduction of p53 deficiency in pre-TCR-deficient mice (RAG−/−, SCID, CD3γ−/−) could alleviate the DN developmental arrest, resulting in increased thymocyte survival and differentiation to the DP stage (5–7). Thus, a p53-mediated checkpoint exists in DN3 pre-T cells.
We have recently identified an antiapoptotic member of the BCL2 family, BCL2A1, as a direct target of the pre-TCR (2). BCL2A1 can bind to and suppress the function of proapoptotic BCL2 proteins and inhibit the execution of apoptosis (8). However, subsequent experiments using in vivo, siRNA-mediated, silencing of BCL2A1 demonstrated that it is not sufficient for the suppression of pre-T cell death (9). These experiments suggested that either other antiapoptotic genes are overexpressed in the absence of BCL2A1, supporting thymocyte survival or that there are additional mechanisms that control life and death at this stage of development. Because we have excluded the former hypothesis, we have turned our attention to the identification of pre-T cell-specific proapoptotic factors.
Cell death is regulated by the members of the BCL-2 family of proteins, as defined by the conservation of 1 to 4 BCL-2 homology domains (BH1–4) (10). From these proteins, Bax and Bak appear to be the direct executioners, because they have the ability to permeabilize the outer mitochondrial membrane (11, 12). A second group of proteins share only the BH3 domain (BH3-only). Among them, BIM and BID and PUMA are “activators,” because they can directly engage the downstream executioners Bax and Bax, whereas others termed “sensitizers” (NOXA, BIK, and BAD) purportedly act only by displacing the activators from the prosurvival proteins (13). Contrary to this “direct-activation model,” Huang and coworkers (14, 15) showed that BH3-only proteins activate Bax or Bak indirectly, by engaging the different antiapoptotic relatives that constrain them. Although the exact mechanisms of apoptosis remains controversial, it is fair to say that the balance between pro and antiapoptotic proteins is a sensitive rheostat that controls life and death in each cell.
We demonstrate here that the BH3-only proapoptotic proteins Bim and Bid are overexpressed in pre-TCR deficient cells. Also, we demonstrate that Bid expression is regulated by the activity of p53 in response to DNA damage, and Bim expression is regulated by the transcription factor FoxO3a. The 2 genes are direct transcriptional targets of the p53 and FoxO3a, because these factors are bound on conserved sited found on the promoters of Bid and Bim, respectively. Our studies also show that the pre-TCR is important for the suppression of Bim expression, because it can phosphorylate Foxo3a through the activation of the PI3K/Akt kinase complex. Last, expression of Bim and Bid is of unique biological significance, because their deletion in pre-TCR deficient thymocytes is able to suppress cell death in vivo and in vitro.
Results
Bim and Bid Are Overexpressed in Proapoptotic, Pre-TCR Deficient T Cell Progenitors.
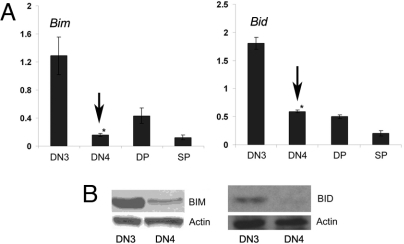
We have initially compared BCL2-family gene expression in pre-TCR nonexpressing, proapoptotic thymocytes, and cells that receive pre-TCR signals, suppress apoptosis, and differentiate. Thus, we have purified “small” (based on FSC axis), pre-TCR-nonexpressing “DN3” (CD25+44-c-kit−) and compared them with “large,” pre-TCR-expressing “DN4” (CD25low44-c-kit−gdTCR−CD19−intracellularTCRb+) cells. Later stages of T cell development (DP: CD4+8+; SP: CD4+ and CD8+ abTCR+) were also included in this analysis. Quantitative, real-time (qRT)-PCR using cDNA from the purified populations revealed that the expression of several proapoptotic genes, including Noxa, Bax, Puma, Hrk, Bad, and Bik (data not shown) was not significantly altered during the transition from the DN3 to the DN4 stage. However, Bim (16) and Bid (17), 2 BH3-only members of the BCL2 family were significantly up-regulated in DN3 cells lacking pre-TCR expression. Indeed, both mRNA (Fig. 1A) and protein (Fig. 1B) levels were high in DN3 cells, and rapidly decreased once cells acquired surface pre-TCR expression (DN4). These observations suggested a pre-TCR-dependent regulation of Bim and Bid expression during early T cell development, and identified these 2 genes as potential executioners of apoptosis in pre-T cells.
Fig. 1.
Proapoptotic, pre-T cells overexpress Bim and Bid. (A) Quantitative RT-PCR of Bim and Bid expression in DN3 pre-TCR−/−, WT DN4, WT DP, and (SP) cells. *, P < 0.001. (B) Bim and Bid protein expression was analyzed pTα−/− DN3 and WT DN4 cells. All data are representative of 4 independent experiments.
p53 Activity Regulates Bid Expression in Developing Thymocytes.
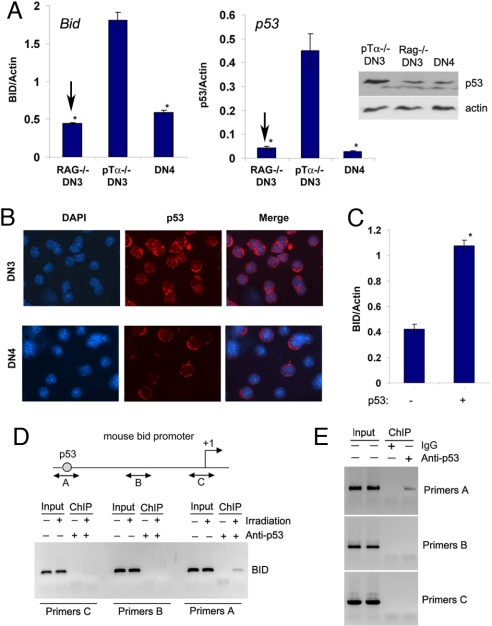
We have initially focused on the regulation of Bid transcription. Although it was suggested previously that Bid is regulated by p53 (18), this regulation was tissue-specific (19). Because in developing lymphocytes p53 can be activated by TCR rearrangement (5), we have compared Bid and p53 expression in DN3 cells purified from either pTα−/− or Rag-2−/− thymi, because the former (but not the latter) are capable of TCR recombination events. Interestingly, p53 mRNA and protein levels were significantly elevated in TCR recombining pTα−/− thymocytes. Bid expression mirrored p53 and was extinguished in p53−/− DN3 cells [Fig. 2A and supporting information (SI) Fig. S1], suggesting that Bid expression is regulated by p53 activation. Immunostaining using p53 antibodies indicated that not only p53 expression was elevated in pTα−/− DN3 cells, but also p53 was localized in the nucleus of these cells suggesting induced transcriptional activity (Fig. 2B).
Fig. 2.
Direct regulation of Bid expression by p53. (A) Quantitative RT-PCRs for Bid and p53 expression; p53 protein expression is also shown (Right, Upper). *, P < 0.001. (B) IF staining showing the p53 expression and localization. (C) Quantitative RT-PCR showing induction of Bid in response to p53 expression. *, P < 0.001. (D) ChIP assay by using gamma-irradiated SCIET27 cells. (E) ChIP assay of purified pTα−/− DN3 cells. All data are representative of 3 independent experiments.
To directly prove that p53 was able to induce Bid expression in pre-T cells, we have overexpressed p53 in a “DN3-like,” pre-TCRneg cell line (SCIET27) (20). As shown in Fig. 2C, p53 expression enhanced significantly Bid transcription. However, because these assays did not prove whether p53 directly activated Bid expression, we have preformed ChIP assays by using a previously characterized p53-binding site identified on the mouse Bid promoter (18). This site is situated ≈5 kb upstream of the first noncoding exon of the Bid locus. We have initially proved that irradiation-induced DNA damage induces p53, Bid expression (data not shown), and the binding of p53 on the Bid promoter in pre-T cells (Fig. 2D). To directly show that p53 is bound on the Bid promoter in Bid overexpressing pTα−/− pre-T cells, we have performed ChIP assays by using purified pTα−/− thymocytes. As shown in Fig. 2E, p53 was indeed bound on the promoter of Bid in these cells. These results strongly suggested that p53 is the main direct regulator of Bid transcription and a potential death trigger for pre-TCR deficient thymocytes.
A Potential Bim Regulator, FoxO3a, Is Controlled by the Pre-TCR.
We have then turned our attention to the regulation of Bim. In different tissues, Bim is a transcriptional target of the Forkhead factors (21). To test the ability of FoxO3a, a pre-T cell expressed Forkhead member, to induce Bim transcription, we have expressed an “active” (nonphosphorylatable) mutant of this factor (FoxO3.AAA) in pre-T cells. FoxO3a activation was inducing rapid cell death (data not shown), and significant up-regulation of Bim gene transcription (Fig. S2A).
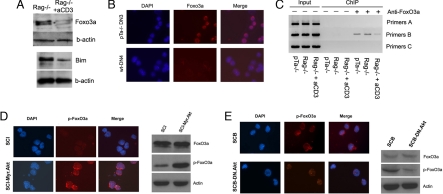
We then studied the expression pattern of FoxO3a during the pre-TCR-regulated early stages of T cell development. We have used anti-CD3 stimulation to dynamically induce pre-TCR signaling (2), and saw that Bim expression was rapidly decreased in response to pre-TCR signals (Fig. S2B). However, FoxO3a mRNA expression was not altered (data not shown). However, because Forkhead transcription factors are mainly regulated by phosphorylation and proteasomal degradation (22), we have traced FoxO3a protein expression and nuclear localization of FoxO3a in thymocytes. We have found that Foxo3a protein expression was sensitive to pre-TCR signals, because it was rapidly down-regulated in pre-TCR+ DN4 cells (Fig. 3A). Bim protein expression was significantly down-regulated 24 h after the induction of pre-TCR signaling. Also, immunofluorescence experiments also proved the presence of more active FoxO3a protein in the nucleus of the pre-TCR deficient cells than that of pre-TCR expressing cells (Fig. 3B). Thus, FoxO3a activity appeared to be directly controlled by pre-TCR signaling.
Fig. 3.
Direct regulation of Bim by the pre-TCR through Foxo3a. (A) FoxO3a and Bim protein expression in the indicated populations. (B) IF staining showing FoxO3a expression and localization in the indicated cell subsets. (C) ChIP assay for Foxo3a binding using DNA from the indicated populations. Rag-1−/− mice were stimulated with aCD3 for 24h. All data are representative of 3 independent experiments. (D) IF staining (Left), demonstrating phospho-FoxO3a expression and localization in SCIET27 and SCIET27 cells expressing with Myr-Akt. Western blot analysis (Right), using protein extracts from the same combination of cells. (E) IF staining (Left) and Western blot analysis (Right) of phosho-FoxO3a expression in SCB29 and SCB29 cells expressing DN-Akt. The data presented are representative of 3 independent experiments.
We have then compared Foxo3a activity by using a parental pre-TCR deficient line SCIET27 and its pre-TCRpos (transfected with TCRb) derivative SCB29. As evident from Fig. S2C, SCIET27 cells expressed high levels of nuclear, active FoxO3a. On the contrary, on pre-TCR expression, in SCB29 cells, total FoxO3a protein expression was down-regulated. Interestingly, the majority of FoxO3a protein was excluded from the nucleus, suggesting protein phosphorylation as this modification controls FoxO3a nuclear shuttling and activation. Indeed, phosphorylation of FoxO3a was induced in response to pre-TCR signaling, and phospho-FoxO3a was excluded from the nucleus of pre-TCRpos (Fig. S2D).
Bim Is a Direct FoxO3a Transcriptional Target.
To address direct Foxo3a involvement in Bim transcription, we have performed ChIP assays by using an identified FoxO binding site on the mouse Bim promoter (23). This site, conserved in the mouse and human genomes, is found ≈160 bp upstream of the transcriptional start. We have initially tested the functionality of our ChIP assay by using serum starvation of thymocytes, and demonstrated that FoxO3a was weekly bound on the promoter of Bim, and that on serum withdrawal, this binding was markedly enhanced (Fig. S3). To directly study FoxO3a binding in ex vivo purified pre-T cells, we have performed a ChIP assay by using pre-TCR deficient DN3 (pTα−/− or Rag-1−/−) and pre-TCR+ DN4 cells. As shown in Fig. 3C, we were able to detect strong binding of FoxO3a in pre-TCR- DN3 cells. On pre-TCR signaling, this binding was significantly attenuated. These results strongly suggest that FoxO3a is a transcriptional regulator of Bim expression during early T cell development.
The Akt Kinase Is Downstream of the Pre-TCR and Upstream of FoxO3a/Bim.
It was previously shown that the Akt/PKB kinase is the major regulator of FoxO3a activation in multiple cell types (24). It was also recently shown that the Akt/PkB kinase complex is essential for early T cell development (25, 26). To directly show that Akt activity is regulated by the pre-TCR, we have initially compared levels of phospho-Akt between SCIET27 (pre-TCRneg) and the TCRb-transfected derivative line SCB29 (pre-TCRpos) cells. As shown in Fig. S4A, pre-TCR expression induced the phosphorylation of Akt. To further prove that Akt is downstream of the pre-TCR in vivo, we have performed a “rescue” experiment, expressing an activated Akt form (Myr-Akt) in pre-TCR-deficient (Rag-2−/−) bone marrow hematopoietic progenitors. Myr-Akt expressing (EGFP+) and “control” EGFP-only expressing progenitors were transplanted in irradiated Rag-2−/−γc−/− alymphoid hosts. Six weeks post transplantation, recipient thymi were analyzed. As shown in Fig. S4B, Myr-Akt expression rescued the pre-TCR imposed developmental block (because 95% of the thymocytes were CD4+8+), suggesting that Akt is indeed a downstream pre-TCR effector. Also, by using nontreated Rag-2−/− and Rag-2−/− treated with anti-CD3 to induce pre-TCR signaling, demonstrated that pre-TCR signaling independently phosphorylate Akt (Fig. S4C).
To further probe this signaling cooperation, we have expressed Myr-Akt in pre-TCR deficient SCIET27 cells. SCIET27 was characterized by nuclear localization of FoxO3a and low levels of phosho-FoxO3a due to the absence of pre-TCR signaling. On Myr-Akt expression, we have noticed a significant reduction in Bim expression (Fig. S5), and induction of FoxO3a phosphorylation and nuclear exclusion (Fig. 3D). Identical results were obtained by using a second pre-TCRneg line (LR2, data not shown). We then expressed in pre-TCRpos SCB29 cells a dominant negative form of Akt (DN-Akt). SCB29 cells demonstrate a distinct pattern of nuclear exclusion of Akt due to pre-TCR signaling. DN-Akt expression was able to out-compete pre-TCR signals, and induce the dephosphorylation and nuclear localization of FoxO3α proteins (Fig. 3E). These experiments were consistent with the hypothesis that survival of pre-T cells depends on pre-TCR/Akt signals that control FoxO3a activation and the expression of proapoptotic Bim.
In Vivo Deletion of Bim and Bid Suppresses Pre-T Cell Death.
Our data suggested that Bim and Bid are essential elements of the mechanism executing apoptosis in pre-T cells. If this hypothesis was correct, deletion of Bim and Bid expression should enhance the survival ability of pre-TCR deficient thymocytes. To test this hypothesis, we have generated double deficient animals crossing pTα−/− (27) to Bid−/− (28), and Bim−/− animals (29).
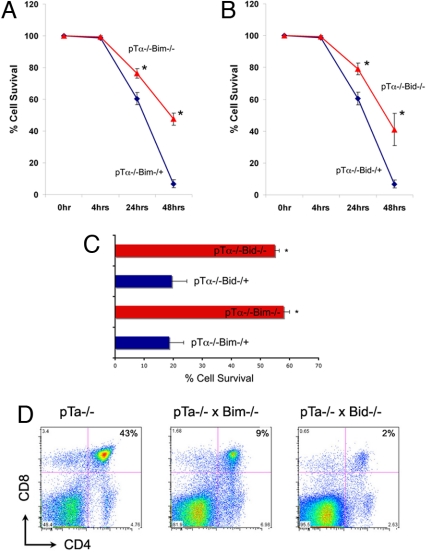
We have initially directly compared survival kinetics of pTα−/−, pTα−/−Bid−/−, and pTα−/−Bim−/− CD25+44− DN3 cells in vitro. Introduction of both Bim and Bid deficiencies significantly delayed the death kinetics of ex vivo cultured DN3 thymocytes (Fig. 4 A and B). At 24 h of culture, the survival of pTα−/−Bid−/− and pTα−/−Bim−/− DN3 thymocytes was significantly enhanced when compared with pTα−/− counterparts. At 48 h in culture, the differences were even more pronounced, because 47% of pTα−/−Bim−/− DN3 thymocytes and 36% of pTα−/−Bid−/− DN3 thymocytes were alive, in comparison with <7% of pTα−/− DN3 thymocytes (Fig. 4 A and B). These in vitro studies provided a support to the notion that these 2 proteins are elements of a pre-T cell apoptotic mechanism.
Fig. 4.
Bim and Bid deficiency suppresses pre-T cell death in vivo. (A and B) DN3 cells from pTα−/−, pTα−/−BIM−/−, and pTα−/−BID−/− mice were sorted for live cells and cultured in 24-well plates. Thus, time 0 was set to 100% to reflect sorted live cells from both populations. Cell viability was analyzed by using Annexin V staining at the indicated time points. Data represent mean ± SD from 4 experimental sets. *, P < 0.001. (C) Quantification of Annexin-V expressing cells in the thymi of the indicated genotypes. n = 5. (D) Phenotypic analysis of total thymocytes used CD4 and CD8 antibodies.
To further prove this hypothesis, we have studied the effect of Bim/Bid deficiency on pre-TCR-deficient thymocyte survival in vivo. We have extracted the thymi from pTα−/−, pTα−/−Bid−/−, and pTα−/−Bim−/− littermate mice and purified DN3 cells, as this population has the highest percentage of proapoptotic cells. Indeed, in pTα−/− thymi, 58 ± 5% of these cells were Annexin-V positive. Introduction of either Bim or Bid deficiency dramatically decreased the percentage of Annexin-V+ cells (to <20% in both genetic backgrounds). These observations (Fig. 4C) were consistent with the in vitro experiments presented previously, and strongly suggest that deletion of either Bim or Bid was sufficient for the suppression of apoptotic death in thymocytes that do not receive pre-TCR signals. Also, to further demonstrate the role of pTα, Bim, and Bid in pre-T cell death programs, triple (pTα−/−, Bim−/−, and Bid−/−) knockouts were generated. These mice do not survive postpartum. However, embryos were harvested at embryonic day 16.5 and DN3 cells were analyzed. The comparison of death kinetics between triple knockouts versus double deletion (pTα−/−, Bid−/−) demonstrated a significant difference (P < 0.01) in the resistance of triple knockouts to apoptosis (Fig. S6).
Because the pre-TCR is able to control proliferation, survival, and differentiation, we wanted to determine whether the decrease in cell death was sufficient to rescue the pre-TCR-induced differentiation arrest at the CD4−8−25+ (DN3) stage. Initially, the total number of thymocytes was significantly elevated in double deficient thymi (Table S1); pTα−/−Bim−/− and pTα−/−Bid−/− mice exhibited 1.9- (P < 0.001) and 1.4-fold (P < 0.01) increase in total number of thymocytes, respectively, compared with pTα−/− mice. This increase was not due to an increase in the absolute number of DP, SP, TCRγδ, or B cells, suggesting a specific effect in the DN compartment. Indeed, pTα−/−Bim−/− and pTα−/−Bid−/− mice had a 2.5- (P < 0.001) and 2.1-fold (P < 0.01) increase in the number of the DN cells, compared with pTα−/− mice (Fig. 4D and Table S1). Most significantly, both pTα−/−xBim−/− and pTα−/−Bid−/− thymi had, in absolute numbers, 2.5–3 times more DN3 cells. This observation was consistent with our previous observations, suggesting enhanced cell survival of pTα−/−Bid−/− and pTα−/−Bim−/− DN thymocytes. Supporting this notion, DN3 cells of all 3 phenotypes showed comparable cell cycle profiles (data not shown). Interestingly, although pTα−/−Bid−/− and pTα−/−Bim−/− thymi contain more DN3 cells that have the ability to survive longer that their pTα−/− counterparts, there were no signs of rescue of the DN3 block and progression to the CD4+8+ stage (Fig. 4D and Fig. S7), uncoupling cell survival from differentiation. Also, to prove the specificity of Bim and Bid function, we have generated pTα−/− mice lacking Bax (30), a proapoptotic gene not differentially regulated during early T cell development; pTα−/−Bax−/− thymocytes were phenotypically identical to pTα−/− cells (Fig. S8), and showed no signs of DN cell accumulation or decreased rates of cell death (data not shown). These results strongly supported the hypothesis that Bim and Bid not only were regulated during pre-TCR-controlled early T cell differentiation, but that they also had a significant role in the execution of pre-T cell death programs.
Discussion
Our studies identify Bim and Bid as important regulators of pre-T cell death. We demonstrate that cells that are unable to express the pre-TCR express high levels of both death inducers. In vivo deletion of either gene in a pre-TCR-deficient background significantly suppresses cell death and expands the progenitor cell pool. We here report the involvement of either of these genes at the initial selection step in the thymus. Our studies also mechanistically explain the overexpression of Bim and Bid in pre-TCR nonexpressing pre-T cells. Bid expression depends on p53 activation, which in turn appears to be related to the ongoing rearrangement of the TCR loci. However, Bim expression depends on FoxO3a nuclear translocation and activation. This event is directly controlled by the pre-TCR and its downstream kinase Akt/PKB.
Our studies and previously published observations shape a previously undescribed understanding of the role of the pre-TCR in the regulation of cell survival (Fig. S9). Cell death appears to be a default pathway at this early developmental checkpoint due to both extrinsic and intrinsic apoptotic stimuli. For example, lack of pre-TCR signals maintains FoxO3a in an active, nuclear-localized state, able to activate Bim gene transcription. Cell intrinsic signals (including DNA damage due to ongoing TCR rearrangement) impinge on p53 activity and sustain high expression of Bid. Also, both Bim and Bid could also be triggered by signaling through death receptors and FADD, in line with the previous findings of Strasser and coworkers (4). The pre-TCR can suppress death by using several mechanisms. It can induce the expression of antiapoptotic factors (BCL2A1) (2), which is able to bind and neutralize the activities of both Bim and Bid (8). It can also activate the Akt/PKB kinase, phosphorylate FoxO3a, and switch-off the expression of Bim. The connection between pre-TCR and p53 remains controversial, as it could be direct and/or indirect. It was shown that pre-TCR signaling can directly phosphorylate and inactivate p53 (31). However, attenuation of p53 activity could be indirect contingent on its ability to block further TCR rearrangement; thus, inactivate the DNA response pathway (32). Our working model is not only based on our observations, but it incorporates and explains important recent studies on the regulation of T cell progenitor death. It is also consistent with recent findings from Zuniga-Pflucker and coworkers (26), suggesting a cooperation between pre-TCR and Notch1 signaling in the regulation of pre-T cell apoptosis. These studies have shown that Notch1 is mainly essential for cellular metabolism of T cell progenitors. However, Notch1 signaling could cooperate with the pre-TCR in the induction of antiapoptotic BCL2A1 (M.M. and I.A., unpublished work) and the activation of the Akt/PKB signaling cascade (M.M. and I.A., unpublished work; and ref. 26).
However, one would notice a major difference between the phenotypes of the pTα−/− × Bim−/− and pTα−/− × Bid−/− mice presented here and previously published rescue experiments. Indeed, thymi of Rag-1−/− × FADD, CD3ε−/− × p53−/−, or SCID × p53−/− animals are larger with the majority of thymocytes being at the CD4+8+ stage (4–6). This discrepancy can be easily explained once we consider the exact function of these molecules; p53 is a well-characterized regulator of cell survival, but also cell proliferation, because it is able to target the expression of several regulators, including the cell cycle inhibitor p21cip1; suggesting that p53 can control both survival and proliferation at this stage of development. Similarly, FADD has also been suggested to be a regulator of cell cycle entry (33) with effects on expression of cell cyle inhibitors (p21), D type cyclins, and cyclin-dependent kinases (34). Thus, although p53 and FADD can affect both survival and proliferation, Bim and Bid control only cell death and are unable to promote cell cycle entry and thymocyte differentiation. This observation is intriguing, because it uncouples progenitor survival from their ability to differentiate. However, we have recently shown that DN3 cells lacking the ability to enter cell cycle due to absence of cyclin D3 cannot differentiate in response to pre-TCR signals (35) coupling pre-T cell proliferation and differentiation. Both these observations suggest a very tightly regulated interplay of cell proliferation and survival that leads to the promotion of differentiation of pre-TCR-expressing T cell progenitors.
Methods
Animals.
All mice were kept in the animal facilities of the University of Chicago and the New York University School of Medicine. All animal experiments were performed in accordance to the guidelines of the Institutional Animal Care and Use Committee of both universities. Genotyping primers are available on request. BID genotyping was previously described (28).
Flow Cytometric Analysis and Cell Sorting.
Anti-CD4(L3T4), CD8(53–6.7), CD25(3C7), CD44(IM7), B220(RA3–6B2), CD11c(HL3), NK1.1(PK136), TER-119, Mac-1(M1/70), Gr-1(RB6–8C5), and TCR-gd and were from BD Biosciences. Surface marker expression by thymocytes and peripheral T cells was visualized by using a flow cytometer (FACSCalibur, Becton Dickinson) and analyzed with Flow-Jo and CELLQuest software, according to standard protocols. Cell sorting was performed by using a Mo-Flo (DakoCytomation).
RT-PCR and qRT-PCR.
Total cellular RNA was isolated by using a Qiagen RNeasy minicolumn. Each cDNA template was made from total RNA with SuperScript II reverse transcriptase kit according to the manufacturer's instructions (Invitrogen). PCRs were performed with RedTaq DNA polymerase (Sigma). Quantitative analysis of cDNA amplification was assessed by incorporation of SYBR Green into double-stranded DNA. Measurement of gene expression was performed by using the ABI PRISM 7700 Sequence Detector (PE Applied Biosystems) and analyzed with ABI Prism Sequence Detection Software version 1.7 (PE Applied Biosystems). All cDNA samples were tested in quadruplicate by using the ABI PRISM 7700 Sequence Detector (PE Applied Biosystems) and analyzed with ABI Prism Sequence Detection Software version 1.7 (PE Applied Biosystems). The specific primers are available on request.
Western Blot Analysis.
Thymocyte protein lysates were prepared as described in Aifantis et al. (20) by using the BIM (B-7929, Sigma), BID (MAB-860, R&D systems), FKHRL1 (Upstate), p-FKHRL-1 (Cell Signaling), or β-Actin (Chemicon) antibodies.
ChIP Assay.
The assays were performed by using the ChIP assay Kit, according to the manufacturer's instructions (Upstate) and as previously described (36). The immunoprecipitated DNA (or the input control) was then purified by using the Qiagen PCR Purification Kit, according to the manufacturer's protocol. PCRs were then performed on the purified DNA by using red taq polymerase, by using specific primers (available on request).
Immunofluorescence Microscopy (IF).
IF was performed as previously described (36), by using the FKHRL1, phospho-FKHRL1, or p53 (sc-6243, Santa Cruz) antibodies.
Retroviral Constructs.
The full length mouse p53 sequence was PCR amplified from cDNA and cloned into the MIGR1 (MSCV-IRES-GFP) vector to make the MIGR1-p53 retroviral vector. The MSCV-based retroviruses expressing Myr-Akt, DN-Akt (37) and FoxO3a.AAA (22) have been previously described. Retroviral supernatants were generated as previously described (38).
Statistical Analysis.
Data were analyzed by unpaired t test and ANOVA, followed by the test of least significant difference for comparisons within and between the groups.
Supplementary Material
Acknowledgments.
We thank A. Brunet (Harvard Medical School, Boston), B. Scheijen (Harvard Medical School, Boston), L. van Parijs (Massachusetts Institute of Technology, Boston), C. W. Brains (Harvard Medical School, Boston), and A. Ruiz-Vela (Harvard Medical School, Boston) for DNA vectors and helpful discussions; and H. von Boehmer and the late S. J. Korsmeyer for mentorship and support during the initial steps of the project. I.A. is a Leukemia and Lymphoma Society Scholar and is supported by the Sidney Kimmel Foundation for Cancer Research, the G&P Foundation for Cancer Research, and National Institutes of Health (NIH)/National Cancer Institute Grant R01CA105129. M.M. is supported by a Leukemia Research Foundation postdoctoral fellowship. K.M.C. is supported by NIH/National Research Service Award Molecular Oncology and Immunology Training Grant 5 T32 CA 009161.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0807557106/DCSupplemental.
References
- 1.Borowski C, et al. On the brink of becoming a T cell. Curr Opin Immunol. 2002;14:200–206. doi: 10.1016/s0952-7915(02)00322-9. [DOI] [PubMed] [Google Scholar]
- 2.Mandal M, et al. The BCL2A1 gene as a pre-T cell receptor-induced regulator of thymocyte survival. J Exp Med. 2005;201:603–614. doi: 10.1084/jem.20041924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aifantis I, Mandal M, Sawai K, Ferrando A, Vilimas T. Regulation of T-cell progenitor survival and cell-cycle entry by the pre-T-cell receptor. Immunol Rev. 2006;209:159–169. doi: 10.1111/j.0105-2896.2006.00343.x. [DOI] [PubMed] [Google Scholar]
- 4.Newton K, Harris AW, Strasser A. FADD/MORT1 regulates the pre-TCR checkpoint and can function as a tumour suppressor. EMBO J. 2000;19:931–941. doi: 10.1093/emboj/19.5.931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Guidos CJ, et al. V(D)J recombination activates a p53-dependent DNA damage checkpoint in scid lymphocyte precursors. Gene Dev. 1996;10:2038–2054. doi: 10.1101/gad.10.16.2038. [DOI] [PubMed] [Google Scholar]
- 6.Haks MC, Krimpenfort P, van den Brakel JH, Kruisbeek AM. Pre-TCR signaling and inactivation of p53 induces crucial cell survival pathways in pre-T cells. Immunity. 1999;11:91–101. doi: 10.1016/s1074-7613(00)80084-9. [DOI] [PubMed] [Google Scholar]
- 7.Jiang D, Lenardo MJ, Zuniga-Pflucker JC. p53 prevents maturation to the CD4+CD8+ stage of thymocyte differentiation in the absence of T cell receptor rearrangement. J Exp Med. 1996;183:1923–1928. doi: 10.1084/jem.183.4.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Certo M, et al. Mitochondria primed by death signals determine cellular addiction to antiapoptotic BCL-2 family members. Cancer Cell. 2006;9:351–365. doi: 10.1016/j.ccr.2006.03.027. [DOI] [PubMed] [Google Scholar]
- 9.Oberdoerffer P, et al. Efficiency of RNA interference in the mouse hematopoietic system varies between cell types and developmental stages. Mol Cell Biol. 2005;25:3896–3905. doi: 10.1128/MCB.25.10.3896-3905.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Scorrano L, Korsmeyer SJ. Mechanisms of cytochrome c release by proapoptotic BCL-2 family members. Biochem Biophys Res Commun. 2003;304:437–444. doi: 10.1016/s0006-291x(03)00615-6. [DOI] [PubMed] [Google Scholar]
- 11.Danial NN, Korsmeyer SJ. Cell death: Critical control points. Cell. 2004;116:205–219. doi: 10.1016/s0092-8674(04)00046-7. [DOI] [PubMed] [Google Scholar]
- 12.Rathmell JC, Lindsten T, Zong WX, Cinalli RM, Thompson CB. Deficiency in Bak and Bax perturbs thymic selection and lymphoid homeostasis. Nat Immunol. 2002;3:932–939. doi: 10.1038/ni834. [DOI] [PubMed] [Google Scholar]
- 13.Letai A, et al. Distinct BH3 domains either sensitize or activate mitochondrial apoptosis, serving as prototype cancer therapeutics. Cancer Cell. 2002;2:183–192. doi: 10.1016/s1535-6108(02)00127-7. [DOI] [PubMed] [Google Scholar]
- 14.Chen L, et al. Differential targeting of prosurvival Bcl-2 proteins by their BH3-only ligands allows complementary apoptotic function. Mol Cell. 2005;17:393–403. doi: 10.1016/j.molcel.2004.12.030. [DOI] [PubMed] [Google Scholar]
- 15.Willis SN, et al. Apoptosis initiated when BH3 ligands engage multiple Bcl-2 homologs, not Bax or Bak. Science. 2007;315:856–859. doi: 10.1126/science.1133289. [DOI] [PubMed] [Google Scholar]
- 16.Strasser A, et al. The role of bim, a proapoptotic BH3-only member of the Bcl-2 family in cell-death control. Ann N Y Acad Sci. 2000;917:541–548. doi: 10.1111/j.1749-6632.2000.tb05419.x. [DOI] [PubMed] [Google Scholar]
- 17.Zinkel SS, et al. A role for proapoptotic BID in the DNA-damage response. Cell. 2005;122:579–591. doi: 10.1016/j.cell.2005.06.022. [DOI] [PubMed] [Google Scholar]
- 18.Sax JK, et al. BID regulation by p53 contributes to chemosensitivity. Nat Cell Biol. 2002;4:842–849. doi: 10.1038/ncb866. [DOI] [PubMed] [Google Scholar]
- 19.Fei P, Bernhard EJ, El-Deiry WS. Tissue-specific induction of p53 targets in vivo. Cancer Res. 2002;62:7316–7327. [PubMed] [Google Scholar]
- 20.Aifantis I, Gounari F, Scorrano L, Borowski C, von Boehmer H. Constitutive pre-TCR signaling promotes differentiation through Ca2+ mobilization and activation of NF-kappaB and NFAT. Nat Immunol. 2001;2:403–409. doi: 10.1038/87704. [DOI] [PubMed] [Google Scholar]
- 21.Dijkers PF, et al. FKHR-L1 can act as a critical effector of cell death induced by cytokine withdrawal: Protein kinase B-enhanced cell survival through maintenance of mitochondrial integrity. J Cell Biol. 2002;156:531–542. doi: 10.1083/jcb.200108084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Brunet A, et al. Akt promotes cell survival by phosphorylating and inhibiting a Forkhead transcription factor. Cell. 1999;96:857–868. doi: 10.1016/s0092-8674(00)80595-4. [DOI] [PubMed] [Google Scholar]
- 23.Essafi A, et al. Direct transcriptional regulation of Bim by FoxO3a mediates STI571-induced apoptosis in Bcr-Abl-expressing cells. Oncogene. 2005;24:2317–2329. doi: 10.1038/sj.onc.1208421. [DOI] [PubMed] [Google Scholar]
- 24.Datta SR, Brunet A, Greenberg ME. Cellular survival: A play in three Akts. Gene Dev. 1999;13:2905–2927. doi: 10.1101/gad.13.22.2905. [DOI] [PubMed] [Google Scholar]
- 25.Mao C, et al. Unequal contribution of Akt isoforms in the double-negative to double-positive thymocyte transition. J Immunol. 2007;178:5443–5453. doi: 10.4049/jimmunol.178.9.5443. [DOI] [PubMed] [Google Scholar]
- 26.Ciofani M, Zuniga-Pflucker JC. Notch promotes survival of pre-T cells at the beta-selection checkpoint by regulating cellular metabolism. Nat Immunol. 2005;6:881–888. doi: 10.1038/ni1234. [DOI] [PubMed] [Google Scholar]
- 27.Fehling HJ, Krotkova A, Saint-Ruf C, von Boehmer H. Crucial role of the pre-T-cell receptor alpha gene in development of alpha beta but not gamma delta T cells. Nature. 1995;375:795–798. doi: 10.1038/375795a0. and erratum (1995) 378:419. [DOI] [PubMed] [Google Scholar]
- 28.Zinkel SS, et al. Proapoptotic BID is required for myeloid homeostasis and tumor suppression. Gene Dev. 2003;17:229–239. doi: 10.1101/gad.1045603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Takeuchi O, et al. Essential role of BAX,BAK in B cell homeostasis and prevention of autoimmune disease. Proc Natl Acad Sci USA. 2005;102:11272–11277. doi: 10.1073/pnas.0504783102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wei MC, et al. Proapoptotic BAX and BAK: A requisite gateway to mitochondrial dysfunction and death. Science. 2001;292:727–730. doi: 10.1126/science.1059108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Murga C, Barber DF. Molecular mechanisms of pre-T cell receptor-induced survival. J Biol Chem. 2002;277:39156–39162. doi: 10.1074/jbc.M203553200. [DOI] [PubMed] [Google Scholar]
- 32.Aifantis I, Buer J, von Boehmer H, Azogui O. Essential role of the pre-T cell receptor in allelic exclusion of the T cell receptor beta locus. Immunity. 1997;7:601–607. doi: 10.1016/s1074-7613(00)80381-7. and erratum (1997) 7:895. [DOI] [PubMed] [Google Scholar]
- 33.Newton K, Kurts C, Harris AW, Strasser A. Effects of a dominant interfering mutant of FADD on signal transduction in activated T cells. Curr Biol. 2001;11:273–276. doi: 10.1016/s0960-9822(01)00067-7. [DOI] [PubMed] [Google Scholar]
- 34.Zhang J, Kabra NH, Cado D, Kang C, Winoto A. FADD-deficient T cells exhibit a disaccord in regulation of the cell cycle machinery. J Biol Chem. 2001;276:29815–29818. doi: 10.1074/jbc.M103838200. [DOI] [PubMed] [Google Scholar]
- 35.Sicinska E, et al. Requirement for cyclin D3 in lymphocyte development and T cell leukemias. Cancer Cell. 2003;4:451–461. doi: 10.1016/s1535-6108(03)00301-5. [DOI] [PubMed] [Google Scholar]
- 36.Vilimas T, et al. Targeting the NF-kappaB signaling pathway in Notch1-induced T-cell leukemia. Nat Med. 2007;13:70–77. doi: 10.1038/nm1524. [DOI] [PubMed] [Google Scholar]
- 37.Van Parijs L, et al. Uncoupling IL-2 signals that regulate T cell proliferation, survival, and Fas-mediated activation-induced cell death. Immunity. 1999;11:281–288. doi: 10.1016/s1074-7613(00)80103-x. [DOI] [PubMed] [Google Scholar]
- 38.Ory DS, Neugeboren BA, Mulligan RC. A stable human-derived packaging cell line for production of high titer retrovirus/vesicular stomatitis virus G pseudotypes. Proc Natl Acad Sci USA. 1996;93:11400–11406. doi: 10.1073/pnas.93.21.11400. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.