Abstract
The formation of skeletal muscle from the epithelial somites involves a series of events triggered by temporally and spatially discrete signals resulting in the generation of muscle fibers which vary in their contractile and metabolic nature. The fiber type composition of muscles varies between individuals and it has now been found that there are differences in fiber type proportions between lean and obese animals and humans. Amongst the possible causes of obesity, it has been suggested that inappropriate prenatal environments may ‘program’ the fetus and may lead to increased risks for disease in adult life. The characteristics of muscle are both heritable and plastic, giving the tissue some ability to adapt to signals and stimuli both pre and postnatally. Given that muscle is a site of fatty acid oxidation and carbohydrate metabolism and that its development can be changed by prenatal events, it is interesting to examine the possible relationship between muscle development and the risk of obesity.
Key words: myogenesis, myogenic regulatory factors, FOXO, PGC-1α, PPAR undernutrition, muscle fibre type specification, obesity, fetal, growth
Introduction
The global epidemic of obesity is generally attributed to individuals combining high energy diets and low levels of physical activity. However, there is now some concern that the habits of the current population may impact upon the health of future generations. These concerns are based on evidence which indicates that if there is a mismatch between the pre and postnatal environments then there may be an increased risk for the offspring of cardiovascular and metabolic diseases in later life.1–3 For example, it is suggested that these mismatches can occur if nutritional conditions are more abundant/favorable postnatally than prenatally.2 In this situation, it appears that the adaptations made by the embryo/fetus to survive in less favorable uterine conditions may not match the postnatal environment and this mismatch may have long term consequences for the health of the individual particularly with respect to the risk of cardiovascular and metabolic diseases.2
It appears that a central tenet of the proposed developmental origin of health and disease is the plasticity of developmental programming.3 Skeletal muscle is a very plastic tissue, responding to signals both pre and postnatally.4 It represents between 40% and 60% of body weight and comprises over 600 individual muscles and not only provides a means of movement but is also a major site of fatty acid oxidation and carbohydrate metabolism. Muscle formation starts early during development and observations that prenatal events can affect the composition, have led to the speculation that under certain conditions the composition of muscle could also be ‘programmed’ so as to increase an individual's propensity to obesity in later life. This review will explore that possibility and will focus on some recently reported pathways to try to provide some new insights as to how muscle fiber composition could be altered in a way that would produce an obesogenic phenotype.
Skeletal muscle fiber types.
Muscle is a heterogenous tissue comprising fibers which vary in their metabolic and contractile nature and which occur in varying proportions in the individual muscles. Muscle fibers are classified on the basis of these various properties into two major ‘types’; type I and type II. Type I or slow twitch and oxidative (SO) fibers have a slow time to peak tension, are rich in mitochondria and derive ATP mainly from oxidative metabolism, rendering them relatively fatigue-resistant. In contrast, type II or fast twitch fibers are subdivided into at least two sub groups type IIa or fast twitch and oxidative and glycolytic (FOG) fibers and type IIb or fast twitch and glycolytic (FG) fibers. Type IIa (FOG) fibers have a fast time to peak tension and have metabolic and morphological properties intermediate to those of type I and type IIb (FG) fibers. Type IIb (FG) fibers have a fast time to peak tension, a low mitochondrial density and are less reliant on oxidative metabolism than either type I or IIa fibers deriving ATP mainly from glycolytic metabolism thus being more fatiguable than the other fiber types.
Heritability.
Fiber type proportions differ between individuals and there is evidence that these proportions may be heritable. In pigs, for example, cross breeding of Duroc and Large white pigs indicates heritability of fiber types, and particularly of the type I (SO) fibers (Table 1). This is consistent with estimates of heritability made in humans and in animals which show that the heritability of type I fibers is higher than that of type II fibers.5,6 In horses5 and in Limousin bulls6 estimates of SO fiber heritability are approximately 30–35%, while human studies, including those conducted using twins, tend to give rather variable data, with estimates ranging from 99.5%,7 to more moderate values of between 20–50%.8
Table 1.
The frequency of the individual fibre types in m longissimus thoracis of pure and cross bred Duroc pigs (from Maltin et al.148)
| Fibre type frequency (%) | |||
| FOG | FG | SO | |
| 0% Duroc | 28.5 (2.1) | 60.6 (3.6) | 10.8 (3.2) |
| 25% Duroc | 32.2 (1.9) | 54.8 (3.7) | 12.8 (2.7) |
| 50% Duroc | 28.2 (1.8) | 56.2 (3.6) | 15.6 (3.6) |
| 100% Duroc | 32.8 (1.9) | 51.5 (3.2) | 15.6 (2.13) |
The values are presented as means with standard errors in brackets.
However, fiber type proportions are not fixed throughout life and the muscle phenotype shows remarkable plasticity and adaptability in response to a range of signals such as exercise/training.9 For example, sprint training tends to increase the proportions of type IIa (FOG),9,10 so tending to increase the time taken to fatigue. In contrast, endurance training tends to convert type IIb (FG) fibers to type IIa (FOG),9 and increase the size of type I (SO) fibers11 so increasing the capacity to use fat as a fuel.12
Muscle fiber type and obesity.
This combination of heritability and plasticity may be particularly relevant in the context of human health and an individual's predisposition to disease, because of the opportunities for modification of the phenotype. The current global rise in obesity has been attributed variously to genetic and environmental factors or a combination of both, however, there is now growing evidence that the specific muscle fiber phenotype may also play a role in obesity. For example, reduced proportions of type I,13 type IIa fibers14 and increased proportions of type IIb fibers13 have been observed in obese individuals, indicating an overall reduction in oxidative capacity. Similarly in the obese, diabetic Zucker rat lower proportions of type IIa (FOG) fibers and reduced expression levels of oxidative metabolism related genes have been identified.15 Lipid oxidation is also reduced in obese individuals16 but can be improved by agents which increase muscle oxidative capacity17 or by exercise, but not by weight loss.16 This observation is important because it points to the central need to change muscle fiber type to change oxidative capacity, and provides clear rationale for the inclusion of exercise as an essential part of programs designed to achieve long term weight loss. Indeed it has been suggested that the type I (SO) phenotype may directly or indirectly protect against both obesity18 and dietary induced insulin resistance,19 and that the oxidative capacity of muscle may predict insulin sensitivity more effectively than the concentrations of muscle triglyceride or long-chain fatty acyl CoA.20 The latter finding is consistent with the observed differences in insulin sensitivity between muscles comprising largely type I (SO) insulin sensitive fibers and those comprising largely type II (FG) insulin resistant fibers.21
The recent finding that a major regulator of fat metabolism in adipose tissues, peroxisome proliferator-activated receptor δ (PPARδ), is highly expressed in muscle and influences the formation of the type I/SO fiber phenotype, provides a link between muscle fiber type, fat metabolism and obesity.18 The peroxisome proliferator-activated receptors (PPARs) are a subfamily of nuclear receptor superfamily comprising three isoforms α, β also known as δ and γ. The PPARs regulate transcription following activation by dietary fatty acids, especially polyunsaturated fatty acids and control a number of metabolic processes. PPARδ appears to play a role in the control of glucose and fatty acid homeostasis and the attenuation of obesity, hyperlipidaemia and insulin resistance,22 and has been identified as a potential target for novel therapeutic treatments of metabolic syndrome.23 PPARδ is the predominant isoform expressed in skeletal muscle and shows differential muscle type expression.18,24 It is most highly expressed in muscles with high proportions of type I (SO) fibers and targeted expression of an activated form PPARδ, or the use of a PPARδ agonist in mice has been shown to increase the expression of type I (SO) fibers, to enhance fatigue resistance and confer resistance to obesity.18 Similarly in humans PPARδ has been found to be more highly expressed oxidative fibers with levels of expression being increased in athletes24 and reduced in patients with diabetes.17
Hence it is clear that there is some relationship between fiber type proportions and obesity, and in particular that oxidative fibers appear to be protective. However, this raises the questions of whether the relationship is causative or consequential, and whether the observed phenotype could be explained by prenatal ‘programming’ of fiber type. In order to explore the possible prenatal ‘programming’ of obesity, it is important to understand the principle processes which drive the formation of muscle in utero.
Myogenesis
The skeletal muscles of the body (and some in the head) are derived from the epithelial somites which, in response to local signals become organized into distinct regions to give rise to the ventral sclerotome which forms the vertebra and ribs, and the dorsal dermomyotome which gives rise to muscles and dermis.25 The process of myogenesis involves a series of morphogenetic changes which are stimulated by a cascade of temporally and spatially discrete signals which trigger various genetic programs leading to cell-specific destiny; these events and the pathways which control them are covered in a number of recent reviews.4,26–30
The myogenic regulatory factors.
One of the key manifestations of myogenesis is the expression of the family of myogenic regulatory factors (MRFs). The family comprises four related but distinct members, MyoD, myf5, myogenin, mrf4 (also known as myf6), all of which are basic helix-loop-helix transcription factors essential for myogenesis, and a number of studies have elucidated the roles, redundancies and cross-regulation of the family members.26,27,31–35 The first studies showed that MyoD had the ability to convert fibroblasts to myoblasts in culture,36 and early in vivo studies indicated that either MyoD or myf5 can specify the skeletal muscle lineage, because muscle appeared to develop normally without one or the other, but did not develop if both genes are absent,34 and in addition, activation of myogenin and mrf4 appeared to be dependant on prior expression of MyoD and/or myf5.34
More recent evidence suggests myf5 is a key determination factor directing cells down the myogenic route33 and that it interacts with muscle gene chromatin to facilitate transcription.31 However, it is also likely that myf5 alone cannot maintain the myogenic program since triple-mutant mice (lacking myogenin, MyoD and mrf4) produce myoblasts but differentiation does not occur.37 Several recent findings indicated that mrf4 plays an important role in early myogenesis. For example, it has been shown that Mrf4 is expressed either before or co-incident with myf5 (hence before MyoD)38 and that in the MyoD:myf5 double-null mouse expression of Mrf4 can direct muscle identity and differentiation,32 whereas in the absence of myf5 and mrf4 the early myotome fails to form from the dermomyotome.27 However, it is also clear that if MyoD, myf5 and mrf4 are all absent, precursor myoblasts are not found and muscle does not form.32,34 Myogenin appears to activate differentiation downstream of MyoD and myf5 expression, since MyoD, in addition to auto-regulation, also activates myogenin expression so that both MyoD and myogenin are expressed during differentiation.
Of all the MRF family members, MyoD is seen as probably being the most important, indeed, in a recent review, Tapscott35 describes MyoD as a ‘master switch’ which is responsible for the orchestration and integration of skeletal muscle gene transcription during myogenesis. One premise for describing MyoD as a master switch is the observation that MyoD can interact with both regulators and chromatin-associated proteins39 to control muscle differentiation.35 In adult muscle, MyoD transcripts accumulate to high levels in muscles which comprise predominantly type II/fast fibers, while myogenin transcripts accumulate in muscle comprising mainly type I/slow fibers,40 and it has been suggested that the ratio of the MRFs may affect fiber type.30
Fiber type specification.
The formation of muscle fibers occurs in a multiphasic process.4 During the first phase, known as primary myogenesis, embryonic myoblasts exit the cell cycle and line up to fuse and form primary myotubes (also called primary fibers) which then increase in size.41,42 Primary myotubes form early during development, around embryonic day 14 in the rat,43 around day 32–38 in the sheep44 and between weeks 6 and 8 in humans.45
Later, between days 17 and 21 in the rat,43 from day 62 in the sheep44 and between weeks 8 and 18 in humans,45 in the second phase known as secondary myogenesis, fetal myoblasts stop proliferating and fuse to form secondary myotubes. Secondary myotubes tend to form on the surface of the primary myotubes, initially at the site of innervation46 and using the primary myotubes as a scaffolding for alignment. The secondary myotubes subsequently elongate along the long axis of the primary fibers and eventually separate to form individual fibers. However, it appears that not all secondary myotubes use the primary myotubes as scaffolding, and there is some evidence particularly from large animals and humans suggesting additional complexities including a tertiary wave of myogenesis and the formation of secondary fibers independently from the primary fiber scaffold.44,47
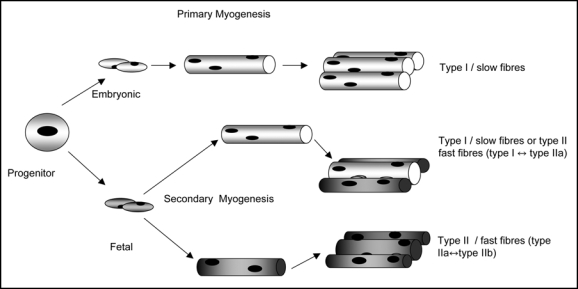
Various models of the contributions made by embryonic and fetal myotubes to fiber type diversity have been proposed41,42,48 but in general there is broad agreement (Fig. 1). Labelling experiments have shown that embryonic myoblasts only contribute to primary myotube formation, whilst fetal or early postnatal myoblast fuse with both primary and later, secondary myotubes.41,49 The findings generally concur that primary fibers, formed from embryonic myoblasts, mature express the slow myosin heavy chain isoform,42 while secondary fibers formed from fetal myoblasts mature to express the fast myosin heavy chain isoforms.42 However, secondary myotubes have been found to show high levels of coexpression of the various myosin heavy chain isoforms found in all the main subpopulations of muscle fibers types42 and consequently it is proposed that although the majority of secondary fibers mature into type II or fast fiber types, there is also a population of fetal myoblasts which are not restricted to either type I or type II phenotype and appear to have an ‘adaptive range’ of type I to type IIa.42 If this finding is correct, then it would suggest that whilst there is an adaptive capacity with regard to both contractile speed and glycolytic capacity, oxidative capacity would be rather less affected.
Figure 1.
A schematic diagram of primary and secondary myogenesis in relation to the formation of fibre type diversity. Type I/slow myotubes are shown as light, type II/ fast myotubes are shown as dark. Muscle progenitors give rise to embryonic myoblasts which fused to form primary myotubes. Subsequently, fetal myoblasts fuse either with primary myotubes or with fetal myoblasts to form secondary fibres. Fibres formed from secondary myogenesis appear to be able to switch between the different fibre types, while type I/slow fibres formed during primary myogenesis do not. Based on Dunglison et al.41 Pin et al.42 and Stockdale.48 The contributions of former colleagues at the Rowett Institute, Aberdeen to this figure are acknowledged.
Although fiber formation occurs from the early stages of development, in the chick, it is suggested that fiber type diversity in the limb muscles may be set as early as embryonic day 4,50 prior to entry of the myoblasts into the limb,51 and possibly within the somite.52 Observations from studies in vivo and in vitro support the contention that the myofibers formed during these waves of development are to a large extent already programmed with a default fiber type. Indeed it is suggested that the myosin heavy chain expression program and fiber type potential is set in myoblasts prior to fusion.42,52 The differences between fiber patterning in chick and quail have been widely exploited to investigate whether myoblasts are intrinsically committed to a particular fiber type fate. Reciprocal transplantation of either somites or lateral plate mesoderm between chick and quail showed that fibers produced in chicks from quail myogenic precursor cells (MPCs) retained quail fiber type patterning and vice versa.52 This and other work indicates that MPCs are not naïve, and that the myoblast lineage provides some level of intrinsic control of fiber type patterning which is independent of extrinsic/environmental factors.
However, it is equally clear local environmental signals also play a central role in fiber type specification in fetal muscle,53–55 and it is very likely that fiber type specification depends on the interaction of both intrinsic and extrinsic controls. Retroviral labelling of MPCs in chick somites showed that neither MPCs in the somites, nor those in the proximal limb were committed to a specific fiber type fate55 and that commitment to the slow or fast fiber type occurred after MPCs had entered the limb bud. The study showed that a considerable proportion of the labelled MPCs gave rise to both fast and slow myotubes, suggesting that they were not intrinsically committed to a fiber type fate and indicating a role for extrinsic signals in fiber type specification. However, the study also showed that there were cells which produced only one type of fiber and that overall, the proportions of slow myotubes were significantly higher from cells labelled at somite stage I compared with those labelled at somite stage VII.55 These findings point to a level of intrinsic control and indicate that slow fiber type fate could be programmed within the somites.55 The findings of Kardon et al.55 are consistent with the general model of primary and secondary myogenesis and could be interpreted as suggesting that embryonic myoblasts are committed to a slow fiber fate early during development possibly in the somites, whereas fetal myoblasts show more plasticity and can form fast or slow fibers depending on extrinsic signals and could be more susceptible to possible ‘programming’ events.
Signalling systems in fiber type specification.
Prenatal events which may program fetal fiber type specification are likely to impact on the molecular and mechanistic factors which regulate fiber type diversity. However, although these factors are not yet fully described, it is becoming clear that pathways such as those involving sonic hedgehog (Shh), Wnt, fibroblast growth factor (FGF) etc.,—extensively described in the early stages of development—may also be important in the later stages of muscle development.29,30 In addition, recent studies have highlighted that a number of pathways known to be important in other systems, such as metabolic responsiveness, are also involved in the regulation of fiber type specification. Such relationships make these new observations of fiber type proportions in relation to obesity intriguing and raise the question as to whether some or any of these pathways provide further insights into the transduction of prenatal events into the postnatal outcomes.
Sonic hedgehog.
Shh has been shown to be a key element in the development of slow fibers in vitro,56 and in zebrafish.57 In avian and mammalian species the role of Shh in differentiation and fiber type development is still a matter of debate. In the mouse limb, Shh is ascribed with the role of acting as a survival and proliferation factor and as being required for the maintenance of MRF expression.58 Similarly in the chick limb, Shh has been shown to inhibit the terminal differentiation of myoblasts destined to become slow fibers, and to act to maintain these cells in a proliferative state.59 However, studies in which Shh has been found to promote the survival and proliferation of myoblasts58 have largely been carried out in limb buds and primary cultures both of which comprise non-myogenic cells in which Shh was also found stimulate proliferation.60 In pure C2 myoblast cell lines, in the absence of non myogenic tissue, Shh appears to promotes myogenic differentiation even in the presence of growth factors which usually inhibit differentiation.60 This would be consistent with the observation that in some but not all myoblasts from chick wing buds in culture exposure to Shh enhanced differentiation and increase the expression of slow myosin.60 It has been suggested that the activity of Hedgehog (Hh) signalling alone is sufficient to induce the slow muscle fiber program.61 However, it seems more likely that while Shh or other members of the Hh family may act directly to affect slow fiber differentiation, the nature and extent of responsiveness to Shh may be dependant on both the intrinsic status of the myoblasts and environmental signals possibly from non myogenic tissues.
Fibroblast growth factor.
In the development of other tissues Hh pathways have been shown to be linked with fibroblast growth factor (FGF) pathways, where FGF signals trigger the activation of Hh family members.62 In muscle, the role of FGF6 in myogenesis is well described63 and it is known that fibroblast growth factor receptor (FREK) expression is seen early during development and that it can be stimulated in myofibers by Shh.64 It remains to be seen whether interactive and/or inductive activities between FGF and HH pathways exist in muscle.
Wnt.
FGF signalling pathways cross-talk with Wnt signalling pathways in a variety of cellular processes and tissues,65 and it has been suggested that in addition to its well described role in early myogenesis, Wnt signalling is also involved in the differentiation of myogenic cells.66,67
Retroviral misexpression studies in the chick show that both Wnt 5a and Wnt 11 signals can alter fiber type in vivo. Expression of Wnt11 decreases the number of slow fibers but increases the number of fast fibers, with expression of Wnt 5a having reciprocal effects consistent with their differential fiber type expression patterns in vivo.66 Overexpression of Wnt4 in MPCs in chick embryos was shown to lead to a significant increase in the area of muscle cells expressing fast myosin heavy chain with a small but non significant reduction in slow myosin area leading to an overall significant increase in the fast:slow myosin heavy chain expressing area.67 These findings were attributed to a Wnt 4-mediated stimulation of proliferation and/or differentiation, particularly in fast myosin heavy chain expressing cells. The similarities between these findings and the observations of secondary myofiber hyperplasia and hypertrophy,68 and increased proportions of type IIb/fast glycolytic fibers69 in myostatin knockout mice, led to the suggestion that Wnt4 may be acting as a antagonist of the negative regulator of muscle growth, myostatin.67
Forkhead box O.
In osteoblasts Wnt signalling pathways have been shown to potentiate transcription mediated by the fork-head box O (FOXO) transcription factors.70 FOXOs are a family of transcription factors which regulate a number of metabolic pathways and response systems,71 and which are expressed in a range of insulin sensitive tissues including muscle. The activity of FOXO depends on insulin/Akt mediated phosphorylation leading to exclusion/export of FOXO from the nucleus71–73 with inactivation by nuclear exclusion preventing the expression of FOXOs target genes such as the insulin receptor, insulin receptor substrate 2, 4EBP171 and peroxisome proliferator-activated receptor gamma coactivator-1 α (PGC-1α).73,74
FOXO1 has been recently implicated as playing an important role in the specification of fiber type and the control of muscle differentiation. Kitamura et al.75 indicated that FOXO1 protein was present in fast and slow muscle, but was more abundant in m. soleus (predominantly type I fibers) than in either m. gastrocnemius or m. vastus lateralis (predominantly type II fibers). Conditional ablation of FOXO1 was shown to lead to an increase in fast fibers with a concomitant two-fold increase in MyoD expression, a decrease in slow fibers with an 80% decrease in myogenin expression and a reduction in treadmill running time.75 This would suggest that FOXO1 was involved in the specification and or maintenance of the type I (SO) fiber program, although this is in contrast to studies in which FOXO1 overexpression is associated with a reduction in the number of type I (SO) fibers and the size of both fast and slow fiber types.76 Interestingly, in pigs and mice the abundance of FOXO1 transcripts has been shown to be highest in type II/fast muscles and lowest in type I/slow muscle.77,78
In proliferating primary myoblasts FOXO1 is localized in the cytoplasm but appears to be translocated to the nucleus in a phosphorylation-independent manner under serum free differentiation conditions and it appears it is continuously exported from the nucleus in proliferating cells.79 However, during terminal differentiation, phosphorylation of FOXO1 appeared to reduce its fusion activity.79 Similarly, in C2C12 myoblasts studies have shown that FOXO1 has a negative effect on myogenesis,80,81 and it appears that the action of FOXO1 to inhibit differentiation in C2C12 occurs at an early stage, probably upstream of myogenin expression.81 In addition, the expression of constitutively active FOXO1 has been shown to increase both the abundance myostatin transcripts and the activity of the myostatin promoter in C2C12 myotubes.78 The association between FOXO1 and myostatin raises the possibility that FOXO1 may be important in determining both fiber type and fiber size, consistent with the finding that overexpression of FOXO1 in mice leads to a marked reduction in the size of both type I and type II fibers.76 Indeed the recent positioning of FOXO1 pathways as being involved in both myogenic differentiation and fiber type specification together with its known roles in the regulation of various metabolic pathways71 is of considerable interest.
Peroxisome proliferator-activated receptor gamma coactivator-1 α.
PGC-1α, as its name suggests, was initially identified through its functional interaction with the nuclear receptor peroxisome proliferator-activated receptor gamma (PPARγ).82 It acts as a transcriptional coactivator of various nuclear receptors and transcription factors83 and has been shown to be important in a wide range of functions in relation to energy metabolism including the induction of mitochondrial biogenesis, the regulation of glucose metabolism and fatty acid oxidation.82 The regulation of PGC-1α expression appears complex. In muscle PGC-1α appears to be regulated by FOXO,74 MRFs, particularly by MyoD84 and by MEF285 and it has been identified as being involved in the regulation of muscle fiber type specification.86
Overexpression of PGC-1α in primary mouse myoblasts leads to a more rapid maturation of the myotubes evidenced by a rapid down regulation of the expression of the immature isoforms of the myosin heavy chain, and an enhancement of the slow oxidative associated myosin isoforms and a reduction of fast glycolytic associated myosin isoforms.87 Loss-of-function studies in vivo showed that although mitochondrial function and density measures appear normal in the fast fibers of PGC-1α deficient mice, exercise capacity was reduced.86 In contrast, forced expression of PGC-1α led to expression of the type I muscle fiber phenotype via a mechanism which involved the co-activation of MEF2 transcription factors and was characterized by increased expression of contractile and mitochondrial elements of the slow fiber type gene program and increased fatigue resistance.86 These findings indicate that PGC-1α expression and down stream signalling is relevant to the regulation of both the contractile and metabolic aspect of fiber type specification, and may be particularly important in the context of prenatal impacts on fiber type and oxidative capacity.
Peroxisome proliferator-activated receptor gamma and delta.
PGC-1α was originally identified through its association with PPARγ. Changes in the expression of PPARγ can lead to changes in muscle, for example rat pups from dams fed a cafeteria diet showed reductions in both muscle cross-sectional area and fiber number concomitant with an increase in muscle levels of IGF-I, IGF-I receptor and PPARγ transcripts.88 These results point to a role for IGF-I and PPARγ in myogenic differentiation and are consistent with the findings that the inhibitory effects of PPARγ on differentiation are mediated via reductions in the expression of myogenin, MyoD and creatine kinase.89
However, it is now known that another PPAR isoform, PPARδ, activates PGC-1α gene transcription in muscle.90 PPARδ (also known as PPARβ) is the most abundant PPAR isoform in muscle,91 it is implicated in the control of muscle development and metabolism, and the determination of fiber type18,92–94 as well as fiber type switching.91 In cell lines PPARδ has been shown to regulate genes involved in fatty acid transport, β-oxidation and mitochondrial respiration.95 Overexpression of PPARδ in mouse muscle leads to increased oxidative capacity mediated via both a hyperplastic increase in oxidative fiber number and by fiber type switching but with no significant effect on muscle mass or fiber size.94 In addition, targeted expression of the activated form of PPARδ not only increases type I fibers but also leads to a significant increase in endurance exercise capacity.18 Hence the data indicate that PPARδ appears to be important for the maintenance of the slow oxidative fiber gene program, and since the PGC-1α promoter contains a PPAR response element, it is possible that the observed effects of PPARδ are mediated to some extent through PGC-1α transcription.
Calcineurin.
Calcineurin is a calcium-calmodulin-regulated serine threonine phosphatase which has been shown to be involved in the regulation of differentiation,96 the determination of fiber type,97–100 regeneration101 and fiber type conversion102,103 and evidence suggests that it may be activated by a range of factors which trigger increases in intracellular calcium. The downstream targets of calcineurin include the MEF2 and the nuclear factor of activated T-cells (NFATs) families transcription factors,97 both of which appear to be activated by increased intracellular calcium.
However, whether and how calcineurin is involved in differentiation is a matter of ongoing debate. Evidence suggests that the mechanism by which calcineurin might regulate differentiation involves calcineurin-induced activation of MEF2 (which in turn may act as a transcription factor for PGC-1α), indirect activation of MyoD via a de-repression of the inhibitory effects of the inhibitor of differentiation (Id) on MyoD, and the induction of myogenin expression.104 It is also suggested that although calcineurin is required for the maintenance of slow fibers in adult muscle, the embryonic development of slow fibers may be calcineurin independent.105 Muscles from young adult transgenic mice in which a calcineurin inhibitor had been expressed from embryonic d9.5, prior to primary myogenesis, were found to have a reduced abundance of the contractile components but not the metabolic components of type I fibers.105 These results were interpreted as indicating that the expression of the contractile elements of slow fibers was dependant on calcineurin activity, whereas the oxidative capacity, mitochondrial content and myoglobin abundance were not.105 However, forced overexpression of constitutively active calcineurin, has been shown to increase the expression of markers of type I fibers,106 suggesting that while fiber type specification during embryogenesis may be calcineurin-independent, there is a requirement for calcineurin signalling postnatally.
Nuclear factors of activated T-cells.
Although the role of calcineurin in differentiation remains to be elucidated further, the argument for its involvement in the process may be strengthened by evidence that nuclear factors of activated T-cells (NFATs) play a role in differentiation. In response to increased intracellular calcium, calcineurin dephosphorylates NFATs which are then translocated to the nucleus107 where they activate transcription. The expression of the NFAT isoforms during the various stages of development implies their role in differential regulation of these developmental stages and appears consistent with the sequential expression and activation of NFATs seen in the differentiation of thymocytes.108 In myogenic tissue, NFATc3 translocates to the nucleus at the onset of differentiation while NFATs c1 and c2 show nuclear translocation in mature myotubes,109 hence suggesting a specific role for NFATc3 in differentiation and primary myogenesis with the remaining isoforms being important in secondary myogenesis and later developmental stages. This role for NFATc3 is further supported by studies in cell cultures showing that NFATc3 promotes differentiation, but it does not appear to be involved in slow myosin heavy chain expression,109 and knockout studies using NFATc3-/- mice which show a reduction in both fiber number and muscle mass due to a reduction in primary myogenesis.110
The involvement of NFATc1 and c2 in secondary myogenesis, fiber type specification and the maintenance of the myogenic state is less clear. For example, in NFATc2-/- mice fiber size is reduced and it is speculated that NFATc2 is involved in fusion and recruitment of myonuclei to newly formed fibers.111 The association of calcineurin with NFATc1 (and GATA-2) in the regeneration of adult muscle101 and the regulation of myf5 expression by a calcineurin and NFAT dependant pathway in reserve or satellite cell cultures supports the role of NFATs in the later stages of myogenesis and/or in regeneration.112
Innervation.
In innervated muscle, intracellular calcium levels change in response to neural activity such that in slow, tonic fibers sustained low levels of calcium exist, while in fast, phasic fibers transiently high levels of calcium are achieved. In the model proposed by Chin et al.98 the intrinsic differences in intracellular calcium levels arising from the differences in neuronal firing pattern were used to explain the observed differences in calcineurin activity and NFAT phosphorylation status, and it was suggested that calcineurin was activated in slow but not fast fibers. The model explained many of the observations in vivo that calcineurin controls the expression of the nerve activity dependant slow fiber gene program, but did not regulate hypertrophic growth.113
However, the model proposed by Chin et al.98 largely relates to mature myofibers and does not necessarily hold true in developing muscle, as pointed out earlier. The onset of myogenesis is accompanied by the arrival of pathfinder motor neurons in the muscle which make contact with the primary myotubes at a point at which acetylcholine receptors (AChR) are clustered. Early studies led to the belief that the clustering of AChR was triggered via motor neuron induced activation of subsynaptic nuclei in developing muscle and that innervation was central to myogenic development. However, soleus muscle development in aneural mice has been shown to largely mirror that of soleus muscles from innervated animals,114 questioning the role of innervation in myogenesis. Moreover, it has also been shown that distribution and de novo formation of AChR clusters can be achieved in the absence of innervation and that calcineurin signalling plays a role in the redistribution of AChR induced by synaptogenic signals,115 this suggests that intrinsic calcineurin activity in muscle fibers rather than that stimulated by innervation is involved in the specification of slow fiber gene expression.
Consequently, these findings suggest that during fiber type specification, mechanisms other than or in addition to neural activity, may be involved. Moreover it appears that, based on the current understanding and the pathways involved, there are several ways in which prenatal ‘programming’ events might impinge leading to changes in fiber type specification.
Muscle fiber number and growth.
The relationship between fiber number and growth has been comprehensively reviewed.116 Briefly muscle fiber number is largely set in utero, and total fiber number shows a similar positive relationship to birth weight as that seen for muscle mass.116 After the MPCs exit the cell cycle no further proliferation occurs, so postnatal growth is hypertrophic rather than hyperplastic.
The Effect of Prenatal Events on Muscle Fiber Type
If it is speculated that the observed changes in fiber type proportions associated with obesity are causative, rather than consequential and that they are established in utero, then this begs the question of how changes myogenesis and in particular in fiber type specification could lead to the observed obesogenic phenotype.
The fiber type phenotype most commonly reported to be associated with obesity shows an increase in the relative proportions of type IIb/FG fibers and a decrease in the relative proportions of the oxidative fiber types, type I/SO and type IIa/FOG fibers, consistent with the observed decrease in oxidative capacity. As outlined in Figure 1 the origin and time of formation of the different fiber types is different. There is also fairly convincing evidence that at least a proportion of the type I (SO) fiber cohort is controlled by intrinsic factors, and is rather resistant to manipulation. Hence, if the observed obesogenic phenotype has prenatal origins, then it would appear that the most likely area for ‘programming’ would be secondary myogenesis which gives rise to a mixture of type I, IIa and IIb fibers (Fig. 1) although impacts on primary myogenesis or both cannot be excluded. The majority of observations are fairly consistent in reporting a decrease in the oxidative capacity of muscle. Hence, proposed differences in the metabolic profiles of the secondary fiber cohorts may be relevant (Fig. 1) and the precise timing of any potential ‘programming’ event to affect fetal myoblast proliferation or differentiation for example, may be critical to the observed outcome.
Although a range of impacts and interventions can affect muscle phenotype4 the majority of studies use models of prenatal maternal undernutrition to create increased susceptibility to obesity in the offspring. It is evident that the observed effects are dependant on the precise nature of the nutritional insult, its extent and its relative gestational timing, and that in general, impacts tend to be mediated via effects on the secondary rather than primary fibers.4 For example periconceptual manipulations, either via 50% nutrient restriction in pregnant ewes117 or via manipulations of the environment of pre-implantation ovine embryos has been shown to have significant impact on fetal fiber proportions.118,119 Nutritional restriction appeared to lead to a reduction in total fiber number together with a reduction in secondary fiber numbers,117 suggestive of precocious fusion. In contrast, manipulation of the pre-implantation environment resulted in an increase in the ratio of secondary to primary fibers following both in vitro culture118 and asynchronous transfer,119 with an increase in total fiber number also being measured following asynchronous transfer.119
Postnatally, observations of the impact of mid-term nutritional restriction on fiber number, type and size can be more difficult to interpret, because of refeeding associated changes in development during gestation or postnatal changes due to maturation. Measurement of fiber number can be informative because fiber number is fixed at birth. However, postnatal muscle size can make quantitative assessment of total fiber number challenging, so in large animals these data are often not collected. Nevertheless the relative proportions (percentage frequency) of the fiber types may be of interest. For example examination of the fiber populations in 8-month-old lambs born to ewes that were subjected to a 50% nutrient restriction during mid gestation only (d28–d70), showed that there was a relative increase in the proportions of type IIb fibers, an increase in the abundance of myosin IIb isoforms and a reduction in carnitine palmitoyltransferase-1 activity (a key enzyme controlling fatty acid oxidation) compared with controls.120 In a similar study, offspring of restricted ewes (50% restriction d28 and d78 of gestation) were slaughtered at slaughter around 9 months old and, despite having a normal birth weight, were heavier, fatter and had reduced muscle weights as a proportion of hot carcase weight than control fed lambs, and showed deregulated glucose uptake.121 These findings suggest that accelerated growth late in gestation, increased postnatal weight gain and a decreased first-phase insulin response may occur in the offspring of restricted ewes.121 Moreover, the observations are seen as being similar to reports of the onset of diabetes mellitus in humans.121
In other studies undernutrition to 50% of nutrient requirements from d30 to d70 of gestation in twin bearing ewes was reported to lead to an apparent reduction in fast fiber density and an increase in slow fiber density and a significant increase in fast fiber but not slow fiber size in lamb muscles at two weeks postnatally.122 However, the apparent increase in fast fiber density may be distorted by the increase in fiber size. A subsequent experiment from the same group and using the same protocol (ewes restricted from either d30–70) reported various changes in fiber densities in several muscles from lambs slaughtered at 24 weeks old.123 The most consistent result was an increase in the density of fast type IIb fibers in both female and male lambs.123 The study also indicated that, consistent with reports from other studies,121 there was a tendency towards an increased fat:lean ratio in the carcases of lamb from restricted ewes.123
IGFs are known to affect myogenic proliferation and differentiation and fiber type specification and to be involved in the activation of pathways which have been implicated as mediating gene transcription in myogenic cells.124–126 Consequently the reports of both maternal and fetal changes in elements of the IGF system127,128 in situations of prenatal undernutrition are of considerable interest in the context of possible fetal ‘programming’ of myogenesis. For example, nutrient restriction to 60% of metabolizable energy (ME) requirements in ewes in mid gestation (d28–d80), led to transient elevation of IGF-II transcript abundance in fetal muscle, such that at the end of the period of nutrient restriction, abundance was higher in restricted fetuses, compared to well fed (150% ME) fetuses, but then fell to below the levels of well fed fetuses following 60 days of refeeding.129 However, there were no differences in fetal body weights or the weight of m. quadriceps at 140 days.129
The IGF signalling system is complex, involving a number of receptors and binding proteins so it is difficult to extrapolate as to exact changes in signalling simply on the basis of alterations in growth factor levels or transcript abundance. Nevertheless, it is interesting to note that in fetal lambs from ewes restricted to 50% of their nutritional requirements during early to mid gestation (d28–d78), protein levels for both IGF-I and IGF-II receptors were elevated in left ventricular tissue at d78, and IGF-I receptor expression remained elevated after late gestational re-alimentation.130 This suggests that undernutrition may have rather wide ranging effects on the IGF system and that if re-alimentation is associated with increased IGF-I receptor levels, this could have consequences for growth.
New Insights?
The evidence gives some support to the contention that nutritional impacts during gestation can affect myogenesis with consequences for muscle fiber composition and possibly for the susceptibility to obesity in later life. As outlined above, in general nutritional insults are associated with impacts on the secondary myogenic cohort with consequences for fast (type II) fibers and total fiber number. Moreover, there is increasing evidence that, at least in the case of sheep, inappropriate nutrition during critical periods of development may lead to changes which can be measured in the fetus,117–119,127–130 but which are also associated with measurable differences postnatally.120–123
While the mechanistic basis by which nutritional impacts in the fetus could lead to fiber type changes postnatally is not clear, there is evidence that restricting maternal nutrition changes the expression of genes which are known to affect the development of muscle. Hence it could be speculated that changes in transcript levels of key genes could entrain a sequence of events that could alter myogenic programming. For example, since IGF-II transcripts are increased in undernourished fetuses it is tempting to speculate that IGF-II may play a role in the aetiology of the observed increase in secondary fibers or fast fibers. Evidence suggests that in normal mouse embryogenesis fast myosin heavy chain expression precedes that of IGF-II,131 suggesting that changes in IGF-II expression might not affect fast myosin heavy chain expression. However, forced expression of IGF-II in myotubes has been shown to be associated with increased proportions of fast myosin heavy chain expressing myotubes,131 indicating that the effects of IGF-II may be directed not at the initiation of secondary myogenesis, but rather at fiber type specification hence establishing the fast myosin heavy chain expressing cohort of fibers. In this context, the findings from maternal nutritional restriction experiments in sheep,129,130 would indicate that changes in elements of the IGF system at around d78–80 would be likely to affect these aspects of both secondary and possibly tertiary myogenesis, hence possibly biasing postnatal fiber type proportions towards a fast phenotype, thereby enhancing the normal postnatal increase in glycolytic fiber types.132 It is also of interest that the expression myostatin, a negative regulator of growth, shows reciprocity with that of IGF-II and IGF-I receptor.133,134
If it is speculated that abnormally increased IGF-II expression at a critical time in gestation could affect myogenic programming in the fetus, leading to the increase in fast fibers and reduced oxidation arising from inappropriate prenatal nutrition, then it is interesting to consider some of the possible downstream targets of IGF-II signalling, and to assess whether such a scenario is tenable. There are a number of interdependent pathways/signalling systems such as calcineurin/NFAT, FOXO1, PGC-1α, PPARδ etc., through which increases in fast fiber or decreases in slow fiber types could be elicited.
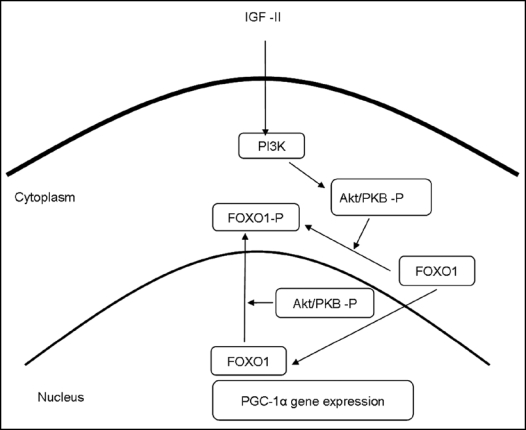
Although a role in differentiation has been proposed for the IGF-II receptor,135 the majority of IGF-II's actions appear to be mediated by binding to the IGF-I receptor in muscle,136 with subsequent downstream activation of pathways involving PI3K and IRS-1and leading to increased Akt activity, or involving the activation of the MAP kinase pathway. IGF-II mediated activation of Akt is particularly interesting in the context of fiber type specification because its downstream effects would be likely to lead to inactivation of FOXO1,137 a reduction in PGC-1α expression, and hence a reduction in both oxidative and slow fiber gene expression (Fig. 2).
Figure 2.
Proposed pathways by which IGF-II signals could affect fibre type. IGF-II signals mediated via PI3K and Akt lead to the phosphorylation and nuclear exclusion of FOXO1, a reduction of PGC-1α expression and hence a reduction in both oxidative and slow fibre gene expression. Based on Machida et al.137
IGF-I signals also affect myogenesis, largely through increasing calcineurin but not NFAT activity 109,138 supporting the concept that in the early stages of differentiation calcineurin induced gene expression is mediated via MEF2 and MyoD rather than NFATs and emphasising the role of IGF-I in early myogenesis. However, the identification of a calcineurin/NFAT responsive element in the IGF-I gene,139 activated by NFATc3 (the NFAT believed to be preferentially involved in primary myogenesis) may suggest a role for NFATc3. The complexities of these pathways are emphasized by the various interactions between FOXO1, Akt and calcineurin which appear to be important for the transduction of signals from the insulin receptor.140
Hence, although the relationships between the pathways controlling the myogenic processes are complex and require further investigation, it is clear that there are a number of ways on which prenatal insults could impinge on myogenic pathways to alter the outcome of fiber type specification.
Muscle Fiber Number and Obesity
Prenatal undernutrition, not only affects muscle fiber type, but also affects muscle fiber number which is set in utero.141,142 Muscle fiber number is determined by the extent of proliferation of the MPCs prior to primary and secondary myogenesis, so that any factor which alters fiber number is likely to have impinged on the proliferative phase of the myogenic program. Studies on maternal undernutrition in guinea pigs and rats show that reductions in fiber number appear to arise largely from impacts on secondary myogenesis.141,142 However, it appears that even very early insults can affect secondary fiber formation. In lambs from ewes subjected to peri conceptual undernutrition, total muscle fiber numbers were reduced by around 20%, apparently due to a reduction in secondary fiber formation.117 This is interesting since it is believed that the embryonic myoblasts which are the precursors of primary myotubes may be committed in the somites, whereas fetal myoblasts which are the secondary myotube precursors form later.
Muscle mass is determined by both hyperplasia and hypertrophy, hence relates to both fiber number and fiber size. Fiber size relates closely to the metabolic demands of the tissue, hence oxidative fibers with a considerable requirement for oxygen tend to be smaller because of the physical limits for diffusion, while fibers which are less dependant on oxidative metabolism have fewer diffusion driven size constraints, and can therefore hypertrophy to a greater extent. The relationship between lean deposition or muscle growth and lipid deposition is well known in livestock production where evidence shows that there is a genetic upper limit to daily protein deposition, with excess dietary protein and non-protein energy being deposited as lipid.143 Consequently once muscle fibers reach their maximum ‘metabolic size’, the limits for protein deposition within the lean tissue may be reached so that excess dietary protein and non-protein energy will be deposited as fat, leading to increased obesity.
The precise effect of low birth weight per se on the risk of disease in later life appears uncertain although the role of maternal under-nutrition appears clearer.144 Nevertheless; studies in animals have demonstrated that there are relationships between birth weight, muscle fiber numbers, rate of growth and, muscle mass. It is suggested that total fiber number and birth weight show a positive linear correlation, and that there is a positive correlation between fiber number and muscle cross-sectional area, whereas there is negative correlation between total muscle fiber number and the rate of postnatal hypertrophy.116 In a study of slaughter pigs, low birth weight individuals were found to have lower total fiber numbers but larger fibers than high birth weight individuals, and also to have higher levels of carcase fat.116 Hence, the relationships between large postnatal fiber size and increased adiposity in low birth weight pigs116,145 show similarities to the relationship between in low birth weight and obesity in later life in humans.146
Based on experimental data from pigs116,145 in situations of low birth weight and reduced fiber number it would appear that the reduced numbers of fibers of both types I and II, grow rapidly in the face of adequate nutrition and reach their maximum ‘metabolic size’ quickly. The signals for this compensatory response is not clear, but re-alimentation in undernourished ewes is associated with an upregulation of IGF-I receptor in the fetal lamb ventricle leading to ventricular enlargement.130 Similarly, postnatal catch up growth in intrauterine growth retarded rat pups appears also to be driven by IGF-I amongst other factors,147 and this type of response might lead to additional fiber hypertrophy in the late fetal or early postnatal period, with consequences for lipid deposition as noted in several studies.116,122,145 However, the available data from human studies of the relationship between low birth weight and muscle mass do not provide enough evidence to test this hypothesis.
Conclusion
A number of studies have linked obesity and muscle fiber characteristics and others have linked prenatal undernutrition with changes in muscle fiber numbers and muscle fiber type proportions. This review has examined whether the two are related and thus whether there is a rational basis for the speculation that prenatal maternal undernutrition can ‘program’ changes in the fetal muscle that will increase the individual's susceptibility to obesity in later life.
Although the process of myogenesis is tightly regulated, it is susceptible to prenatal perturbation. The evidence suggests that some muscle fibers, particularly primary fibers formed very early in development, may be more resistant to insults than the secondary cohort of fibers formed later. Moreover, it does appear possible that changes in the secondary myogenesis could be sufficient to account for some of the features of obesity-prone phenotype, and consideration of the recently described pathways implicated in fiber type specification provides a speculative mechanism for fetal adaptation to undernutrition in utero.
If the apparent relationships between muscle fiber characteristics and susceptibility to obesity are fully substantiated, then a greater understanding of the precise processes underpinning myogenesis and fiber type specification may be central to the ability to target advice, practices and therapeutics aimed at reducing obesity and its consequences.
Abbreviations
- SO
slow twitch oxidative
- FOG
fast twitch oxidative glycolytic fibre
- FG
fast twitch glycolytic
- PPAR
peroxisome proliferator-activated receptor
- PPARδ
peroxisome proliferator-activated receptor δ
- PPARγ
peroxisome proliferator-activated receptor γ
- MRFs
myogenic regulatory factors
- MPCs
myogenic precursor cells
- Hh
hedgehog
- Shh
sonic hedgehog
- FGF
fibroblast growth factor
- FREK
fibroblast growth factor receptor
- FOXO
Forkhead box O
- PGC-1α
peroxisome proliferator-activated receptor gamma coactivator 1-α
- NFAT
nuclear factor of activated T-cells
- AChRs
acetylcholine receptors
- MEF2
myocyte enhancing factor 2
Footnotes
Previously published online as an Organogenesis E-publication: http://www.landesbioscience.com/journals/organogenesis/article/6312
References
- 1.McMillen IC, MacLaughlin SM, Muhlhausler BS, Gentili S, Duffield JL, Morrison JL. Developmental origins of adult health and disease: the role of periconceptional and foetal nutrition. Basic Clin Pharmacol Toxicol. 2008;102:82–89. doi: 10.1111/j.1742-7843.2007.00188.x. [DOI] [PubMed] [Google Scholar]
- 2.Hanson MA, Gluckman PD. Developmental origins of health and disease: new insights. Basic Clin Pharmacol Toxicol. 2008;102:90–93. doi: 10.1111/j.1742-7843.2007.00186.x. [DOI] [PubMed] [Google Scholar]
- 3.Gluckman PD, Lillycrop KA, Vickers MH, Pleasants AB, Phillips ES, Beedle AS, Burdge GC, Hanson MA. Metabolic plasticity during mammalian development is directionally dependent on early nutritional status. Proc Natl Acad Sci USA. 2007;104:12796–12800. doi: 10.1073/pnas.0705667104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Maltin CA, Delday MI, Sinclair KD, Steven J, Sneddon AA. Impact of manipulations of myogenesis in utero in the performance of adult muscle. Reproduction. 2001;122:359–374. doi: 10.1530/rep.0.1220359. [DOI] [PubMed] [Google Scholar]
- 5.Rivero JLL, Barrey E. Heritabilities and genetic and phenotypic parameters for gluteus medius muscle fiber type composition, fiber size and capillaries in purebred Spanish horses. Livestock Production Science. 2001;72:233–241. [Google Scholar]
- 6.Renard G, Jurie C, Robelin J, Picard B, Geay Y, Menissier F. Genetic variability of muscle biological characteristics of young Limousin bulls. Genet Sel Evol. 1995;27:287–298. [Google Scholar]
- 7.Komi PV, Viitasalo JH, Havu M, Thorstensson A, Sjodin B, Karlsson J. Skeletal muscle fibers and muscle enzyme activites in monozygous and dizyogous twins of both sexes. Acta Physiol Scand. 1977;100:385–392. doi: 10.1111/j.1365-201X.1977.tb00001.x. [DOI] [PubMed] [Google Scholar]
- 8.Bouchard C, Simoneau JA, Lortie G, Boulay MR, Marcotte M, Thibault MC. Genetic effects in human skeletal muscle fiber type distribution and enzyme activities. Can J Physiol Pharmacol. 1986;64:1245–1251. doi: 10.1139/y86-210. [DOI] [PubMed] [Google Scholar]
- 9.Abernethy PJ, Thayer R, Taylor AW. Acute and chronic responses of skeletal muscle to endurance and sprint exercise. A review. Sports Med. 1990;10:365–389. doi: 10.2165/00007256-199010060-00004. [DOI] [PubMed] [Google Scholar]
- 10.Ross A, Leveritt M. Long term metabolic and skeletal muscle adaptations to short-sprint training: implications for sprint training and tapering. Sports Med. 2001;31:1063–1082. doi: 10.2165/00007256-200131150-00003. [DOI] [PubMed] [Google Scholar]
- 11.Costill DL, Fink WJ, Pollock ML. Muscle fiber composition and enzyme activities of elite distance runners. Med Sci Sports. 1976;8:96–100. [PubMed] [Google Scholar]
- 12.Martin WH., 3rd Effects of acute and chronic exercise on fat metabolism. Exerc Sport Sci Rev. 1996;24:203–231. [PubMed] [Google Scholar]
- 13.Tanner CJ, Barakat HA, Lynis Dohm G, Pories WJ, MacDonald KG, Cunningham PRG, Swanson MS, Houmard JA. Muscle fiber type is associated with obesity and weight loss. Am J Physiol Endocrinol Metab. 2002;282:1191–1196. doi: 10.1152/ajpendo.00416.2001. [DOI] [PubMed] [Google Scholar]
- 14.Oberbach A, Bossenz Y, Lehmann S, Niebauer J, Adams V, Paschke R, Schon MR, Bluher M, Punkt K. Altered fiber distribution and fiber specific glycolytic and oxidative enzyme activity in skeletal muscle of patients with type two diabetes. Diabetes Care. 2006;29:895–900. doi: 10.2337/diacare.29.04.06.dc05-1854. [DOI] [PubMed] [Google Scholar]
- 15.Adachi T, Kikuchi N, Yasuda K, Anahara R, Gu N, Matsunaga T, Yamamura T, Mori C, Tsujimoto G, Tsuda K, Ishihara A. Fiber type distribution and gene expression levels of both succinate dehydrogenase and peroxisome proliferator-activated receptor-gamma coactivator-1alpha of fibers in the soleus muscle of Zucker diabetic fatty rats. Exp Physiol. 2007;92:449–455. doi: 10.1113/expphysiol.2006.035451. [DOI] [PubMed] [Google Scholar]
- 16.Berggren J, Boyle KE, Chapman WH, Houmard JA. Skeletal muscle lipid oxidation and obesity: influence of weight loss and exercise. Am J Physiol Endocrinol Metab. 2008 doi: 10.1152/ajpendo.00354.2007. [E pub before print] [DOI] [PubMed] [Google Scholar]
- 17.Mensink M, Hesselink MK, Russell AP, Schaart G, Sels JP, Schrauwen P. Improved skeletal muscle oxidative enzyme activity and restoration of PGC-1alpha and PPAR beta/delta gene expression upon rosiglitazone treatment in obese patients with type 2 diabetes mellitus. Int J Obes (Lond) 2007;8:1302–1310. doi: 10.1038/sj.ijo.0803567. [DOI] [PubMed] [Google Scholar]
- 18.Wang YX, Zhang CL, Yu RT, Cho HK, Nelson MC, Bayuga-Ocampo CR, Ham J, Kang H, Evans RM. Regulation of muscle fiber type and running endurance by PPARδ. PLoS Biology. 2004;2:294. doi: 10.1371/journal.pbio.0020294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ryder JW, Bassel-Duby R, Olsen EN, Zierath JR. Skeletal muscle reprogramming by activation of calcineurin improves insulin action on metabolite pathways. J Biol Chem. 2003;278:44298–44304. doi: 10.1074/jbc.M304510200. [DOI] [PubMed] [Google Scholar]
- 20.Bruce CR, Anderson MJ, Carey AL, Newman DG, Bonen A, Kriketos AD, Cooney GJ, Hawley JA. Muscle oxidative capacity is a better predictor of insulin sensitivity that lipid status. J Clin Endocrinol Metab. 2003;88:5444–5451. doi: 10.1210/jc.2003-030791. [DOI] [PubMed] [Google Scholar]
- 21.Lillioja S, Young AA, Culter CL, Ivy JL, Abbott WG, Zawadzki JK, Yki-Järvinen H, Christin L, Secomb TW, Bogardus C. Skeletal muscle capillary density and fiber type are possible determinants of in vivo insulin resistance in man. J Clin Invest. 1987;80:415–424. doi: 10.1172/JCI113088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Reilly SM, Lee CH. PPARδ as a therapeutic target in metabolic disease. FEBS letts. 2008;582:26–31. doi: 10.1016/j.febslet.2007.11.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Luquet S, Lopez-Soriano J, Holst D, Fredenrich A, Melki J, Rassoulzadegan M, Grimaldi PA. Peroxisome proliferator-activated receptor delta controls muscle development and oxidative capability. FASEB J. 2003;17:2299–2301. doi: 10.1096/fj.03-0269fje. [DOI] [PubMed] [Google Scholar]
- 24.Krämer DK, Ahlsén M, Norrbom J, Jansson E, Hjeltnes N, Gustafsson T, Krook A. Human skeletal muscle fiber type variations correlate with PPAR alpha, PPAR delta and PGC-1 alpha mRNA. Acta Physiol. 2006;188:207–216. doi: 10.1111/j.1748-1716.2006.01620.x. [DOI] [PubMed] [Google Scholar]
- 25.Christ B, Huang R, Scaal M. Amniote somite derivatives. Dev Dyn. 2007;236:2382–2396. doi: 10.1002/dvdy.21189. [DOI] [PubMed] [Google Scholar]
- 26.Buckingham M, Bajard L, Chang T, Daubas P, Hadchouel J, Meilhac S, Montarras D, Rocancourt D, Relaix F. The formation of skeletal muscle from somite to limb. J Anat. 2003;202:59–68. doi: 10.1046/j.1469-7580.2003.00139.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Buckingham M. Myogenic progenitor cells and skeletal myogenesis in vertebrates. Curr Opin Genet Dev. 2006;16:525–532. doi: 10.1016/j.gde.2006.08.008. [DOI] [PubMed] [Google Scholar]
- 28.Kalcheim C, Ben-Yair R. Cell rearrangements during development of the somite and its derivatives. Curr Opin Genet Dev. 2005;15:371–380. doi: 10.1016/j.gde.2005.05.004. [DOI] [PubMed] [Google Scholar]
- 29.Francis-West PH, Antoni L, Anakwe K. Regulation of myogenic differentiation in the developing limb bud. J Anat. 2003;202:69–81. doi: 10.1046/j.1469-7580.2003.00136.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Te Kronnie G, Reggiani C. Skeletal muscle fiber type specification during embryonic development. J Muscle Res Cell Motil. 2002;23:65–69. doi: 10.1023/a:1019940932275. [DOI] [PubMed] [Google Scholar]
- 31.Bergstrom DA, Tapscott SJ. Molecular distinction between specification and differentiation in the myogenic basic helix-loop-helix transcription factor family. Mol Cell Biol. 2001;21:2404–2412. doi: 10.1128/MCB.21.7.2404-2412.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kassar Duchossoy L, Gayraud-Morel B, Gomes D, Rocancourt D, Buckingham M, Shinin V, Tajbakhsh S. Mrf 4 determines skeletal muscle identity in Myf5:MyoD double-mutant mice. Nature. 2004;431:466–471. doi: 10.1038/nature02876. [DOI] [PubMed] [Google Scholar]
- 33.Hadchouel J, Carvajal JJ, Daubas P, Bajard L, Chang T, Rocancourt D, Cox D, Summerbell D, Tajbakhsh S, Rigby PW, Buckingham M. Analysis of a key regulatory region upstream of the Myf5 gene reveals multiple phases of myogenesis, orchestrated at each site by a combination of elements dispersed throughout the locus. Development. 2003;130:3415–3426. doi: 10.1242/dev.00552. [DOI] [PubMed] [Google Scholar]
- 34.Rudnicki MA, Schnegelsberg PN, Stead RH, Braun T, Arnold HH, Jaenisch R. MyoD or Myf-5 is required for the formation of skeletal muscle. Cell. 1993;75:1351–1359. doi: 10.1016/0092-8674(93)90621-v. [DOI] [PubMed] [Google Scholar]
- 35.Tapscott SJ. The circuitry of a master switch: MyoD and the regulation of skeletal muscle gene transcription. Development. 2005;132:2685–2695. doi: 10.1242/dev.01874. [DOI] [PubMed] [Google Scholar]
- 36.Davis RL, Weintraub H, Lassar AB. Expression of a single transfected cDNA converts fibroblasts to myoblasts. Cell. 1987;51:987–1000. doi: 10.1016/0092-8674(87)90585-x. [DOI] [PubMed] [Google Scholar]
- 37.Valdez MR, Richardson JA, Klein WH, Olson EN. Failure of Myf5 to support myogenic differentiation without myogenin, MyoD, and MRF4. Dev Biol. 2000;219:287–298. doi: 10.1006/dbio.2000.9621. [DOI] [PubMed] [Google Scholar]
- 38.Summerbell D, Halai C, Rigby PWJ. Expression of the myogenic regulatory factor Mrf4 precedes or is contemporaneous with that of Myf5 in the somatic bud. Mech Dev. 2002;117:331–335. doi: 10.1016/s0925-4773(02)00208-3. [DOI] [PubMed] [Google Scholar]
- 39.McKinsey TA, Zhang CL, Olsen EN. Signaling chromatin to make muscle. Curr Opin Cell Biol. 2002;14:763–772. doi: 10.1016/s0955-0674(02)00389-7. [DOI] [PubMed] [Google Scholar]
- 40.Hughes SM, Taylor JM, Tapscott SJ, Gurley CM, Carter WJ, Peterson CA. Selective accumulation of MyoD and myogenin mRNAs in fast and slow adult skeletal muscle is controlled by innervation and hormones. Development. 1993;118:1137–1147. doi: 10.1242/dev.118.4.1137. [DOI] [PubMed] [Google Scholar]
- 41.Dunglisson GF, Scotting PJ, Wigmore PM. Rat embryonic myoblasts are restricted to forming primary fibers while later myogenic populations are pluripotent. Mech Dev. 1999;87:11–19. doi: 10.1016/s0925-4773(99)00134-3. [DOI] [PubMed] [Google Scholar]
- 42.Pin CL, Hrycyshyn AW, Rogers KA, Rushlow WJ, Merrifield PA. Embryonic and fetal rat myoblasts form different muscle fiber types in an ectopic in vivo environment. Dev Dyn. 2002;224:253–266. doi: 10.1002/dvdy.10106. [DOI] [PubMed] [Google Scholar]
- 43.Wigmore PM, Dunglisson GF. The generation of fiber diversity during myogenesis. Int J Dev Biol. 1998;42:117–125. [PubMed] [Google Scholar]
- 44.Wilson SJ, McEwan JC, Sheard PW, Harris AJ. Early stages of myogenesis in a large mammal: formation of successive generations of myotubes in sheep tibialis cranialis muscle. J Muscle Res Cell Motil. 1992;13:534–550. doi: 10.1007/BF01737996. [DOI] [PubMed] [Google Scholar]
- 45.Barbet JP, Thornell LE, Butler-Browne GS. Immunocytochemical characterisation of two generations of fibers during the development of the human quadriceps muscle. Mech Dev. 1991;35:3–11. doi: 10.1016/0925-4773(91)90036-6. [DOI] [PubMed] [Google Scholar]
- 46.Duxson MJ, Sheard PW. Formation of new myoblasts occurs exclusively at the multiple innervations zones of and embryonic large muscle. Dev Dyn. 1995;204:391–405. doi: 10.1002/aja.1002040406. [DOI] [PubMed] [Google Scholar]
- 47.Drager A, Wees AG, Fitzsimons RB. Primary secondary and tertiary myotubes in developing skeletal muscle a new approach to the analysis of human myogenesis. J Neurol Sci. 1987;81:19–43. doi: 10.1016/0022-510x(87)90181-x. [DOI] [PubMed] [Google Scholar]
- 48.Stockdale FE. Mechanisms of formation of muscle fiber types. Cell Struct Funct. 1997;22:37–43. doi: 10.1247/csf.22.37. [DOI] [PubMed] [Google Scholar]
- 49.Hughes SM, Blau HM. Muscle fiber pattern is independent of cell lineage in postnatal rodent development. Cell. 1992;68:659–671. doi: 10.1016/0092-8674(92)90142-y. [DOI] [PubMed] [Google Scholar]
- 50.DiMario JX, Fernyak SE, Stoackdale FE. Myoblasts transferred to the limbs of embryos are committed to specific fiber fates. Nature. 1993;362:165–167. doi: 10.1038/362165a0. [DOI] [PubMed] [Google Scholar]
- 51.Van Swearingen J, Lance-Jones C. Slow and fast muscle fibers are preferentially derived from myoblasts migrating into the chick limb bud at different developmental times. Dev Biol. 1995;170:321–337. doi: 10.1006/dbio.1995.1218. [DOI] [PubMed] [Google Scholar]
- 52.Nikovits W, Cann GM, Huang R, Christ B, Stockdale FE. Patterning of fast and slow fibers within embryonic muscles is established independantly of signals from the surrounding mesenchyme. Development. 2001;128:2537–2544. doi: 10.1242/dev.128.13.2537. [DOI] [PubMed] [Google Scholar]
- 53.Cho M, Webster SG, Blau HM. Evidence for myoblast-extrinsic regulation of slow myosin heavy chain expression during fiber formation in embryonic development. J Cell Biol. 1993;121:795–810. doi: 10.1083/jcb.121.4.795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Robson LG, Hughes SM. Local signals in the chick limb bud can override myoblast lineage commitment; induction of slow myosin heavy chain in fast myoblasts. Mech Dev. 1999;85:59–71. doi: 10.1016/s0925-4773(99)00060-x. [DOI] [PubMed] [Google Scholar]
- 55.Kardon G, Campbell JK, Tabin CJ. Local extrinsic signals determine muscle and endothelial cell fate and patterning in the vertebrate limb. Dev Cell. 2002;3:533–545. doi: 10.1016/s1534-5807(02)00291-5. [DOI] [PubMed] [Google Scholar]
- 56.Norris W, Neyt C, Ingham PW, Currie PD. Slow muscle induction by Hedgehog signalling in vitro. J Cell Sci. 2000;113:2695–2703. doi: 10.1242/jcs.113.15.2695. [DOI] [PubMed] [Google Scholar]
- 57.Blagden CS, Currie PD, Ingham PW, Hughes SM. Hedgehog induction of zebrafish slow muscle is mediated by Sonic hedgehog. Genes Dev. 1997;11:2163–2175. doi: 10.1101/gad.11.17.2163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kruger M, Mennerich D, Fees S, Schafer R, Mundios S, Braun T. Sonic hedgehog is a survival factor for hypaxial muscle during mouse development. Development. 2001;128:743–752. doi: 10.1242/dev.128.5.743. [DOI] [PubMed] [Google Scholar]
- 59.Bren-Mattison Y, Olwin BB. Sonic hedgehog inhibits the terminal differentiation of limb myoblasts committed to the slow lineage. Dev Biol. 2002;242:130–148. doi: 10.1006/dbio.2001.0528. [DOI] [PubMed] [Google Scholar]
- 60.Li X, Blagden CS, Bildsoe H, Bonnin MA, Duprez D, Hughes SM. Hedgehog can derive terminal differentiation of amniote slow skeletal muscle. BMC Dev Biol. 2004;4:9. doi: 10.1186/1471-213X-4-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Baxendale S, Davison C, Muxworthy C, Wolff C, Ingham PW, Roy S. The B-cell maturation factor Blimp-1 specifies vertebrate slow-twitch muscle fiber identity in response to Hedgehog signalling. Nature Genetics. 2004;36:88–93. doi: 10.1038/ng1280. [DOI] [PubMed] [Google Scholar]
- 62.Lavine KJ, White AC, Park C, Smith CS, Choi K, Long F, Hui C, Ornitz DM. Fibroblast growth factor signals regulate a wave of Hedgehog activation that is essential for coronary vascular development. Genes Dev. 2006;20:1651–1666. doi: 10.1101/gad.1411406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Armand AS, Laziz I, Chanoine C. FGF6 in myogenesis. Biochim Biophys Acta. 2006;1763:773–778. doi: 10.1016/j.bbamcr.2006.06.005. [DOI] [PubMed] [Google Scholar]
- 64.Kahane N, Cinnamon Y, Bachelet I, Kalcheim C. The third wave of myotome colonization by mitotically competent progenitors: regulating the balance between differentiation and proliferation during muscle development. Development. 2001;128:2187–2198. doi: 10.1242/dev.128.12.2187. [DOI] [PubMed] [Google Scholar]
- 65.Katoh M, Katoh M. Cross-talk of WNT and FGF signalling pathways at GSK3beta to regulate beta catenin and SNAIL signalling cascades. Cancer Biol Ther. 2006;5:1059–1064. doi: 10.4161/cbt.5.9.3151. [DOI] [PubMed] [Google Scholar]
- 66.Anakwe K, Robson L, Hadley J, Buxton P, Church V, Allen S, Hartmann C, Harfe B, Nohno T, Brown AM, Evans DJ, Francis-West P. Wnt signalling regulates myogenic differentiation in the developing avian wing. Development. 2003;130:3503–3514. doi: 10.1242/dev.00538. [DOI] [PubMed] [Google Scholar]
- 67.Takata H, Trada K, Oka H, Sunada Y, Moriguchi T, Nohno T. Involvement of Wnt4 signaling during myogenic proliferation and differentiation of skeletal muscle. Dev Dyn. 2007;236:2800–2807. doi: 10.1002/dvdy.21327. [DOI] [PubMed] [Google Scholar]
- 68.Lee SJ. Quadrupling muscle mass in mice by targeting TGFbeta signaling pathways. PLoS ONE. 2007;2:789. doi: 10.1371/journal.pone.0000789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Amthor H, Macharia R, Navarrete R, SChuelke M, Brown SC, Otto A, Voit T, Muntoni F, Vbrova G, Partridge T, Zammit P, Bunger L, Patel K. Lack of myostatin results in excessive muscle growth but impaired force generation. PNAS. 2007;104:1835–1840. doi: 10.1073/pnas.0604893104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Almeida M, Han L, Martin-Millan M, O'Brien CA, Manolagas SC. Oxidative stress antagonizes Wnt signalling in osteoblast precursors by diverting β-catenin from TCF- to FOXO-mediated transcription. J Biol Chem. 2007;282:27298–27305. doi: 10.1074/jbc.M702811200. [DOI] [PubMed] [Google Scholar]
- 71.Nakae J, Oki M, Cao Y. The FOXO transcription factors and metabolic regulation. FEBS letts. 2008;582:54–67. doi: 10.1016/j.febslet.2007.11.025. [DOI] [PubMed] [Google Scholar]
- 72.Dowell P, Otto T, Adi S, Lane DM. Convergence of the peroxisome proliferator-activated receptor γ and Foxo1 signaling pathways. J Biol Chem. 2003;278:45485–45491. doi: 10.1074/jbc.M309069200. [DOI] [PubMed] [Google Scholar]
- 73.Southgate RJ, Bruce CR, Carey AL, Steinberg GR, Walder K, Monks R, Watt MJ, Hawley JA, Birnbaum MJ, Febbraio MA. PGC-1alpha gene expression is downregulated by Akt-mediated phosphorylation and nuclear exclusion of FoxO1 in insulin-stimulated skeletal muscle. FASEB J. 2005;19:2072–2074. doi: 10.1096/fj.05-3993fje. [DOI] [PubMed] [Google Scholar]
- 74.Daitoku H, Yamagata K, Matsuzakai H, Hatta M, Fukamizu A. Regulation of PGC-1 promoter activity by protein kinase B and the forkhead transcription factor FKHR. Diabetes. 2003;52:642–649. doi: 10.2337/diabetes.52.3.642. [DOI] [PubMed] [Google Scholar]
- 75.Kitamura T, Kitamura YI, Funahashi Y, Shawber CJ, Castrillon DH, Kollipara R, DePinho RA, Kitajewski J, Accli D. A Foxo/Notch pathway control myogenic differentiation and fiber type specification. J Clin Invest. 2007;117:2477–2485. doi: 10.1172/JCI32054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Kamei Y, Miura S, Suzuki M, Kai Y, Mizukami J, Taniguchi T, Mochida K, Hata T, Matsuda J, Aburatani H, Nishino I, Ezaki O. Skeletal muscle FOXO1 (FKHR) transgenic mice have less skeletal muscle mass, downregulated Type I (slow twitch/red muscle) fiber genes, and impaired glycemic control. J Biol Chem. 2004;279:41114–41123. doi: 10.1074/jbc.M400674200. [DOI] [PubMed] [Google Scholar]
- 77.Yang YJ, Pang WJ, Bai L, Yang GS. Expression of FoxO1 mRNA in muscle tissue of Bamei, Landrace and Landrace x Bamei. Yi Chuan. 2008;30:185–189. doi: 10.3724/sp.j.1005.2008.00185. [DOI] [PubMed] [Google Scholar]
- 78.Allen DL, Unterman TG. Regulation of myostatin expression and myoblast differentiation by FoxO and SMAD transcription factors. Am J Physiol Cell Physiol. 2007;292:188–199. doi: 10.1152/ajpcell.00542.2005. [DOI] [PubMed] [Google Scholar]
- 79.Bois PRJ, Grosveld GC. FKHR (FOXO1a) is required for myotubes fusion of primary mouse myoblasts. EMBO J. 2003;22:1147–1157. doi: 10.1093/emboj/cdg116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Hribal ML, Nakae J, Kitamura T, Shutter JR, Accili D. Regulation of insulin-like growth factor-dependent myoblast differentiation by Foxo forkhead transcription factors. J Cell Biol. 2003;162:535–541. doi: 10.1083/jcb.200212107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Wu AL, Kim JH, Zhang C, Unterman TG, Chen J. Forkhead Box Protein O1 Negatively Regulates Skeletal Myocyte Differentiation through Degradation of Mammalian Target of Rapamycin Pathway Components. Endocrinology. 2008;149:1407–1414. doi: 10.1210/en.2007-1470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Finck BN, Kelly DP. PGC-1 coactivators: inducible regulators of energy metabolism in health and disease. J Clin Invest. 2006;116:615–622. doi: 10.1172/JCI27794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Knutti D, Kralli A. PGC-1, a versatile coactivator. Trends Endocrinol Metab. 2001;12:360–365. doi: 10.1016/s1043-2760(01)00457-x. [DOI] [PubMed] [Google Scholar]
- 84.Chang JH, Lin KH, Shih CH, Chang YJ, Chi HC, Chen SL. Myogenic basic helix-loop-helix proteins regulate the expression of peroxisomal proliferator-activated receptor-γ coactivator-1α. Endocrinology. 2006;147:3093–3106. doi: 10.1210/en.2005-1317. [DOI] [PubMed] [Google Scholar]
- 85.Czbryt MP, McAnally J, Fishman GI, Olson EN. Regulation of peroxisome proliferatoractivated receptor γ coactivator 1α (PGC-1α) and mitochondrial function by MEF2 and HDAC5. PNAS. 2003;100:1711–1716. doi: 10.1073/pnas.0337639100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Lin J, Wu H, Tarr PT, Zhang CY, Wu Z, Boss O, Michael LF, Puigserver P, Isotani E, Olson EN, Lowell BB, Bassel-Duby R, Spiegelman BM. Transcriptional co-activator PGC-1 alpha drives the formation of slow-twitch muscle fibers. Nature. 2002;418:797–801. doi: 10.1038/nature00904. [DOI] [PubMed] [Google Scholar]
- 87.Mortensen OH, Frandsen L, Schjerling P, Nishimura E, Grunnet N. PGC-1alpha and PGC-1beta have both similar and distinct effects on myofiber switching toward an oxidative phenotype. Am J Physiol Endocrinol Metab. 2006;291:807–816. doi: 10.1152/ajpendo.00591.2005. [DOI] [PubMed] [Google Scholar]
- 88.Bayol SA, Simbi BH, Stickland NC. A maternal cafeteria diet during gestation and lactation promotes adiposity and impairs skeletal muscle development and metabolism in rat offspring at weaning. J Physiol. 2005;567:951–961. doi: 10.1113/jphysiol.2005.088989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Singh J, Verma NK, Kansagra SM, Kate BN, Dey CS. Altered PPARgamma expression inhibits myogenic differentiation in C2C12 skeletal muscle cells. Mol Cell Biochem. 2007;294:163–171. doi: 10.1007/s11010-006-9256-x. [DOI] [PubMed] [Google Scholar]
- 90.Hondares E, Pineda-Torra I, Iglesias R, Staels B, Villarroya F, Giralt M. PPARdelta, but not PPARalpha, activates PGC-1alpha gene transcription in muscle. Biochem Biophys Res Commun. 2007;354:1021–1027. doi: 10.1016/j.bbrc.2007.01.092. [DOI] [PubMed] [Google Scholar]
- 91.Schuler M, Ali F, Chambon C, Duteil D, Bornert JM, Tardivel A, Desvergne B, Wahli W, Chambon P, Metzger D. PGC1alpha expression is controlled in skeletal muscles by PPARbeta, whose ablation results in fiber-type switching, obesity, and type 2 diabetes. Cell Metab. 2006;4:407–414. doi: 10.1016/j.cmet.2006.10.003. [DOI] [PubMed] [Google Scholar]
- 92.Gaudel C, Grimaldi PA. Metabolic Functions of Peroxisome Proliferator-Activated Receptor beta/delta in Skeletal Muscle. PPAR Res. 2007;2007:86394. doi: 10.1155/2007/86394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Grimaldi PA. Roles of PPARdelta in the control of muscle development and metabolism. Biochem Soc Trans. 2003;31:1130–1132. doi: 10.1042/bst0311130. [DOI] [PubMed] [Google Scholar]
- 94.Luquet S, Lopez-Soriano J, Holst D, Fredenrich A, Melki J, Rassoulzadegan M, Grimaldi PA. Peroxisome proliferator-activated receptor delta controls muscle development and oxidative capability. FASEB J. 2003;17:2299–2301. doi: 10.1096/fj.03-0269fje. [DOI] [PubMed] [Google Scholar]
- 95.Tanaka T, Yamamoto J, Iwasaki S, Asaba H, Hamura H, Ikeda Y, Watanabe M, Magoori K, Ioka RX, Tachibana K, Watanabe Y, Uchiyama Y, Sumi K, Iguchi H, Ito S, Doi T, Hamakubo T, Naito M, Auwerx J, Yanagisawa M, Kodama T, Sakai J. Activation of peroxisome proliferator-activated receptor delta induces fatty acid beta-oxidation in skeletal muscle and attenuates metabolic syndrome. Proc Natl Acad Sci USA. 2003;100:15924–15929. doi: 10.1073/pnas.0306981100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Friday BB, Horsley V, Pavlath GK. Calcineurin activity is required for the initiation of skeletal muscle differentiation. J Cell Biol. 2000;149:657–666. doi: 10.1083/jcb.149.3.657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Schulz RA, Yutzey KE. Calcineurin signaling and NFAT activation in cardiovascular and skeletal muscle development. Dev Biol. 2004;266:1–16. doi: 10.1016/j.ydbio.2003.10.008. [DOI] [PubMed] [Google Scholar]
- 98.Chin ER, Olson EN, Richardson JA, Yang Q, Humphries C, Shelton JM, Wu H, Zhu W, Bassel-Duby R, Williams RS. A calcineurin-dependent transcriptional pathway controls skeletal muscle fiber type. Genes Dev. 1998;12:2499–2509. doi: 10.1101/gad.12.16.2499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Swoap SJ, Hunter RB, Stevenson EJ, Felton HM, Kansagra NV, Lang JM, Esser KA, Kandarian SC. The calcineurin-NFAT pathway and muscle fiber-type gene expression. Am J Physiol Cell Physiol. 2000;279:915–924. doi: 10.1152/ajpcell.2000.279.4.C915. [DOI] [PubMed] [Google Scholar]
- 100.Talmadge RJ, Otis JS, Rittler MR, Garcia ND, Spencer SR, Lees SJ, Naya FJ. Calcineurin activation influences muscle phenotype in a muscle-specific fashion. BMC Cell Biol. 2004;28:5–28. doi: 10.1186/1471-2121-5-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Sakuma K, Nishikawa J, Nakao R, Watanabe K, Totsuka T, Nakano H, Sano M, Yasuhara M. Calcineurin is a potent regulator for skeletal muscle regeneration by association with NFATc1 and GATA-2. Acta Neuropathol. 2003;105:271–280. doi: 10.1007/s00401-002-0647-0. [DOI] [PubMed] [Google Scholar]
- 102.Bigard X, Sanchez H, Zoll J, Mateo P, Rousseau V, Veksler V, Ventura-Clapier R. Calcineurin Co-regulates contractile and metabolic components of slow muscle phenotype. J Biol Chem. 2000;275:19653–19660. doi: 10.1074/jbc.M000430200. [DOI] [PubMed] [Google Scholar]
- 103.da Costa N, Edgar J, Ooi PT, Su Y, Meissner JD, Chang KC. Calcineurin differentially regulates fast myosin heavy chain genes in oxidative muscle fiber type conversion. Cell Tissue Res. 2007;329:515–527. doi: 10.1007/s00441-007-0441-3. [DOI] [PubMed] [Google Scholar]
- 104.Friday BB, Mitchell PO, Kegley KM, Pavlath GK. Calcineurin initiates skeletal muscle differentiation by activating MEF2 and MyoD. Differentiation. 2003;71:217–227. doi: 10.1046/j.1432-0436.2003.710303.x. [DOI] [PubMed] [Google Scholar]
- 105.Oh M, Rybkin II, Copeland V, Czubryt MP, Shelton JM, van Rooij E, Richardson JA, Hill JA, De Windt LJ, Bassel-Duby R, Olson EN, Rothermel BA. Calcineurin is necessary for the maintenance but not embryonic development of slow muscle fibers. Mol Cell Biol. 2005;25:6629–6638. doi: 10.1128/MCB.25.15.6629-6638.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Wu H, Rothermel B, Kanatous S, Rosenberg P, Naya FJ, Shelton JM, Hutcheson KA, DiMaio JM, Olson EN, Bassel-Duby R, Williams RS. Activation of MEF2 by muscle activity is mediated through a calcineurin-dependent pathway. EMBO J. 2001;20:6414–6423. doi: 10.1093/emboj/20.22.6414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Beals CR, Sheridan CM, Turek CW, Gardner P, Crabtree GR. Nuclear export of NF-ATc enhanced by glycogen synthase kinase-3. Science. 1997;275:1930–1934. doi: 10.1126/science.275.5308.1930. [DOI] [PubMed] [Google Scholar]
- 108.Adachi S, Amasaki Y, Miyatake S, Arai N, Iwata M. Successive expression and activation of NFAT family members during thymocyte differentiation. J Biol Chem. 2000;275:14708–14716. doi: 10.1074/jbc.275.19.14708. [DOI] [PubMed] [Google Scholar]
- 109.Delling U, Tureckova J, Lim HW, De Windt LJ, Rotwein P, Molkentin JD. A calcineurin-NFATc3-dependent pathway regulates skeletal muscle differentiation and slow myosin heavy-chain expression. Mol Cell Biol. 2000;20:6600–6611. doi: 10.1128/mcb.20.17.6600-6611.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Kegley KM, Gephart J, Warren GL, Pavlath GK. Altered primary myogenesis in NFATC3(-/-) mice leads to decreased muscle size in the adult. Dev Biol. 2001;232:115–126. doi: 10.1006/dbio.2001.0179. [DOI] [PubMed] [Google Scholar]
- 111.Horsley V, Friday BB, Matteson S, Kegley KM, Gephart J, Pavlath GK. Regulation of the growth of multinucleated muscle cells by an NFATC2-dependent pathway. J Cell Biol. 2001;153:329–338. doi: 10.1083/jcb.153.2.329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Friday BB, Pavlath GK. A calcineurin- and NFAT-dependent pathway regulates Myf5 gene expression in skeletal muscle reserve cells. J Cell Sci. 2000;114:303–310. doi: 10.1242/jcs.114.2.303. [DOI] [PubMed] [Google Scholar]
- 113.Serrano AL, Murgia M, Pallafacchina G, Calabria E, Coniglio P, Lømo T, Schiaffino S. Calcineurin controls nerve activity-dependent specification of slow skeletal muscle fibers but not muscle growth. Proc Natl Acad Sci USA. 2001;98:13108–13113. doi: 10.1073/pnas.231148598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Hughes DS, Schade RR, Ontell M. Ablation of the fetal mouse spinal cord: the effect on soleus muscle cytoarchitecture. Dev Dyn. 1992;193:164–174. doi: 10.1002/aja.1001930208. [DOI] [PubMed] [Google Scholar]
- 115.Madhavan R, Zhao XT, Chan F, Wu Z, Peng HB. The involvement of calcineurin in acetylcholine receptor redistribution in muscle. Mol Cell Neurosci. 2003;23:587–599. doi: 10.1016/s1044-7431(03)00101-5. [DOI] [PubMed] [Google Scholar]
- 116.Rehfeldt C, Kuhn G. Consequences of birth weight for postnatal growth performance and carcass quality in pigs as related to myogenesis. J Anim Sci. 2006;84:113–123. doi: 10.2527/2006.8413_supple113x. [DOI] [PubMed] [Google Scholar]
- 117.Quigley SP, Kleemann DO, Kakar MA, Owens JA, Nattrass GS, Maddocks S, Walker SK. Myogenesis in sheep is altered by maternal feed intake during the peri-conception period. Anim Reprod Sci. 2005;87:241–251. doi: 10.1016/j.anireprosci.2004.11.005. [DOI] [PubMed] [Google Scholar]
- 118.Maxfield EK, Sinclair KD, Broadbent PJ, McEvoy TG, Robinson JJ, Maltin CA. Short-term culture of ovine embryos modifies fetal myogenesis. Am J Physiol. 1998;274:1121–1123. doi: 10.1152/ajpendo.1998.274.6.E1121. [DOI] [PubMed] [Google Scholar]
- 119.Maxfield EK, Sinclair KD, Dunne LD, Broadbent PJ, Robinson JJ, Stewart E, Kyle DG, Maltin CA. Temporary exposure of ovine embryos to an advanced uterine environment does not affect fetal weight but alters fetal muscle development. Biol Reprod. 1998;59:321–325. doi: 10.1095/biolreprod59.2.321. [DOI] [PubMed] [Google Scholar]
- 120.Zhu MJ, Ford SP, Means WJ, Hess BW, Nathanielsz PW, Du M. Maternal nutrient restriction affects properties of skeletal muscle in offspring. J Physiol. 2006;575:241–250. doi: 10.1113/jphysiol.2006.112110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Ford SP, Hess BW, Schwope MM, Nijland MJ, Gilbert JS, Vonnahme KA, Means WJ, Han H, Nathanielsz PW. Maternal undernutrition during early to mid-gestation in the ewe results in altered growth, adiposity, and glucose tolerance in male offspring. J Anim Sci. 2007;85:1285–1294. doi: 10.2527/jas.2005-624. [DOI] [PubMed] [Google Scholar]
- 122.Fahey AJ, Brameld JM, Parr T, Buttery PJ. The effect of maternal undernutrition before muscle differentiation on the muscle fiber development of the newborn lamb. J Anim Sci. 2005;83:2564–2571. doi: 10.2527/2005.83112564x. [DOI] [PubMed] [Google Scholar]
- 123.Daniel ZC, Brameld JM, Craigon J, Scollan ND, Buttery PJ. Effect of maternal dietary restriction during pregnancy on lamb carcass characteristics and muscle fiber composition. J Anim Sci. 2007;85:1565–1576. doi: 10.2527/jas.2006-743. [DOI] [PubMed] [Google Scholar]
- 124.Tollefsen SE, Lajara R, McCusker RH, Clemmons DR, Rotwein P. Insulin-like growth factors (IGF) in muscle development. Expression of IGF-I, the IGF-I receptor, and an IGF binding protein during myoblast differentiation. J Biol Chem. 1989;264:13810–13817. [PubMed] [Google Scholar]
- 125.Semsarian C, Wu MJ, Ju YK, Marciniec T, Yeoh T, Allen DG, Harvey RP, Graham RM. Skeletal muscle hypertrophy is mediated by a Ca2+-dependent calcineurin signalling pathway. Nature. 1999;400:576–581. doi: 10.1038/23054. [DOI] [PubMed] [Google Scholar]
- 126.Espinosa A, Estrada M, Jaimovich E. IGF-I and insulin induce different intracellular calcium signals in skeletal muscle cells. J Endocrinol. 2004;182:339–352. doi: 10.1677/joe.0.1820339. [DOI] [PubMed] [Google Scholar]
- 127.Osgerby JC, Wathes DC, Howard D, Gadd TS. The effect of maternal undernutrition on the placental growth trajectory and the uterine insulin-like growth factor axis in the pregnant ewe. J Endocrinol. 2004;182:89–103. doi: 10.1677/joe.0.1820089. [DOI] [PubMed] [Google Scholar]
- 128.Heasman L, Brameld J, Mostvn A, Budge H, Dawson J, Buttery P, Stephenson T, Symonds ME. Maternal nutrient restriction during early to mid gestation alters the relationship between insulin-like growth factor I and bodyweight at term in fetal sheep. Reprod Fertil Dev. 2000;12:345–350. doi: 10.1071/rd00115. [DOI] [PubMed] [Google Scholar]
- 129.Brameld JM, Mostyn A, Dandrea J, Stephenson TJ, Dawson JM, Buttery PJ, Symonds ME. Maternal nutrition alters the expression of insulin-like growth factors in fetal sheep liver and skeletal muscle. J Endocrinol. 2000;167:429–437. doi: 10.1677/joe.0.1670429. [DOI] [PubMed] [Google Scholar]
- 130.Dong F, Ford SP, Fang CX, Nijland MJ, Nathanielsz PW, Ren J. Maternal nutrient restriction during early to mid gestation upregulates cardiac insulin-like growth factor (IGF) receptors associated with enlarged ventricular size in fetal sheep. Growth Horm IGF Res. 2005;15:291–299. doi: 10.1016/j.ghir.2005.05.003. [DOI] [PubMed] [Google Scholar]
- 131.Merrick D, Ting T, Stadler LK, Smith J. A role for Insulin-like growth factor 2 in specification of the fast skeletal muscle fiber. BMC Dev Biol. 2007;7:65. doi: 10.1186/1471-213X-7-65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Whipple G, Koohmaraie M. Effects of lamb age, muscle type and 24 hour activity of endogenous proteinases on post-mortem proteolysis. J. Anim Sci. 1992;70:798–804. doi: 10.2527/1992.703798x. [DOI] [PubMed] [Google Scholar]
- 133.Kocamis H, Gahr SA, Batelli L, Hubbs AF, Killefer J. IGF-I, IGF-II, and IGF-receptor-1 transcript and IGF-II protein expression in myostatin knockout mice tissues. Muscle Nerve. 2002;26:55–63. doi: 10.1002/mus.10160. [DOI] [PubMed] [Google Scholar]
- 134.Yamaguchi A, Fujikawa T, Tateoka M, Soya H, Sakuma K, Sugiura T, Morita I, Ikeda Y, Hirai T. The expression of IGF-I and myostatin mRNAs in skeletal muscle of hypophysectomized and underfed rats during postnatal growth. Acta Physiol (Oxf) 2006;186:291–300. doi: 10.1111/j.1748-1716.2006.01569.x. [DOI] [PubMed] [Google Scholar]
- 135.Rosenthal SM, Hsiao D, Silverman LA. An insulin-like growth factor-II (IGF-II) analog with highly selective affinity for IGF-II receptors stimulates differentiation, but not IGF-I receptor downregulation in muscle cells. Endocrinology. 1994;135:38–44. doi: 10.1210/endo.135.1.8013373. [DOI] [PubMed] [Google Scholar]
- 136.Florini JR, Ewton DZ, Coolican SA. Growth hormone and the insulin-like growth factor system in myogenesis. Endocr Rev. 1996;17:481–517. doi: 10.1210/edrv-17-5-481. [DOI] [PubMed] [Google Scholar]
- 137.Machida S, Spangenburg EE, Booth FW. Forkhead transcription factor FoxO1 transduces insulin-like growth factor's signal to p27Kip1 in primary skeletal muscle satellite cells. J Cell Physiol. 2003;196:523–531. doi: 10.1002/jcp.10339. [DOI] [PubMed] [Google Scholar]
- 138.Spangenburg EE, Bowles DK, Booth FW. Insulin-like growth factor-induced transcriptional activity of the skeletal alpha-actin gene is regulated by signaling mechanisms linked to voltage-gated calcium channels during myoblast differentiation. Endocrinology. 2004;145:2054–2063. doi: 10.1210/en.2003-1476. [DOI] [PubMed] [Google Scholar]
- 139.Alfieri CM, Evans-Anderson HJ, Yutzey KE. Developmental regulation of the mouse IGF-I exon 1 promoter region by calcineurin activation of NFAT in skeletal muscle. Am J Physiol Cell Physiol. 2007;292:1887–1894. doi: 10.1152/ajpcell.00506.2006. [DOI] [PubMed] [Google Scholar]
- 140.Ni YG, Wang N, Cao DJ, Sachan N, Morris DJ, Gerard RD, Kuro-O M, Rothermel BA, Hill JA. FoxO transcription factors activate Akt and attenuate insulin signaling in heart by inhibiting protein phosphatases. Proc Natl Acad Sci USA. 2007;104:20517–20522. doi: 10.1073/pnas.0610290104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Dwyer CM, Madgwick AJ, Ward SS, Stickland NC. Effect of maternal undernutrition in early gestation on the development of fetal myofibers in the guinea-pig. Reprod Fertil Dev. 1995;7:1285–1292. doi: 10.1071/rd9951285. [DOI] [PubMed] [Google Scholar]
- 142.Wilson SJ, Ross JJ, Harris AJ. A critical period for formation of secondary myotubes defined by prenatal undernourishment in rats. Development. 1988;102:815–821. doi: 10.1242/dev.102.4.815. [DOI] [PubMed] [Google Scholar]
- 143.Moughan PJ, Jacobson LH, Morel PC. A genetic upper limit to whole-body protein deposition in a strain of growing pigs. J Anim Sci. 2006;84:3301–3309. doi: 10.2527/jas.2005-277. [DOI] [PubMed] [Google Scholar]
- 144.Gardner DS, Buttery PJ, Daniel Z, Symonds ME. Factors affecting birth weight in sheep: maternal environment. Reproduction. 2007;133:297–307. doi: 10.1530/REP-06-0042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Bee G. Effect of early gestation feeding, birth weight, and gender of progeny on muscle fiber characteristics of pigs at slaughter. J Anim Sci. 2004;82:826–836. doi: 10.2527/2004.823826x. [DOI] [PubMed] [Google Scholar]
- 146.Wells JC, Chomtho S, Fewtrell MS. Programming of body composition by early growth and nutrition. Proc Nutr Soc. 2007;66:423–434. doi: 10.1017/S0029665107005691. [DOI] [PubMed] [Google Scholar]
- 147.Muaku SM, Thissen JP, Gerard G, Ketelslegers JM, Maiter D. Postnatal catch-up growth induced by growth hormone and insulin-like growth factor-I in rats with intrauterine growth retardation caused by maternal protein malnutrition. Pediatr Res. 1997;42:370–377. doi: 10.1203/00006450-199709000-00019. [DOI] [PubMed] [Google Scholar]
- 148.Maltin CA, Steven J, Warkup CC. Assay for Duroc Muscle fiber type. 1998. International application published under the patent cooperation treaty (PCT) WO 98/15837. [Google Scholar]