Abstract
Removing the function of a specific gene from a developing organ, by making a ‘knockout’ mouse, is a powerful method for analyzing the molecular pathways that control organogenesis. The technique is expensive, though, in terms of time and money, and complex strategies for producing conditional knockouts are needed for genes that are essential for early development of the embryo, for which an unconditional knockout would be lethal before the organ of interest begins to form. Small interfering RNAs (siRNAs) offer a method of knocking down the expression of specific genes with no need for genomic manipulation. Almost as soon as they had been discovered, siRNAs began to be used to explore the molecular biology of mammalian cells in conventional, two-dimensional culture. They have now also been applied successfully, by several groups, to knock down specific genes in various organ rudiments developing in organ culture. This article reviews the basic technique of siRNA-mediated gene knockdown and how it is being applied to organ culture. It also reviews some of the current problems and challenges in the field, and the ways in which these problems are likely to be overcome.
Key words: siRNA, RNAi, organ culture, organogenesis, organ development, 3D culture
Introduction
One of the standard methods of investigating the functions of a gene or protein in development is to remove it from a developing system and to see what happens. This general technique has proved to be one of the most powerful in modern biology, and is largely responsible for our current understanding of the molecular basis of organogenesis.
Removal of gene function is normally achieved by mutation, nowadays usually achieved by artificial gene targeting. In mice, now the most frequently used organism in developmental biology research,1 the most common method of artificial gene targeting is by homologous recombination in ES cells.2–6 When done properly and after breeding of the engineered mice, this technique succeeds in completely removing the gene from the organism. In some contexts this is an advantage—it helps to produce, for example, unambiguous results—but in other contexts it is a decided disadvantage. Organogenesis begins, by definition, right at the end of the embryonic period of development (the presence of growing organs defines the developing animal as a fetus). Deletion of any gene that is required for embryonic development can mean that the foetal stage is never reached and the role of that gene in organ development cannot be addressed in a simple knockout animal.
The problem of early deleterious effects of a knockout on embryonic development can be solved by making conditional knockout animals. The dominant method for this is the cre-lox system.7 In this technique, the gene of interest is flanked with lox-P sites in a manner that does not interfere with the function of that gene. Separately (and usually in a different colony of animals, that can later be crossed with those carrying the lox-P-flanked gene), the open reading frame of a ‘driver’ gene is replaced by cre recombinase. As the animal carrying this develops, cre recombinase will be expressed in any cell in which the driver gene is normally expressed. When both of these systems are present in the same animal, the cre recombinase causes recombination of the lox-P sites and causes deletion of the gene of interest. By choosing a driver gene that is expressed only in the organ of interest at the time of interest, an experimenter can arrange for the targeted gene to be deleted only where the driver gene is activated, and therefore avoid the problem of embryonic lethality.
Conditional knockouts have proved very useful in studies of organogenesis8–12 but even this powerful technique has its problems. Fundamentally, as the real complexity of the genome due to alternative splicing events is beginning to be realized thanks to the different genome and transcriptome sequencing programs, identifying and generating the mutation most likely to result in complete deletion of all functional isoforms becomes more problematic. This issue affects conventional and conditional knockouts alike. Technically, there is the problem that it can be used only when a suitable driver gene can be identified; at later stages of organogenesis many tissue-specific genes tend to be activated but at earlier stages there are few genes that are expressed in one organ and that are not also expressed elsewhere in the body and earlier in embryogenesis. Alternatively, tissue-specific enhancers can sometimes be identified in the regulatory sequences of driver genes, which can be used in combination with a minimal promoter to express cre as an ectopic transgenic construct. However, taking enhancer elements outside their normal context can result in unexpected results and big differences between individual lines, and too often this approach results in a ‘hit-or-miss’ project. Practically, there is also the problem of the expense, in terms of both time and money, of creating two strains of engineered mice and of breeding them together.
Non-genetic methods for removing the function of specific genes are also available, at least for some genes and stages of development. Many organ rudiments will develop in organ culture, at least through their early stages, and culture techniques have been used with great effect to study kidneys, salivary glands, lungs, prostates etc. Organ culture renders the rudiment accessible to drugs, antibodies and enzymes and these can be used to inhibit the actions of gene products at any time of an experimenter's choosing. Their use is, however, limited to a fairly narrow range of targets. Antibodies and enzymes can target only extracellular molecules. Many drugs can act on intracellular targets but they suffer from the problems of relative, rather than absolute, specificities and drugs are available to inhibit only a tiny subset of the more than 30,000 genes in the genome.
What is urgently needed is a method of inhibiting the expression of specific genes that can be used in organ culture and that can be applied to a very large range of possible targets. The properties of small interfering RNAs (siRNAs) seem, at least at first sight, to meet this requirement and they have already been used successfully in some systems. They are not yet, however, living up to their full apparent promise and this review is intended to provide a balanced evaluation of their current potential and an overview of the problems still to be addressed.
siRNAs and RNA Interference in Conventional (Monolayer) Culture
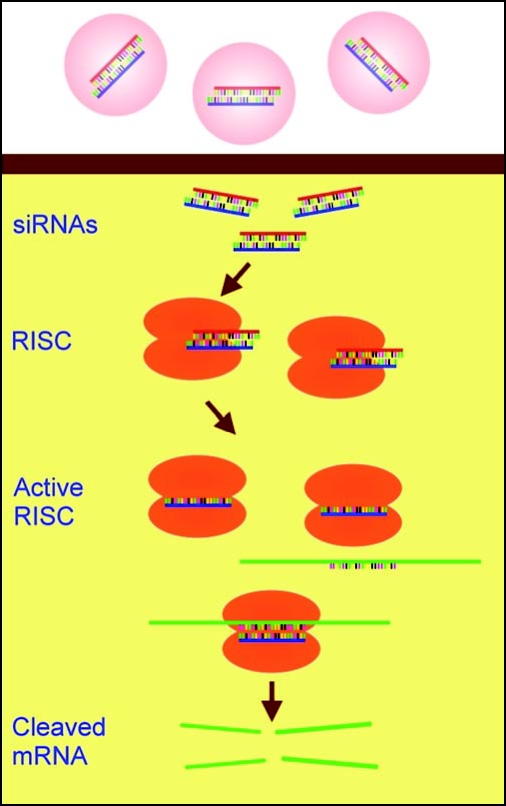
RNA interference is a system for the post-transcriptional repression of gene expression and it has been reviewed extensively elsewhere.13 Briefly, siRNAs consist of a duplex of ≈22-nt RNAs, the 5′ 20 bases of which are complementary so that the duplex formed has two projecting nucleotides at each 3′ end.14,15 If introduced into the cytoplasm of a cell, a siRNA is bound by an RNA-induced silencing complex (RISC).16 Unwinding of the siRNA duplex when it binds to the RISC creates an active form of the RISC complex, carrying one strand of the siRNA, which can bind to any mRNA with a sequence complementary to that strand. Once bound, the RISC complex cleaves the mRNA (Fig. 1). The reaction does not destroy the original siRNA strand bound in the RISC, so the complex can go on to destroy other complementary mRNAs. In this way, low concentrations of siRNA can be effective in silencing large numbers of mRNA molecules, at least in principle. Also, with careful selection of sequences, siRNA can be used to knock down only mRNAs encoding specific splice forms of a gene.
Figure 1.
The basic mechanism of siRNA-mediated RNA interference. siRNA duplexes complexed with a transfection vehicle (pink) are taken across the cell membrane by the vehicle. There, they are bound by the RISC complexes and become single-stranded and active. The siRNA strand in an active RISC complex binds to complementary bases of its target mRNA and the RISC complex then cleaves that mRNA.
From an experimenter's point of view, siRNAs can be synthesized chemically, can be made by RNase III cleavage of longer double-stranded RNAs made from plasmids, or from a palindromic sequence downstream of a polymerase III promoter in a plasmid or virus that is transfected into the cell (the palindrome results in the formation of a double-stranded RNA hairpin loop that is cut by the cell into siRNA). The choice of method is dictated largely by considerations of economics (chemical synthesis is convenient but expensive, yet it might in the end be advantageous to compare siRNAs from different companies to get optimal knockdown efficiencies), accessibility (viruses may diffuse less well than small siRNAs) and the cell type involved (some are difficult to transfect). It also depends on the desired duration of gene silencing, transient transfection with siRNA usually providing a few days' silencing but permanent integration of a plasmid or virus providing permanent knockdown. Inducible systems for stably transfected siRNA expression constructs have been described that are regulated by tetracycline17,18 using modified RNA polymerase III promoters. More versatile control of knockdown is to be expected from the observation that custom siRNA target sequences can be placed in the backbone of an endogenous micro RNA (miRNA) which can be expressed from any RNA polymerase II promoter.19,20
Of these common methods of transfection, only the viral method allows efficient uptake in mammalian cells without the need for transfection reagents: mammalian cells do not, in general, show the ability of some invertebrates, such as Caenorhabditis elegans, to take up dsRNA without transfection reagents.21 Naked siRNAs (whether synthesized chemically or made from dsRNAs) and siRNA-encoding plasmids have to be complexed with transfection reagents before they will cross cell membranes. A variety of such reagents is available from commercial suppliers. The efficiency of each seems to vary between cell types, and must be determined empirically. In general, siRNA is made into a complex with its transfection reagent and is then applied in a suspension on cells in subconfluent two-dimensional culture, left for a few hours, then washed away. The inertness of the transfection reagents used should not be taken for granted, however; at the very least, some can modulate the stabilities of RNAs to which they are bound and apparently target them to particles in the cytoplasm where they may not fulfil their intended functions.22
Supported by adequate controls,23 the use of siRNA in conventional two-dimensional cell culture has proved a very valuable technique in a range of cell biological and physiological investigations.24–28 It is therefore natural to think of extending it to three-dimensional organ culture.
Successful Application of siRNA to Organ Culture: The Promise
In principle, the combination of exogenously-applied siRNAs and organs growing in culture should be a powerful method for knocking down the expression of any gene of interest at any time of an experimenter's choosing. The first published practical demonstration of this was produced by Takayoshi Sakai and colleagues, who used oligofectamine to transfect cultured salivary glands from E12.5 mouse embryos with 500 nM synthetic siRNA duplexes.29 Control duplexes had no significant effect on either morphogenesis or on gene expression, but siRNAs targeting fibronectin caused decrease in fibronectin expression, as judged by immunostaining, and a dramatic, dose-dependent and reversible inhibition of epithelial branching. A similar phenotype was also produced by function-blocking anti-fibronectin antibodies. In principle, the entire experiment could have been done using the antibodies, but what made the study so powerful from the point of view of introducing siRNA as a technique for organogenesis was the demonstration that it would replicate the effect of established techniques, such as antibody inhibition, so exactly. In the same paper (or at least, in the supplementary information provided alongside it), the authors used the same siRNAs to knock down the expression of fibronectin in both cultured embryonic kidneys and cultured embryonic lungs, and obtained a very similar general effect.
In the next application of siRNA technology to cultured kidneys, another group used an siRNA technique similar to that described above to target a protein that was intracellular and therefore unreachable by antibodies.30 The protein, WT1, had already been identified as being a tumor suppressor; the mutation of which was implicated in the development of the childhood nephroblastoma, Wilms' tumor.31,32 It was known to be expressed in a complex pattern that changed as kidney development proceeded33 and a simple transgenic deletion of the gene in mice turned out to result in renal agenesis rather than development of the tumor.34 A technique for removing Wt1 function at different stages of renal development was therefore required to find the stage at which Wt1 loss might be connected with tumorigenesis rather than causing renal agenesis.
Synthetic siRNA duplexes, again complexed with oligofectamine, were first applied to cultured E9.5 urogenital ridges, which are the parts of the embryos in which kidneys would form approximately one day later. The siRNA prevented the expression of WT1 protein in most cells, as judged by immunofluorescence, and produced a renal agenesis phenocopy of the Wt1 knockout mouse. Application of the siRNAs at later stages of development identified a stage at which knockdown of WT1 resulted not in renal agenesis, but rather in too much proliferation and a failure of differentiation, an effect with potential relevance to the development of a tumor. Immunostaining for WT1 protein showed that the knockdown was not absolute; low levels of WT1 (typical of early stages of renal development) were present in many cells, and a very few showed high levels of expression.
In the same paper, two other proteins were targeted, the transcription factor Pax2 and the signalling molecule Wnt4. Both showed knockdown, at least in mesenchymal tissues, and study of the expression of Wnt4, WT1 and Pax2 when each was knocked down was used to establish a likely hierarchy of their interactions.
The range of organs for which siRNA has been successfully used has steadily expanded since these early papers (Table 1). Some of these experiments have used intact organ rudiments, but others have used rudiments reconstructed from separated cells. The reason for this will be discussed in section 5 below.
Table 1.
Examples of siRNA in organ culture
| Organ | Target of siRNA | Reference |
| Atrioventricular canal | Endoglin | 35 |
| Lung | Fibronectin | 29 |
| Lung | Hoxb5 | 36 |
| Lung | Fak | 37 |
| Kidney | Fibronectin | 29 |
| Kidney | Wt1, Wnt4, Pax2 | 30 |
| Kidney | GDNF | 38 |
| Ovary | GDF-9 | 39 |
| Palatal shelves | Smad2 | 40 |
| Re-constructed Ovary | Wee-1 | 41 |
| Salivary gland | Fibronectin | 29 |
| Re-constructed Skin | VEGF | 42 |
| Re-constructed Tooth | Msx1, Dlx2 | 43 |
| Re-constructed Thymus | various | 44 |
| Urogenital Ridge | Alks and Smads | 45 |
| Wolffian Duct | Activin | 46 |
siRNA in Organ Culture: The Pitfalls
Although the use of siRNA in organ culture has provided very valuable information in a number of systems, as recounted above and summarized in Table 1, it does not always work so well and several researchers involved in this field—and we include ourselves in this statement—have had to endure much frustration as well as occasional success. If this were not so, there would have been far more siRNA organ culture papers published by now. The pitfalls of the technique fall into two categories—problems inherent in any siRNA experiment and problems caused specifically by organ culture.
Problems inherent in any siRNA approach include the selection of effective and specific siRNA sequences, selection of a transfection method, monitoring the efficiency of knockdown and establishing the half-life of the protein. Much has been written elsewhere about design and selection of effective siRNA sequences and siRNAs for a large fraction of the genome can now be bought pre-tested off the shelf. In general, those that work in one cell will work in another, although there is always a small risk that one may be destroyed by cell-specific RNA editing systems.47 Once a siRNA has been designed, it has to be delivered across the plasma membrane into the cell. Some mammalian cells will take up naked siRNA but inefficiently, so that uneconomical amounts are needed. For this reason, siRNA is normally complexed to lipophilic carriers, which are available from a number of rival companies, each of which seems to claim that its own products are best. The ability of those companies to each make substantiated claims underlines the fact that different approaches suit different cells. Viruses can also be used to transfect siRNA (encoded as a hairpin loop or in miRNA backbone) into cells, although this is less likely to work in organ culture for reasons discussed below. Good advice on the choice of transfection method is available in the literature.48,49 It remains a feature of almost all siRNA experiments, though, that not all cells are transfected. A “70% knockdown,” for example, tends to reflect not all of the cells suffering a 70% reduction in expression but rather deep knockdown in about 70% of cells and 30% of cells being unaffected. This may make a real difference to the interpretation of an experiment.
Once an effective siRNA has been transfected into cells, it should lead to the destruction of its target mRNA (and no other). This will block protein synthesis, but the disappearance of the protein itself will depend on its half-life. The half-life of a protein is determined partially by the intrinsic properties of the protein but generally much more by the environment in which it finds itself, particularly with respect to proteases and ubiquitination mechanisms. Long-lived proteins can make siRNA experiments difficult to perform and to interpret, especially if the timing of the knockdown is important. Of course, conditional knockout models will suffer of this problem as well.
Protein half-life is not the only potential problem with knockdown. The amount of a specific protein that is expressed by a cell is often under the control of feedback, and this is especially true of proteins whose concentration is critical to cellular and developmental events. These are the very types of proteins in which investigators tend to be interested. Feedback loops can be driven directly by a protein's concentration or indirectly by consequences of the protein's function and they can operate at the levels of transcription, translation, export and degradation.50–54 Attempts to knock down a protein by siRNA may therefore be thwarted, at least partially, by the cell ‘fighting back’ and using post-translational methods to maintain a constant level of the protein.
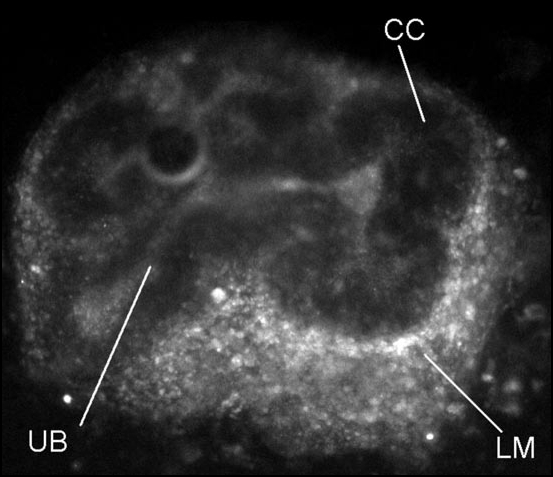
The problems listed above can affect any siRNA experiment. The extra problems associated particularly with organ culture arise mainly from the three-dimensional nature of the tissue involved. In simple, two-dimensional culture of cells on the bottom of a flask, siRNA complexes or siRNA-encoding viruses can gain direct access to the surface of the cells. In three dimensional tissues or in artificial collagen gels, most cells are separated from the source of complexed siRNAs by diffusion pathways packed with extracellular matrix and sometimes a fully-formed basement membrane. These elements can act as barriers to diffusion or they can act as ‘traps’ that bind and concentrate siRNAs due to multiple low-affinity charge interactions. The theoretical possibility of organ rudiments containing these barriers and traps can be tested empirically by using siRNA duplexes that are labelled with fluorophores such as Cy3. Such experiments typically show the distribution to be markedly uneven, even after days of culture (Fig. 2). The patterns can vary from organ to organ and also between developmental stages within one organ. In early salivary glands, for example, Cy3-labelled siRNA tends to be associated more with epithelial tissues than with mesenchymal29 whereas in early kidneys it penetrates mesenchyme better than it does epithelia.30 A little later in kidney development, at the stage of mesenchymal condensation,55 an intra-mesenchymal barrier seems to form that interferes with penetration of siRNA into the condensate (Fig 2).
Figure 2.
Uneven penetration of labelled siRNAs into a cultured kidney rudiment. An E12.5 kidney rudiment was cultured on a polycarbonate filter supported on a Trowell-type grid, and fluorescently-labelled siRNA, complexed to oligofectamine, was added to the medium. The siRNA has penetrated the loose mesenchyme (‘LM’) well and some seems to have reached at least the lumen of the ureteric bud (‘UB’), but it has substantially failed to enter the cap condensates (‘CC’). This is disappointing because the cap condensates give rise to the nephrons of the kidney and studying the genetic mechanisms of nephron development is a priority in renal research.
These diffusion barriers have created serious problems to our own work in kidney and we have had to abandon, at least temporarily, attempts to knock down genes at late stages of renal development precisely because we see effective knockdown only in the most peripheral cells. Even at younger stages, we have found some genes easier to knock down than others—those genes that are not already switched on seem to be, in general, easier to knock down, presumably because the problem of protein half life is greatly reduced when there is none of that protein present at time zero.
Approaches to More Reliable siRNA Techniques for Organ Culture
There are two, essentially opposite though not mutually exclusive, approaches to making siRNA-based gene knockdown work better in organ culture. The first is to adapt existing siRNA methods to use in organ culture while the second is to adapt existing organ culture methods for better susceptibility to siRNA treatment.
Many adaptations to the use of siRNA-mediated knockdown have already been described for two-dimensional cultures, but combinations of all these approaches may be needed to fully utilize the potential of siRNA in organ cultures. For transient knockdown experiments, the use of chemically synthesized siRNA molecules will usually be sufficient. However, when developmental processes take longer than transient knockdown will last, stable integration of siRNA expression constructs will be essential. In contrast to normal cell culture, simple selection for antibiotic resistance is not an option. Viral systems, especially Lentiviruses that infect both dividing and non-dividing cells with high efficiency, will be necessary. Unfortunately, at least in kidney organ cultures, Lentiviruses are not capable of deeply penetrating the tissue (Berry and Hohenstein, unpublished data). If this problem can be solved (see below), inducible systems will be needed to express the stably integrated siRNA cassette at the right time and place. The observation that RNA polymerase II-driven miRNA based vectors show improved knockdown at low copy numbers20 on top of the possibility of using a variety of regulatable systems as mentioned before would make this a likely system of choice.
Adaptation of organ culture techniques for siRNA generally involves disaggregating the organ rudiment so that cells are exposed to siRNA and then re-aggregating it so that it is still capable of organotypic development.
An example of this approach is that developed by Park and colleagues, who wished to use siRNA for analysis of the development of ovarian follicles.41 To render ovarian cells susceptible to siRNA, these authors dissected ovaries and used enzymes to dissociate them into cell suspensions. They then used FuGene6 (Roche) to transfect either oocytes or somatic cells with siRNA, achieving transfection efficiencies of 40–50% and 50–60% for each cell type respectively and achieving knockdown of Wee1 expression to 30–40% of wild type levels. The transfected cells (and untransfected controls) were then aggregated with phytohaemagglutinin and coated with sodium alginate, which was then converted to a calcium alginate gel that encapsulated the reconstructed ‘ovaries’. These reconstructed ‘ovaries’ were then cultured for several days; even controls grew more slowly than unmolested organs, but the ones with Wee1 knockdown showed marked and specific developmental effects.
Song and colleagues used a similar aggregation technique to knock down gene expression in tooth development.43 They removed molar tooth rudiments from E13.5 mouse embryos, separated their epithelial and mesenchymal components and reduced the mesenchyme to a suspension of single cells with the aid of low calcium-medium. They then transfected them with Lentiviruses that encoded hairpin loops that would be processed by cells to yield siRNAs that targeted Msx1, Dlx2 and Barx1. They then centrifuged the cells to make a firm pellet and recombined this pellet with the epithelial component of tooth rudiments (either as a complete epithelium or as suspended cells, which sorted spontaneously from the mesenchyme and re-formed its epithelial tissue). The recombination was cultured overnight in vitro and then in subrenal culture in vivo and controls developed into recognizable molars. Knockdown of Msx1 and Dlx2 produced a phenocopy of their transgenic knockout phenotypes (arrest at tooth bud stage in the case of Msx1, normal development in the case of Dlx2). Knockdown of Barx1, for which no tooth data exist from conventional knockout mice, revealed a novel degenerative phenotype. The detailed experiments in this report also revealed an interesting restriction to the re-aggregation approach: in the case of tooth rudiments, at least, it seems to be important that the time that mesenchymes spend separated from the epithelium must be minimized, because their ability to generate teeth in re-aggregated rudiments decreases rapidly with the time they spend alone.
Reaggregation techniques connect strongly with the cutting edge of tissue engineering, and new methods for producing organotypic tissues from dispersed cells are being reported at an increasing rate in the tissue engineering literature. Depending on what question is being asked, many of these systems seem to offer a real potential for rendering a developing organ system accessible to siRNA. Examples include brain,56 cartilage,57 liver,58 tooth,59 thymus60 and vascular tissues.61
Conclusions
This is a fast-moving field, so any conclusions written at this stage must be regarded as both tentative and temporary. The use of siRNA has established itself in ordinary two-dimensional cell culture and it seems to us that with over a dozen research papers already published, from a variety of unconnected laboratories, the use of siRNA is also becoming established in organ culture. Its use here is not straightforward, however. The problems of ensuring adequate penetration are particularly acute and it seems that recent developments in disaggregation, transfection and reconstitution of organ rudiments offer a promising way forward and the combination of this with improved transfection methods offers most promise of all. The speed and flexibility of siRNA-based approaches offer substantial advantages over the production of transgenic mice, particularly for preliminary experiments to gain enough information to know that a full transgenic (which has 100% knockout rather than a difficult-to-interpret knock-down) is worth making. In cases where it is difficult to generate a true loss-of-function knock-out model that inactivates all isoforms of a protein, siRNA-mediated knockdown may instead provide the means of testing the phenotypes of loss of specific isoforms. Other advantages of knockdown over knockout, as the possibility of generating allelic series of models to study dosage effects of genes,62 improving knockdown efficiencies by linking several target molecules in a single miRNA-based vector or even the possibility of linking target sequences against different proteins in the same construct for combined knockdown63 make knockdown experiments in organ cultures in many cases a complementation to knockout studies rather than a replacement. For this reason, we expect that the future will bring more applications in which siRNAs are used for high-throughout screens for genes that control the development of organs, in much the same way that they are already being used to identify genes that control the development of invertebrate embryos.64
Footnotes
Previously published online as an Organogenesis E-publication: www.landesbioscience.com/journals/organogenesis/article/6642
References
- 1.Davies JA. Developmental biologists choice of subjects approximates to a power law, with no evidence for the existence of a special group of model organisms. BMC Dev Biol. 2007;7:40. doi: 10.1186/1471-213X-7-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Doetschman T, Gregg RG, Maeda N, Hooper ML, Melton DW, Thompson S, et al. Targetted correction of a mutant HPRT gene in mouse embryonic stem cells. Nature. 1987;330:576–578. doi: 10.1038/330576a0. [DOI] [PubMed] [Google Scholar]
- 3.Gossler A, Doetschman T, Korn R, Serfling E, Kemler R. Transgenesis by means of blastocyst-derived embryonic stem cell lines. Proc Natl Acad Sci USA. 1986;83:9065–9069. doi: 10.1073/pnas.83.23.9065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Stanton BR, Reid SW, Parada LF. Germ line transmission of an inactive N-myc allele generated by homologous recombination in mouse embryonic stem cells. Mol Cell Biol. 1990;10:6755–6758. doi: 10.1128/mcb.10.12.6755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Robertson EJ. Using embryonic stem cells to introduce mutations into the mouse germ line. Biol Reprod. 1991;44:238–245. doi: 10.1095/biolreprod44.2.238. [DOI] [PubMed] [Google Scholar]
- 6.Houdebine LM. Transgenic animal models in biomedical research. Methods Mol Biol. 2007;360:163–202. doi: 10.1385/1-59745-165-7:163. [DOI] [PubMed] [Google Scholar]
- 7.Sauer B. Inducible gene targeting in mice using the Cre/lox system. Methods. 1998;14:381–392. doi: 10.1006/meth.1998.0593. [DOI] [PubMed] [Google Scholar]
- 8.Dworniczak B, Skryabin B, Tchinda J, Heuck S, Seesing FJ, Metzger D, et al. Inducible Cre/loxP recombination in the mouse proximal tubule. Nephron Exp Nephrol. 2007;106:e11–e20. doi: 10.1159/000100554. [DOI] [PubMed] [Google Scholar]
- 9.Wells JM, Esni F, Boivin GP, Aronow BJ, Stuart W, Combs C, et al. Wnt/beta-catenin signaling is required for development of the exocrine pancreas. BMC Dev Biol. 2007;7:4. doi: 10.1186/1471-213X-7-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Loomes KM, Russo P, Ryan M, Nelson A, Underkoffler L, Glover C, et al. Bile duct proliferation in liver-specific Jag1 conditional knockout mice: effects of gene dosage. Hepatology 2007. 45:323–330. doi: 10.1002/hep.21460. [DOI] [PubMed] [Google Scholar]
- 11.Ma X, Ziel-van der Made AC, Autar B, van der Korput HA, Vermeij M, van Duijn P, et al. Targeted biallelic inactivation of Pten in the mouse prostate leads to prostate cancer accompanied by increased epithelial cell proliferation but not by reduced apoptosis. Cancer Res. 2005;65:5730–5739. doi: 10.1158/0008-5472.CAN-04-4519. [DOI] [PubMed] [Google Scholar]
- 12.Rojek A, Fuchtbauer EM, Kwon TH, Frokiaer J, Nielsen S. Severe urinary concentrating defect in renal collecting duct-selective AQP2 conditional-knockout mice. Proc Natl Acad Sci USA. 2006;103:6037–6042. doi: 10.1073/pnas.0511324103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hannon GJ. RNA interference. Nature. 2002;418:244–251. doi: 10.1038/418244a. [DOI] [PubMed] [Google Scholar]
- 14.Elbashir SM, Lendeckel W, Tuschl T. RNA interference is mediated by 21- and 22-nucleotide RNAs. Genes Dev. 2001;15:188–200. doi: 10.1101/gad.862301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zamore PD, Tuschl T, Sharp PA, Bartel DP. RNAi: double-stranded RNA directs the ATP-dependent cleavage of mRNA at 21 to 23 nucleotide intervals. Cell. 2000;101:25–33. doi: 10.1016/S0092-8674(00)80620-0. [DOI] [PubMed] [Google Scholar]
- 16.Paroo Z, Liu Q, Wang X. Biochemical mechanisms of the RNA-induced silencing complex. Cell Res. 2007;17:187–194. doi: 10.1038/sj.cr.7310148. [DOI] [PubMed] [Google Scholar]
- 17.van de WM, Oving I, Muncan V, Pon Fong MT, Brantjes H, van Leenen D, et al. Specific inhibition of gene expression using a stably integrated, inducible small-interfering-RNA vector. EMBO Rep. 2003;4:609–615. doi: 10.1038/sj.embor.embor865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ventura A, Meissner A, Dillon CP, McManus M, Sharp PA, van Parijs L, et al. Cre-lox-regulated conditional RNA interference from transgenes. Proc Natl Acad Sci USA. 2004;101:10380–10385. doi: 10.1073/pnas.0403954101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zeng Y, Wagner EJ, Cullen BR. Both natural and designed micro RNAs can inhibit the expression of cognate mRNAs when expressed in human cells. Mol Cell. 2002;9:1327–1333. doi: 10.1016/s1097-2765(02)00541-5. [DOI] [PubMed] [Google Scholar]
- 20.Stegmeier F, Hu G, Rickles RJ, Hannon GJ, Elledge SJ. A lentiviral microRNA-based system for single-copy polymerase II-regulated RNA interference in mammalian cells. Proc Natl Acad Sci USA. 2005;102:13212–13217. doi: 10.1073/pnas.0506306102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Timmons L, Fire A. Specific interference by ingested dsRNA. Nature. 1998;395:854. doi: 10.1038/27579. [DOI] [PubMed] [Google Scholar]
- 22.Barreau C, Dutertre S, Paillard L, Osborne HB. Liposome-mediated RNA transfection should be used with caution. RNA. 2006;12:1790–1793. doi: 10.1261/rna.191706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Whither RNAi? Nat Cell Biol. 2003;5:489–490. doi: 10.1038/ncb0603-490. [DOI] [PubMed] [Google Scholar]
- 24.Elbashir SM, Harborth J, Lendeckel W, Yalcin A, Weber K, Tuschl T. Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature. 2001;411:494–498. doi: 10.1038/35078107. [DOI] [PubMed] [Google Scholar]
- 25.Harborth J, Elbashir SM, Bechert K, Tuschl T, Weber K. Identification of essential genes in cultured mammalian cells using small interfering RNAs. J Cell Sci. 2001;114:4557–4565. doi: 10.1242/jcs.114.24.4557. [DOI] [PubMed] [Google Scholar]
- 26.Marker PC. Datta MW, Hernandez AM, Schlicht MJ, Kahler AJ, DeGueme AM, Dhir R, Shah RB, Farach-Carson C, Barrett A, Datta S. Urol Onco. Vol. 25. Atlanta, GA.: Department of Pathology, Emory University; 2007. Perlecan a candidate gene for the CAPB locus, regulates prostate cancer cell growth via the Sonic Hedgehog pathway; p. 280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.De F, V, Castellone MD, De Vita G, Cirafici AM, Hershman JM, Guerrero C, et al. RET/papillary thyroid carcinoma oncogenic signaling through the Rap1 small GTPase. Cancer Res. 2007;67:381–390. doi: 10.1158/0008-5472.CAN-06-0981. [DOI] [PubMed] [Google Scholar]
- 28.Shaw RM, Fay AJ, Puthenveedu MA, von Zastrow M, Jan YN, Jan LY. Microtubule plusend-tracking proteins target gap junctions directly from the cell interior to adherens junctions. Cell. 2007;128:547–560. doi: 10.1016/j.cell.2006.12.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sakai T, Larsen M, Yamada KM. Fibronectin requirement in branching morphogenesis. Nature. 2003;423:876–881. doi: 10.1038/nature01712. [DOI] [PubMed] [Google Scholar]
- 30.Davies JA, Ladomery M, Hohenstein P, Michael L, Shafe A, Spraggon L, et al. Development of an siRNA-based method for repressing specific genes in renal organ culture and its use to show that the Wt1 tumor suppressor is required for nephron differentiation. Hum Mol Genet. 2004;13:235–246. doi: 10.1093/hmg/ddh015. [DOI] [PubMed] [Google Scholar]
- 31.McLorie GA. Wilms' tumor (nephroblastoma) Curr Opin Urol. 2001;11:567–570. doi: 10.1097/00042307-200111000-00002. [DOI] [PubMed] [Google Scholar]
- 32.Hohenstein P, Hastie ND. The many facets of the Wilms' tumor gene, WT1. Hum Mol Genet. 2006;15:R196–R201. doi: 10.1093/hmg/ddl196. Spec No 2. [DOI] [PubMed] [Google Scholar]
- 33.Armstrong JF, Pritchard-Jones K, Bickmore WA, Hastie ND, Bard JB. The expression of the Wilms' tumor gene, WT1, in the developing mammalian embryo. Mech Dev. 1993;40:85–97. doi: 10.1016/0925-4773(93)90090-k. [DOI] [PubMed] [Google Scholar]
- 34.Kreidberg JA, Sariola H, Loring JM, Maeda M, Pelletier J, Housman D, et al. WT-1 is required for early kidney development. Cell. 1993;74:679–691. doi: 10.1016/0092-8674(93)90515-r. [DOI] [PubMed] [Google Scholar]
- 35.Mercado-Pimentel ME, Hubbard AD, Runyan RB. Endoglin and Alk5 regulate epithelialmesenchymal transformation during cardiac valve formation. Dev Biol. 2007;304:420–432. doi: 10.1016/j.ydbio.2006.12.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Volpe MV, Ramadurai SM, Pham LD, Nielsen HC. Hoxb-5 down regulation alters Tenascin-C, FGF10 and Hoxb gene expression patterns in pseudoglandular period fetal mouse lung. Front Biosci. 2007;12:860–873. doi: 10.2741/2108. [DOI] [PubMed] [Google Scholar]
- 37.Gill SE, Pape MC, Leco KJ. Tissue inhibitor of metalloproteinases 3 regulates extracellular matrix—cell signaling during bronchiole branching morphogenesis. Dev Biol. 2006;298:540–554. doi: 10.1016/j.ydbio.2006.07.004. [DOI] [PubMed] [Google Scholar]
- 38.Michael L, Sweeney DE, Davies JA. The lectin Dolichos biflorus agglutinin is a sensitive indicator of branching morphogenetic activity in the developing mouse metanephric collecting duct system. J Anat. 2007;210:89–97. doi: 10.1111/j.1469-7580.2006.00670.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang C, Roy SK. Expression of growth differentiation factor 9 in the oocytes is essential for the development of primordial follicles in the hamster ovary. Endocrinology. 2006;147:1725–1734. doi: 10.1210/en.2005-1208. [DOI] [PubMed] [Google Scholar]
- 40.Shiomi N, Cui XM, Yamamoto T, Saito T, Shuler CF. Inhibition of SMAD2 expression prevents murine palatal fusion. Dev Dyn. 2006;235:1785–1793. doi: 10.1002/dvdy.20819. [DOI] [PubMed] [Google Scholar]
- 41.Park CE, Lee D, Kim KH, Lee KA. Establishment of ovarian reconstruction system in culture for functional genomic analysis. J Biosci Bioeng. 2006;102:396–401. doi: 10.1263/jbb.102.396. [DOI] [PubMed] [Google Scholar]
- 42.Mildner M, Ballaun C, Stichenwirth M, Bauer R, Gmeiner R, Buchberger M, et al. Gene silencing in a human organotypic skin model. Biochem Biophys Res Commun. 2006;348:76–82. doi: 10.1016/j.bbrc.2006.07.035. [DOI] [PubMed] [Google Scholar]
- 43.Song Y, Zhang Z, Yu X, Yan M, Zhang X, Gu S, et al. Application of lentivirus-mediated RNAi in studying gene function in mammalian tooth development. Dev Dyn. 2006;235:1334–1344. doi: 10.1002/dvdy.20706. [DOI] [PubMed] [Google Scholar]
- 44.Hernandez-Hoyos G, Alberola-Ila J. Analysis of T-cell development by using short interfering RNA to knock down protein expression. Methods Enzymol. 2005;392:199–217. doi: 10.1016/S0076-6879(04)92012-5. [DOI] [PubMed] [Google Scholar]
- 45.Zhan Y, Fujino A, MacLaughlin DT, Manganaro TF, Szotek PP, Arango NA, et al. Mullerian inhibiting substance regulates its receptor/SMAD signaling and causes mesenchymal transition of the coelomic epithelial cells early in Mullerian duct regression. Development. 2006;133:2359–2369. doi: 10.1242/dev.02383. [DOI] [PubMed] [Google Scholar]
- 46.Maeshima A, Vaughn DA, Choi Y, Nigam SK. Activin A is an endogenous inhibitor of ureteric bud outgrowth from the Wolffian duct. Dev Biol. 2006;295:473–485. doi: 10.1016/j.ydbio.2006.03.011. [DOI] [PubMed] [Google Scholar]
- 47.Nishikura K. Editor meets silencer: crosstalk between RNA editing and RNA interference. Nat Rev Mol Cell Biol. 2006;7:919–931. doi: 10.1038/nrm2061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Li CX, Parker A, Menocal E, Xiang S, Borodyansky L, Fruehauf JH. Delivery of RNA interference. Cell Cycle. 2006;5:2103–2109. doi: 10.4161/cc.5.18.3192. [DOI] [PubMed] [Google Scholar]
- 49.Gilmore IR, Fox SP, Hollins AJ, Akhtar S. Delivery strategies for siRNA-mediated gene silencing. Curr Drug Deliv. 2006;3:147–155. doi: 10.2174/156720106776359159. [DOI] [PubMed] [Google Scholar]
- 50.Hasler U, Nielsen S, Feraille E, Martin PY. Posttranscriptional control of aquaporin-2 abundance by vasopressin in renal collecting duct principal cells. Am J Physiol Renal Physiol. 2006;290:F177–F187. doi: 10.1152/ajprenal.00056.2005. [DOI] [PubMed] [Google Scholar]
- 51.Schmidt TJ, Meyer AS. Autoregulation of corticosteroid receptors. How, when, where, and why? Receptor. 1994;4:229–257. [PubMed] [Google Scholar]
- 52.Ladurner AG. Rheostat control of gene expression by metabolites. Mol Cell. 2006;24:1–11. doi: 10.1016/j.molcel.2006.09.002. [DOI] [PubMed] [Google Scholar]
- 53.Ju D, Wang L, Mao X, Xie Y. Homeostatic regulation of the proteasome via an Rpn4-dependent feedback circuit. Biochem Biophys Res Commun. 2004;321:51–57. doi: 10.1016/j.bbrc.2004.06.105. [DOI] [PubMed] [Google Scholar]
- 54.Gulow K, Bienert D, Haas IG. BiP is feed-back regulated by control of protein translation efficiency. J Cell Sci. 2002;115:2443–2452. doi: 10.1242/jcs.115.11.2443. [DOI] [PubMed] [Google Scholar]
- 55.Davies JA, Bard JB. The development of the kidney. Curr Top Dev Biol. 1998;39:245–301. doi: 10.1016/S0070-2153(08)60458-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ajioka I, Nakajima K. Birth-date-dependent segregation of the mouse cerebral cortical neurons in reaggregation cultures. Eur J Neurosci. 2005;22:331–342. doi: 10.1111/j.1460-9568.2005.04214.x. [DOI] [PubMed] [Google Scholar]
- 57.Rosowski M, Falb M, Tschirschmann M, Lauster R. Initiation of mesenchymal condensation in alginate hollow spheres—a useful model for understanding cartilage repair? Artif Organs. 2006;30:775–784. doi: 10.1111/j.1525-1594.2006.00300.x. [DOI] [PubMed] [Google Scholar]
- 58.Zhu XH, Wang CH, Tong YW. Growing tissue-like constructs with Hep3B/HepG2 liver cells on PHBV microspheres of different sizes. J Biomed Mater Res B Appl Biomater. 2006 doi: 10.1002/jbm.b.30698. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 59.Yamamoto H, Kim EJ, Cho SW, Jung HS. Analysis of tooth formation by reaggregated dental mesenchyme from mouse embryo. J Electron Microsc (Tokyo) 2003;52:559–566. doi: 10.1093/jmicro/52.6.559. [DOI] [PubMed] [Google Scholar]
- 60.Ueno T, Liu C, Nitta T, Takahama Y. Development of T-lymphocytes in mouse fetal thymus organ culture. Methods Mol Biol. 2005;(290):117–133. doi: 10.1385/1-59259-838-2:117. [DOI] [PubMed] [Google Scholar]
- 61.Perez-Pomares JM, Mironov V, Guadix JA, Macias D, Markwald RR, Munoz-Chapuli R. In vitro self assembly of proepicardial cell aggregates, an embryonic vasculogenic model for vascular tissue engineering. Anat Rec A Discov Mol Cell Evol Biol. 2006;288:700–713. doi: 10.1002/ar.a.20338. [DOI] [PubMed] [Google Scholar]
- 62.Hemann MT, Fridman JS, Zilfou JT, Hernando E, Paddison PJ, Cordon-Cardo C, et al. An epi-allelic series of p53 hypomorphs created by stable RNAi produces distinct tumor phenotypes in vivo. Nat Genet. 2003;33:396–400. doi: 10.1038/ng1091. [DOI] [PubMed] [Google Scholar]
- 63.Chung KH, Hart CC, Al Bassam S, Avery A, Taylor J, Patel PD, et al. Polycistronic RNA polymerase II expression vectors for RNA interference based on BIC/miR-155. Nucleic Acids Res. 2006;34:e53. doi: 10.1093/nar/gkl143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Fraser AG, Kamath RS, Zipperlen P, Martinez Campos M, Sohrmann M, Ahringer J. Functional genomic analysis of C. elegans chromosome I by systematic RNA interference. Nature. 2000;408:325–330. doi: 10.1038/35042517. [DOI] [PubMed] [Google Scholar]