Abstract
The proteolytic processing of amyloid β precursor protein (APP) has long been studied because of its association with the pathology of Alzheimer's disease (AD). The ectodomain of APP is shed by α- or β-secretase cleavage. The remaining membrane bound stub can then undergo regulated intramembrane proteolysis (RIP) by γ-secretase. This cleavage can release amyloid β (Aβ) from the stub left by β-secretase cleavage but also releases the APP intracellular domain (AICD) after α- or β-secretase cleavage. The physiological functions of this proteolytic processing are not well understood. We compare the proteolytic processing of APP to the ligand-dependent RIP of Notch. In this review, we discuss recent evidence suggesting that TAG1 is a functional ligand for APP. The interaction between TAG1 and APP triggers γ-secretase-dependent release of AICD. TAG1, APP and Fe65 colocalise in the neurogenic ventricular zone and in fetal neural progenitor cells in vitro. Experiments in TAG1, APP and Fe65 null mice as well as TAG1 and APP double-null mice demonstrate that TAG1 induces a γ-secretase- and Fe65-dependent suppression of neurogenesis.
Key words: Amyloid β precursor protein, APP, TAG1, AICD, Fe65, neurogenesis, Alzheimer's disease
Shedding and RIPping of APP
Amyloid β precursor protein (APP) has been the subject of intensive study because of its association with the pathology of Alzheimer's disease (AD). APP is a type I transmembrane glycoprotein cleaved by specific proteases, sheddases, to shed a large, secreted, soluble luminal or extracellular domain (APPs) leaving a membrane-bound stub1 (see Fig. 1). The remaining membrane-bound stub then undergoes regulated intramembrane proteolysis (RIP)2 by an intramembrane cleaving protease (I-CLIP), the γ-secretase complex.3
Figure 1.
A schematic diagram of amyloid β precursor protein (APP) proteolytic processing (not drawn to scale). APP is a large transmembrane molecule (unboxed at top). There is a large extracellular or luminal domain with β- and α-secretase cleavage sites (β and α, respectively) close to the membrane. The γ-secretase sites (γ) occur within the transmembrane region. On proteolytic processing the ectodomain is first shed by either α- or β-secretase mediated cleavage (middle, left and right boxes, respectively). Cleavage by α-secretase (box at middle left) releases APPsα extracellularily or luminally and leaves a C83 (also known as CTFα) stub in the membrane. Cleavage by β-secretase (box at middle right) releases APPsβ extracellularly or luminally and leaves a C99 (also known as CTFβ) stub in the membrane. Regulated intramembrane proteolysis (RIP) by γ-secretase subsequently cleaves the stubs remaining in the membrane (bottom). Cleavage of C83 (box on bottom left) liberates p3 and the APP intracellular domain (AICD). Cleavage of C99 (box on the bottom right) liberates amyloid β peptide (Aβ) and the APP intracellular domain (AICD).
Initial ectodomain shedding is a common feature of the proteolytic processing of many type I and type II transmembrane proteins. In the case of APP, the sheddases responsible for ectodomain secretion are α- or β-secretases. Cleavage by α-secretase, which is mediated by members of the ADAM (a disintegrin and metalloproteinase) family of proteinases, including in particular ADAM 10 and ADAM 17,4–8 results in secretion of APPsα and retention of a membrane-tethered C83 stub, also known as C-terminal fragment α (CTFα). Cleavage by β-amyloid-cleaving enzyme-1 (BACE) or β-secretase, results in secretion of APPsβ and retention of a membrane-tethered C99 stub, also known as C-terminal fragment α (CTFβ).9
After ectodomain shedding of the APPs, the remaining C-terminal fragment still tethered to the membrane can undergo RIP by the γ-secretase complex.2,10 Cleavage by the γ-secretase complex releases the C-terminal APP intracellular domain (AICD, also sometimes known as AID or C-terminal fragment γ, CTFγ) and simultaneously a small N-terminal fragment. Following cleavage by α-secretase the N-terminal fragment is the nonamyloidogenic, p3. However, following cleavage by β-secretase the N-terminal fragment is the potentially amyloidogenic, Aβ. The γ-secretase complex minimally contains a quartet of proteins:11,12 presenilin 1 or 2 (PS1 or PS2), nicastrin, anterior pharynx-defective phenotype-1 (Aph-1) and presenilin enhancer (Pen-2).13–16 Nicastrin appears to act as a receptor ensuring that the luminal or extracellular N-terminal of the sheddase cleaved substrate is of the correct length,17 which means that ectodomain shedding is required for γ-secretase-dependent RIP to proceed.3,12 Aph-1 appears to act as a scaffold to which first nicastrin and then presenilin and Pen-2 are bound. Pen-2 then triggers endoproteolysis of presenilin into an active heterodimer.3,12 The γ-secretase complex is thought to cleave the membrane bound C-terminal fragment of APP at multiple sites: referred to as γ, ζ and ε. The γ site is variable and can occur at amino acids 38, 40 and 42 in the C99 stub left by BACE-mediated ectodomain shedding via cleavage at the β-secretase site.18 The generation of Aβ40 and Aβ42 are most common and an increase in the ratio of Aβ42 to Aβ40 can increase the risk of polymerization and amyloid deposition.18 Cutting at the ε-site at amino acid 49, also known as the S3-like site by analogy with Notch, leads to release of AICD.3,19 It appears the γ-secretase complex may cut first at the ε-site and then cut back every helical turn of the Aβ substrate to generate the γ-site cleavage products.20,21 Although in theory the cleavage of the APP substrate should always generate equimolar amounts of Aβ and AICD,20 it appears that Aβ and AICD generation are not directly linked and can be independently modulated by mutations of APP22 or knockdown of TMP21 another protein that may serve as an additional modulatory subunit in the γ-secretase complex.23
Physiological Functions of Proteolytic Processing of APP
As the secretase-mediated proteolytic processing of APP can lead to potentially harmful production of Aβ, it is a puzzle as to why it exists. It may in part be that selection pressure against production of Aβ42 has not been great since its deleterious effects are usually not seen until late in life and so may have little impact on reproductive success. However, it has been speculated that proteolytic processing of APP may serve important physiological functions in inter- and intracellular signaling, which unfortunately have the potential to produce Aβ42 as a byproduct. Although most of the literature on APP has focused on its potential pathological roles in the development of AD, in recent years there has been increasing interest in the physiological functions of APP.
APPs has been implicated in diverse cellular processes involved in cell proliferation, cell survival, neuroprotection, enhancement of memory, neuronal excitability and regulation of synaptic plasticity.24–29 Expression of APPsα was sufficient to recover anatomical, behavioral and electrophysiological abnormalities of APP-deficient mice30 suggesting that many of the physiological functions of APP are served by the secretion of APPsα. APPs also controls neural stem cell division in the adult subventricular zone.31 The N-terminal region of APPs was reported to stimulate neural stem cell proliferation32,33 and APPsα was found to bind to EGF expressing neural stem cells in the subventricular zone nd in combination with epidermal growth factor (EGF), to stimulate proliferation of neural stem cells in vitro.34
One view of the role of the γ-secretase cleavage has been that it serves to facilitate protein turnover after secretion of the APPs.3,12 The release of the AICD allows removal of the protein stubs from the membrane and subsequent intracellular proteosomal degradation. Indeed, the γ-secretase complex has even been dubbed the “proteosome of the membrane.”35,36 An alternative view is that the γ-secretase proteolytic products of the N-terminal stubs left in the membrane may also serve physiological functions in inter- and intracellular signaling. In addition to its pathological role in amyloid deposition in AD, the Aβ generated by γ-secretase RIP following β-secretase cleavage appears to play more direct roles in regulation of cell death. It had been reported that binding of the antibody 22C11 to the extracellular domain of APP activated G protein Go-dependent signaling37 and that the familial AD (FAD) mutations of APP that constitutively activated Go triggered apoptosis via the Gβγ subunit complex.38 It has now been reported that Aβ, which binds to APP causing clustering of APP in the membrane,39 acts as an APP ligand triggering Go protein activation-mediated neuronal degeneration.40
Physiological Functions of AICD
The hypothesis that AICD may also serve signaling functions has been more controversial. Investigation of the functions of AICD was long overshadowed by the widely recognized pathological importance of Aβ. One of the reasons why AICD received little attention may be that it is unstable and short-lived and so was difficult to detect. However, the transient nature of AICD is itself consistent with the notion that AICD may serve an intracellular signaling function. γ-secretase complex-mediated RIP cleaves not only APP but also many other Type I transmembrane proteins, including Notch.3,10,41–43 In the case of Notch, RIP clearly serves an important role in regulation of intracellular signaling by the Notch intracellular domain (NICD). Notch, like APP, is first cleaved by a metalloprotease sheddase, ADAM 17 or tumor necrosis factor (TNF) α-converting enzyme (TACE), just outside the membrane to shed the transmembrane stub.10,44,45 Then, the transmembrane stub of Notch undergoes RIP mediated by presenilin-dependent γ-secretase46 cleaving the protein at an intramembrane site (S3) to release the NICD, which translocates to the nucleus.47 In the nucleus, NICD acts as a second messenger to modulate target gene expression.48,49 Depending upon the factors and cofactors recruited, NICD can act by at least two distinct pathways in the nucleus. In one pathway, NICD can bind to a complex containing CSL (CBF1/RBP-J in vertebrates, suppressor of hairless in Drosophila and Lag-1 in C. elegans) DNA-binding proteins and other proteins, including the coactivator Mastermind (Mam) and Ski-interacting protein (SKIP),50–54 converting this CLS protein complex from a repressor of transcription to an activator of transcription.55 As yet, surprisingly few immediate target genes have been identified for this ubiquitous Notch signalling pathway.56 Two targets are the hairy/enhancer of split (HES) and HES-related (HERP) repressor protein families of transcription regulators.57–60 HES represses the expression of a group of proneural differentiation, basic helix-loop-helix (bHLH) genes, including NeuroD, Mash, Math and Neurogenins.61–63 In the other distinct pathway, cytosolic NICD recruits Deltex1 and translocates it to the nucleus where, by binding to transcriptional activator p300, the NICD-Deltex1 complex inhibits transcriptional activation by Mash1.64,65 NICD-dependent transcriptional activity is now known to play an important role in many cellular functions including cell proliferation, differentiation and apoptosis.54,60,66
By analogy to NICD, it might be hypothesized that AICD could serve as an intracellular signaling molecule, perhaps even modulating transcription.67 One mechanism by which AICD could achieve this may be by binding to Numb and thus inhibiting Notch-mediated signaling.68 Alternatively, AICD might more directly acts as a regulator of transcription or a modulator of another regulator of transcription. The hypothesis that AICD can modulate transcriptional activity has been highly controversial. Several studies have suggested that AICD can regulate transcription of various endogenous genes, including KAI1, GSK-3β, APP, BACE, neprilysin, α2-actin, transgelin and EGFR.69–74 Yet others did not find any evidence for AICD-mediated regulation of the transcription of KAI1, GSK-3β, APP and neprilysin.75–77 It has also been highly controversial as to whether AICD is translocated to the nucleus78,79 or modulates transcription while still bound to the membrane through interaction with scaffolding or transcriptional regulatory proteins.80,81 Initially, it appeared that AICD is stabilized by Fe65, interacts with the transcriptional factor Tip60 and translocates to the nucleus.78 In experiments using an artificial reporter system in which Gal4 was fused to the N-terminal of AICD such that it could be released by γ-secretase-dependent cleavage and expression of a luciferase reporter driven by the interaction of the Gal4 with the Gal4 response element, AICD appeared to have transcriptional activity in complex with Fe65 and histone acetyltransferase Tip60.79 However, this study using an artificial reporter system does not demonstrate whether AICD itself is involved in endogenous transcriptional activation. Even the question of whether endogenous AICD translocates to the nucleus remains unresolved. Later studies indicated that membrane-tethered AICD recruits and activates Fe65 allowing its translocation to the nucleus but that it is not essential for Fe65-dependent transcriptional transactivation.81 Moreover, a subsequent study confirmed that Fe65 alone was sufficient for transcriptional transactivation and that APP and Tip60 play positive and negative modulatory roles, respectively.82 While, yet another study suggested that APP stabilizes Tip60 through CDK-dependent phosphorylation allowing APP to signal to the nucleus by a γ-secretase-independent mechanism.83 Investigation of the phosphorylation of APP at Thr688 (residue numbering for the APP695 form), which reduces or prevents Fe65 binding to APP84 and disrupts the stabilization of AICD by Fe65 binding,85 suggested that activation of Fe65 may involve liberation from membrane bound APP on phosphorylation and that unphosphorylated, but not phosphorylated, AICD translocates to the nucleus independently of Fe65.80 Thus the role of AICD in Fe65/Tip60-mediated transcriptional activity remains unclear. However, in the case of the epidermal growth factor receptor (EGFR), direct binding of endogenous AICD to the EGFR promoter is reported.74 It has also been suggested that AICD can enhance the transcriptional activation of another transcription factor, p53.86 The evidence falls short of conclusively demonstrating that AICD is in itself a transcription factor or a nuclearly translocated modulator of transcription factors. Nevertheless, together the literature suggests the possibility that intracellular release of ACID could serve as an intracellular signal, perhaps even playing a role in the modulation of the expression of certain proteins.
TAG1-APP Ligand-Receptor Triggered Release of AICD
An important aspect of the cellular function of the RIP of Notch is that the γ-secretase mediated cleavage of NICD is stimulated by ligand binding to Notch. At the cell surface, the Notch receptor responds to ligand binding to its extracellular epidermal growth factor (EGF)-like repeats.87 Delta, Serrate and Lag (DSL) family proteins can bind to Notch and stimulate ectodomain shedding.45,61,88 Consistent with the notion that the γ-secretase complex senses whether the N-terminal stub has been sufficiently cleaved via nicastrin,17 it appears that ligand-induced extracellular cleavage regulates γ-secretase-mediated proteolytic activation of Notch1.89 Recently, we have shown that F3/contactin and its homologue NB-3 are functional ligands for the Notch receptor. Activation of Notch by these F3/contactin family proteins modulates oligodendrocyte differentiation via the transcriptional factor Deltex.56,90–92
By analogy with the RIP of Notch, one might predict that ligand interaction with APP might stimulate release of AICD. It was reported that f-spondin binds to the extracellular domain of APP and inhibits β-secretase cleavage.93 Could a ligand bind to APP promoting, rather than inhibiting, ectodomain shedding of APP and thus facilitating γ-secretase cleavage in a manner similar to ligand-activation of Notch RIP? We investigated the interaction between TAG1, a member of the F3/Contactin family of glycophosphatidylinositol (GPI)-linked proteins, and APP.94 Cell adhesion, coimmunoprecipitation and pull-down assays all suggested that APP and TAG1 bind to each other. Using the artificial luciferase reporter system in which Gal4 is fused to the N-terminal end of AICD adjacent to the γ-secretase ε cleavage site,79 we investigated whether the TAG1-APP interaction could trigger intracellular release of AICD and hence activate the Gal4 promoter driven luciferase reporter. We found that TAG1 stimulated activation of the luciferase reporter and that this activation was dose-dependently blocked by two different γ-secretase inhibitors indicating that TAG1 triggers AICD release in a γ-secretase-dependent manner. Moreover, Western blotting confirmed that TAG1 dose-dependently increased expression of AICD. Thus, it appears that TAG1 is a novel ligand for APP capable of facilitating γ-secretase-dependent release of AICD. This suggests that APP may function as a receptor for TAG1 in a manner similar to that in which Notch functions as a receptor for DSL and F3/NB3 (Fig. 2).
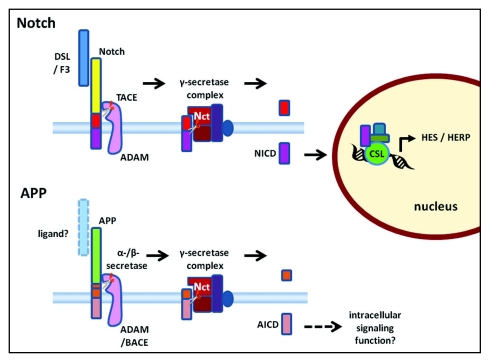
Figure 2.
A schematic diagram of ligand-dependent γ-secretase cleavage of Notch (top) and associated intracellular signaling compared with proteolytic processing of APP (bottom) (not drawn to scale). A Delta, Serrate and Lag (DSL) or F3/contactin family protein (F3) acts as a ligand binding to the extracellular domain of Notch. Binding of the ligand stimulates ectodomain shedding by tumour necrosis factor (TNF) α-converting enzyme (TACE), a disintegrin and metalloproteinase (ADAM). Similarly, the extracellular portion of APP is cleaved by α-secretase, an ADAM, or β-secretase, β-amyloid-cleaving enzyme-1 (BACE). Nicastrin (Nct) in the γ-secretase complex acts as a receptor interacting with the extracellular N-terminal of the protein stub left in the membrane. Only when the ectodomain has been shed can γ-secretase cleavage of the membrane bound stub proceed. The γ-secretase-dependent cleavage releases the Notch intracellular domain (NICD) in the case of Notch and the APP intracellular domain (AICD) in the case of APP. NICD is known to serve as an intracellular signaling molecule regulating protein expression on translocation to the nucleus. In the example illustrated in the diagram, NICD complexes with CSL and other proteins to regulate expression of the hairy/enhancer of split (HES) and HES-related (HERP) repressor protein families of transcription regulators.
Physiological Function of TAG1-APP Interaction in Neurogenesis
If TAG1 is a ligand for APP, what physiological functions does this ligand-receptor interaction mediate? In the central nervous system (CNS), GPI-linked recognition molecules, such as TAG1, NB-3 and F3/contactin, have been implicated in key developmental events, including selective axonal fasciculation, neural cell adhesion and migration, and neurite outgrowth.95 TAG1 is expressed from early in development, for example on the cell bodies of motor neurons in spinal cord at E10.5 and during their lateral migration from the ventricular zone at E13.95 Likewise, APP is expressed early in neural development. APP mRNA transcripts are reported in mouse oocytes and early in mouse embryogenesis.96 In the mouse neural tube, APP is expressed as early as E9.5, when the neural stem cells and RC2-positive radial glia are actively dividing.97 APP is expressed by neuroepithelial cells in the cortical ventricular zone, particularly in the apical portion where mitosis takes place at E14 to E16.98 APPsα binds to EGF-positive fetal neural stem cells from the cortical subventricular zone and, together with EGF, APP stimulates proliferation of the cells from embryonic neurospheres in vitro.34 A recent in utero RNA interference study indicated that Aβ plays a critical role in neural migration.99 It has also been reported that neurogenesis is increased in an AD mouse model100 and in the adult human hippocampus in AD,101 although the changes in neurogenesis in AD and their implications are controversial.102 Together these data suggest that TAG1 and APP may both be involved in neural development.
We found that TAG1 and APP colocalize in the neurogenic niche of the ventricular zone in the developing mouse brain and within neural progenitor cells (NPCs) isolated from this region.94 NPCs isolated from TAG1 null and APP null mice showed abnormally enhanced neurogenesis, suggesting that TAG1 and APP are involved in negative regulation of neurogenesis. Moreover, consistent with the notion that TAG1 and APP act by a common pathway rather than by two separate and additive pathways, NPCs from TAG1/APP double null mice showed a similar enhancement in neurogenesis to NPCs from single TAG1 and APP null mice. Treatment with soluble TAG1 protein during differentiation normalized neurogenesis in NPCs isolated from TAG1 null mice but not in NPCs isolated from TAG1/APP double null mice. Moreover, transfection of NPCs isolated from TAG1 null mice with AICD rescued the suppression of neurogenesis. Thus, it appears that binding of TAG1 to APP can trigger an AICD-dependent suppression of neurogenesis. As Fe65 has been proposed as a partner of AICD in intracellular signaling mechanisms,78,79,81,84 we also investigated the localization of Fe65. Fe65 colocalized with TAG1 and APP in the fetal cortical ventricular zone and NPCs isolated from fetal brain.94 NPC's isolated from Fe65 null mice also showed a similar enhancement of neurogenesis to that observed in NPCs isolated from TAG1, APP and TAG1/APP null mice. Using a Gal4 promoter-driven luciferase reporter system in which Gal4 was fused to the N-terminal of Fe65 we showed that cotransfection of TAG1-transfected cells with APP led to increased reporter activity indicative of mobilization of Fe65. But cotransfection of TAG1-transfected cells with an NPTY to NATA mutation that abolishes Fe65 binding84 did not increase reporter activity. Transfection of NPCs derived from TAG1 null mice with AICD with the same NPTY to NATA mutation abolishing Fe65 binding did not rescue the suppression of neurogenesis. Thus these data suggest that Fe65 is required for the AICD-dependent suppression of neurogenesis by TAG1.
Conclusions
Together, our data94 are consistent with the hypothesis that TAG1 binding to APP stimulates γ-secretase-dependent cleavage of APP to release AICD which suppresses neurogenesis by a mechanism involving Fe65 (Fig. 3). At this stage the mechanisms by which AICD suppresses neurogenesis are unknown. It may be interesting to investigate whether AICD acting in concert with or through modulation of Fe65 influences gene transcription. The discovery that TAG1 can act as an AICD-releasing ligand for APP suggests the possibility that other molecules may likewise act as ligands of APP. It may be interesting to investigate whether ligand-stimulated cleavage of APP is involved in physiological regulation of other cellular functions. Our finding that TAG1 binding to APP triggers AICD cleavage may also have implications in AD. It may be of interest to investigate whether ligand-stimulated cleavage influences the probability of amyloidogenic cleavage of APP. Conversely, abnormal proteolytic processing of APP in AD may have consequences for intracellular signaling.
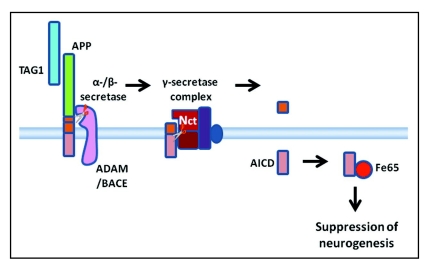
Figure 3.
A diagrammatic summary of a working model of TAG1-APP signaling. TAG1 is a ligand for APP. When TAG1 binds to APP this stimulates or facilitates ectodomain shedding by α- or β-secretases. Once the ectodomain has been shed, γ-secretase cleavage of the membrane bound stub can proceed. Thus, TAG1 binding leads to γ-secretase-dependent cleavage releasing ACID intracellularly. AICD interacts with the scaffolding protein, Fe65. This results in an Fe65-dependent suppression of neurogenesis.
Footnotes
Previously published online as a Cell Adhesion & Migration E-publication: www.landesbioscience.com/journals/celladhesion/article/5790
References
- 1.Lichtenthaler SF, Steiner H. Sheddases and intramembrane-cleaving proteases: RIPpers of the membrane. Symposium on regulated intramembrane proteolysis. EMBO Rep. 2007;8:537–541. doi: 10.1038/sj.embor.7400978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brown MS, Ye J, Rawson RB, Goldstein JL. Regulated intramembrane proteolysis: A control mechanism conserved from bacteria to humans. Cell. 2000;100:391–398. doi: 10.1016/s0092-8674(00)80675-3. [DOI] [PubMed] [Google Scholar]
- 3.Wolfe MS, Kopan R. Intramembrane proteolysis: Theme and variations. Science. 2004;305:1119–1123. doi: 10.1126/science.1096187. [DOI] [PubMed] [Google Scholar]
- 4.Allinson TM, Parkin ET, Turner AJ, Hooper NM. ADAMs family members as amyloid precursor protein alpha-secretases. J Neurosci Res. 2003;74:342–352. doi: 10.1002/jnr.10737. [DOI] [PubMed] [Google Scholar]
- 5.Asai M, Hattori C, Szabo B, Sasagawa N, Maruyama K, Tanuma S, Ishiura S. Putative function of ADAM9, ADAM10, and ADAM17 as APP alpha-secretase. Biochem Biophys Res Commun. 2003;301:231–235. doi: 10.1016/s0006-291x(02)02999-6. [DOI] [PubMed] [Google Scholar]
- 6.Lammich S, Kojro E, Postina R, Gilbert S, Pfeiffer R, Jasionowski M, Haass C, Fahrenholz F. Constitutive and regulated alpha-secretase cleavage of Alzheimer's amyloid precursor protein by a disintegrin metalloprotease. Proc Natl Acad Sci USA. 1999;96:3922–3927. doi: 10.1073/pnas.96.7.3922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Parvathy S, Karran EH, Turner AJ, Hooper NM. The secretases that cleave angiotensin converting enzyme and the amyloid precursor protein are distinct from tumour necrosis factor-alpha convertase. FEBS Lett. 1998;431:63–65. doi: 10.1016/s0014-5793(98)00726-1. [DOI] [PubMed] [Google Scholar]
- 8.Slack BE, Ma LK, Seah CC. Constitutive shedding of the amyloid precursor protein ectodomain is up-regulated by tumour necrosis factor-alpha converting enzyme. Biochem J. 2001;357:787–794. doi: 10.1042/0264-6021:3570787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Walsh DM, Minogue AM, Sala FC, Fadeeva JV, Wasco W, Selkoe DJ. The APP family of proteins: Similarities and differences. Biochem Soc Trans. 2007;35:416–420. doi: 10.1042/BST0350416. [DOI] [PubMed] [Google Scholar]
- 10.Parks AL, Curtis D. Presenilin diversifies its portfolio. Trends Genet. 2007;23:140–150. doi: 10.1016/j.tig.2007.01.008. [DOI] [PubMed] [Google Scholar]
- 11.Haass C. Take five—BACE and the gamma-secretase quartet conduct Alzheimer's amyloid beta-peptide generation. EMBO J. 2004;23:483–488. doi: 10.1038/sj.emboj.7600061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Selkoe DJ, Wolfe MS. Presenilin: Running with scissors in the membrane. Cell. 2007;131:215–221. doi: 10.1016/j.cell.2007.10.012. [DOI] [PubMed] [Google Scholar]
- 13.Yu G, Nishimura M, Arawaka S, Levitan D, Zhang L, Tandon A, Song YQ, Rogaeva E, Chen F, Kawarai T, Supala A, Levesque L, Yu H, Yang DS, Holmes E, Milman P, Liang Y, Zhang DM, Xu DH, Sato C, Rogaev E, Smith M, Janus C, Zhang Y, Aebersold R, Farrer LS, Sorbi S, Bruni A, Fraser P, St George-Hyslop P. Nicastrin modulates presenilin-mediated notch/glp-1 signal transduction and betaAPP processing. Nature. 2000;407:48–54. doi: 10.1038/35024009. [DOI] [PubMed] [Google Scholar]
- 14.Francis R, McGrath G, Zhang J, Ruddy DA, Sym M, Apfeld J, Nicoll M, Maxwell M, Hai B, Ellis MC, Parks AL, Xu W, Li J, Gurney M, Myers RL, Himes CS, Hiebsch R, Ruble C, Nye JS, Curtis D. aph-1 and pen-2 are required for Notch pathway signaling, gamma-secretase cleavage of betaAPP, and presenilin protein accumulation. Dev Cell. 2002;3:85–97. doi: 10.1016/s1534-5807(02)00189-2. [DOI] [PubMed] [Google Scholar]
- 15.Goutte C, Tsunozaki M, Hale VA, Priess JR. APH-1 is a multipass membrane protein essential for the Notch signaling pathway in Caenorhabditis elegans embryos. Proc Natl Acad Sci USA. 2002;99:775–779. doi: 10.1073/pnas.022523499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.De Strooper B. Aph-1, Pen-2, and Nicastrin with Presenilin generate an active gamma-Secretase complex. Neuron. 2003;38:9–12. doi: 10.1016/s0896-6273(03)00205-8. [DOI] [PubMed] [Google Scholar]
- 17.Shah S, Lee SF, Tabuchi K, Hao YH, Yu C, LaPlant Q, Ball H, Dann CE, IIIrd, Sudhof T, Yu G. Nicastrin functions as a gamma-secretase-substrate receptor. Cell. 2005;122:435–447. doi: 10.1016/j.cell.2005.05.022. [DOI] [PubMed] [Google Scholar]
- 18.Haass C, Selkoe DJ. Soluble protein oligomers in neurodegeneration: Lessons from the Alzheimer's amyloid beta-peptide. Nat Rev Mol Cell Biol. 2007;8:101–112. doi: 10.1038/nrm2101. [DOI] [PubMed] [Google Scholar]
- 19.De Strooper B, Annaert W, Cupers P, Saftig P, Craessaerts K, Mumm JS, Schroeter EH, Schrijvers V, Wolfe MS, Ray WJ, Goate A, Kopan R. A presenilin-1-dependent gamma-secretase-like protease mediates release of Notch intracellular domain. Nature. 1999;398:518–522. doi: 10.1038/19083. [DOI] [PubMed] [Google Scholar]
- 20.Kakuda N, Funamoto S, Yagishita S, Takami M, Osawa S, Dohmae N, Ihara Y. Equimolar production of amyloid beta-protein and amyloid precursor protein intracellular domain from beta-carboxyl-terminal fragment by gamma-secretase. J Biol Chem. 2006;281:14776–14786. doi: 10.1074/jbc.M513453200. [DOI] [PubMed] [Google Scholar]
- 21.Wolfe MS. When loss is gain: Reduced presenilin proteolytic function leads to increased Abeta42/Abeta40. Talking Point on the role of presenilin mutations in Alzheimer disease. EMBO Rep. 2007;8:136–140. doi: 10.1038/sj.embor.7400896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hecimovic S, Wang J, Dolios G, Martinez M, Wang R, Goate AM. Mutations in APP have independent effects on Abeta and CTFgamma generation. Neurobiol Dis. 2004;17:205–218. doi: 10.1016/j.nbd.2004.04.018. [DOI] [PubMed] [Google Scholar]
- 23.Chen F, Hasegawa H, Schmitt-Ulms G, Kawarai T, Bohm C, Katayama T, Gu Y, Sanjo N, Glista M, Rogaeva E, Wakutani Y, Pardossi-Piquard R, Ruan X, Tandon A, Checler F, Marambaud P, Hansen K, Westaway D, St George-Hyslop P, Fraser P. TMP21 is a presenilin complex component that modulates gamma-secretase but not epsilon-secretase activity. Nature. 2006;440:1208–1212. doi: 10.1038/nature04667. [DOI] [PubMed] [Google Scholar]
- 24.Mattson MP. Cellular actions of beta-amyloid precursor protein and its soluble and fibrillogenic derivatives. Physiol Rev. 1997;77:1081–1132. doi: 10.1152/physrev.1997.77.4.1081. [DOI] [PubMed] [Google Scholar]
- 25.Turner PR, O'Connor K, Tate WP, Abraham WC. Roles of amyloid precursor protein and its fragments in regulating neural activity, plasticity and memory. Prog Neurobiol. 2003;70:1–32. doi: 10.1016/s0301-0082(03)00089-3. [DOI] [PubMed] [Google Scholar]
- 26.Reinhard C, Hebert SS, De SB. The amyloid-beta precursor protein: Integrating structure with biological function. EMBO J. 2005;24:3996–4006. doi: 10.1038/sj.emboj.7600860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Meziane H, Dodart JC, Mathis C, Little S, Clemens J, Paul SM, Ungerer A. Memory-enhancing effects of secreted forms of the beta-amyloid precursor protein in normal and amnestic mice. Proc Natl Acad Sci USA. 1998;95:12683–12688. doi: 10.1073/pnas.95.21.12683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Roch JM, Masliah E, Roch-Levecq AC, Sundsmo MP, Otero DA, Veinbergs I, Saitoh T. Increase of synaptic density and memory retention by a peptide representing the trophic domain of the amyloid beta/A4 protein precursor. Proc Natl Acad Sci USA. 1994;91:7450–7454. doi: 10.1073/pnas.91.16.7450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zheng H, Koo EH. The amyloid precursor protein: Beyond amyloid. Mol Neurodegener. 2006;1:5. doi: 10.1186/1750-1326-1-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ring S, Weyer SW, Kilian SB, Waldron E, Pietrzik CU, Filippov MA, Herms J, Buchholz C, Eckman CB, Korte M, Wolfer DP, Muller UC. The secreted beta-amyloid precursor protein ectodomain APPs alpha is sufficient to rescue the anatomical, behavioral, and electrophysiological abnormalities of APP-deficient mice. J Neurosci. 2007;27:7817–7826. doi: 10.1523/JNEUROSCI.1026-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Conti L, Cattaneo E. Controlling neural stem cell division within the adult subventricular zone: An APPealing job. Trends Neurosci. 2005;28:57–59. doi: 10.1016/j.tins.2004.11.005. [DOI] [PubMed] [Google Scholar]
- 32.Hayashi Y, Kashiwagi K, Ohta J, Nakajima M, Kawashima T, Yoshikawa K. Alzheimer amyloid protein precursor enhances proliferation of neural stem cells from fetal rat brain. Biochem Biophys Res Commun. 1994;205:936–943. doi: 10.1006/bbrc.1994.2755. [DOI] [PubMed] [Google Scholar]
- 33.Ohsawa I, Takamura C, Morimoto T, Ishiguro M, Kohsaka S. Amino-terminal region of secreted form of amyloid precursor protein stimulates proliferation of neural stem cells. Eur J Neurosci. 1999;11:1907–1913. doi: 10.1046/j.1460-9568.1999.00601.x. [DOI] [PubMed] [Google Scholar]
- 34.Caille I, Allinquant B, Dupont E, Bouillot C, Langer A, Muller U, Prochiantz A. Soluble form of amyloid precursor protein regulates proliferation of progenitors in the adult sub-ventricular zone. Development. 2004;131:2173–2181. doi: 10.1242/dev.01103. [DOI] [PubMed] [Google Scholar]
- 35.Kopan R, Ilagan MX. Gamma-secretase: Proteasome of the membrane? Nat Rev Mol Cell Biol. 2004;5:499–504. doi: 10.1038/nrm1406. [DOI] [PubMed] [Google Scholar]
- 36.Small DH. Is gamma-secretase a multienzyme complex for membrane protein degradation? Models and speculations. Peptides. 2002;23:1317–1321. doi: 10.1016/s0196-9781(02)00072-4. [DOI] [PubMed] [Google Scholar]
- 37.Okamoto T, Takeda S, Murayama Y, Ogata E, Nishimoto I. Ligand-dependent G protein coupling function of amyloid transmembrane precursor. J Biol Chem. 1995;270:4205–4208. doi: 10.1074/jbc.270.9.4205. [DOI] [PubMed] [Google Scholar]
- 38.Giambarella U, Yamatsuji T, Okamoto T, Matsui T, Ikezu T, Murayama Y, Levine MA, Katz A, Gautam N, Nishimoto I. G protein betagamma complex-mediated apoptosis by familial Alzheimer's disease mutant of APP. EMBO J. 1997;16:4897–4907. doi: 10.1093/emboj/16.16.4897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Heredia L, Lin R, Vigo FS, Kedikian G, Busciglio J, Lorenzo A. Deposition of amyloid fibrils promotes cell-surface accumulation of amyloid beta precursor protein. Neurobiol Dis. 2004;16:617–629. doi: 10.1016/j.nbd.2004.04.015. [DOI] [PubMed] [Google Scholar]
- 40.Sola Vigo F, Kedikian G, Heredia L, Heredia F, Anel AD, Rosa AL, Lorenzo A. Amyloid-beta precursor protein mediates neuronal toxicity of amyloid beta through Go protein activation. Neurobiol Aging. 2008 doi: 10.1016/j.neurobiolaging.2007.11.017. [DOI] [PubMed] [Google Scholar]
- 41.Hartmann D, Tournoy J, Saftig P, Annaert W, De SB. Implication of APP secretases in notch signaling. J Mol Neurosci. 2001;17:171–181. doi: 10.1385/JMN:17:2:171. [DOI] [PubMed] [Google Scholar]
- 42.Selkoe D, Kopan R. Notch and Presenilin: Regulated intramembrane proteolysis links development and degeneration. Annu Rev Neurosci. 2003;26:565–597. doi: 10.1146/annurev.neuro.26.041002.131334. [DOI] [PubMed] [Google Scholar]
- 43.Ebinu JO, Yankner BA. A RIP tide in neuronal signal transduction. Neuron. 2002;34:499–502. doi: 10.1016/s0896-6273(02)00704-3. [DOI] [PubMed] [Google Scholar]
- 44.Brou C, Logeat F, Gupta N, Bessia C, LeBail O, Doedens JR, Cumano A, Roux P, Black RA, Israel A. A novel proteolytic cleavage involved in Notch signaling: The role of the disintegrin-metalloprotease TACE. Mol Cell. 2000;5:207–216. doi: 10.1016/s1097-2765(00)80417-7. [DOI] [PubMed] [Google Scholar]
- 45.Bianchi S, Dotti MT, Federico A. Physiology and pathology of notch signalling system. J Cell Physiol. 2006;207:300–308. doi: 10.1002/jcp.20542. [DOI] [PubMed] [Google Scholar]
- 46.Kopan R, Goate A. A common enzyme connects notch signaling and Alzheimer's disease. Genes Dev. 2000;14:2799–2806. doi: 10.1101/gad.836900. [DOI] [PubMed] [Google Scholar]
- 47.Schroeter EH, Kisslinger JA, Kopan R. Notch-1 signalling requires ligand-induced proteolytic release of intracellular domain. Nature. 1998;393:382–386. doi: 10.1038/30756. [DOI] [PubMed] [Google Scholar]
- 48.Jarriault S, Le BO, Hirsinger E, Pourquie O, Logeat F, Strong CF, Brou C, Seidah NG, Isra I. A Delta-1 activation of notch-1 signaling results in HES-1 transactivation. Mol Cell Biol. 1998;18:7423–7431. doi: 10.1128/mcb.18.12.7423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jarriault S, Brou C, Logeat F, Schroeter EH, Kopan R, Israel A. Signalling downstream of activated mammalian Notch. Nature. 1995;377:355–358. doi: 10.1038/377355a0. [DOI] [PubMed] [Google Scholar]
- 50.Kao HY, Ordentlich P, Koyano-Nakagawa N, Tang Z, Downes M, Kintner CR, Evans RM, Kadesch T. A histone deacetylase corepressor complex regulates the Notch signal transduction pathway. Genes Dev. 1998;12:2269–2277. doi: 10.1101/gad.12.15.2269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Taniguchi Y, Furukawa T, Tun T, Han H, Honjo T. LIM protein KyoT2 negatively regulates transcription by association with the RBP-J DNA-binding protein. Mol Cell Biol. 1998;18:644–654. doi: 10.1128/mcb.18.1.644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hsieh JJ, Zhou S, Chen L, Young DB, Hayward SD. CIR, a corepressor linking the DNA binding factor CBF1 to the histone deacetylase complex. Proc Natl Acad Sci USA. 1999;96:23–28. doi: 10.1073/pnas.96.1.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhou S, Fujimuro M, Hsieh JJ, Chen L, Miyamoto A, Weinmaster G, Hayward SD. SKIP, a CBF1-associated protein, interacts with the ankyrin repeat domain of NotchIC To facilitate NotchIC function. Mol Cell Biol. 2000;20:2400–2410. doi: 10.1128/mcb.20.7.2400-2410.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bray SJ. Notch signalling: A simple pathway becomes complex. Nat Rev Mol Cell Biol. 2006;7:678–689. doi: 10.1038/nrm2009. [DOI] [PubMed] [Google Scholar]
- 55.Kopan R. Notch: A membrane-bound transcription factor. J Cell Sci. 2002;115:1095–1097. doi: 10.1242/jcs.115.6.1095. [DOI] [PubMed] [Google Scholar]
- 56.Ma QH, Yang WL, Dawe GS, Xiao ZC. Notch signalling and oligodendrocyte development. In: Dheen ST, Ling EA, editors. Trends in Glial Research - Basic and Applied. Kerala: Research Signpost; 2007. pp. 107–118. [Google Scholar]
- 57.Egan SE, St-Pierre B, Leow CC. Notch receptors, partners and regulators: From conserved domains to powerful functions. Curr Top Microbiol Immunol. 1998;228:273–324. doi: 10.1007/978-3-642-80481-6_11. [DOI] [PubMed] [Google Scholar]
- 58.Iso T, Kedes L, Hamamori Y. HES and HERP families: Multiple effectors of the Notch signaling pathway. J Cell Physiol. 2003;194:237–255. doi: 10.1002/jcp.10208. [DOI] [PubMed] [Google Scholar]
- 59.Iso T, Chung G, Hamamori Y, Kedes L. HERP1 is a cell type-specific primary target of Notch. J Biol Chem. 2002;277:6598–6607. doi: 10.1074/jbc.M110495200. [DOI] [PubMed] [Google Scholar]
- 60.Artavanis-Tsakonas S, Rand MD, Lake RJ. Notch signaling: Cell fate control and signal integration in development. Science. 1999;284:770–776. doi: 10.1126/science.284.5415.770. [DOI] [PubMed] [Google Scholar]
- 61.Bray SJ. Expression and function of Enhancer of split bHLH proteins during Drosophila neurogenesis. Perspect Dev Neurobiol. 1997;4:313–323. [PubMed] [Google Scholar]
- 62.Cau E, Gradwohl G, Fode C, Guillemot F. Mash1 activates a cascade of bHLH regulators in olfactory neuron progenitors. Development. 1997;124:1611–1621. doi: 10.1242/dev.124.8.1611. [DOI] [PubMed] [Google Scholar]
- 63.Fisher A, Caudy M. The function of hairy-related bHLH repressor proteins in cell fate decisions. Bioessays. 1998;20:298–306. doi: 10.1002/(SICI)1521-1878(199804)20:4<298::AID-BIES6>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- 64.Yamamoto N, Yamamoto S, Inagaki F, Kawaichi M, Fukamizu A, Kishi N, Matsuno K, Nakamura K, Weinmaster G, Okano H, Nakafuku M. Role of Deltex-1 as a transcriptional regulator downstream of the Notch receptor. J Biol Chem. 2001;276:45031–45040. doi: 10.1074/jbc.M105245200. [DOI] [PubMed] [Google Scholar]
- 65.Ramain P, Khechumian K, Seugnet L, Arbogast N, Ackermann C, Heitzler P. Novel Notch alleles reveal a Deltex-dependent pathway repressing neural fate. Curr Biol. 2001;11:1729–1738. doi: 10.1016/s0960-9822(01)00562-0. [DOI] [PubMed] [Google Scholar]
- 66.Nelson BR, Hartman BH, Georgi SA, Lan MS, Reh TA. Transient inactivation of Notch signaling synchronizes differentiation of neural progenitor cells. Dev Biol. 2007;304:479–498. doi: 10.1016/j.ydbio.2007.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Steiner H, Haass C. Nuclear signaling: A common function of presenilin substrates? J Mol Neurosci. 2001;17:193–198. doi: 10.1385/JMN:17:2:193. [DOI] [PubMed] [Google Scholar]
- 68.Roncarati R, Sestan N, Scheinfeld MH, Berechid BE, Lopez PA, Meucci O, McGlade JC, Rakic P, D'Adamio L. The gamma-secretase-generated intracellular domain of beta-amyloid precursor protein binds Numb and inhibits Notch signaling. Proc Natl Acad Sci USA. 2002;99:7102–7107. doi: 10.1073/pnas.102192599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Baek SH, Ohgi KA, Rose DW, Koo EH, Glass CK, Rosenfeld MG. Exchange of N-CoR corepressor and Tip60 coactivator complexes links gene expression by NF-kappaB and beta-amyloid precursor protein. Cell. 2002;110:55–67. doi: 10.1016/s0092-8674(02)00809-7. [DOI] [PubMed] [Google Scholar]
- 70.Kim HS, Kim EM, Lee JP, Park CH, Kim S, Seo JH, Chang KA, Yu E, Jeong SJ, Chong YH, Suh YH. C-terminal fragments of amyloid precursor protein exert neurotoxicity by inducing glycogen synthase kinase-3beta expression. FASEB J. 2003;17:1951–1953. doi: 10.1096/fj.03-0106fje. [DOI] [PubMed] [Google Scholar]
- 71.Muller T, Concannon CG, Ward MW, Walsh CM, Tirniceriu AL, Tribl F, Kogel D, Prehn JH, Egensperger R. Modulation of gene expression and cytoskeletal dynamics by the amyloid precursor protein intracellular domain (AICD) Mol Biol Cell. 2007;18:201–210. doi: 10.1091/mbc.E06-04-0283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Pardossi-Piquard R, Petit A, Kawarai T, Sunyach C, ves da CC, Vincent B, Ring S, D'Adamio L, Shen J, Muller U, St George HP, Checler F. Presenilin-dependent transcriptional control of the Abeta-degrading enzyme neprilysin by intracellular domains of betaAPP and APLP. Neuron. 2005;46:541–554. doi: 10.1016/j.neuron.2005.04.008. [DOI] [PubMed] [Google Scholar]
- 73.von Rotz RC, Kohli BM, Bosset J, Meier M, Suzuki T, Nitsch RM, Konietzko U. The APP intracellular domain forms nuclear multiprotein complexes and regulates the transcription of its own precursor. J Cell Sci. 2004;117:4435–4448. doi: 10.1242/jcs.01323. [DOI] [PubMed] [Google Scholar]
- 74.Zhang YW, Wang R, Liu Q, Zhang H, Liao FF, Xu H. Presenilin/gamma-secretase-dependent processing of beta-amyloid precursor protein regulates EGF receptor expression. Proc Natl Acad Sci USA. 2007;104:10613–10618. doi: 10.1073/pnas.0703903104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Chen AC, Selkoe DJ. Response to: Pardossi-Piquard et al., “Presenilin-Dependent Transcriptional Control of the Abeta-Degrading Enzyme Neprilysin by Intracellular Domains of betaAPP and APLP.” Neuron 46, 541–554. Neuron. 2007;53:479–483. doi: 10.1016/j.neuron.2007.01.023. [DOI] [PubMed] [Google Scholar]
- 76.Hebert SS, Serneels L, Tolia A, Craessaerts K, Derks C, Filippov MA, Muller U, De SB. Regulated intramembrane proteolysis of amyloid precursor protein and regulation of expression of putative target genes. EMBO Rep. 2006;7:739–745. doi: 10.1038/sj.embor.7400704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Pardossi-Piquard R, Dunys J, Kawarai T, Sunyach C, ves da CC, Vincent B, Sevalle J, Pimplikar S, St George-Hyslop P, Checler F. Response to Correspondence: Pardossi-Piquard et al., “Presenilin-Dependent Transcriptional Control of the Abeta-Degrading Enzyme Neprilysin by Intracellular Domains of betaAPP and APLP.” Neuron 46, 541–554. Neuron. 2007;53:483–486. doi: 10.1016/j.neuron.2007.01.024. [DOI] [PubMed] [Google Scholar]
- 78.Kimberly WT, Zheng JB, Guenette SY, Selkoe DJ. The intracellular domain of the beta-amyloid precursor protein is stabilized by Fe65 and translocates to the nucleus in a notch-like manner. J Biol Chem. 2001;276:40288–40292. doi: 10.1074/jbc.C100447200. [DOI] [PubMed] [Google Scholar]
- 79.Cao X, Sudhof TC. A transcriptionally [correction of transcriptively] active complex of APP with Fe65 and histone acetyltransferase Tip60. Science. 2001;293:115–120. doi: 10.1126/science.1058783. [DOI] [PubMed] [Google Scholar]
- 80.Nakaya T, Suzuki T. Role of APP phosphorylation in FE65-dependent gene transactivation mediated by AICD. Genes Cells. 2006;11:633–645. doi: 10.1111/j.1365-2443.2006.00968.x. [DOI] [PubMed] [Google Scholar]
- 81.Cao X, Sudhof TC. Dissection of amyloid-beta precursor protein-dependent transcriptional transactivation. J Biol Chem. 2004;279:24601–24611. doi: 10.1074/jbc.M402248200. [DOI] [PubMed] [Google Scholar]
- 82.Yang Z, Cool BH, Martin GM, Hu Q. A dominant role for FE65 (APBB1) in nuclear signaling. J Biol Chem. 2006;281:4207–4214. doi: 10.1074/jbc.M508445200. [DOI] [PubMed] [Google Scholar]
- 83.Hass MR, Yankner BA. A {gamma}-secretase-independent mechanism of signal transduction by the amyloid precursor protein. J Biol Chem. 2005;280:36895–36904. doi: 10.1074/jbc.M502861200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Ando K, Iijima KI, Elliott JI, Kirino Y, Suzuki T. Phosphorylation-dependent regulation of the interaction of amyloid precursor protein with Fe65 affects the production of beta-amyloid. J Biol Chem. 2001;276:40353–40361. doi: 10.1074/jbc.M104059200. [DOI] [PubMed] [Google Scholar]
- 85.Kimberly WT, Zheng JB, Town T, Flavell RA, Selkoe DJ. Physiological regulation of the beta-amyloid precursor protein signaling domain by c-Jun N-terminal kinase JNK3 during neuronal differentiation. J Neurosci. 2005;25:5533–5543. doi: 10.1523/JNEUROSCI.4883-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Ozaki T, Li Y, Kikuchi H, Tomita T, Iwatsubo T, Nakagawara A. The intracellular domain of the amyloid precursor protein (AICD) enhances the p53-mediated apoptosis. Biochem Biophys Res Commun. 2006;351:57–63. doi: 10.1016/j.bbrc.2006.09.162. [DOI] [PubMed] [Google Scholar]
- 87.Rebay I, Fleming RJ, Fehon RG, Cherbas L, Cherbas P, Artavanis-Tsakonas S. Specific EGF repeats of Notch mediate interactions with Delta and Serrate: Implications for Notch as a multifunctional receptor. Cell. 1991;67:687–699. doi: 10.1016/0092-8674(91)90064-6. [DOI] [PubMed] [Google Scholar]
- 88.Baron M. An overview of the Notch signalling pathway. Semin Cell Dev Biol. 2003;14:113–119. doi: 10.1016/s1084-9521(02)00179-9. [DOI] [PubMed] [Google Scholar]
- 89.Mumm JS, Schroeter EH, Saxena MT, Griesemer A, Tian X, Pan DJ, Ray WJ, Kopan R. A ligand-induced extracellular cleavage regulates gamma-secretase-like proteolytic activation of Notch1. Mol Cell. 2000;5:197–206. doi: 10.1016/s1097-2765(00)80416-5. [DOI] [PubMed] [Google Scholar]
- 90.Hu QD, Ang BT, Karsak M, Hu WP, Cui XY, Duka T, Takeda Y, Chia W, Sankar N, Ng YK, Ling EA, Maciag T, Small D, Trifonova R, Kopan R, Okano H, Nakafuku M, Chiba S, Hirai H, Aster JC, Schachner M, Pallen CJ, Watanabe K, Xiao ZC. F3/contactin acts as a functional ligand for Notch during oligodendrocyte maturation. Cell. 2003;115:163–175. doi: 10.1016/s0092-8674(03)00810-9. [DOI] [PubMed] [Google Scholar]
- 91.Hu QD, Ma QH, Gennarini G, Xiao ZC. Cross-talk between F3/contactin and Notch at axoglial interface: A role in oligodendrocyte development. Dev Neurosci. 2006;28:25–33. doi: 10.1159/000090750. [DOI] [PubMed] [Google Scholar]
- 92.Cui XY, Hu QD, Tekaya M, Shimoda Y, Ang BT, Nie DY, Sun L, Hu WP, Karsak M, Duka T, Takeda Y, Ou LY, Dawe GS, Yu FG, Ahmed S, Jin LH, Schachner M, Watanabe K, Arsenijevic Y, Xiao ZC. NB-3/Notch1 pathway via Deltex1 promotes neural progenitor cell differentiation into oligodendrocytes. J Biol Chem. 2004;279:25858–25865. doi: 10.1074/jbc.M313505200. [DOI] [PubMed] [Google Scholar]
- 93.Ho A, Sudhof TC. Binding of F-spondin to amyloid-beta precursor protein: A candidate amyloid-beta precursor protein ligand that modulates amyloid-beta precursor protein cleavage. Proc Natl Acad Sci USA. 2004;101:2548–2553. doi: 10.1073/pnas.0308655100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Ma QH, Futagawa T, Yang WL, Jiang XD, Zeng L, Takeda Y, Xu RX, Bagnard D, Schachner M, Furley AJ, Karagogeos D, Watanabe K, Dawe GS, Xiao ZC. A TAG1-APP signalling pathway through Fe65 negatively modulates neurogenesis. Nat Cell Biol. 2008;10:283–294. doi: 10.1038/ncb1690. [DOI] [PubMed] [Google Scholar]
- 95.Karagogeos D. Neural GPI-anchored cell adhesion molecules. Front Biosci. 2003;8:1304–1320. doi: 10.2741/1214. [DOI] [PubMed] [Google Scholar]
- 96.Fisher S, Gearhart JD, Oster-Granite ML. Expression of the amyloid precursor protein gene in mouse oocytes and embryos. Proc Natl Acad Sci USA. 1991;88:1779–1782. doi: 10.1073/pnas.88.5.1779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Salbaum JM, Ruddle FH. Embryonic expression pattern of amyloid protein precursor suggests a role in differentiation of specific subsets of neurons. J Exp Zool. 1994;269:116–127. doi: 10.1002/jez.1402690205. [DOI] [PubMed] [Google Scholar]
- 98.Lopez-Sanchez N, Muller U, Frade JM. Lengthening of G2/mitosis in cortical precursors from mice lacking beta-amyloid precursor protein. Neuroscience. 2005;130:51–60. doi: 10.1016/j.neuroscience.2004.09.020. [DOI] [PubMed] [Google Scholar]
- 99.Young-Pearse TL, Bai J, Chang R, Zheng JB, LoTurco JJ, Selkoe DJ. A critical function for beta-amyloid precursor protein in neuronal migration revealed by in utero RNA interference. J Neurosci. 2007;27:14459–14469. doi: 10.1523/JNEUROSCI.4701-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Jin K, Galvan V, Xie L, Mao XO, Gorostiza OF, Bredesen DE, Greenberg DA. Enhanced neurogenesis in Alzheimer's disease transgenic (PDGF-APPSw,Ind) mice. Proc Natl Acad Sci USA. 2004;101:13363–13367. doi: 10.1073/pnas.0403678101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Jin K, Peel AL, Mao XO, Xie L, Cottrell BA, Henshall DC, Greenberg DA. Increased hippocampal neurogenesis in Alzheimer's disease. Proc Natl Acad Sci USA. 2004;101:343–347. doi: 10.1073/pnas.2634794100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Kuhn HG, Cooper-Kuhn M, Boekhoorn K, Lucassen PJ. Changes in neurogenesis in dementia and Alzheimer mouse models: Are they functionally relevant? Eur Arch Psychiatry Clin Neurosci. 2007;257:281–289. doi: 10.1007/s00406-007-0732-4. [DOI] [PubMed] [Google Scholar]