Abstract
Chronic inflammation and inflammatory cytokines have recently been implicated in the development and progression of various types of cancer. In the brain, neuroinflammatory cytokines affect the growth and differentiation of both normal and malignant glial cells, with interleukin 1 (IL-1) shown to be secreted by the majority of glioblastoma cells. Recently, elevated levels of sphingosine kinase 1 (SphK1), but not SphK2, were correlated with a shorter survival prognosis for patients with glioblastoma multiforme. SphK1 is a lipid kinase that produces the pro-growth, anti-apoptotic sphingosine 1-phosphate, which can induce invasion of glioblastoma cells. Here, we show that the expression of IL-1 correlates with the expression of SphK1 in glioblastoma cells, and neutralizing anti-IL-1 antibodies inhibit both the growth and invasion of glioblastoma cells. Furthermore, IL-1 up-regulates SphK1 mRNA levels, protein expression, and activity in both primary human astrocytes and various glioblastoma cell lines; however, it does not affect SphK2 expression. The IL-1-induced SphK1 up-regulation can be blocked by the inhibition of JNK, the overexpression of the dominant-negative c-Jun(TAM67), and the down-regulation of c-Jun expression by small interference RNA. Activation of SphK1 expression by IL-1 occurs on the level of transcription and is mediated via a novel AP-1 element located within the first intron of the sphk1 gene. In summary, our results suggest that SphK1 expression is transcriptionally regulated by IL-1 in glioblastoma cells, and this pathway may be important in regulating survival and invasiveness of glioblastoma cells.
Glioblastoma multiforme (GBM)3 is the most common malignant glioma, which has an extremely invasive phenotype (1, 2). The ability of glioblastoma cells to diffusely infiltrate deeply into healthy brain tissue is the major obstacle for successful treatments, including surgery and radiotherapy, thus leading to the death of the patients within 10–12 months (3, 4). Over the years, ample studies have aimed at understanding the molecular mechanisms of invasion of glioblastoma cells; however, they still remain elusive. Recently, the enhanced expression of several GBM biomarkers, such as EGFR, uPAR, uPA, PAI-1, and MMP-9, attracted significant attention (5–8). More recently, a shorter prognosis for survival of patients with GBM was correlated with the elevated expression of SphK1, an enzyme that generates S1P by phosphorylating sphingosine (9). S1P is a potent lipid mediator of various cell processes, including cell proliferation, differentiation, survival, and migration (10). In the brain, S1P is present at high concentrations (11), and it has been shown to stimulate the migration and invasion of glioblastoma cells (12). S1P can either be secreted by the cells to activate G-coupled S1P receptors (S1P1–5), or act via unknown intracellular processes (10, 13). In addition to SphK1, S1P can also be generated by SphK2, nevertheless the expression of SphK2 does not correlate with prognosis of GBM patients.
In the past decade, it has been shown that the activity of SphK1 is stimulated by various growth factors and cytokines, including epidermal growth factor, tumor necrosis factor-α, IL-1, vascular endothelial growth factor, transforming growth factor-β, and nerve growth factor (10). These factors induce rapid and transient activation of SphK1 and its subsequent translocation to the plasma membrane. In contrast to these observations, knowledge about the transcriptional regulation of the sphk1 gene is limited. The intrinsic expression of SphK1 depends on the specificity protein 1 binding elements located within the 5′-flanking region of the sphk1 gene (14). In addition, the expression of SphK1 is enhanced by epidermal growth factor, IL-1, tumor necrosis factor-α, histamine, prolactin, and glial cell-derived neurotrophic factor in different cell types (15–19); however, molecular mechanisms controlling the transcription of sphk1 gene in response to these factors are not known.
Accordingly, the factors responsible for the enhanced SphK1 expression in GBM patients have not been identified. However, it recently became apparent that inflammation can be directly linked to the development and progression of many cancers (20, 21), with inflammatory cytokines involved in these processes. IL-1 is one of the major regulators of inflammation, immune responses, and is a major neuroinflammatory cytokine in the brain that is released in response to injury or a growing tumor (22, 23). It can induce the secretion of other proinflammatory cytokines, such as IL-6 and IL-8, as well as promote proliferation of glioblastoma cells and astrocytes (24–27). Moreover, IL-1 has been implicated in tumorigenesis, tumor invasiveness, and metastasis of various cancer cells (23, 28, 29). More importantly, glioblastoma cells have recently been shown to secrete substantial, physiologically relevant amounts of IL-1 (30).
We report here that the expression level of IL-1 correlates with the levels of SphK1, but not SphK2 in glioblastoma cell lines. IL-1 also activates the expression of SphK1, but not SphK2 in both glioblastoma cells and primary human astrocytes. We show that the response to IL-1 is mediated by the activation of the JNK-c-jun pathway, and subsequent binding of c-Jun to a novel AP-1 element located within the first intron of the sphk1 gene. Moreover, depletion of IL-1 or SphK1 levels decreases the growth and invasiveness of glioblastoma cells. Therefore, both IL-1 and SphK1 may be considered as future therapeutic targets for GBM.
EXPERIMENTAL PROCEDURES
Cell Culture—Human glioblastoma U373-MG cells were obtained from American Type Culture Collection (Rockville, MD), whereas human glioblastoma A172, U87, U251, and T98G cells were obtained from Dr. Jaharul Haque (Cleveland Clinic Foundation, Cleveland, OH). The U373-TAM67 cells expressing the dominant-negative c-Jun(TAM67) were described previously (31). Human cortical astrocyte cultures were established using dissociated human cerebral tissue established exactly as previously described (32). Cortical tissue was provided by Advanced Bioscience Resources (Alameda, CA), and the protocol for obtaining postmortem fetal neural tissue complied with the federal guidelines for fetal research, and with the Uniformed Anatomical Gift Act. Cells were cultured in Dulbecco's modified Eagle's medium supplemented with 10% fetal calf serum, antibiotics, sodium pyruvate, and non-essential amino acids.
Cytokines and Cell Stimulation—Cells were stimulated with 10 ng/ml IL-1 (a gift from Immunex Corp., Seattle, WA), 5 μm S1P (Biomol Research Laboratories, Plymouth Meeting, PA) or 100 nm phorbol 12-myristate 13-acetate (PMA, Fisher Scientific Inc., Waltham, MA). For inhibitor studies, cells were pretreated with 1 μm SP600125, 10 μm BAY-117082, 10 μm SB202190, 5 μm parthenolide, 5 μg/ml Actinomycin D (Sigma), 1 μm CAY10470, 10 μm JNK peptide inhibitor I, 10 μm, JNK negative control peptide inhibitor I (EMD Biosciences, Inc., San Diego, CA), 10 μm LY294002, 1 μm U0126 (Cell Signaling Technology, Inc., Beverly, MA), 500 nm TAK-1 inhibitor (AnalytiCon Discovery, GmbH, Potsdam, Germany), 10 μm sphingosine kinase-1 inhibitor SK1-I (33), and then stimulated with IL-1 as described in the figure legends.
RNA Preparation and Quantitative PCR—Total RNA was prepared by phenol extraction, exactly as described previously (34). SphK1, SphK2, IL-1β, α1-antichymotrypsin (ACT), c-Jun, and 18 S mRNA levels were measured using TaqMan technology (Applied Biosystems, Foster City, CA), according to the supplier's instructions. Briefly, 1 μg of total RNA was reverse-transcribed using the High-Capacity cDNA archive kit (Applied Biosystems). Subsequently, the cDNAs were diluted 10-fold (Sphk1, SphK2, IL-1β, ACT, and c-jun), or 10,000-fold (18 S rRNA). For real-time PCR, pre-mixed primer-probe sets, and TaqMan Universal PCR Master Mix were purchased from Applied Biosystems and cDNAs were amplified using ABI 7900HT cycler. SphK1a, SphK1c, and glyceraldehyde-3-phosphate dehydrogenase mRNA levels were measured using DyNAmo™ SYBR Green quantitative PCR Kit (New England Biolabs, Inc., Ipswich, MA). To measure the expression of mRNAs for SphK1-spliced isoforms, specific primers located on the boundaries of the exons were designed and cDNA was amplified using ABI 7900HT cycler.
Semiquantitative PCR—The expression of SphK1 isoforms were analyzed by PCR using isoform specific primers, shown below. The PCR products were analyzed by agarose gel electrophoresis after staining with ethidium bromide.
Synthetic Oligonucleotides—The following oligonucleotides were synthesized to amplify the DNA fragments from the 5′-flanking region of the SphK1 gene: (300), 5′-GGGGGTCGACGCAGCTCGTCCCAAGCTC-3′ and 5′-GCTTTCTAGAACCCGGGCGGGAACCAGCTC-3′; (1200), 5′-TAACGTCGACCCGCGGCTCCCGC-3′ and 5′-GCTTTCTAGAACCCGGGCGGGAACCAGCTC-3′; (2400), 5′-GGGGGTCGACGCAGCTCGTCCCAAGCTC-3′ and 5′-GGCATCTAGAGAGATCCAAGTGGCCCGCC-3′; (L3R3), 5′-TCCCTCTAGATCACACCTTGGCAAG-3′ and 5′-CTGGGGATCCAGGTCTCTTCTGGGC-3′; (L4R4), 5′-GGCTTCTAGAAGCCAGAAAAG-3′ and 5′-GTAGGATCCTAAGGGTACAGGAGG-3′; (L5R5), 5′-GAGGTCTAGAAGAGACCTGGGTCC-3′ and 5′-GATGGATCCCCTTCCCACGGTTGC-3′. The mutations within the AP-1 and CRE elements were generated using the following primers: AP-1 mutant, 5′-GCTCTCTAGAATCCGTCGGGCCGGAACC-3′ and 5′-GGATTCTAGAGAGCCCCGTGGCTCCC-3′; CRE mutant, 5′-GGAAGGATCCCGGTGCTCCTGCAGCCAC-3′ and 5′-ACCGGGATCCTTCCCTGCGGCGGCTGG-3′. The AP-1 double-stranded oligonucleotides used in EMSA had the following sequence: 5′-GATCTGGGGCTCTGACTCATCCGTCGA-3′ and 5′-GATCTCGACGGATGAGTCAGAGCCCCA-3′. The expression of SphK1 isoforms was assayed using the following isoform specific primers: (AB-R), 5′-CCTGCCTTCAGCTCCTTATC-3′; (AB-F), 5′-CCGGACCGACTGGGTCCAG-3′; (R), 5′-CCTTGCCGCCGCGCGGG-3′; (A-R), 5′-GGCCGCCCGCTGGATCC-3′; (B-R), 5′-TTCCGCCGCTCAGTGAGCA-3′; (B-F), 5′-GGTTATGGATCCAGTGGTCG-3′; (C-F), 5′-GGGATTTTTACGCAGCTGGAC-3′; and (AC-F), 5′-GGTTATGGATCCAGCGGG CG-3′.
Plasmid Construction—Plasmids pSphK1(300)CAT, pSphK1(1200)CAT, and pSphK1(2400)CAT were generated by cloning the XbaI/SalI-digested PCR products into the XbaI/XhoI sites of ptkCATΔEH. pSphK1(L3R3)CAT, pSphK1(L4R4)CAT, and pSphK1(L5R5)CAT plasmids were generated by cloning the XbaI/BamHI-digested PCR products into the XbaI/BamHI sites of pSphK1(1200)CAT. pSphK1(L4R4)CAT plasmid was utilized as the template to construct plasmids with mutated AP-1 and CRE sites using the QuikChange XL site-directed mutagenesis kit (Stratagene, La Jolla, CA) according to the manufacturer's instructions.
Transient Transfections—Primary human astrocytes were transfected in 12-well clusters using FuGENE6 transfection reagent (Roche Applied Science), with 300 ng of the reporter CAT plasmid and 100 ng of the expression plasmid encoding β-galactosidase. One day after transfection, the cells were stimulated with IL-1 or PMA, cultured another 24 h, and harvested. Extracts were prepared by freeze thawing. Chloramphenicol acetyltransferase (CAT) and β-galactosidase assays were performed as described (35, 36). CAT activities were normalized to β-galactosidase activity and are means ± S.E. (3–5 determinations).
EMSA—Nuclear extracts were prepared as described (37). The oligonucleotides used for EMSA were designed to contain four base-long single-stranded 5′-overhangs at each end after annealing. Double-stranded DNA fragments were labeled by filling in the 5′-protruding ends with Klenow enzyme using [α-32P]dCTP (3000 Ci/mmol). EMSA was carried out according to the published procedures (38). Briefly, 5 μg of nuclear extracts and ∼10 fmol (10,000 cpm) of probe were used. The competition experiment was performed in the presence of a 100-fold excess of the cold oligonucleotides. Polyclonal anti-c-Fos, anti-FosB, anti-Fra1, anti-Fra2, anti-c-Jun, anti-JunB, and anti-JunD antisera were purchased from Santa Cruz Biotechnology (Santa Cruz, CA) and used for the supershift studies.
Western Blotting and Antibodies—Cells were lysed in 10 mm Tris, pH 7.4, 150 mm sodium chloride, 1 mm EDTA, 0.5% Nonidet P-40, 1% Triton X-100, 1 mm sodium orthovanadate, 0.2 mm phenylmethylsulfonyl fluoride, and protease inhibitor mixture (Roche Applied Science). Equal amounts of proteins were resolved using SDS-PAGE and electroblotted onto nitrocellulose membranes (Schleicher & Schuell, Keene, NH). Polyclonal anti-IκBε, anti-c-fos, anti-phospho-c-jun, and anti-tubulin antisera were purchased from Santa Cruz Biotechnology, whereas anti-phospho-p38, anti-phospho-ERK, anti-phospho-JNK, anti-phospho-MKK3/6, anti-IκBα, anti-phospho-IκBα, anti-phospho-IKK, anti-phospho-p65, anti-phospho-ATF-2, and anti-phospho-Akt antisera were purchased from Cell Signaling Technology, Inc. (Beverly, MA). Polyclonal anti-phospho-CREB/anti-phospho-ATF-1 antibodies were purchased from Upstate (Charlottesville, VA). Anti-sphingosine kinase 1 antibodies were described previously (39). Antigen-antibody complexes were visualized by enhanced chemiluminescence according to the manufacturer's instructions (Pierce). The neutralizing anti-IL-1β antibodies were purchased from R&D Systems (Minneapolis, MN).
Down-regulation with siRNA—The expression of p65 and c-jun was down-regulated using SMARTpool siRNAs purchased from Dharmacon, Inc. (Lafayette, CO). SphK1 mRNA was down-regulated with siRNA targeted to a unique hSphK1 sequence as described previously (40). siRNAs were transfected into cells using Dharmafect 1 according to the manufacturer's instructions (Dharmacon, Inc., Lafayette, CO).
Sphingosine Kinase Activity Assay—SphK activity was measured using 10-μg cell lysates with 50 μm sphingosine, in the presence of 0.25% Triton X-100 and [γ-32P]ATP (10 μCi and 1 mm) containing MgCl2 (10 mm) in buffer containing 20 mm Tris (pH 7.4), 10% glycerol, 1 mm 2-mercaptoethanol, 1 mm EDTA, 5 mm sodium orthovanadate, 40 mm β-glycerophosphate, 15 mm NaF, 10 μg/ml leupeptin, aprotinin, and soybean trypsin inhibitor, 1 mm phenylmethylsulfonyl fluoride, and 0.5 mm 4-deoxypyridoxine in a final volume of 200 μl, as described previously (41, 42). S1P was separated by TLC on silica gel G-60 with chloroform/acetone/methanol/acetic acid/water (10:4:3: 2:1, v/v) as a solvent, and the radioactive spots corresponding to S1P were quantified with a FX Molecular Imager (Bio-Rad). Background was determined in the absence of substrate. Activity is expressed as picomoles of S1P formed per min per mg of protein.
Invasion Assay—The invasion of the cells was measured in a modified Boyden chamber, using polycarbonate filters (25 by 80 mm, 12-μm pore size) coated with Matrigel (BD Biosciences). IL-1 was added to both the upper and lower chamber, and the cells were also added to the upper chamber at 5 × 104 cells/well. After 7 h, non-migratory cells on the upper membrane surface were mechanically removed, and the cells that traversed and spread on the lower surface of the filter were fixed and stained with Diff-Quik (Fisher Scientific). The migrated cells were counted with a microscope and a 10× objective. Each data point is the average number of cells in five random fields, and is the average ± S.D. of three individual wells.
Proliferation Assay—The number of viable cells was measured using the WST-1 assay (Roche Applied Science), according to the supplier's instructions.
RESULTS
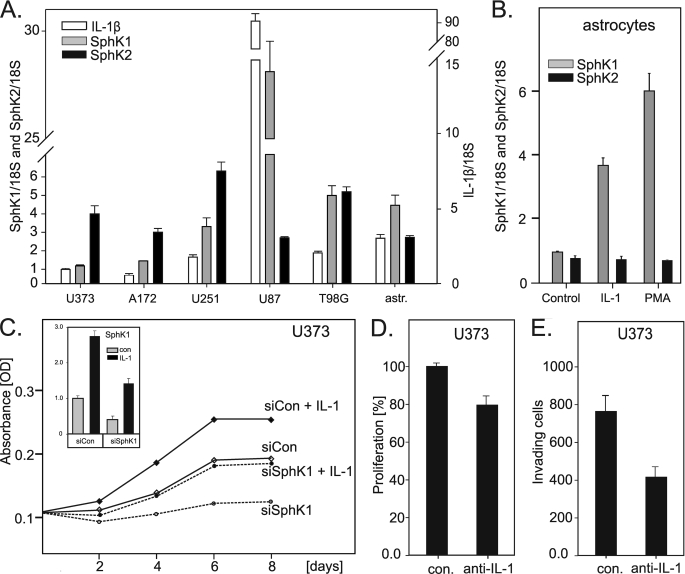
Coexpression of SphK1 and IL-1β in Glioblastoma Cells; Effect of IL-1 on Proliferation and Invasion—Enhanced expression of SphK1, but not SphK2, has been correlated with the shorter survival of glioblastoma patients (9); nevertheless, factors that enhance SphK1 expression in the tumors of these patients have not been identified. Because glioblastoma cells secrete IL-1 and chronic inflammation has been linked to cancer, the expression of SphK1 could be regulated by proinflammatory cytokines, including IL-1. To examine whether IL-1 is responsible for the enhanced expression of SphK1, we analyzed the expression of SphK1, SphK2, and IL-1β in several glioblastoma cell lines, and found drastic differences in their expression (Fig. 1A). More importantly, the enhanced expression of IL-1β correlated with high expression levels of SphK1, but not SphK2. Of note, U87 cells expressed the highest levels of SphK1 and IL-1β, and these cells are fairly invasive in comparison to the other cell lines (43).
FIGURE 1.
Effect of IL-1 on proliferation, invasion, and expression of SphK1. RNA was isolated from untreated cultures of the indicated glioblastoma cell lines or primary human astrocytes, reverse transcribed, and the expression of IL-1β, SphK1, and SphK2 was analyzed by quantitative PCR (A). Data are expressed as a -fold induction after normalization to 18 S RNA. B, primary human astrocytes were stimulated with 10 ng/ml IL-1 or 100 nm PMA for 18 h, and the expression of SphK1 and SphK2 was analyzed as described above. C, U373 cells were transfected with either control or SphK1 siRNA. Subsequently, cells were treated with 10 ng/ml IL-1 as indicated, and the number of viable cells was monitored using a WST-1 assay as described under “Experimental Procedures.” The efficiency of SphK1 down-regulation was analyzed by quantitative PCR (inset). D and E, U373 cells were incubated with 5 μg/ml control or anti-IL-1 neutralizing antibodies for 24 h. The proliferation (D) and the in vitro invasion into Matrigel (E) were analyzed as described under “Experimental Procedures.”
To determine if IL-1 can also induce the expression of SphK1 in primary cells, we analyzed the expression of both SphK1 and SphK2 in primary human astrocytes. As a control, we stimulated cells with PMA, which has previously been shown to activate SphK1 expression in other cell types (44). Similarly to the coexpression of SphK1 and IL-1 in glioblastoma cells, the expression of SphK1 was significantly up-regulated by IL-1 (and PMA) in astrocytes, while the expression of SphK2 was not changed (Fig. 1B); thus suggesting that the activation of SphK1 expression by IL-1 may occur in the brain during inflammation. We conclude that IL-1 is an important regulator of SphK1 expression in human astrocytes and glioblastoma cells.
Because IL-1 is secreted by glioblastoma cells and induces the expression of SphK1, whose product S1P stimulates their growth and invasion, we analyzed whether inhibition of IL-1 and SphK1 affects growth and invasiveness of these cells. We used U373 cells as a model, because their response to S1P is well characterized (12). In fact, IL-1 increased the number of viable U373 cells, while the knockdown of SphK1 had an opposite effect (Fig. 1C). Furthermore, neutralizing antibodies to IL-1β significantly decreased both the number of U373 cells and their invasion (Fig. 1, D and E). Thus, IL-1 is an important factor that can influence glioblastoma cell number and invasion, likely via the activation of Sphk1 expression.
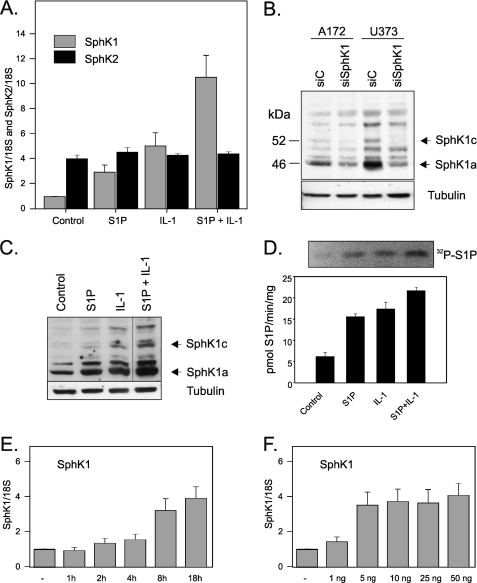
IL-1 Up-regulates SphK1 Expression in Glioblastoma Cells—Subsequently, we used U373 and A172 cells, which express low levels of SphK1 and IL-1β (Fig. 1A), and analyzed the expression of SphK1 and SphK2 in response to IL-1. In addition, we stimulated these cells with S1P to determine if this product of SphK1 can modulate SphK1 expression. We found that both IL-1 and S1P efficiently activated the expression of SphK1 on the mRNA level in U373, and they had an additive effect when used together (Fig. 2A). In contrast to SphK1, SphK2 expression was not affected by IL-1 or S1P. The up-regulation of SphK1 expression by IL-1 was time- and dose-dependent (Fig. 2, E and F, respectively), with a maximum stimulation at 8–18 h. The up-regulation of SphK1 expression on the mRNA level was paralleled by similar changes on the protein level (Fig. 2C) and in the enzymatic activity (Fig. 2D). Because of the presence of nonspecific bands detected by SphK1 antibodies, the identity of SphK1 bands was established by the down-regulation of SphK1 expression using specific siRNA (Fig. 2B). We found that both U373 and A172 cells express two SphK1 isoforms, migrating at 46 and 52 kDa, which likely represent SphK1a and Sphk1c splice variants described previously (45, 46). These data indicate that (i) SphK1 expression is tightly regulated by IL-1, which likely provides a sustained pool of S1P, and (ii) S1P provides additional positive feedback loop, which further enhances expression of SphK1.
FIGURE 2.
IL-1 up-regulates SphK1 expression and activity in glioblastoma cells. U373 cells were stimulated with 10 ng/ml IL-1 or 5 μm S1P for 18 h. A, SphK1 or SphK2 mRNA expression was analyzed using real-time PCR. B, A172 and U373 cells were transfected with either control or SphK1 siRNA. 48 h later, protein lysates were prepared and down-regulation of SphK1 protein expression was measured by Western blotting. C, lysates were prepared, and SphK1 protein level was determined by Western blotting. D, sphingosine kinase assays were performed in whole cell lysates, and bands corresponding to [32P]S1P were separated and quantitated. E, U373 cells were treated with 10 ng/ml IL-1 for the indicated times or for 18 h with indicated concentration of IL-1. F, subsequently, RNA was isolated, reverse-transcribed, and SphK1 mRNA expression was analyzed using real-time PCR (TaqMan). Data shown in the A, E, and F are expressed as -fold induction after normalization to 18 S rRNA.
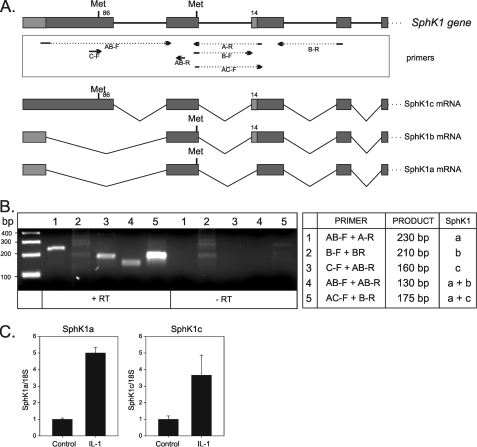
Identification of the SphK1 Isoforms Regulated by IL-1—Multiple isoforms of SphK1 have previously been shown to exist in rats, mice, and humans (45, 46). The gene encoding SphK1 contains five exons and can be alternatively spliced producing SphK1 isoforms: SphK1a, SphK1b, and SphK1c. SphK1a lacks a fragment of the third exon encoding 14 amino acids, which is present in SphK1b and SphK1c, whereas SphK1c possesses an additional 86 amino acids at the N terminus (Fig. 3A). To determine which of the SphK1 isoforms are expressed in U373 cells, isoform-specific primers were designed on the boundaries of the exons to specifically detect different mRNA splice variants (Fig. 3A). We found that both SphK1a and SphK1c isoforms are expressed in U373 cells, whereas the SphK1b was not detected (Fig. 3B). Subsequently, using isoform-specific quantitative PCR, we tested whether IL-1 regulates the expression of both isoforms. In fact, the expression of both SphK1a and SphK1c was similarly up-regulated by IL-1 (Fig. 3C).
FIGURE 3.
IL-1 regulates expression of both isoforms SphK1a and SphK1c expressed in U373 cells. A, structure of the human sphk1 gene and putative spliced isoforms: SphK1a, SphK1b, and SphK1c. Specific primers, located on the boundaries of the exons, were designed, and the expression of SphK1 isoforms was analyzed by PCR. B, ethidium bromide-stained PCR products are shown with the table indicating the primers used, and the expected size of the products. C, U373 cells were stimulated with 10 ng/ml IL-1 for 18 h, and expression of both SphK1a and SphK1c isoforms was determined by real-time PCR using isoform-specific primer sets (numbers 1 and 3, respectively). Data are expressed as -fold induction after normalization to glyceraldehyde-3-phosphate dehydrogenase mRNA.
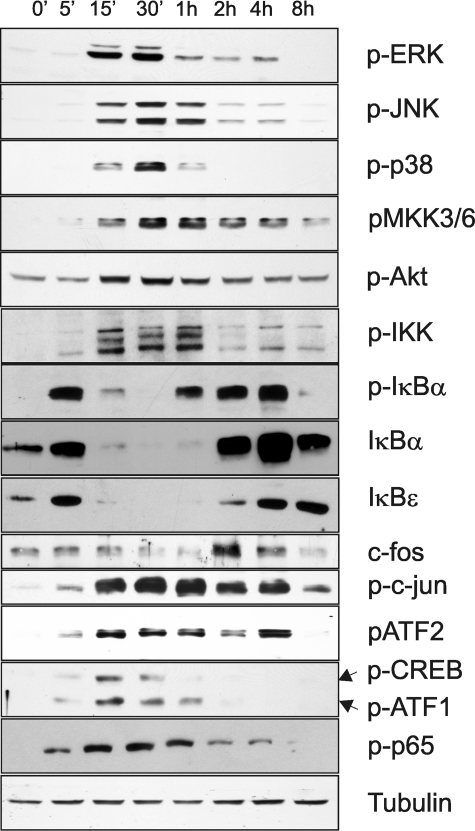
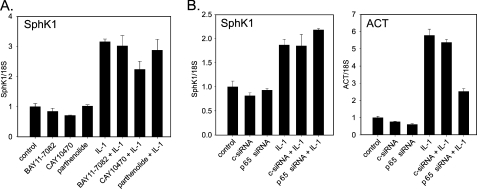
IL-1 Regulates SphK1 Expression via an NF-κB-independent Pathway—IL-1 has been shown to activate multiple signaling cascades in a variety of cell types (32, 47, 48). As expected, IL-1 activated a broad spectrum of signaling molecules and transcription factors in glioblastoma cells (Fig. 4). We found that IL-1 caused rapid phosphorylation of ERK1/2, JNK, p38, MKK3/6, Akt, IKK, c-Jun, ATF-2, ATF-1, CREB, p65, and IκBα, induced the degradation of IκBα and IκBε, and activated the synthesis of c-fos (Fig. 4). Rapid phosphorylation of IKK and IκB proteins, followed by IκBs degradation and phosphorylation of p65 suggested that IKK/NF-κB activation may regulate the expression of SphK1 in response to IL-1. Surprisingly, pharmacological inhibitors of NF-κB activation (BAY11–7082, CAY10470, and parthenolide) did not abolish the induction of SphK1 expression by IL-1 (Fig. 5A). Moreover, the knockdown of p65 expression did not abolish the activation of SphK1 expression by IL-1; however, it diminished the activation of the NF-κB-dependent act gene (Fig. 5B) (47). We conclude that the activation of SphK1 expression by IL-1 is NF-κB-independent.
FIGURE 4.
IL-1 activates multiple signaling pathways in glioblastoma cells. U373 cells were stimulated with 10 ng/ml IL-1 for the indicated time periods, and protein lysates were prepared. The activation of signaling molecules was analyzed by Western blotting using specific antibodies. Tubulin is included as a loading control.
FIGURE 5.
IL-1 regulates SphK1 expression via NF-κB-independent pathway. A, U373 cells were pretreated with 5 μm BAY11–7082, 1 μm CAY10470, or 5 μm parthenolide for 1 h, and subsequently stimulated with 10 ng/ml IL-1 for 18 h. SphK1 mRNA expression was measured using real-time PCR. Data are expressed as -fold induction after normalization to 18 S rRNA. B, U373 cells were transfected with either control siRNA or SMART-pool siRNA to p65, incubated for 48 h, and then stimulated with 10 ng/ml IL-1 for 18 h. RNA was isolated and SphK1 mRNA expression was analyzed as described above. ACT mRNA expression is shown as a positive control for p65 regulation.
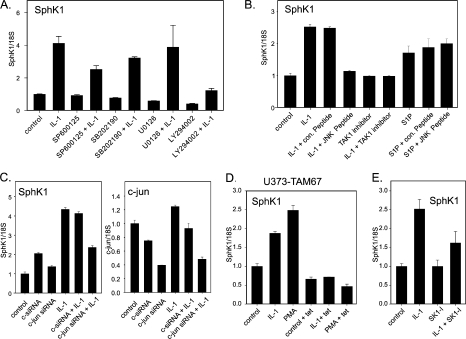
IL-1, but Not S1P, Regulates SphK1 Expression via JNK-c-Jun—To identify the signaling pathway(s) regulating SphK1 expression in response to IL-1, we employed several pharmacological inhibitors to block the signaling pathways activated by IL-1 in U373 cells (Fig. 6A). Inhibition of p38, MEK1/2, and phosphatidylinositol 3-kinase (using SB202190, U0126, and LY294002, respectively) diminished the intrinsic expression of SphK1, but did not affect the relative fold of activation by IL-1. However, the inhibition of JNK (using SP600125) blocked SphK1 up-regulation by IL-1 by ∼45% (Fig. 6A), which was similar to the efficiency of blocking c-Jun phosphorylation (50%, data not shown). We further confirmed the importance of the JNK pathway in the regulation of SphK1 expression using a specific JNK peptide inhibitor, which blocks the activation domain of JNK and prevents phosphorylation of c-Jun. In fact, IL-1 no longer activated SphK1 expression in the presence of this specific peptide, whereas the control peptide was ineffective (Fig. 6B). In contrast, S1P-activated expression of SphK1 was not blocked by JNK peptide inhibitor (Fig. 6B), suggesting an independent mechanism of SphK1 activation by S1P. In addition, we used a TAK-1 inhibitor, which has recently been shown to efficiently block JNK phosphorylation, but not NF-κB activation (49). Inhibition of TAK-1 abolished activation of SphK1 expression by IL-1 (Fig. 6B). Because c-Jun is a target of activated JNK, and it controls the expression of many IL-1-regulated genes (50), we down-regulated c-Jun expression and analyzed activation of SphK1 by IL-1 (Fig. 6C). In agreement with our inhibitor studies, the knockdown of c-Jun expression (by 60%) decreased the IL-1-mediated SphK1 up-regulation (by 50%) (Fig. 6C). These results suggest that the JNK-c-Jun pathway mediates the activation of SphK1 expression in response to IL-1. Subsequently, we used U373-TAM67 cells, which express dominant-negative c-Jun-(TAM67) under the control of tetracycline-regulated promoter (31). In these cells, overexpressed dominant-negative c-Jun(TAM67) efficiently quenches AP-1 and blocks the expression of AP-1-dependent genes (51). Overexpression of c-Jun(TAM67) completely blocked both IL-1- and PMA-induced SphK1 expression, suggesting that AP-1 complex may regulate activation of SphK1 (Fig. 6D).
FIGURE 6.
IL-1 activates SphK1 expression via JNK and c-Jun. A, U373 cells were pretreated with 1 μm SP600125, 10 μm SB202190, 1 μm U0126, or 10μm LY294002 for 1 h, and subsequently stimulated with 10 ng/ml IL-1 for 18 h. SphK1 mRNA expression was measured using real-time PCR. Data are expressed as -fold induction after normalization to 18S rRNA. B, cells were pretreated with either 10 μm JNK peptide inhibitor or 10 μm control peptide for 1 h, and then stimulated with 10 ng/ml IL-1 or 5 μm S1P for 18 h. SphK1 mRNA expression was analyzed as described above. C, cells were transfected with either control or c-jun SMART pool siRNA, incubated for 48 h, and then stimulated with 10 ng/ml IL-1 for 18 h. SphK1 mRNA expression was analyzed as described above. D, U373-TAM67 cells expressing inducible dominant-negative c-Jun(TAM-67) were cultured in the presence of tetracycline for 24 h and then stimulated with 10 ng/ml IL-1 or 100 nm PMA. SphK1 mRNA expression was analyzed as described above. E, U373 cells were pretreated with 10 μm SK1-I for 1 h, and then stimulated with 10 ng/ml IL-1 for 18 h. SphK1 mRNA expression was analyzed as described above.
IL-1 induces rapid phosphorylation of SphK1, leading to the increased enzymatic activity and production of S1P (52). Therefore, we tested whether activation of SphK1 expression is mediated via generated S1P. However, IL-1-induced SphK1 expression was only partially affected by the SK1-I (33), which is a specific SphK1 inhibitor (Fig. 6E). Moreover, S1P activates SphK1 expression independently of JNK (Fig. 6B), and it does not activate the IL-1-responsive SphK1 reporter (Fig. 7C). Thus, IL-1 activates SphK1 expression via a mechanism independent of S1P generation.
FIGURE 7.
Identification of the critical AP-1 binding element within the first intron of the sphk1 gene. A, U373 cells were left untreated or preincubated with 10 ng/ml IL-1 for 18 h, and then treated with 5μg/ml actinomycin D. RNA was isolated at indicated times, reverse-transcribed, and SphK1 mRNA expression was analyzed using real-time PCR. Data are expressed as -fold induction after normalization to 18 S rRNA. B, primary human astrocytes were transiently transfected with the indicated reporter plasmids, and aβ-galactosiordase expression vector. One day after transfection, cells were stimulated with 10 ng/ml IL-1, 5μm S1P, or 100 nm PMA, cultured for an additional 24 h, and harvested. CAT activities were normalized to β-galactosidase activities to account for transfection efficiency. Results are expressed as -fold induction. C, point mutation were introduced into AP-1 and CRE binding sites located in the promoter of SphK1. Primary human astrocytes were transfected with the obtained plasmids and analyzed as described above. D, nuclear extracts were prepared from control and IL-1-treated U373 cells as indicated. DNA binding activity of AP-1 was than analyzed by EMSA using the 32P-labeled oligonucleotide probe (left panel). Specific antibodies or normal rabbit serum (NRS) were added to the binding reaction, and binding was analyzed as described above in control nuclear extract (middle panel) or extract from 4-h IL-1-treated cells (right panel).
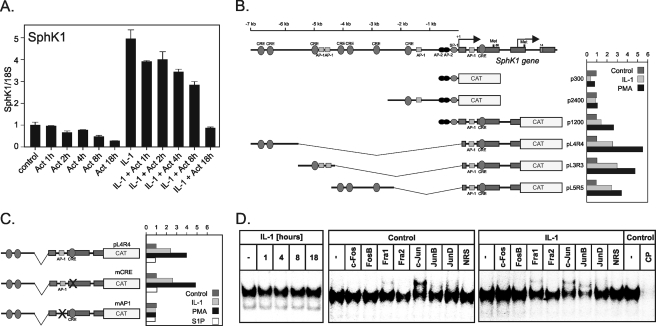
Identification of a Critical AP-1 Element within the First Intron of the sphk1 Gene—SphK1 expression is regulated on the level of transcription by several stimuli, including PMA and nerve growth factor (16, 44). To verify that IL-1 activates SphK1 transcription, we analyzed the time-dependent degradation of SphK1 mRNA in control and IL-1-treated cells exposed to Actinomycin D, which blocks ongoing transcription. Interestingly, the rate of SphK1 mRNA degradation was similar in control and IL-1-treated cells (Fig. 7A), indicating that mRNA stability was not affected by exposure to IL-1.
Because the activation of SphK1 expression by IL-1 was abolished in the presence of c-Jun(TAM67) (Fig. 6D), we screened the 8-kb-long 5′-flanking region of the sphk1 gene for the presence of putative binding sites for the complexes containing c-Jun using the Mat Inspector Program. This analysis resulted in the identification of several putative AP-1 and CREB regulatory elements (Fig. 7B). Subsequently, we generated several reporter constructs covering the entire 8-kb-long 5′-flanking region of the sphk1 gene and the first three exons, and analyzed the responsiveness of these reporters to IL-1 (and PMA) in primary human astrocytes. This analysis yielded the identification of the fragment that was necessary for both IL-1 and PMA response (Fig. 7B). This fragment harbors putative binding sites for AP-1 located in the first intron (+587 to +593), and CREB located in the second exon (+850 to +857). Because both c-Jun and CREB were phosphorylated in response to IL-1 (Fig. 4), we mutated both of these putative regulatory elements to asses their contribution to the IL-1, S1P, and PMA responsiveness (Fig. 7C). The reporter containing mutated AP-1 element was no longer responsive to IL-1 (and PMA), whereas mutation of the CREB element had no effect. These results indicate that the AP-1 element located in the first intron at +587 to +593 is critical for the IL-1-mediated activation of SphK1 expression. Furthermore, the analyzed reporters were not responsive to S1P, suggesting that S1P activates SphK1 expression by different regulatory elements. Importantly, the AP-1 element is constitutively bound by AP-1; although, binding of AP-1 does not increase in response to IL-1 (Fig. 7D). In control cells, the AP-1 complex contains mostly c-Jun, whereas c-Jun and JunB are the major component of the AP-1 complex in the IL-1-treated cells.
DISCUSSION
Sphingosine kinases and S1P have been strongly correlated with progression of many human cancers. Specifically, the expression of SphK1 mRNA is up-regulated in numerous solid tumors, including breast, colon, lung, ovary, stomach, uterus, kidney, rectum (53), and various cell lines such as glioma cells (9). In addition, high levels of SphK1, but not SphK2 correlate with a poor prognosis for patients with glioblastoma multiforme (9). In agreement, high concentrations of S1P have been reported in the brain (11), and S1P has been described to stimulate proliferation, motility, and invasiveness of glioblastoma cells (12, 54).
Many growth factors, cytokines, and other external stimuli have been described to increase SphK1 activity and intracellular concentration of S1P (10). However, little is known about the transcriptional regulation of the SphK1 gene. To date, the induction of SphK1 mRNA expression has been shown to be stimulated by PMA (44), epidermal growth factor (15), nerve growth factor (16), histamine (17), platelet-derived growth factor (18), and glial cell-derived neurotrophic factor (19). In our study, we report for the first time that IL-1 and S1P up-regulate the expression of SphK1 in U373 glioblastoma cell line (Fig. 1A) and primary human astrocytes (Fig. 1B).
Because the mechanism of PMA-induced transcriptional regulation of SphK1 has been described previously, we used PMA as a positive control for our study of the IL-1-mediated regulation of SphK1. Previously, PMA had been shown to regulate SphK1 gene expression in the MEG-O1 cell line (44). These studies indicated that the minimal PMA-responsive element of the sphk1 gene was the specificity protein 1 and activator protein-2-binding sites in the 5′-flanking region. In contrast, in human primary astrocytes PMA-induced SphK1 expression is mediated via AP-1-binding site located in the first intron of the sphk1 gene, and did not require the previously described specificity protein 1 and AP-2-binding sites. The apparent difference in the transcriptional regulation of the sphk1 gene, in response to PMA treatment, may be explained by the different cell types used in these studies.
Gliomas are characterized by extensive areas of necrosis, which are surrounded by highly anaplastic cells. The necrotic cell death triggers inflammation and release of inflammatory mediators, including IL-1, which is one of the major regulators of inflammation. It seems probable that IL-1 available at the site of inflammatory in the tumor, could enhance SphK1 expression and activity; therefore, increasing local production of S1P, which boosts glioma cell proliferation, migration, and invasion. Moreover, it has also been recently published that glioblastoma cells are able to secrete IL-1 (30). In our studies, we show that the high level of SphK1 mRNA in various glioblastoma cell lines is correlated with high level of IL-1 expression (Fig. 3, B and C).
IL-1 has been shown to induce NF-κB signaling in many cell types, and the NF-κB pathway has been shown to be constitutively active in numerous cancers, including gliomas (55). In agreement, we found that IL-1 treatment of U373 activates the NF-κB pathway, manifested by IKK, p65, and IκBα phosphorylation, as well as IκBα and IκBε degradation (Fig. 4). Surprisingly, pharmacological inhibition of the NF-κB pathway by BAY11–7082, CAY10470, and parthenolide had no effect on the IL-1-induced SphK1 expression (Fig. 5A). In agreement, siRNA directed toward p65 also did not ablate the IL-1-induced SphK1 transcriptional regulation (Fig. 5B).
Several lines of evidence suggest that JNK/c-Jun may mediate the IL-1-induced SphK1 transcription in glioma cells: (i) blockade of JNK signaling by a JNK peptide inhibitor blocked SphK1 expression by >50% (Fig. 6B); (ii) inhibition of JNK activity by SP600125 attenuated SphK1 induction (Fig. 6A); (iii) overexpression of dominant-negative c-Jun(TAM67), which lacks the transactivation domain, also blocked the IL-1-induced effects on SphK1 expression (Fig. 6D); (iv) molecular targeting of c-Jun by siRNA blocked SphK1 induction (Fig. 6C); and (v) mutation of the AP-1-binding site in the first intron of the sphk1 gene totally abolished basal, and IL-1 stimulated SphK1 expression (Fig. 7C). EMSA assays confirmed binding of the AP-1 complex to the AP-1 element located at +587 to +593 bp from the transcriptional start site (Fig. 7D). Furthermore, c-Jun is the main component of the AP-1 binding complex in control cells, whereas c-Jun and JunB complexes occupy the AP-1 element of the sphk1 gene after IL-1 stimulation (Fig. 7D). Collectively, our data suggest that SphK1 transcription is a downstream target of IL-1 signaling, and that JNK/c-Jun/AP-1 pathway is indispensable for this transcriptional activation.
Because most GBM therapies remain ineffective, and enhanced SphK1 expression correlates with a poor prognosis for survival, targeting of SphK1 maybe an important additional therapy to combine with existing treatments. Down-regulation of SphK1 expression with siRNA or pharmacological inhibition in glioblastoma cell lines has been shown to significantly decrease the rate of proliferation of glioblastoma cells (9). In agreement, overexpression of dominant-negative SphK1 in MCF7 cells decreases their tumor forming ability in nude mice (56). Additionally, an antibody to S1P has been recently shown to negatively contribute to tumor growth (57). To our knowledge, this is the first demonstration of the molecular mechanism regulating transcription of SphK1 in gliomas, which may provide new clues for possible future therapy targets.
This work was supported, in whole or in part, by National Institutes of Health (NIH) Grant R21NS063283 (to T. K.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
The abbreviations used are: GBM, glioblastoma multiforme; AP-1, -2, activating proteins 1 and 2; CAT, chloramphenicol acetyltransferase; EMSA, electrophoretic mobility shift assay; IL-1, interleukin-1; PMA, phorbol 12-myristate 13 acetate; S1P, sphingosine 1-phosphate; SphK1, sphingosine kinase 1; JNK, c-Jun N-terminal kinase; ACT, α1-antichymotrypsin; siRNA, small interference RNA; CREB, cAMP-response element-binding protein.
References
- 1.Belda-Iniesta, C., de Castro Carpeno, J., Casado Saenz, E., Cejas Guerrero, P., Perona, R., and Gonzalez Baron, M. (2006) Clin. Transl. Oncol. 8 635–641 [DOI] [PubMed] [Google Scholar]
- 2.Gilbertson, R. J., and Rich, J. N. (2007) Nat. Rev. Cancer 7 733–736 [DOI] [PubMed] [Google Scholar]
- 3.Hill, C., Hunter, S. B., and Brat, D. J. (2003) Adv. Anat. Pathol. 10 212–217 [DOI] [PubMed] [Google Scholar]
- 4.Deb, P., Sharma, M. C., Mahapatra, A. K., Agarwal, D., and Sarkar, C. (2005) Neurol. India 53 329–332 [PubMed] [Google Scholar]
- 5.Gondi, C. S., Lakka, S. S., Dinh, D. H., Olivero, W. C., Gujrati, M., and Rao, J. S. (2004) Neuron Glia Biol. 1 165–176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lakka, S. S., Gondi, C. S., Dinh, D. H., Olivero, W. C., Gujrati, M., Rao, V. H., Sioka, C., and Rao, J. S. (2005) J. Biol. Chem. 280 21882–21892 [DOI] [PubMed] [Google Scholar]
- 7.Muracciole, X., Romain, S., Dufour, H., Palmari, J., Chinot, O., Ouafik, L., Grisoli, F., Branger, D. F., and Martin, P. M. (2002) Int. J. Radiat. Oncol. Biol. Phys. 52 592–598 [DOI] [PubMed] [Google Scholar]
- 8.Laerum, O. D., Nygaar, S. J., Steine, S., Mork, S. J., Engebraaten, O., Peraud, A., Kleihues, P., and Ohgaki, H. (2001) J. Neurooncol. 54 1–8 [DOI] [PubMed] [Google Scholar]
- 9.Van Brocklyn, J. R., Jackson, C. A., Pearl, D. K., Kotur, M. S., Snyder, P. J., and Prior, T. W. (2005) J. Neuropathol. Exp. Neurol. 64 695–705 [DOI] [PubMed] [Google Scholar]
- 10.Spiegel, S., and Milstien, S. (2003) Nat. Rev. Mol. Cell. Biol. 4 397–407 [DOI] [PubMed] [Google Scholar]
- 11.Edsall, L. C., and Spiegel, S. (1999) Anal. Biochem. 272 80–86 [DOI] [PubMed] [Google Scholar]
- 12.Van Brocklyn, J. R., Young, N., and Roof, R. (2003) Cancer Lett 199 53–60 [DOI] [PubMed] [Google Scholar]
- 13.Sanchez, T., and Hla, T. (2004) J. Cell. Biochem. 92 913–922 [DOI] [PubMed] [Google Scholar]
- 14.Imamura, T., Miyauchi-Senda, N., Tanaka, S., and Shiota, K. (2004) J. Vet. Med. Sci. 66 1387–1393 [DOI] [PubMed] [Google Scholar]
- 15.Doll, F., Pfeilschifter, J., and Huwiler, A. (2005) Biochim. Biophys. Acta 1738 72–81 [DOI] [PubMed] [Google Scholar]
- 16.Sobue, S., Hagiwara, K., Banno, Y., Tamiya-Koizumi, K., Suzuki, M., Takagi, A., Kojima, T., Asano, H., Nozawa, Y., and Murate, T. (2005) J. Neurochem. 95 940–949 [DOI] [PubMed] [Google Scholar]
- 17.Huwiler, A., Doll, F., Ren, S., Klawitter, S., Greening, A., Romer, I., Bubnova, S., Reinsberg, L., and Pfeilschifter, J. (2006) Biochim. Biophys. Acta 1761 367–376 [DOI] [PubMed] [Google Scholar]
- 18.Francy, J. M., Nag, A., Conroy, E. J., Hengst, J. A., and Yun, J. K. (2007) Biochim. Biophys. Acta 1769 253–265 [DOI] [PubMed] [Google Scholar]
- 19.Murakami, M., Ichihara, M., Sobue, S., Kikuchi, R., Ito, H., Kimura, A., Iwasaki, T., Takagi, A., Kojima, T., Takahashi, M., Suzuki, M., Banno, Y., Nozawa, Y., and Murate, T. (2007) J. Neurochem. 102 1585–1594 [DOI] [PubMed] [Google Scholar]
- 20.Coussens, L. M., and Werb, Z. (2002) Nature 420 860–867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Karin, M., and Greten, F. R. (2005) Nat. Rev. Immunol. 5 749–759 [DOI] [PubMed] [Google Scholar]
- 22.Rothwell, N. J., and Luheshi, G. N. (2000) Trends Neurosci. 23 618–625 [DOI] [PubMed] [Google Scholar]
- 23.Basu, A., Krady, J. K., and Levison, S. W. (2004) J. Neurosci. Res. 78 151–156 [DOI] [PubMed] [Google Scholar]
- 24.Tada, M., and de Tribolet, N. (1993) J. Neurooncol. 17 261–271 [DOI] [PubMed] [Google Scholar]
- 25.Cinque, S., Willems, J., Depraetere, S., Vermeire, L., and Joniau, M. (1992) Immunol. Lett. 34 267–271 [DOI] [PubMed] [Google Scholar]
- 26.Giulian, D., and Lachman, L. B. (1985) Science 228 497–499 [DOI] [PubMed] [Google Scholar]
- 27.Giulian, D., Young, D. G., Woodward, J., Brown, D. C., and Lachman, L. B. (1988) J. Neurosci. 8 709–714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Apte, R. N., Dotan, S., Elkabets, M., White, M. R., Reich, E., Carmi, Y., Song, X., Dvozkin, T., Krelin, Y., and Voronov, E. (2006) Cancer Metastasis Rev. 25 387–408 [DOI] [PubMed] [Google Scholar]
- 29.Apte, R. N., Krelin, Y., Song, X., Dotan, S., Recih, E., Elkabets, M., Carmi, Y., Dvorkin, T., White, R. M., Gayvoronsky, L., Segal, S., and Voronov, E. (2006) Eur. J. Cancer 42 751–759 [DOI] [PubMed] [Google Scholar]
- 30.Lu, T., Tian, L., Han, Y., Vogelbaum, M., and Stark, G. R. (2007) Proc. Natl. Acad. Sci. U. S. A. 104 4365–4370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gopalan, S. M., Wilczynska, K. M., Konik, B. S., Bryan, L., and Kordula, T. (2006) J. Biol. Chem. 281 1956–1963 [DOI] [PubMed] [Google Scholar]
- 32.Wilczynska, K. M., Gopalan, S. M., Bugno, M., Kasza, A., Konik, B. S., Bryan, L., Wright, S., Griswold-Prenner, I., and Kordula, T. (2006) J. Biol. Chem. 281 34955–34964 [DOI] [PubMed] [Google Scholar]
- 33.Paugh, S. W., Paugh, B. S., Rahmani, M., Kapitonov, D., Almenara, J. A., Kordula, T., Milstien, S., Adams, J. K., Zipkin, R. E., Grant, S., and Spiegel, S. (2008) Blood 112 1382–1391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gopalan, S., Kasza, A., Xu, W., Kiss, D. L., Wilczynska, K. M., Rydel, R. E., and Kordula, T. (2005) J. Neurochem. 94 763–773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Seed, B., and Sheen, J. Y. (1988) Gene (Amst.) 67 271–277 [DOI] [PubMed] [Google Scholar]
- 36.Delegeane, A. M., Ferland, L. H., and Mellon, P. L. (1987) Mol. Cell. Biol. 7 3994–4002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Baeuerle, P. A., and Baltimore, D. (1988) Science 242 540–546 [DOI] [PubMed] [Google Scholar]
- 38.Sawadogo, M., Van Dyke, M. W., Gregor, P. D., and Roeder, R. G. (1988) J. Biol. Chem. 263 11985–11993 [PubMed] [Google Scholar]
- 39.Hait, N. C., Sarkar, S., Le Stunff, H., Mikami, A., Maceyka, M., Milstien, S., and Spiegel, S. (2005) J. Biol. Chem. 280 29462–29469 [DOI] [PubMed] [Google Scholar]
- 40.Bektas, M., Jolly, P. S., Muller, C., Eberle, J., Spiegel, S., and Geilen, C. C. (2005) Oncogene 24 178–187 [DOI] [PubMed] [Google Scholar]
- 41.Kohama, T., Olivera, A., Edsall, L., Nagiec, M. M., Dickson, R., and Spiegel, S. (1998) J. Biol. Chem. 273 23722–23728 [DOI] [PubMed] [Google Scholar]
- 42.Paugh, S. W., Payne, S. G., Barbour, S. E., Milstien, S., and Spiegel, S. (2003) FEBS Lett. 554 189–193 [DOI] [PubMed] [Google Scholar]
- 43.Schichor, C., Kerkau, S., Visted, T., Martini, R., Bjerkvig, R., Tonn, J. C., and Goldbrunner, R. (2005) J. Neurooncol. 73 9–18 [DOI] [PubMed] [Google Scholar]
- 44.Nakade, Y., Banno, Y. K. T. K., Hagiwara, K., Sobue, S., Koda, M., Suzuki, M., Kojima, T., Takagi, A., Asano, H., Nozawa, Y., and Murate, T. (2003) Biochim. Biophys. Acta 1635 104–116 [DOI] [PubMed] [Google Scholar]
- 45.Imamura, T., Ohgane, J., Ito, S., Ogawa, T., Hattori, N., Tanaka, S., and Shiota, K. (2001) Genomics 76 117–125 [DOI] [PubMed] [Google Scholar]
- 46.Venkataraman, K., Thangada, S., Michaud, J., Oo, M. L., Ai, Y., Lee, Y. M., Wu, M., Parikh, N. S., Khan, F., Proia, R. L., and Hla, T. (2006) Biochem. J. 397 461–471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kordula, T., Bugno, M., Rydel, R. E., and Travis, J. (2000) J. Neurosci. 20 7510–7516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Eberhardt, W., Huwiler, A., Beck, K. F., Walpen, S., and Pfeilschifter, J. (2000) J. Immunol. 165 5788–5797 [DOI] [PubMed] [Google Scholar]
- 49.Yao, J., Kim, T. W., Qin, J., Jiang, Z., Qian, Y., Xiao, H., Lu, Y., Qian, W., Gulen, M. F., Sizemore, N., DiDonato, J., Sato, S., Akira, S., Su, B., and Li, X. (2007) J. Biol. Chem. 282 6075–6089 [DOI] [PubMed] [Google Scholar]
- 50.Saklatvala, J., Dean, J., and Finch, A. (1999) Biochem. Soc. Symp. 64 63–77 [PubMed] [Google Scholar]
- 51.Gopalan, S. M., Wilczynska, K. M., Konik, B. S., Bryan, L., and Kordula, T. (2006) J. Biol. Chem. 281 13126–13133 [DOI] [PubMed] [Google Scholar]
- 52.Billich, A., Bornancin, F., Mechtcheriakova, D., Natt, F., Huesken, D., and Baumruker, T. (2005) Cell Signal. 17 1203–1217 [DOI] [PubMed] [Google Scholar]
- 53.French, K. J., Schrecengost, R. S., Lee, B. D., Zhuang, Y., Smith, S. N., Eberly, J. L., Yun, J. K., and Smith, C. D. (2003) Cancer Res. 63 5962–5969 [PubMed] [Google Scholar]
- 54.Van Brocklyn, J., Letterle, C., Snyder, P., and Prior, T. (2002) Cancer Lett. 181 195–204 [DOI] [PubMed] [Google Scholar]
- 55.Kiss, D. L., Xu, W., Gopalan, S., Buzanowska, K., Wilczynska, K. M., Rydel, R. E., and Kordula, T. (2005) J. Neurochem. 92 730–738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sukocheva, O. A., Wang, L., Albanese, N., Pitson, S. M., Vadas, M. A., and Xia, P. (2003) Mol. Endocrinol. 17 2002–2012 [DOI] [PubMed] [Google Scholar]
- 57.Visentin, B., Vekich, J. A., Sibbald, B. J., Cavalli, A. L., Moreno, K. M., Matteo, R. G., Garland, W. A., Lu, Y., Yu, S., Hall, H. S., Kundra, V., Mills, G. B., and Sabbadini, R. A. (2006) Cancer Cell 9 225–238 [DOI] [PubMed] [Google Scholar]