Abstract
The secreted trefoil factor family 2 (TFF2) protein contributes to the protection of the gastrointestinal mucosa from injury by strengthening and stabilizing mucin gels, stimulating epithelial restitution, and restraining the associated inflammation. Although trefoil factors have been shown to activate signaling pathways, no cell surface receptor has been directly linked to trefoil peptide signaling. Here we demonstrate the ability of TFF2 peptide to activate signaling via the CXCR4 chemokine receptor in cancer cell lines. We found that both mouse and human TFF2 proteins (at ∼0.5 μm) activate Ca2+ signaling in lymphoblastic Jurkat cells that could be abrogated by receptor desensitization (with SDF-1α) or pretreatment with the specific antagonist AMD3100 or an anti-CXCR4 antibody. TFF2 pretreatment of Jurkat cells decreased Ca2+ rise and chemotactic response to SDF-1α. In addition, the CXCR4-negative gastric epithelial cell line AGS became highly responsive to TFF2 treatment upon expression of the CXCR4 receptor. TFF2-induced activation of mitogen-activated protein kinases in gastric and pancreatic cancer cells, KATO III and AsPC-1, respectively, was also dependent on the presence of the CXCR4 receptor. Finally we demonstrate a distinct proliferative effect of TFF2 protein on an AGS gastric cancer cell line that expresses CXCR4. Overall these data identify CXCR4 as a bona fide signaling receptor for TFF2 and suggest a mechanism through which TFF2 may modulate immune and tumorigenic responses in vivo.
Trefoil factor 2 (TFF2),2 previously known as spasmolytic polypeptide, is a unique member of the trefoil family that is expressed primarily in gastric mucous neck cells and is up-regulated in the setting of chronic inflammation. Experimental induction of ulceration in the rat stomach leads to rapid up-regulation of TFF2 expression with high levels observed 30 min after ulceration with persistence for up to 10 days (1). TFF2 is secreted into the mucus layer of the gastrointestinal tract of mammals where it stabilizes the mucin gel layer and stimulates migration of epithelial cells (2–4), suggesting an important role in restitution and in maintenance of the integrity of the gut. Exogenous administration of recombinant TFF2, either orally or intravenously, provides mucosal protection in several rodent models of acute gastric or intestinal injury (5, 6). A TFF2-/- knock-out mouse model has confirmed the importance of TFF2 in the protection of gastrointestinal mucosa against chronic injury (7).
It is widely accepted that trefoil factors exert their biological action through a cell surface receptor. This suggestion comes from studies on binding of 125I-labeled TFF2 that demonstrated specific binding sites in the gastric glands, intestine, and colon that could be displaced by non-radioactive TFF2 (6, 8–10). Structural studies have revealed potential binding sites for receptors for all members of the trefoil factor family (11, 12). In concordance with this hypothesis, several membrane proteins were found to interact with TFF2. First it was shown that recombinant human TFF2 (and TFF3) could bind to a 28-kDa peptide from membrane fractions of rat jejunum and two human adenocarcinoma cell lines, MCF-7 and Colony-29 (13). Later it was found that recombinant TFF3 fused with biotin selectively bound with a 50-kDa protein from the membrane of rat small intestinal cells (14). However, these 28- and 50-kDa proteins were characterized only by their molecular size without further identification. Two TFF2-binding proteins that have been characterized include a 140-kDa protein, the β subunit of the fibronectin receptor, and a 224-kDa protein called muclin (15). Another TFF2-binding protein was isolated by probing two-dimensional blots of mouse stomach with a murine TFF2 fusion protein, leading to the identification of the gastric foveolar protein blottin, a murine homolog of the human peptide TFIZ1(16). Although these three proteins have now been well characterized, none of them has been shown to mediate responses to TFF2, and no activated signaling cascades have been shown.
Despite the absence of an identified cell surface receptor for TFF2, there is nevertheless clear evidence that TFF2 and TFF3 rapidly activate signal transduction pathways (17, 18). TFF3 prevents cell death via activation of the serine/threonine kinase AKT in colon cancer cell lines (19). The TFF3 protein also activates STAT3 signaling in human colorectal cancer cells, thus providing cells with invasion potential (20). TFF3 treatment leads to EGF receptor activation and β-catenin phosphorylation in HT-29 cells (21) and to transient phosphorylation of ERK1/2 in oral keratinocytes (22). With respect to TFF2, recombinant peptide enhances the migration of human bronchial epithelial cell line BEAS-2B (4). TFF2 has been shown to induce phosphorylation of c-Jun NH2-terminal kinase (JNK) and ERK1/2. Consistent with this observation, the motogenic effect of TFF2 is significantly inhibited by antagonists of ERK kinases and protein kinase C but not by inhibitors of p38 mitogen-activated protein kinase (MAPK). It is believed that the motogenic effect of trefoil factors and of TFF2 in particular, could contribute to in vivo restitution of gastric epithelium by enhancing cell migration.
Although previous studies have suggested that TFF2 functions primarily in cytoprotection, accumulating evidence now suggests that TFF2 may also play a role in the regulation of host immunity. For example, recombinant TFF2 reduces inflammation in rat and mouse models of colitis (23, 24). In addition, TFF2 was detected in rat lymphoid tissues (spleen, lymph nodes, and bone marrow) (25). Recently we and others found TFF2 mRNA expression in primary and secondary lymphopoietic organs (26, 27). These data suggest that TFF2 may play some function in the immune system. In concordance with these findings, we detected an exacerbated inflammatory response to acute injury in TFF2 knock-out animals (27, 28). These observations prompted us to look at the possible function of TFF2 in immune cells. Unexpectedly we found that TFF2 modulates Ca2+ and AKT signaling in lymphoblastic Jurkat cells and that these effects appear to be mediated through the CXCR4 receptor.
EXPERIMENTAL PROCEDURES
Chemicals, Chemokines, Peptides, and Antibodies—The CXCR4 receptor antagonist AMD3100, was purchased from Sigma. The Ca2+-sensing dye Indo-1 and the transfection reagent Lipofectamine 2000 were obtained from Molecular Probes/Invitrogen. The recombinant human chemokine stromal cell-derived factor (SDF)-1α was purchased from R&D Systems (Minneapolis, MN). Unlabeled (azide-free) and phycoerythrin-conjugated anti-human CXCR4 (12G5) and anti-mouse CXCR4 (clone 2B11) antibodies were purchased from BD Pharmingen. 12G5 antibodies recognize conformation-dependent epitope including amino acid 28 in the NH2 terminus, amino acids 179, 181, 182, and 190 in the second extracellular loop (ECL2); and amino acid 274 in the ECL3 (29). Antibody 2B11 was raised to 63 amino acids of the NH2 terminus of CXCR4 receptor (30). Isotypic mouse IgG2a antibodies were from eBioscience (San Diego, CA). Antibodies to total human AKT, Ser-473 (or Thr-308)-phosphorylated AKT, and total and phosphorylated ERK1/2 were purchased from Cell Signaling Technology (Beverly, MA). Antibodies to α-tubulin were purchased from Oncogene Science, Inc. (Cambridge, MA). Rabbit anti-TFF2 antibodies to a carboxyl-terminal peptide (16 amino acids) of hTFF2 (which also recognize mTFF2) were developed previously and characterized in our laboratory (31).
Cell Lines—KATO III, AsPC-1, AGS, NIH3T3, HEK293T/17, and Jurkat E6-1 cancer cell lines were purchased from the American Type Culture Collection (Manassas, VA). AGS, NIH3T3, HEK293T/17, and AsPC-1 cells were cultured in DMEM supplemented with heat-inactivated 10% fetal bovine serum (Invitrogen), 100 units/ml penicillin, and 100 units/ml streptomycin. KATO III and Jurkat cells were cultivated in RPMI 1640 medium with 10% fetal bovine serum. All cell cultures were incubated at 37 °C in a humidified atmosphere (5% CO2). Where indicated, cells were starved in the respective medium with bovine serum albumin (0.5%) but lacking serum overnight.
Cloning and Expression of mTFF2 in Eukaryotic Cells—To express recombinant mouse TFF2 in CHO-K1 and Jurkat cells, we used a retroviral vector, pMIG, which was kindly provided by Dr. David Baltimore (Caltech, Pasadena, CA). This vector contains a multiple cloning site followed by an internal ribosome binding site and the green fluorescent protein (GFP) gene. A strong viral long terminal repeat promoter controls transcription of both the cloned and GFP genes. The coding region of immature mouse TFF2 along with the noncoding 5′ flanking region and EcoRI/SalI flanking restriction sites was amplified from MGC clone (BC050086; Open Biosystems, Baltimore, MD) by PCR and introduced into corresponding restriction sites of the pMIG polylinker. The integrity of the resultant construct, pMIG-mTFF2 (GFP), was verified by sequencing.
The stable CHO-K1 cell line secreting recombinant mouse TFF2 was generated by infection with the pMIG-mTFF2 retrovirus in the presence of Polybrene (5 μg/ml). The GFP-positive pool of cells was collected by flow cytometric sorting. Stable pools of Jurkat cells bearing empty pMIG or pMIG-mTFF2 were selected in identical fashion.
Recombinant TFF2—Recombinant human TFF2 purified from Escherichia coli (>98% pure by SDS-PAGE and high pressure liquid chromatography analysis) was obtained from Peprotech (Rocky Hills, NJ). Human glycosylated TFF2 purified from yeast that have been used in the majority of the published studies on the biological function of TFF2 peptide was kindly provided by Dr. Lars Thim (“Novo Nordisk,” Maaloev, Denmark). For murine TFF2 production, CHO/pMIG-mTFF2 cells were expanded, and the TFF2 was purified from the supernatant of adherent cells by a combination of gel filtration on Sephadex G-50 and ion-exchange chromatography. Homogeneity of the final product was validated through electrophoresis in an 18% polyacrylamide gel under both reducing and nonreducing conditions. The presence of the monomeric form of secreted recombinant TFF2 was confirmed in Western blot analysis by using antibodies developed to the carboxyl end of the human counterpart.
Chemotaxis Assay—For the migration assay we used Jurkat T cells transfected with retroviral vector pMIG (control) or pMIG-TFF2. The latter expresses both TFF2 and GFP proteins from a single bicistronic transcription unit. For the migration assay, control or TFF2-expressing cells were washed, and 3 × 106 cells/ml were suspended in medium containing RPMI 1640 medium with 1% bovine serum albumin with low endotoxin content (Sigma, CAS 9048-46-8). 100-μl aliquots of cell suspensions were applied to the upper chambers of 24-well Transwell plates (Boyden chamber, Costar 3422, 5-μm-diameter pore size). 600 μl of RPMI 1640 medium/BSA supplemented with the indicated concentrations of SDF-1α (R&D Systems) were loaded into the lower chambers. Cells were allowed to migrate for 3 h at 37 °C. After incubation, the porous inserts were removed carefully, and the viable cells were counted using a hemocytometer. The results are expressed as the percentage of cells that migrated to the bottom chamber. Each experiment was performed seven to eight times in triplicate. In experiments with recombinant TFF2, Jurkat cells were applied to the upper chambers with different concentrations of recombinant protein (75–1000 nm) in the upper and lower chamber. 600 μl of RPMI 1640 medium/BSA supplemented with 100 ng/ml SDF-1 were loaded into the lower chambers.
Generation of Stably Transfected AGS Cells Bearing the CXCR4-GFP Chimeric Receptor—The retroviral construct LZRS-CXCR4-GFP-IRES-Zeocin encoding simian CXCR4 was kindly provided by Drs. Eloise Anthony and Peter L. Hordijk (32). Amphotrophic retrovirus was obtained, and virgin AGS cells (5 × 106) were infected with this retrovirus (at a multiplicity of 1:10) as described above. Infected GFP-positive cells (∼3%) were collected by flow cytometric sorting (twice), expanded, and stored in working aliquots in vapors of liquid nitrogen. AGS cells expressing GFP comprised 96% of the final purified cell population.
Measurement of Ca2+ Level by Flow Cytometry—Jurkat cells (2.5 × 106 cells/ml) were resuspended in RPMI 1640 medium containing 0.5% BSA and incubated with the Ca2+-binding dye Indo-1 AM at a final concentration of 5 μm for 1 h at 37 °C in the dark with agitation. Loaded cells were washed, resuspended in Hanks' balanced salt solution medium containing 2 mm CaCl2 and 1 mm MgCl2, and left for 20 min at room temperature. Cells were aliquoted into fluorescence-activated cell sorter tubes that were immediately transferred into a 37 °C water bath for an additional 5 min prior to measurements. Equilibrated cells were then used for flow cytometric analysis of the Ca2+ level using an LSRII machine (BD Biosciences). The base-line intracellular Ca2+ level was recorded for an initial 25–30 s followed by a stimulation with the indicated concentrations of SDF-1α, human or murine TFF2, gastrin, ionomycin, or diluent (phosphate-buffered saline). Data collection was continued at the speed of 2000 events/s for an additional 4–10 min. An increase in binding of cytosolic Ca2+ to Indo-1 results in a change of the emission spectrum of Indo-1 from 510 nm (free form) to 420 nm (Ca2+-bound form). Thus, blue (4′,6-diamidino-2-phenylindole channel, 420 nm) and violet (Indo channel, 510 nm) cell fluorescence was measured, and data were plotted using FlowJo software (version 6.4; Tree Star, Inc.). Intracellular calcium mobilization in response to SDF-1α or recombinant mouse/human TFF2 in the presence of AMD3100 or anti-CXCR antibody was measured after Jurkat cells were preincubated for 40 min at 37 °C with AMD3100 at a concentration of 0.5 μg/ml or with 13 μg/ml 12G5 antibody (eBioscience) accordingly.
Immunodetection of Phosphorylation Status of Proteins—Jurkat cells were washed twice in RPMI 1640 medium, suspended at 10 × 106 cells/ml in the same medium with 0.5% BSA, and starved for 20 h at 37 °C in 5% CO2. Cancer lines AGS, KATO III, or AsPC-1 or recombinant cells AGS/CXCR4 were seeded in 60-mm dishes (5 × 105 cells/dish), grown for 24 h in DMEM supplemented with 10% fetal bovine serum, and starved overnight in DMEM supplemented with 0.5% BSA. Cells were stimulated with of SDF-1 (100 nm) or TFF2 (500 nm) at different time points, washed with ice-cold phosphate-buffered saline, and resuspended in lysis buffer (50 mm Tris-HCl, pH 7.4, 1% Nonidet P-40, 150 mm NaCl, 0.5% sodium deoxycholate, 1 mm phenylmethylsulfonyl fluoride, 10 μg/ml aprotinin, 1 μg/ml leupeptin, 1 μg/ml pepstatin, 2 mm sodium vanadate, 2 mm sodium fluoride, and 0.25 mm sodium pyrophosphate) for 20 min. After centrifugation (15 min at 13,000 rpm), the soluble fractions were kept at -80 °C until use. Protein concentration was detected with a kit (Bio-Rad). Equivalent amounts of protein from each sample were run on 10% Tricine-glycine SDS-PAGE (Invitrogen) and transferred to 0.45-μm nitrocellulose membranes (Bio-Rad) according to standard protocols. The blots were probed with anti-phospho-AKT (Thr-308) or -AKT (Ser-473) and then anti-total AKT or with anti-phospho-ERK1/2 (Thr-202/Tyr-204) mouse monoclonal antibodies followed by anti-total ERK1/2 rabbit polyclonal antibodies. Bound primary antibodies were visualized with appropriate peroxidase-coupled secondary antibodies (Santa Cruz Biotechnology) using Enhanced Luminol Chemiluminescence Reagent (Amersham Biosciences).
Proliferation Assay—To assess a proliferative effect of cytokines on epithelial cells, AGS and AGS/CXCR4 cells were plated at density 12 × 103 cells/well in a 96-well plate in complete DMEM for 24 h. The next day this medium was replaced with DMEM supplemented with 0.1% BSA. The following day various concentrations of cytokines were added to quiescent cells in triplicates for an additional 72 h. At the end cell growth was analyzed using either Cell Counting Kit-8 (WST-8, Dojindo Laboratories, Rockville, MD) or the BrdU Cell Proliferation Assay (Chemicon, Temecula, CA) according to the manufacturer's instructions.
Studies of Binding of Recombinant TFF2 to Jurkat Cells—Binding experiments were performed with 3 × 105 Jurkat cells resuspended in 50 μl of phosphate-buffered saline, 3% BSA containing anti-CXCR4-phycoerythrin monoclonal antibody 2B11 (10 μg/ml) or 12G5 (10 μg/ml) in the presence of SDF-1 or TFF2 (in the concentrations indicated in the figure legends) at 4 °C for 1 h. The cells were washed twice in phosphate-buffered saline, 3% BSA buffer. As a negative control, cells were stained with phycoerythrin-labeled isotype-matched antibodies. Finally the cells were analyzed on an LSRII flow cytometer.
Statistical Analysis—If not specially stated, each experiment was repeated three to eight times. Statistical significance of the differences observed between experimental groups was determined by a two-tailed t test. p values less than 0.05 were considered to be significant.
RESULTS
TFF2 Attenuates SDF-1α/CXCR4-mediated Chemotaxis and Ca2+ Signaling in Jurkat Cells—We and others have previously demonstrated TFF2 mRNA expression in the secondary mouse lymphoid organs, namely the spleen and thymus (25–27). To evaluate possible TFF2 regulation in splenocytes by mitogens, we stimulated mouse splenocytes with T cell and B cell mitogens, concanavalin A and lipopolysaccharide, respectively. Total mRNA was isolated from activated splenocytes, and the level of TFF2 mRNA was analyzed by quantitative real time reverse transcription PCR. This analysis revealed a distinct increase in TFF2 mRNA abundance in response to concanavalin A (40-fold up-regulation), whereas lipopolysaccharide induced a more modest 2.5-fold increase compared with unstimulated cells (data not shown). To study the possibility of TFF2 function in T cells, we used as a model the well characterized Jurkat clone E6-1 T cell line, which does not express endogenous TFF2 mRNA (data not shown). TFF2 gene expression has been shown in a number of malignancies (33), but the absence of TFF2 expression in Jurkat cells provided us with an opportunity to study TFF2 function through stable overexpression. We cloned the TFF2 cDNA (including its own secretory signal) into a retroviral vector (pMIG) that contained an additional marker, GFP, for easier selection of the infected Jurkat cells. Jurkat cells were infected with recombinant retrovirus pMIG-TFF2 (GFP) or empty retrovirus pMIG (GFP). Respective pools of GFP-positive stable clones (>99%; not shown) were collected after sorting by flow cytometry. Western blot analysis revealed secretion of TFF2 protein into the culture medium by the pooled Jurkat clones containing the pMIG-TFF2 retrovirus but not the empty pMIG retrovirus (supplemental Fig. 1A). Although GFP expression was strictly localized to the cytoplasm, the majority of TFF2 protein was found in the culture medium. Under reducing conditions, the secreted recombinant mouse TFF2 migrated in concordance with the expected calculated molecular mass of around 12 kDa for the secreted monomer form. Nonreduced recombinant murine TFF2 migrates slightly slower than the reduced form, and both migrated as a single band (supplemental Fig. 1B). Because no oligomeric forms of recombinant murine TFF2 were detected we suggested that the arrangement of disulfide bonds at the positions of Cys residues and that folding for the recombinant protein were likely to be identical to that of the native protein.
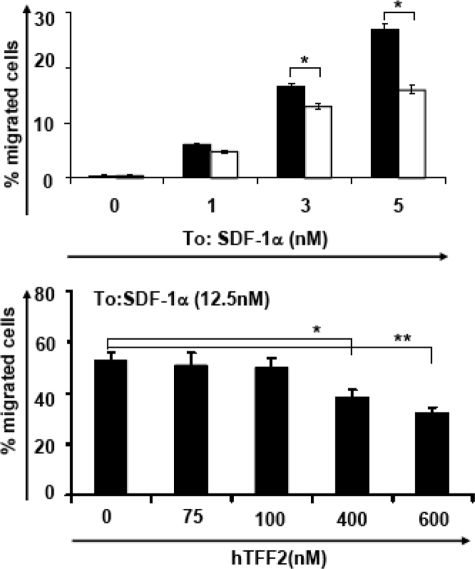
A common and important property of hematopoietic cells is chemotaxis (34). The directional movement of many hematopoietic cells, including T cells, is regulated in part by the chemokine SDF-1α/CXCL12 through the CXCR4 receptor. Therefore, we tested whether TFF2 expression affects SDF-1α/CXCR4-mediated chemotaxis of Jurkat cells. We quantified the migration of Jurkat pMIG and pMIG-mTFF2 cells in response to various concentrations of SDF-1α. As shown in Fig. 1 (top), the pool of TFF2-expressing Jurkat cells migrated ∼30–40% less efficiently than Jurkat cells containing the empty pMIG retrovirus. This inhibitory effect of TFF2 on SDF-1α-mediated migration was statistically significant for all ligand concentrations tested (p < 0.003).
FIGURE 1.
Endogenous and exogenous TFF2 inhibits chemotaxis Jurkat cells to SDF-1α. Top, Jurkat cells bearing empty retrovirus (pMIG; filled bars) or cells expressing mTFF2 (pMIG-mTFF2; empty bars) were tested in chemotactic assays to the indicated concentrations of SDF-1 for 3 h. The relative number of migrated cells versus input cells was calculated (mean ± S. E.) and is shown on the ordinate. Statistically significant differences (p < 0.003) between strains is depicted by asterisks (*). Bottom, parental Jurkat cells were tested in a chemotactic assay to SDF-1α (12.5 nm) as above but in the presence of various concentrations of recombinant mTFF2 prior to (15 min) and during migration in both chambers. Statistically significant differences (*, p < 0.05; **, p < 0.002) between treated and untreated cells are indicated.
To support the notion that TFF2 modulates chemokine-dependent cellular migration through a cell surface mechanism, we tested the effect of exogenous recombinant TFF2 on SDF-1α-dependent chemotaxis of non-transfected, parental Jurkat cells. Purified murine TFF2 was added to Jurkat cells in the upper chamber of a Boyden chamber assay, and Jurkat cells were allowed to migrate toward SDF-1α. A significant (up to 30%) inhibition of SDF-1α-dependent Jurkat cell migration was observed when recombinant murine TFF2 was applied at a concentration 500–600 nm (Fig. 1, bottom).
TFF2 did not affect growth or proliferation of Jurkat cell (not shown), reducing the likelihood of a nonspecific toxic effect. However, we could not further discriminate whether TFF2 affects chemotaxis and/or chemokinesis of Jurkat cells because other chemotactic compounds (like IGF-1, serum, etc.) did not work in this assay. Recombinant TFF2 on its own was unable to stimulate chemotaxis at concentrations ranging from 10 to 1000 nm (data not shown). The inhibitory effect of TFF2 on SDF-1α-dependent chemotaxis suggested the existence of some sort of interaction between TFF2 signaling and CXCR4 signaling in Jurkat cells. One explanation for the interaction might include down-regulation of CXCR4 receptor expression on the surface of Jurkat cells in response to TFF2 stimulation. However, flow cytometry did not reveal any variations in the surface expression of the CXCR4 receptor in Jurkat pMIG-mTFF2 compared with Jurkat pMIG cells (data not shown).
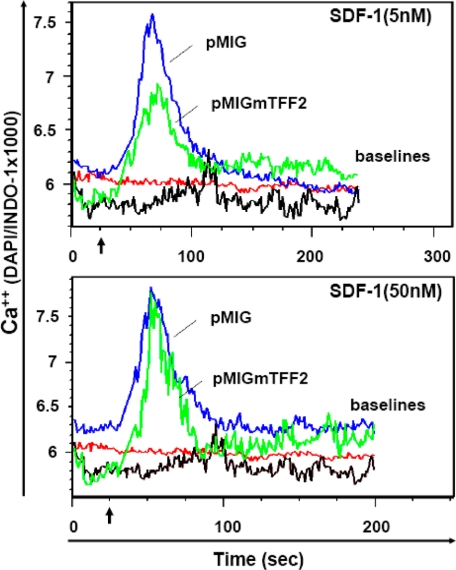
The SDF-1α-induced migration of Jurkat cells is accompanied by the intracellular mobilization of Ca2+ ions (35). Therefore, we tested for a Ca2+ spike in Jurkat cells with or without TFF2 expression in response to SDF-1α stimulation (Fig. 2). TFF2 expression (in pMIG-mTFF2 Jurkat cells) resulted in a marked decrease in the magnitude of calcium flux after stimulation with 5 nm SDF-1α (Fig. 2, top panel). However, this inhibitory effect on calcium signaling was mostly abrogated when a much higher dose of SDF-1α (50 nm) was applied (Fig. 2, bottom panel). The latter observation raises the possibility of competition between two ligands for a common receptor, although heterologous desensitization of CXCR4 signaling by TFF2 through a distinct receptor could not be excluded.
FIGURE 2.
Endogenous TFF2 decreases Ca2+ flux induced by the low but not high dose of SDF-1α in Jurkat cells. Jurkat pMIG (blue line) and Jurkat pMIG-mTFF2 (green line) cells were loaded with Indo-1 (Ca2+-sensing dye) and tested for changes in the intracellular Ca2+ level (blue/violet fluorescence ratio) upon stimulation with 5 nm (top) or 50 nm (bottom) SDF-1α for 4 min by flow cytometry. The same cells challenged with buffer are shown as red (pMIG) or black (pMIG-mTFF2) lines (base lines). Chemokine (or buffer) was added at the time point indicated by arrows. The top and bottom figures represent data from the same experiment. DAPI, 4′,6-diamidino-2-phenylindole.
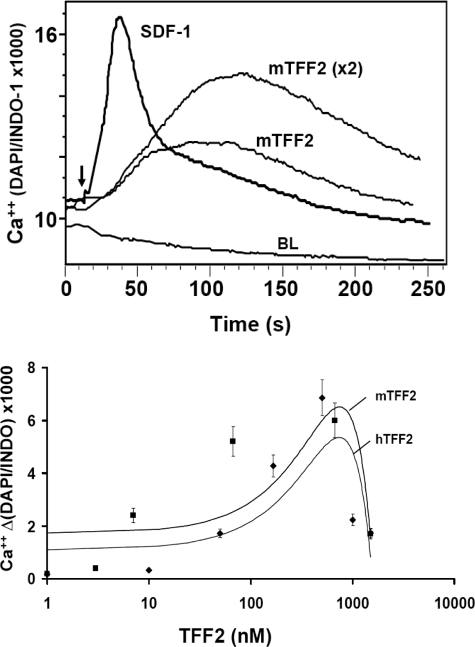
TFF2 Stimulates Calcium Mobilization and Protein Kinase B/AKT Activation in Jurkat Cells—We next tested recombinant TFF2 protein for the ability to directly stimulate signaling pathways in Jurkat cells. To this end, we measured Ca2+ flux in Jurkat cells after exposure to different concentrations of highly purified recombinant mouse TFF2. As shown in Fig. 3 (top), murine TFF2 was able to induce a prominent calcium rise in Jurkat cells with a magnitude comparable to that induced with 12.5 nm SDF-1α. However, to achieve the maximum calcium response, the effective concentration of TFF2 required (500–600 nm) was at least 50 times higher than needed for SDF-1α.In addition, the kinetics of the responses were quite different. The amplitude of calcium release induced by TFF2 was broader than that seen in SDF-1α-treated cells, and the peak calcium rise was achieved in ∼100 s after TFF2 stimulation, whereas it was reached in 40 s after SDF-1α treatment. Additional experiments showed that the TFF2 (but not SDF-1α)-induced Ca2+ spike was completely abrogated when free extracellular Ca2+ ions were depleted with EDTA in testing buffer (not shown). Dose-response studies with recombinant TFF2 show that no significant calcium release was detected at concentrations below 50 nm; a maximum response was observed at 500–600 nm mTFF2, whereas higher peptide doses possessed an inhibitory effect (Fig. 3, bottom).
FIGURE 3.
Recombinant mouse TFF2 stimulates Ca2+ flux in Jurkat cells. Top, cells were loaded with Indo-1 dye and tested for Ca2+ flux by flow cytometry upon stimulation (arrow) with SDF-1α (12.5 nm) or mTFF2 (300 and 600 nm) or buffer (BL) for 4 min. Bottom, Ca2+ flux stimulated with various doses of mouse (rhombus) or human (square) recombinant TFF2 peptides was measured in Jurkat cells by flow cytometry as above. The maximal increases of Ca2+ level above the respective basal levels versus the tested concentrations are shown. The drawn trend lines were created using Microsoft Excel. DAPI, 4′,6-diamidino-2-phenylindole.
Although the recombinant murine TFF2 used in the above studies was homogenous based on analysis in gel electrophoresis (supplemental Fig. 2), we could not completely exclude the possibility that tiny impurities could produce the responses attributed to TFF2. Therefore, as additional controls, we tested two preparations of recombinant human TFF2 that were purified from bacteria or yeast by others. We found that both proteins were able to induce a calcium mobilization in Jurkat cells with the same optimal effective concentration (∼500–600 nm) as recombinant mouse TFF2 (Fig. 3, bottom, and data not shown). In both cases of human TFF2 proteins, we observed the same inhibitory effect at high TFF2 concentrations (>1 μm) on calcium mobilization in Jurkat cells.
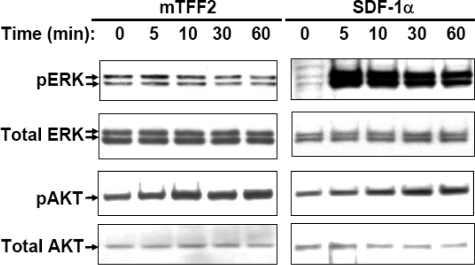
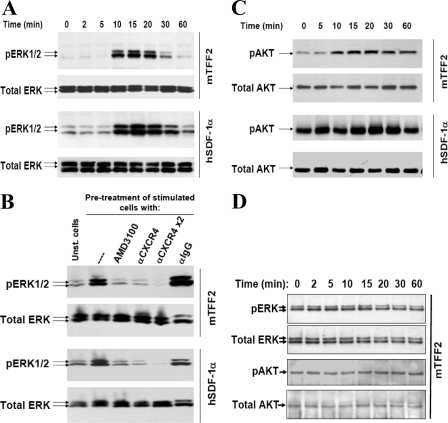
A change of cytoplasmic calcium concentration following a specific stimulus is often considered as primary evidence for receptor-mediated activation of signaling pathways in T cells (36). To further examine downstream TFF2 signaling, we investigated possible activation of protein kinase B/AKT and ERK1/2 kinases in Jurkat cells at various time points following treatment with 600 nm mTFF2. We found that TFF2 stimulated AKT but not ERK1/2 phosphorylation, whereas SDF-1α activated both kinases (Fig. 4). The protein kinase B/AKT (Thr-308) phosphorylation was detected as early as 5 min after TFF2 or SDF-1α challenge in agreement with previous observations (37, 38). Thus, the pattern of kinase activation by TFF2 in Jurkat cells was different from that for SDF-1α. Overall taken together our data indicate some similarities but also important differences in the signaling pathways activated by TFF2 and SDF-1α in Jurkat cells.
FIGURE 4.
Trefoil peptide and SDF-1 activate different signaling pathways in Jurkat cells. Cells were starved in serum-free medium overnight and then stimulated with mTFF2 (600 nm) or SDF-1α (12.5 nm) for the indicated times. Protein extracts were prepared and analyzed for activation/phosphorylation of ERK1/2 or AKT (Thr-308) kinases by Western blot analysis using the corresponding antibodies. Final detection with anti-total ERK1/2 or anti-total AKT antibodies was performed with respective membrane to ensure uniform sample loading. pAKT, phospho-AKT; pERK, phospho-ERK.
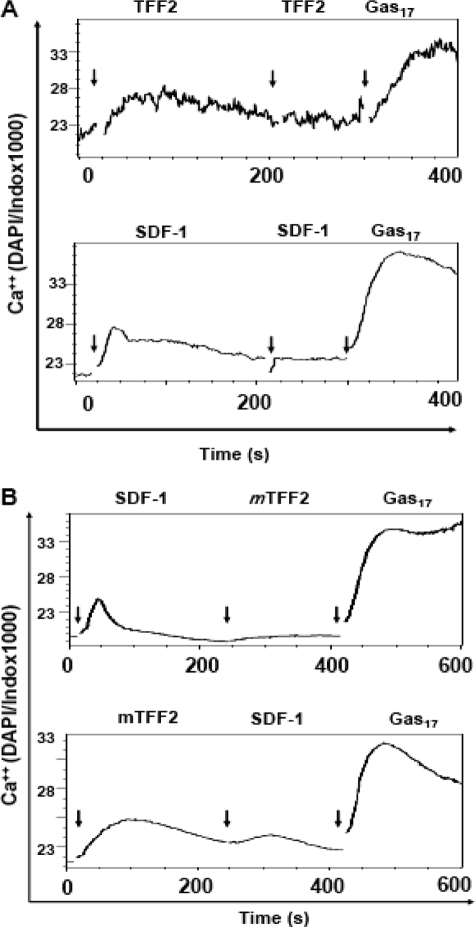
CXCR4 Receptor Is Necessary for TFF2-stimulated Ca2+ Flux in Jurkat Cells—The finding of a TFF2-induced Ca2+ flux in Jurkat cells strongly indicated the presence of a cell surface receptor(s) for the trefoil peptide. Interestingly the finding of an inhibitory effect on calcium rise at high TFF2 concentrations resembles a response pattern described previously for many chemokine family receptor(s) upon ligand stimulation. In addition, the observed competition between exogenous SDF-1α and endogenously expressed TFF2 on Ca2+ flux assay induced by the former (Fig. 3) raised the possibility that CXCR4 receptor might actually be part of the putative TFF2 receptor complex in Jurkat cells. The nine amino-terminal amino acid residues of SDF-1α have been shown to be absolutely required for its chemotactic properties (39). Curiously the amino terminus of the mature TFF2 peptide shows a weak similarity to that of SDF-1α.3 To further establish a link between the putative TFF2 receptor and CXCR4, we measured Ca2+ flux in competition and receptor desensitization experiments. First we treated Jurkat cells repeatedly with SDF-1α or TFF2 at optimal concentrations (Fig. 5A). As expected, we did not detect aCa2+ flux after the second SDF-1α treatment because of receptor desensitization (Fig. 5A, bottom). A similar effect was found for TFF2, strengthening a receptor-dependent mechanism for the observed Ca2+ signaling. Next we treated Jurkat cells with sequential doses of TFF2 (600 nm) and SDF-1α (12.5 nm) and vice versa. In this experiment we also introduced a third (control) treatment of amidated gastrin (Gas17) because the CCK-2 G-protein-coupled receptor is expressed on Jurkat cells (40). Pretreatment of Jurkat cells with TFF2 decreased the Ca2+ flux induced by SDF-1α by 40–60% (Fig. 5B, bottom, and data not shown). In contrast, SDF-1α pretreatment completely abrogated the calcium spike observed in response to TFF2 treatment (Fig. 5B, bottom). Importantly neither TFF2 nor SDF-1 stimulation blocked the Ca2+ rise induced by amidated gastrin, indicating the specificity of the inhibition. Taken together, these data suggest that SDF-1α completely desensitizes TFF2 receptors, whereas TFF2 only partially desensitizes SDF-1α receptor (CXCR4) in Jurkat cells.
FIGURE 5.
Homologous and heterologous desensitization of TFF2- and SDF-1-dependent calcium signaling in Jurkat cells. A, cells were loaded with Ca2+-sensing dye and repeatedly treated with mTFF2 (600 nm; top) or SDF-1 (12.5 nm; bottom) with a final treatment with gastrin (Gas17; 100 nm) at the indicated time points (arrowheads) and in each case followed by the measuring of cellular Ca2+ level by flow cytometry (∼2000 cells/s) for 10 min. B, Ca2+ flux was measured as in A, but cells were successively treated with 12.5 nm SDF-1α and 600 nm mTFF2 (top) or vice versa (bottom) with final stimulation by gastrin (100 nm). Data are shown as curves created by FlowJo software. DAPI, 4′,6-diamidino-2-phenylindole.
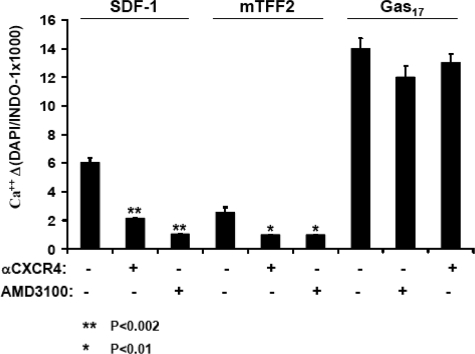
To ascertain a direct involvement of the CXCR4 receptor in TFF2-induced Ca2+ flux, we pretreated Jurkat cells with an anti-CXCR4 antibody (12G5) or AMD3100, a specific small molecule antagonist of the CXCR4 receptor (41). As expected, both pretreatments significantly decreased the magnitude of the Ca2+ response to SDF-1α but not to gastrin stimulation (Fig. 6, left, and data not shown). Curiously neutralization of the CXCR4 receptor also prominently decreased the Ca2+ rise observed in TFF2-challenged cells (Fig. 6, right). However, both pretreatments did not affect calcium mobilization stimulated by gastrin. Overall the simplest explanation for the above results is that TFF2, similar to SDF-1α, interacts directly with the CXCR4 receptor.
FIGURE 6.
Neutralization of the CXCR4 receptor blocks TFF2-induced Ca2+ flux in Jurkat cells. Cells were loaded with Ca2+-sensing dye and treated or not with 12G5 antibodies (5 γ/ml) or antagonist AMD3100 (0.6 μm) for 15 min prior to stimulation with SDF-1α (12.5 nm) or mTFF2 (600 nm). The measurements of calcium ion level were performed by flow cytometry for 4 min. The maximal Ca2+ level was determined for each stimulus with/without treatment and is shown on the abscissa. The indicated treatments did not affect a robust Ca2+ response to gastrin (Gas17). The data shown (mean ± S. E.) are representative of similar data obtained in two additional experiments. DAPI, 4′,6-diamidino-2-phenylindole.
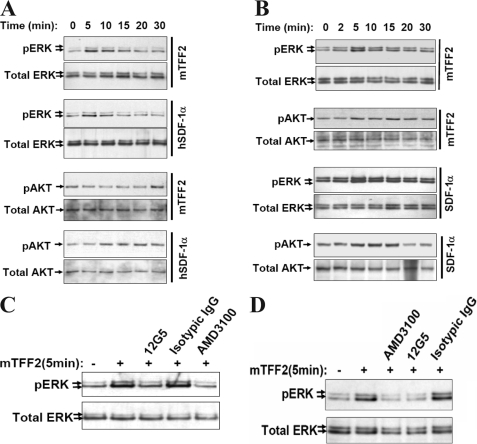
TFF2 Stimulates ERK1/2 and AKT Activation in AGS Cells Engineered to Express CXCR4-GFP Receptor—To demonstrate that CXCR4 receptor expression is sufficient to mediate TFF2 signaling, we generated gastric epithelial AGS cells stably expressing the recombinant CXCR4 receptor fused at its carboxyl terminus to GFP (see “Experimental Procedures”). Previous studies have shown that the CXCR4-GFP fusion product results in a fully functional CXCR4 receptor (32, 42). We chose AGS cells in part because they are unique among a tested panel of gastric cancer cell lines in that they do not endogenously express CXCR4 mRNA.3 In addition, TFF2 is abundantly expressed in stomach and is known to promote restitution of the gastric epithelium. As expected, the unmodified AGS cells did not respond to mTFF2 challenge with any detectable activation of mitogen-activated protein or AKT kinases (Fig. 7D). In contrast, AGS/CXCR4-GFP cells responded to TFF2 treatment with robust phosphorylation of ERK1/2 and protein kinase B/AKT (Fig. 7, A and C, top panels). Interestingly in contrast to our findings in Jurkat cells, the pattern and kinetics of kinase activation by TFF2 in AGS cells closely resembled that seen with SDF-1α treatment (Fig. 7, A and C, bottom panels).
FIGURE 7.
TFF2 activates CXCR4-dependent signaling in AGS cells engineered to express the chemokine receptor. AGS (D) or AGS/CXCR4-GFP (A–C) cells were serum-deprived overnight and tested for phosphorylation of ERK1/2 (A, B, and D) or AKT (C and D) kinases by Western blot with respective phosphospecific antibodies upon stimulation with SDF-1α (12.5 nm) or mTFF2 (600 nm). At the end of the assay, membranes were incubated with correspondent antibodies to total kinases to ensure uniform sample loading. In B, AGS/CXCR4 cells were pretreated with AMD3100 (0.6 μm), anti-CXCR4 antibodies (5 or 10 γ/ml), or isotype matched antibodies (IgG; 10 γ/ml) for 15 min prior to stimulation. pAKT, phospho-AKT; pERK, phospho-ERK; Unst., unstimulated.
To confirm that TFF2 does trigger intracellular signaling in AGS cells through the CXCR4 receptor, we preincubated AGS/CXCR4-GFP cells with the specific CXCR4 inhibitor AMD3100 (0.6 μm for 15 min) or the CXCR4-neutralizing antibody (12G5; 5 and 10 μg/ml for 15 min) prior to mTFF2 or SDF-1α stimulation. We found that CXCR4 receptor neutralization abrogated ERK1/2 activation upon TFF2 and SDF-1α stimulation, whereas the isotype-matched antibody did not affect ERK1/2 activation (Fig. 7B). Taken together, these results clearly show that, similar to SDF-1α, TFF2 activates ERK1/2 and AKT kinases through the CXCR4 receptor.
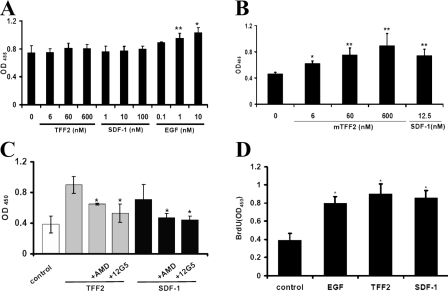
TFF2 Stimulates Proliferation of AGS/CXCR4—Activation of the SDF-1/CXCR4 axis has been shown to stimulate proliferation in several cell lines that is dependent on ERK1/2 activation (43–46). Therefore, we investigated whether the TFF2/CXCR4 axis promotes proliferation of AGS/CXCR4 cells. Initially we evaluated the TFF2 effect on the number of viable AGS or AGS/CXCR4 cells. Cells were starved for 24 h in serum-free medium followed by incubation with either mTFF2, SDF-1α, or EGF for 72 h. The number of viable cells was then assessed using the WST-8 proliferation assay kit according to the manufacturer's instructions. TFF2 and SDF-1α clearly increased the number of viable AGS/CXCR4 cells in a dose-depended manner (Fig. 8B). Importantly pretreatment with AMD3100 or the anti-CXCR4-neutralizing antibody decreased the number of viable AGS/CXCR4-GFP cells following TFF2 or SDF-1 stimulation, implying direct involvement of the CXCR4 receptor in this proliferative/survival effect (Fig. 8C). In contrast, EGF but not TFF2 or SDF-1α increased in a dose-dependent manner the number of viable AGS cells, which lack the CXCR4 receptor (Fig. 8A). These results are in agreement with previous reports that did not observe any effect of trefoil peptides on the growth or [3H]thymidine incorporation in AGS cells (47). To independently determine whether TFF2 does stimulate AGS/CXCR4 cell proliferation we measured BrdUrd incorporation into DNA of these cells in the above experimental settings. We found that TFF2 significantly increased DNA synthesis in AGS/CXCR4 cells (Fig. 8D). Taken together our results showed that TFF2 similarly to SDF-1α promotes proliferation of AGS/CXCR4 cells through the CXCR4 receptor.
FIGURE 8.
TFF2 stimulates proliferation of AGS/CXCR4 cells through the CXCR4 receptor. AGS (A) or AGS/CXCR4 (B) cells (12 × 103 cells/well) were cultivated in triplicate in serum-free DMEM (+0.1% BSA) with or without TFF2, SDF-1, or EGF at the indicated concentrations. The number of viable cells was determined with the WST-8 growth assay kit 72 h later. Each bar represents the mean ± S.E. of three independent measurements. Statistically significant stimulation of growth by cytokines (versus control) is indicated by asterisks (*) above the respective bars (*, p < 0.05; **, p < 0.01). C, neutralization of the CXCR4 receptor with the antagonist AMD3100 (AMD; 0.6 μm) or anti-CXCR4 antibodies (12G5;10 γ/ml) abrogates the growth stimulatory effect of TFF2 (600 nm) and SDF-1 (12.5 nm) in AGS/CXCR4 cells. The growth effect was measured as in A.An asterisk marks a statistically significant blocking effect (p < 0.05) in comparison with untreated but stimulated cells. D, TFF2 (600 nm), SDF-1 (12.5 nm), and EGF (10 ng/ml) stimulate incorporation of BrdUrd (BrdU) in AGS/CXCR4 cells. Cells were plated and grown as above, but BrdUrd incorporation in newly synthesized DNA was assessed 21 h later as described under “Experimental Procedures.” Each bar represents the mean ± S.E. of three replicate determinations. (*, p < 0.05 versus control value).
TFF2 Activates CXCR4 Signaling Pathways in AsPC-1 and KATO III Cancer Cell Lines—Under normal physiologic conditions, TFF2 peptide is most abundant in the stomach and is found at minimal levels in the pancreas (48). Therefore, we next investigated whether TFF2 is able to activate CXCR4 signaling in relevant cancer cell lines that naturally express the CXCR4 chemokine receptor. We selected the human pancreatic cell line AsPC-1 and the gastric carcinoma cell line KATO III for further study. It has been reported that the CXCR4 receptor is functional in the AsPC-1 cell line (49, 50). We were also able to show by flow cytometry that KATO III cells also display CXCR4 receptors on their cell surface (data not shown). Activation of ERK1/2 kinases was observed in both KATO III and AsPC-1 cells treated with mTFF2 (600 nm) or SDF-1 (12.5 nm) (Fig. 9, A and B). In KATO III cells, SDF-1α but not TFF2 was able to induce AKT kinase activation (Fig. 9A). In contrast, both TFF2 and SDF-1α clearly stimulated AKT kinase activity in AsPC-1 cells (Fig. 9B). The CXCR4 antagonist AMD3100 and the neutralizing anti-CXCR4 12G5 antibody completely abrogated ERK1/2 activation after TFF2 and SDF-1α stimulation, whereas the control isotopic antibody did not affect phosphorylation of ERK1/2 in either cell line (Fig. 9, C and D). Thus, TFF2 was able to activate CXCR4 signaling in native epithelial cancer cell lines expressing the endogenous CXCR4 chemokine receptor.
FIGURE 9.
TFF2 activates CXCR4 signaling in cancer cells naturally expressing the chemokine receptor. Gastric KATO III (A and C) and pancreatic AsPC-1 (B and D) cells were tested for ERK1/2 and AKT (Thr-308) phosphorylation upon stimulation with SDF-1 (12.5 nm) or mTFF2 (600 nm) as indicated on the right of each panel by Western blot. C and D, neutralization of the CXCR4 receptor with AMD3100 (0.6 μm) or specific (12G5; 10 γ/ml) but not isotypic antibodies (IgG; 10 γ/ml) blocks ERK1/2 activation by mTFF2 (600 nm; 5 min) in KATO III (C) or AsPC-1 (D) cells. pAKT, phospho-AKT; pERK, phospho-ERK.
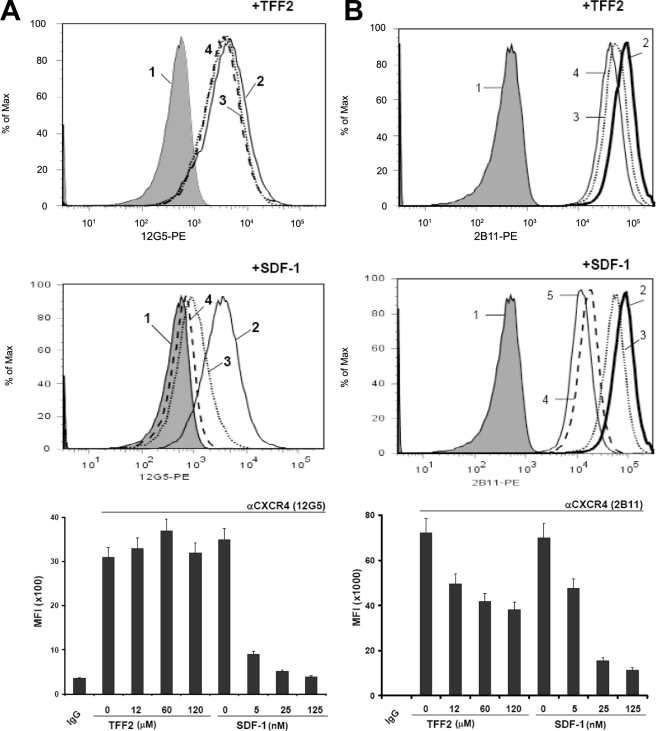
TFF2 Inhibits Binding of the Anti-CXCR4 2B11 Antibody to Jurkat Cells—The ability of TFF2 to recognize the CXCR4 receptor on Jurkat cells was assessed in a binding assay with monoclonal anti-CXCR4 antibodies specific to different portions of the receptor. The 2B11 antibody was raised to the NH2 terminus of the CXCR4 receptor (30), whereas the 12G5 antibody is specific to the conformation-dependent receptor epitopes (29) (see “Experimental Procedures” for details). TFF2 at a concentration of 12 μm significantly (by 40%) inhibited the ability of the 2B11 (phycoerythrin-labeled) antibody to recognize the CXCR4 receptor (Fig. 10B, top panel). In the same assay, SDF-1 at lower (nm) concentrations produced a similar inhibitory effect (Fig. 10B, middle panel). In both cases, the effect was dose-dependent, consistent with a specific interaction between the ligands and the CXCR4 receptor (Fig. 10B, bottom panel). However, TFF2 did not inhibit binding of the CXCR4 receptor with the 12G5 antibody (Fig. 10A, top and bottom panels), which recognizes conformation-dependent epitopes, including primarily amino acids in the second and third extracellular loops (29). However, binding of the 12G5 antibody could be easily inhibited by SDF-1 (Fig. 10A, middle and bottom panels). These data strongly suggest that TFF2 binds with the amino terminus of CXCR4 in contrast to SDF-1, which has multiple binding sites within the CXCR4 receptor. The data also raise the possibility that the observed inhibition of TFF2/CXCR4 signaling by the 12G5 antibody may not necessarily be linked to interference with ligand binding to the CXCR4 receptor.
FIGURE 10.
TFF2 inhibits binding of the anti-CXCR4 2B11 antibody to Jurkat cells. Cells were incubated with phycoerythrin-labeled anti-CXCR4 (lines 2–5), 12G5 (A), or 2B11 (B) or isotype-matched (line 1, tinted area) antibody in the presence of various concentrations of TFF2 (top panels) or SDF-1 (middle panels) for 1 h at 4 °C. At the end of the incubation period, cells were washed and analyzed with flow cytometry. Antibody binding in the presence of the following concentrations of ligands are shown: in A and B (top panels): line 2, no TFF2; line 3, 12 μm TFF2; line 4, 60 μm TFF2; in A and B (middle panels): line 2, no SDF-1; line 3, 5 nm SDF-1; line 4, 25 nm SDF-1; line 5, 125 nm SDF-1. For the purpose of clarity, not all of the obtained curves are shown in the top and middle panels. Bottom panels (A and B) summarize the median fluorescence intensities (MFI) of cell labeling that were calculated using data from the top and middle panels. The data shown (mean ± S. E.) are representative of three independent experiments.
DISCUSSION
In this study, we present several lines of evidence to demonstrate interaction of TFF2 with the CXCR4 receptor. The serendipitous finding of a TFF2 inhibitory effect on SDF-1α-mediated chemotaxis prompted us to further investigate the mechanism for this effect in Jurkat cells. We additionally found that TFF2 itself can stimulate Ca2+ flux and AKT kinase phosphorylation in Jurkat cells. Because neutralization of the CXCR4 receptor strongly dampened the effect of TFF2 on signaling, we reasoned that this chemokine receptor is a necessary component of the TFF2 signaling apparatus in lymphoblastic cells. To further support a direct interaction, we generated the AGS/CXCR4 cell line that stably expresses the CXCR4 receptor. In the CXCR4-expressing AGS cell line, but not the original parent AGS cell line, TFF2 treatment strongly activated phosphorylation of mitogen-activated protein kinase and AKT kinase, promoting proliferation of these cells. Importantly we showed that ERK1/2 activation by TFF2 could be abrogated by antagonism or neutralization of the CXCR4 receptor. In addition, we demonstrated that TFF2 stimulated CXCR4-dependent signaling in gastric and pancreatic cancer cells that naturally express the chemokine receptor. Thus, the CXCR4 receptor is a primary target of TFF2 in both lymphocytic and epithelial cancer cell lines.
Until now, efforts to identify a TFF2 receptor have used primarily biochemical approaches, and findings from these studies have been limited by the absence of evidence linking TFF2-interacting proteins to any downstream signaling pathway that would be consistent with a receptor mechanism (13–15). Because TFF2 is able to activate, through the CXCR4 receptor, signaling cascades that play an essential role in cell migration, proliferation, and survival, we believe that some of the previously described biological effects of TFF2 can be attributed to CXCR4 receptor signaling. First, the motogenic effect of recombinant TFF2 was shown on the human bronchial epithelial cell line BEAS-2B (4), IEC-6 cells (47), breast cancer MCF-7 cells, and MDA-MB-231 cells (33, 51, 52). Curiously all of these cell lines express a functional CXCR4 receptor (53–55). However, signaling pathways were investigated only in BEAS-2B cells, and it was shown that BEAS-2B cells responded to TFF2 by ERK1/2 activation.
Second, several recent studies support the idea of an essential role for CXCR4 in the preservation of mucosal integrity. A functional CXCR4 receptor was found to be expressed in epithelial cell lines and human gastric epithelium (56–58). Our data show that in epithelial cells TFF2 stimulation of CXCR4-expressing cells leads to activation of AKT and mitogen-activated protein kinase signaling cascades, which are generally accepted to be two important survival pathways. Indeed recombinant human TFF2 has been shown to be cytoprotective through in vivo models of gastric injury (23, 59, 60). In one study, during gastric ulcer healing, epithelial CXCR4 expression levels were significantly increased, suggesting that CXCR4 might participate in the maintenance and renewal of the gastric epithelium (61). In the same study, SDF-1 levels were significantly down-regulated during ulcer healing, suggesting that another ligand (e.g. TFF2) might account for CXCR4 stimulation in this setting (58). Thus, both CXCR4 and TFF2 have been shown to be up-regulated during regeneration and repair of the gastric mucosa. In light of our current finding, we would speculate that TFF2 contributes in vivo to the healing of the gastrointestinal epithelium through CXCR4 signaling.
High concentrations of recombinant TFF2 were required to activate CXCR4 receptor signaling in our experiments. However, this is consistent with earlier published work that indicated that trefoil factors activate signaling cascades at high concentrations. Notably in the earlier study by Graness et al. (4), TFF2 protein was able to activate ERK1/2 signaling cascade in BEAS-2B cells but only at high concentration (800 nm). Another member of the trefoil factor family, TFF3 protein, was able to stimulate STAT3 signaling pathway at a similar order (10-7 m) of concentration (20). In more recent studies, ERK1/2 was activated with recombinant TFF3 at a concentration of 0.5 μm (22). Previous reports have indicated that normal physiological levels of TFF2 in vivo are quite high and thus sufficient to stimulate signaling. Indeed levels of TFF2 in the intestinal lumen are significant (around 700 and 500 nm in samples from jejunostomy and ileostomy, respectively) and consistent with high levels found in the gastric lumen (up to 10 μm local concentration) in previous reports (62, 63). These observations suggest that the TFF2-receptor interaction in vivo likely occurs with a somewhat lower affinity than the SDF-1α-CXCR4 interaction. Earlier studies on trefoil factor receptor with radiolabeled native porcine TFF2 and mucosal cell membranes/intestinal cells suggested a low affinity binding with Kd of ∼10-6–10-7 m (64).
In our in vitro binding experiments, TFF2 showed an inhibitory effect on the binding of a monoclonal anti-CXCR4 antibody with specificity for the amino terminus of the CXCR4 receptor, whereas no inhibition of binding was found with an antibody specific for the second and third extracellular loops of CXCR4. This suggests that TFF2 recognizes at a minimum the amino terminus of the CXCR4 receptor, although the TFF2 concentration for the antibody displacement effect (12 μm) appears higher than the concentration required for activation of signaling pathways (600 nm). Interestingly a similar concentration discrepancy in functional and binding assays has been noted previously for antagonist AMD3100, which inhibits only 30% of binding by radiolabeled SDF-1 to CXCR4 (65). It was later shown that AMD3100 inhibits with high affinity functional responses (chemotaxis and calcium mobilization), whereas it inhibits with much lower (∼3000-fold) affinity the binding of radiolabeled SDF-1 to CXCR4 (66).
Although most chemokine receptors bind more than one chemokine, and most chemokines bind to more than one receptor, SDF-1α was until very recently considered as a unique ligand for the CXCR4 receptor (67). Because the SDF-1/CXCR4 signaling axis is essential for chemotaxis, angiogenesis, and hematopoiesis, one would expect tight regulation of this pathway and thus multiple receptors or ligands. Recently several studies have demonstrated that other proteins (human defensin hBD3 and migration inhibitory factor) can function as non-cognate ligands for the CXCR4 receptor (68, 69) despite the fact that their primary structures are quite different from SDF-1 and other chemokines (68, 69). Similarly to TFF2, human defensin hBD3 (abundant intestinal secreted protein) at high (∼3 μm) concentrations inhibits SDF-1-stimulated Ca2+ flux and chemotaxis through the CXCR4 receptor (69). However, in contrast to TFF2, hBD3 inhibits SDF-1-mediated ERK1/2 activation and does not stimulate Ca2+ flux itself while stimulating CXCR4 internalization. Thus, hBD3 acts more as an antagonist than an agonist of the CXCR4 receptor.
TFF2 also differed in several respects from SDF-1 in its signaling through the CXCR4 receptor, and overall TFF2/CXCR4 signaling was found to be cell type-dependent. In gastric epithelial cells, TFF2/CXCR4 signaling was quite similar to SDF-1α/CXCR4, but in the Jurkat lymphoid cell line, TFF2/CXCR4 behaved quite differently. In Jurkat cells, TFF2 stimulated Ca2+ signaling and AKT but not ERK1/2 phosphorylation, inhibited SDF-1-dependent Ca2+ flux and migration, and on its own was unable to stimulate Jurkat migration or proliferation. The fact that TFF2 inhibition of SDF-1 Ca2+ signaling could be overcome at high doses strongly suggests the possibility of a competitive interaction between TFF2 and SDF-1, and overall the data are consistent with TFF2 functioning as a partial agonist at the CXCR4 receptor. The role of TFF2 in the regulation of lymphoid function is not yet clear, although recent work has suggested a role for TFF2 in the modulation of immune responses. The application of recombinant TFF2 has been shown to reduce inflammation in experimental animal models, and TFF2 knock-out mice show increased inflammatory responses. TFF2 is up-regulated in response to inflammation and injury, and thus increased levels might control CXCR4 immune cells that are recruited to the inflamed gut. Although the interaction between SDF-1 and CXCR4 has been thought to exert a homeostatic role for the hematopoietic system, recent data have suggested that the SDF1/CXCR4 axis modulates inflammation and T cell survival (70). It has been shown that SDF-1α recruits leukocytes to sites of inflammation in animal models of arthritis, peritonitis, and acute lung injury (71–75). Recent clinical and experimental data on models of colitis suggest that T cells are involved in sustained colonic inflammation (76). In light of recent published data on the role of the CXCR4/SDF-1 axis in the modulation of immune responses, we suspect that the ameliorating effect of TFF2 on inflammation may be due to the inhibitory effect on T cells. Thus, it is tempting to speculate that TFF2 may inhibit SDF-1-dependent immune cell recruitment or survival and thus suppress or dampen inflammation in the gastrointestinal mucosa.
In conclusion, we have for the first time identified a signaling receptor for the trefoil family peptide TFF2. We show here that TFF2 activated the CXCR4 chemokine receptor and affected proliferation of receptor-expressing epithelial cancer cells. In addition, TFF2 also induced Ca2+ signaling in Jurkat cells through the CXCR4 receptor and modulated responses to SDF-1α. Thus, TFF2 represents a novel addition to the growing class of chemokine-like molecules (68).
Supplementary Material
Acknowledgments
We greatly appreciate the gift of recombinant human TFF2 from Dr. Lars Thim (Novo Nordisk). We also thank Russell Ericksen for careful reading of the manuscript and helpful suggestions.
This work was supported, in whole or in part, by National Institutes of Health Grant 2RO1DK060758 (to T. C. W.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1 and 2.
Footnotes
The abbreviations used are: TFF, trefoil factor family; EGF, epidermal growth factor; ERK, extracellular signal-regulated kinase; DMEM, Dulbecco's modified Eagle's medium; m, mouse; h, human; GFP, green fluorescent protein; CHO, Chinese hamster ovary; BSA, bovine serum albumin; Tricine, N-[2-hydroxy-1,1-bis(hydroxymethyl)ethyl]glycine; SDF, stromal cell-derived factor; PKB/AKT, serine/threonin protein kinase B.
Z. Dubeykovskaya, A. Dubeykovskiy, J. Solal-Cohen, and T. C. Wang, unpublished observation.
References
- 1.Alison, M. R., Chinery, R., Poulsom, R., Ashwood, P., Longcroft, J. M., and Wright, N. A. (1995) J. Pathol. 175 405-414 [DOI] [PubMed] [Google Scholar]
- 2.Hoffmann, W., and Jagla, W. (2002) Int. Rev. Cytol. 213 147-181 [DOI] [PubMed] [Google Scholar]
- 3.Oertel, M., Graness, A., Thim, L., Buhling, F., Kalbacher, H., and Hoffmann, W. (2001) Am. J. Respir. Cell Mol. Biol. 25 418-424 [DOI] [PubMed] [Google Scholar]
- 4.Graness, A., Chwieralski, C. E., Reinhold, D., Thim, L., and Hoffmann, W. (2002) J. Biol. Chem. 277 18440-18446 [DOI] [PubMed] [Google Scholar]
- 5.Babyatsky, M. W., deBeaumont, M., Thim, L., and Podolsky, D. K. (1996) Gastroenterology 110 489-497 [DOI] [PubMed] [Google Scholar]
- 6.Poulsen, S. S., Kissow, H., Hare, K., Hartmann, B., and Thim, L. (2005) Regul. Pept. 126 163-171 [DOI] [PubMed] [Google Scholar]
- 7.Farrell, J. J., Taupin, D., Koh, T. J., Chen, D., Zhao, C. M., Podolsky, D. K., and Wang, T. C. (2002) J. Clin. Investig. 109 193-204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Frandsen, E. K. (1988) Regul. Pept. 20 45-52 [DOI] [PubMed] [Google Scholar]
- 9.Poulsen, S. S., Thulesen, J., Nexo, E., and Thim, L. (1998) Gut 43 240-247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Poulsen, S. S., Thulesen, J., Hartmann, B., Kissow, H. L., Nexo, E., and Thim, L. (2003) Regul. Pept. 115 91-99 [DOI] [PubMed] [Google Scholar]
- 11.Carr, M. D., Bauer, C. J., Gradwell, M. J., and Feeney, J. (1994) Proc. Natl. Acad. Sci. U. S. A. 91 2206-2210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.De, A., Brown, D. G., Gorman, M. A., Carr, M., Sanderson, M. R., and Freemont, P. S. (1994) Proc. Natl. Acad. Sci. U. S. A. 91 1084-1088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chinery, R., and Cox, H. M. (1995) Peptides 16 749-755 [DOI] [PubMed] [Google Scholar]
- 14.Tan, X. D., Hsueh, W., Chang, H., Wei, K. R., and Gonzalez-Crussi, F. (1997) Biochem. Biophys. Res. Commun. 237 673-677 [DOI] [PubMed] [Google Scholar]
- 15.Thim, L., and Mortz, E. (2000) Regul. Pept. 90 61-68 [DOI] [PubMed] [Google Scholar]
- 16.Otto, W. R., Patel, K., McKinnell, I., Evans, M. D., Lee, C. Y., Frith, D., Hanrahan, S., Blight, K., Blin, N., Kayademir, T., Poulsom, R., Jeffery, R., Hunt, T., Wright, N. A., McGregor, F., and Oien, K. A. (2006) Proteomics 6 4235-4245 [DOI] [PubMed] [Google Scholar]
- 17.Emami, S., Rodrigues, S., Rodrigue, C. M., Le Floch, N., Rivat, C., Attoub, S., Bruyneel, E., and Gespach, C. (2004) Peptides 25 885-898 [DOI] [PubMed] [Google Scholar]
- 18.Hoffmann, W. (2005) CMLS Cell. Mol. Life Sci. 62 2932-2938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Taupin, D. R., Kinoshita, K., and Podolsky, D. K. (2000) Proc. Natl. Acad. Sci. U. S. A. 97 799-804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rivat, C., Rodrigues, S., Bruyneel, E., Pietu, G., Robert, A., Redeuilh, G., Bracke, M., Gespach, C., and Attoub, S. (2005) Cancer Res. 65 195-202 [PubMed] [Google Scholar]
- 21.Liu, D., el-Hariry, I., Karayiannakis, A. J., Wilding, J., Chinery, R., Kmiot, W., McCrea, P. D., Gullick, W. J., and Pignatelli, M. (1997) Lab. Investig. 77 557-563 [PubMed] [Google Scholar]
- 22.Storesund, T., Hayashi, K., Kolltveit, K. M., Bryne, M., and Schenck, K. (2008) Eur. J. Oral Sci. 116 135-140 [DOI] [PubMed] [Google Scholar]
- 23.Tran, C. P., Cook, G. A., Yeomans, N. D., Thim, L., and Giraud, A. S. (1999) Gut 44 636-642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Soriano-Izquierdo, A., Gironella, M., Massaguer, A., May, F. E., Salas, A., Sans, M., Poulsom, R., Thim, L., Pique, J. M., and Panes, J. (2004) J. Leukoc. Biol. 75 214-223 [DOI] [PubMed] [Google Scholar]
- 25.Cook, G. A., Familari, M., Thim, L., and Giraud, A. S. (1999) FEBS Lett. 456 155-159 [DOI] [PubMed] [Google Scholar]
- 26.Baus-Loncar, M., Kayademir, T., Takaishi, S., and Wang, T. (2005) CMLS Cell. Mol. Life Sci. 62 2947-2955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kurt-Jones, E. A., Cao, L., Sandor, F., Rogers, A. B., Whary, M. T., Nambiar, P. R., Cerny, A., Bowen, G., Yan, J., Takaishi, S., Chi, A. L., Reed, G., Houghton, J., Fox, J. G., and Wang, T. C. (2007) Infect. Immun. 75 471-480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fox, J. G., Rogers, A. B., Whary, M. T., Ge, Z., Ohtani, M., Jones, E. K., and Wang, T. C. (2007) Am. J. Pathol. 171 1520-1528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kalinkovich, A., Tavor, S., Avigdor, A., Kahn, J., Brill, A., Petit, I., Goichberg, P., Tesio, M., Netzer, N., Naparstek, E., Hardan, I., Nagler, A., Resnick, I., Tsimanis, A., and Lapidot, T. (2006) Cancer Res. 66 11013-11020 [DOI] [PubMed] [Google Scholar]
- 30.Forster, R., Kremmer, E., Schubel, A., Breitfeld, D., Kleinschmidt, A., Nerl, C., Bernhardt, G., and Lipp, M. (1998) J. Immunol. 160 1522-1531 [PubMed] [Google Scholar]
- 31.Tu, S., Chi, A. L., Lim, S., Cui, G., Dubeykovskaya, Z., Ai, W., Fleming, J. V., Takaishi, S., and Wang, T. C. (2007) Am. J. Physiol. 292 G1726-G1737 [DOI] [PubMed] [Google Scholar]
- 32.van Buul, J. D., Voermans, C., van Gelderen, J., Anthony, E. C., van der Schoot, C. E., and Hordijk, P. L. (2003) J. Biol. Chem. 278 30302-30310 [DOI] [PubMed] [Google Scholar]
- 33.May, F. E., Semple, J. I., Prest, S. J., and Westley, B. R. (2004) Peptides 25 865-872 [DOI] [PubMed] [Google Scholar]
- 34.Johnston, B., and Butcher, E. C. (2002) Semin. Immunol. 14 83-92 [DOI] [PubMed] [Google Scholar]
- 35.Majka, M., Ratajczak, J., Kowalska, M. A., and Ratajczak, M. Z. (2000) Eur. J. Haematol. 64 164-172 [DOI] [PubMed] [Google Scholar]
- 36.Quintana, A., Griesemer, D., Schwarz, E. C., and Hoth, M. (2005) Pfluegers Arch. Eur. J. Physiol. 450 1-12 [DOI] [PubMed] [Google Scholar]
- 37.Cherla, R. P., and Ganju, R. K. (2001) J. Immunol. 166 3067-3074 [DOI] [PubMed] [Google Scholar]
- 38.Weber, K. S., Ostermann, G., Zernecke, A., Schroder, A., Klickstein, L. B., and Weber, C. (2001) Mol. Biol. Cell 12 3074-3086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Loetscher, P., Gong, J. H., Dewald, B., Baggiolini, M., and Clark-Lewis, I. (1998) J. Biol. Chem. 273 22279-22283 [DOI] [PubMed] [Google Scholar]
- 40.Pannequin, J., Tantiongco, J. P., Kovac, S., Shulkes, A., and Baldwin, G. S. (2004) J. Endocrinol. 181 315-325 [DOI] [PubMed] [Google Scholar]
- 41.Schols, D., Struyf, S., Van Damme, J., Este, J. A., Henson, G., and De Clercq, E. (1997) J. Exp. Med. 186 1383-1388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tarasova, N. I., Stauber, R. H., and Michejda, C. J. (1998) J. Biol. Chem. 273 15883-15886 [DOI] [PubMed] [Google Scholar]
- 43.Bajetto, A., Barbero, S., Bonavia, R., Piccioli, P., Pirani, P., Florio, T., and Schettini, G. (2001) J. Neurochem. 77 1226-1236 [DOI] [PubMed] [Google Scholar]
- 44.Bajetto, A., Barbieri, F., Dorcaratto, A., Barbero, S., Daga, A., Porcile, C., Ravetti, J. L., Zona, G., Spaziante, R., Corte, G., Schettini, G., and Florio, T. (2006) Neurochem. Int. 49 423-432 [DOI] [PubMed] [Google Scholar]
- 45.Barbero, S., Bonavia, R., Bajetto, A., Porcile, C., Pirani, P., Ravetti, J. L., Zona, G. L., Spaziante, R., Florio, T., and Schettini, G. (2003) Cancer Res. 63 1969-1974 [PubMed] [Google Scholar]
- 46.Porcile, C., Bajetto, A., Barbieri, F., Barbero, S., Bonavia, R., Biglieri, M., Pirani, P., Florio, T., and Schettini, G. (2005) Exp. Cell Res. 308 241-253 [DOI] [PubMed] [Google Scholar]
- 47.Dignass, A., Lynch-Devaney, K., Kindon, H., Thim, L., and Podolsky, D. K. (1994) J. Clin. Investig. 94 376-383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Madsen, J., Nielsen, O., Tornoe, I., Thim, L., and Holmskov, U. (2007) J. Histochem. Cytochem. 55 505-513 [DOI] [PubMed] [Google Scholar]
- 49.Koshiba, T., Hosotani, R., Miyamoto, Y., Ida, J., Tsuji, S., Nakajima, S., Kawaguchi, M., Kobayashi, H., Doi, R., Hori, T., Fujii, N., and Imamura, M. (2000) Clin. Cancer Res. 6 3530-3535 [PubMed] [Google Scholar]
- 50.Marchesi, F., Monti, P., Leone, B. E., Zerbi, A., Vecchi, A., Piemonti, L., Mantovani, A., and Allavena, P. (2004) Cancer Res. 64 8420-8427 [DOI] [PubMed] [Google Scholar]
- 51.Zhao, M., Mueller, B. M., Discipio, R. G., and Schraufstatter, I. U. (2008) Breast Cancer Res. Treat. 110 211-222 [DOI] [PubMed] [Google Scholar]
- 52.Ueda, Y., Neel, N. F., Schutyser, E., Raman, D., and Richmond, A. (2006) Cancer Res. 66 5665-5675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lalani, E. N., Williams, R., Jayaram, Y., Gilbert, C., Chaudhary, K. S., Siu, L. S., Koumarianou, A., Playford, R., and Stamp, G. W. (1999) Lab. Investig. 79 537-546 [PubMed] [Google Scholar]
- 54.Dewan, M. Z., Ahmed, S., Iwasaki, Y., Ohba, K., Toi, M., and Yamamoto, N. (2006) Biomed. Pharmacother. 60 273-276 [DOI] [PubMed] [Google Scholar]
- 55.Eddleston, J., Christiansen, S. C., and Zuraw, B. L. (2002) J. Immunol. 169 6445-6451 [DOI] [PubMed] [Google Scholar]
- 56.Dwinell, M. B., Eckmann, L., Leopard, J. D., Varki, N. M., and Kagnoff, M. F. (1999) Gastroenterology 117 359-367 [DOI] [PubMed] [Google Scholar]
- 57.Jordan, N. J., Kolios, G., Abbot, S. E., Sinai, M. A., Thompson, D. A., Petraki, K., and Westwick, J. (1999) J. Clin. Investig. 104 1061-1069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Norsett, K. G., Laegreid, A., Langaas, M., Worlund, S., Fossmark, R., Waldum, H. L., and Sandvik, A. K. (2005) Physiol. Genomics 22 24-32 [DOI] [PubMed] [Google Scholar]
- 59.Playford, R. J., Marchbank, T., Calnan, D. P., Calam, J., Royston, P., Batten, J. J., and Hansen, H. F. (1995) Gastroenterology 108 92-101 [DOI] [PubMed] [Google Scholar]
- 60.Cook, G. A., Thim, L., Yeomans, N. D., and Giraud, A. S. (1998) J. Gastroenterol. Hepatol. 13 363-370 [DOI] [PubMed] [Google Scholar]
- 61.Akimoto, M., Hashimoto, H., Maeda, A., Shigemoto, M., and Yamashita, K. (2002) Clin. Sci. (Lond.) 103 Suppl. 48, 450S-454S [DOI] [PubMed] [Google Scholar]
- 62.Jeffrey, G. P., Oates, P. S., Wang, T. C., Babyatsky, M. W., and Brand, S. J. (1994) Gastroenterology 106 336-345 [DOI] [PubMed] [Google Scholar]
- 63.Kjellev, S., Vestergaard, E. M., Nexo, E., Thygesen, P., Eghoj, M. S., Jeppesen, P. B., Thim, L., Pedersen, N. B., and Poulsen, S. S. (2007) Peptides 28 1197-1206 [DOI] [PubMed] [Google Scholar]
- 64.Frandsen, E. K., Jorgensen, K. H., and Thim, L. (1986) Regul. Pept. 16 291-297 [DOI] [PubMed] [Google Scholar]
- 65.Banisadr, G., Dicou, E., Berbar, T., Rostene, W., Lombet, A., and Haour, F. (2000) J. Neuroimmunol. 110 151-160 [DOI] [PubMed] [Google Scholar]
- 66.Gupta, S. K., Pillarisetti, K., Thomas, R. A., and Aiyar, N. (2001) Immunol. Lett. 78 29-34 [DOI] [PubMed] [Google Scholar]
- 67.Loetscher, M., Geiser, T., O'Reilly, T., Zwahlen, R., Baggiolini, M., and Moser, B. (1994) J. Biol. Chem. 269 232-237 [PubMed] [Google Scholar]
- 68.Bernhagen, J., Krohn, R., Lue, H., Gregory, J. L., Zernecke, A., Koenen, R. R., Dewor, M., Georgiev, I., Schober, A., Leng, L., Kooistra, T., Fingerle-Rowson, G., Ghezzi, P., Kleemann, R., McColl, S. R., Bucala, R., Hickey, M. J., and Weber, C. (2007) Nat. Med. 13 587-596 [DOI] [PubMed] [Google Scholar]
- 69.Feng, Z., Dubyak, G. R., Lederman, M. M., and Weinberg, A. (2006) J. Immunol. 177 782-786 [DOI] [PubMed] [Google Scholar]
- 70.Nanki, T., Hayashida, K., El-Gabalawy, H. S., Suson, S., Shi, K., Girschick, H. J., Yavuz, S., and Lipsky, P. E. (2000) J. Immunol. 165 6590-6598 [DOI] [PubMed] [Google Scholar]
- 71.Matthys, P., Hatse, S., Vermeire, K., Wuyts, A., Bridger, G., Henson, G. W., De Clercq, E., Billiau, A., and Schols, D. (2001) J. Immunol. 167 4686-4692 [DOI] [PubMed] [Google Scholar]
- 72.Buckley, C. D., Amft, N., Bradfield, P. F., Pilling, D., Ross, E., Arenzana-Seisdedos, F., Amara, A., Curnow, S. J., Lord, J. M., Scheel-Toellner, D., and Salmon, M. (2000) J. Immunol. 165 3423-3429 [DOI] [PubMed] [Google Scholar]
- 73.Kanbe, K., Takagishi, K., and Chen, Q. (2002) Arthritis Rheum. 46 130-137 [DOI] [PubMed] [Google Scholar]
- 74.Blades, M. C., Ingegnoli, F., Wheller, S. K., Manzo, A., Wahid, S., Panayi, G. S., Perretti, M., and Pitzalis, C. (2002) Arthritis Rheum. 46 824-836 [DOI] [PubMed] [Google Scholar]
- 75.Petty, J. M., Sueblinvong, V., Lenox, C. C., Jones, C. C., Cosgrove, G. P., Cool, C. D., Rai, P. R., Brown, K. K., Weiss, D. J., Poynter, M. E., and Suratt, B. T. (2007) J. Immunol. 178 8148-8157 [DOI] [PubMed] [Google Scholar]
- 76.Mikami, S., Nakase, H., Yamamoto, S., Takeda, Y., Yoshino, T., Kasahara, K., Ueno, S., Uza, N., Oishi, S., Fuji, N., Nagasawa, T., and Chiba, T. (2008) J. Pharmacol. Exp. Ther. 327 383-392 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.