Abstract
Studies from budding yeast and ciliates have suggested that telomerase extension of telomeres requires the conventional DNA replication machinery, yet little is known about how DNA replication proteins regulate telomerase action in higher eukaryotic cells. Here we investigate the role of one of the DNA replication factors, flap endonuclease I (FEN1), in regulating telomerase activity in mammalian cells. FEN1 is a nuclease that plays an important role in DNA replication, repair, and recombination. We show that FEN1 is in complex with telomerase in vivo via telomeric DNA. We further demonstrate that FEN1 deficiency in mouse embryonic fibroblasts leads to an increase in telomere end-to-end fusions. In cancer cells, FEN1 deficiency induces gradual shortening of telomeres but does not alter the single-stranded G-overhangs. This is, to our knowledge, the first evidence that FEN1 and telomerase physically co-exist as a complex and that FEN1 can regulate telomerase activity at telomeres in mammalian cells.
Telomeres are distinct DNA-protein structures that protect eukaryotic chromosome ends from degradation and inappropriate recombination or fusions. Maintenance of functional telomeres is essential for long term cell proliferation and stem cell self-renewal. In normal human somatic cells, telomeres progressively shorten each time a cell divides (1). When a subset of telomeres becomes critically short, these short telomeres are recognized as damaged DNA (1–6). Activation of the DNA damage response pathway then induces cellular senescence, impairing cell proliferation (7–9).
To counteract telomere shortening, the cells activate a special reverse transcriptase, telomerase, to elongate short telomeres. Telomerase adds telomeric DNA repeats to chromosome ends, thus keeping telomeres functional (10, 11). Indeed, telomerase is up-regulated in cells that need long term proliferation potential such as embryonic stem cells, germline cells, cancer stem cells, activated lymphocytes, and the majority of human cancer cells (12–16). The critical roles of telomerase in tumor proliferation and stem cell behavior underscore the importance of understanding the regulatory mechanisms for telomerase action at telomeres.
Telomerase elongation of telomeres is a highly coordinated and tightly regulated process, so that the length of the telomeric repeats is kept within a cell type-specific narrow range from 3 to 20 kb in human cells (17). Telomere homeostasis is maintained by a number of proteins associated with the telomere and/or telomerase. These proteins control the recruitment and accessibility of telomerase to telomeres and regulate telomerase activities at telomeres. Defects in certain proteins, among which are DNA metabolic proteins, have been shown to positively or negatively influence telomere length (18).
Several studies in yeast and ciliates have suggested that telomerase-dependent telomere extension is coupled with conventional DNA replication and requires certain DNA replication proteins. For example, inactivation of components of budding yeast DNA replication machinery such as polymerase α (polα),2 primase, and polymerase δ (polδ) abolishes de novo addition of telomeric DNA by telomerase (19). Certain temperature-sensitive mutations in budding yeast polα or the large subunit of replication factor C cause uncontrolled telomerase-dependent telomere elongation (20–22). Consistent with these observations, budding yeast polα physically interacts with Cdc13p, a protein that directly interacts with Est1p and regulates telomerase activity at yeast telomeres (23). Similarly, mutations in polα/primase and polδ in fission yeast lead to abnormal lengthening of telomeres, and polα interacts with the telomerase catalytic subunit, Trt1 (24). In ciliate Euplotes crassus, it has been demonstrated that telomerase physically interacts with primase and proliferating cell nuclear antigen (25). Additionally, partial inhibition of polα and polδ by aphidicolin causes C-strand and G-strand telomere heterogeneity in Euplotes (26). Together, these results suggest that in lower eukaryotes, telomerase action at telomeres is tightly regulated by activities of DNA replication proteins. However, research in higher eukaryotes on how telomerase couples with conventional DNA replication to maintain telomeres is lacking.
The flap endonuclease 1 (FEN1) is an evolutionarily conserved component of the DNA replication machinery from archaebacteria to humans (27, 28). It is a multifunctional structure-specific nuclease containing flap endonuclease activity (29), 5′ → 3′ exonuclease activity (29, 30), and gap-dependent endonuclease activity (31, 32). FEN1 is required for Okazaki fragment processing and maturation during lagging strand DNA synthesis and long patch DNA base excision repair. Its exonuclease activity is important for processing DNA ends during homologous recombination (31). Deficiency in FEN1 leads to an increase in genome instability and tumorigenesis (33, 34). Many cancers have been found to carry somatic mutations in the FEN1 nuclease domain (33). Therefore, FEN1 plays a vital role in maintaining genome stability.
Several studies have revealed an important role for FEN1 in maintaining telomere integrity. Deletion of FEN1 in yeast results in high telomere instability, characterized by heterogeneous telomere length and a sharp increase in the amount of single-stranded G-overhangs (35, 36). In telomerase-negative normal human somatic cells, FEN1 has been shown to associate with telomeres (37, 38). Deficiency in FEN1 leads to loss of telomeres replicated by the lagging strand synthesis (38), suggesting that FEN1 is important for a faithful replication of lagging strand telomere DNA. However, it is unknown whether FEN1 is important for regulating telomerase-mediated telomere maintenance in cancer cells.
Considering the role of FEN1 in DNA replication and repair, we hypothesized that functional FEN1 might be required for telomerase-mediated telomere maintenance. We now provide the first evidence that FEN1 is in complex with the telomerase catalytic subunit, hTERT. Mammalian cells expressing nuclease-deficient FEN1 displayed increased telomere instability, and FEN1 deficiency induced gradual shortening of telomeres in cancer cells. We propose that FEN1 plays an important role in regulating telomerase activity at telomeres.
EXPERIMENTAL PROCEDURES
Cell Culture—Mouse embryonic fibroblasts (MEFs) were cultured at 37 °C under 5% CO2 in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum and 2-mercaptoethanol. All other cells were cultured under similar conditions in a 4:1 mixture of Dulbecco's modified Eagle's medium and medium 199 supplemented with 10% cosmic calf serum (Hyclone).
Antibodies—The following primary antibodies were used: monoclonal anti-FEN1 and mouse anti-actin (BD Biosciences), rabbit polyclonal anti-FEN1 (Bethyl), rabbit polyclonal anti-hTERT (Rockland Inc., it only recognizes exogenously overexpressed hTERT), monoclonal anti-Myc 9E10 (Santa Cruz), monoclonal anti-FLAG (Sigma). Secondary antibodies were: horseradish peroxidase-conjugated anti-mouse (BD Biosciences) or anti-rabbit IgG (Bethyl).
Construction of Plasmids—Myc tag was added at the N terminus of FEN1 cDNA by PCR cloning. The primers used to amplify FEN1 were 5′-ATC GAT GCT AGC ATG GAA CAA AAA CTC ATC TCA GAA GAG GAT CTG AAT ATG GGA ATT CAA GGC CTG GCC AA-3′ and 5′-CCC GGG TTA TTT TCC CCT TTT AAA CTT C-3′. PCR was performed at 94 °C for 2 min, 30 cycles of 94 °C for 30 s, 55 °C for 30 s, and 72 °C for 1 min followed by extension at 72 °C for 10 min. The DNA product was gel-purified, subjected to NheI and SmaI digestion (sites are underlined), and then cloned into an NheI- and SmaI-digested pCI-neo vector (Promega). The cloned sequences were subjected to DNA sequencing to ensure that no mutations were introduced.
Telomere Repeat Amplification Protocol (TRAP) Analysis—Nonradioactive TRAP was performed as described (39) except that Cy-3-labeled primer was used to replace Cy-5-labeled primer and 28 cycles of PCR (95 °C for 30 s, 52 °C for 30 s, and 72 °C for 30 s) were used to amplify telomerase-extended product. The products were separated on 10% polyacrylamide gels and visualized with an FX Molecular Imager (Bio-Rad) or a Storm PhosphorImager 860 (GE Healthcare) using a blue laser. Relative telomerase activity was estimated using ImageQuant software (GE Healthcare) by determining the ratio of the 36-bp internal standard to the 6-bp telomerase-specific ladder.
Co-immunoprecipitation-TRAP—Co-IP-TRAP was performed as described (39) except that 3 μg of each antibody was coupled to protein A/G+-agarose beads (Santa Cruz Biotechnology) by incubating overnight at 4 °C with constant rotation. Antibodies were used for IP of proteins from HeLa cell lysate corresponding to 500,000 cells. After IP, the agarose bead pellets were resuspended with lysis buffer in a final volume of 40 μl, and 2 μl (equivalent to 25,000 cells) were used for nonradioactive TRAP analysis to detect telomerase activity. To test the specificity of FEN1·telomerase co-immunoprecipitation, 3 μg of anti-FEN1 blocking peptide (Bethyl) or an irrelevant blocking peptide (which blocks anti-GAPDH and GAPDH interaction but does not block interaction between anti-FEN1 antibody and FEN1 protein) was added to lysate during IP.
Co-immunoprecipitation—HeLa cells were transfected with a total of 10 μg of plasmid DNA using FuGENE HD transfection reagent (Roche Applied Science) according to the manufacturer's instructions as follows: 5 μg of pCI-neo vector + 5 μg of pcDNA3.1-hTERT, 5 μg of pCI-neo-Myc-FEN1 + 5 μg of pcDNA3.1-hTERT, 5 μg of pCI-neo vector + 5 μg of pCR3-FLAG-hTERT, or 5 μg of pCI-neo-Myc-FEN1 + 5 μg of pCR3-FLAG-hTERT. The cells were harvested 18 h after transfection, and the cell pellets were washed once with cold phosphate-buffered saline, resuspended in lysis buffer (50 mm Tris-HCl, pH 7.4, 50 mm NaCl, 0.1% Nonidet P-40, 2 mm dithiothreitol) supplemented with EDTA-free complete protease inhibitor (Roche Applied Science), sonicated on ice (50 J/watt s, three 5-s pulses), and centrifuged (13,000 rpm, 15 min, 4 °C). The resulting supernatants were first precleared by incubating with 20 μl of 50% slurry of protein G-agarose beads for 2 h at 4 °C with constant rotation. After brief centrifugation, precleared lysates were transferred to new tubes and used for IP. Anti-Myc (3 μg) antibody was coupled to 20 μl of the 50% slurry of protein G-agarose beads by incubating for 1 h at 4 °C with constant rotation. Antibody-coated beads were washed once with lysis buffer prior to use in IP reactions. Precleared lysates were treated with or without DNase I (136 units/ml, RNase-free; Roche Applied Science) or RNase A (100 μg/ml, DNase-free; Roche Applied Science) and incubated with antibody-coupled beads in the presence of 50 μg of bovine serum albumin for overnight at 4 °C with constant rotation. The bead pellets were then washed five times with lysis buffer (400 μl for 10 min with rotation at 4 °C), then resuspended in Laemmli buffer, boiled for 5 min, and used immediately on 7% SDS-PAGE for immunoblotting.
RNA Interference—Small interfering RNAs (siRNAs) were synthesized for the target sequences of FEN1: GCAGCACAAUGAUGAGUGCTT (siRNA-1) and GCUGCCAAUCCAGGAAUUCTT (siRNA-2). Control siRNA targeted luciferase, and the sequence was CGUACGCGGAAUACUUCGATT. HeLa cells were transfected with 5 nm siRNA with X-tremeGENE transfection reagent (Roche Applied Science) according to the manufacturer's instructions. The cells were harvested 72 h after transfection to isolate DNA for TRF analysis and telomere overhang measurement and to isolate protein for immunoblotting. For long term treatment of siRNA, siRNA was transfected to HeLa cells, and transfection was repeated at 3-day intervals.
Terminal Restriction Fragment Analysis—Telomere length was determined by TRF analysis as described (40).
Fluorescence in Situ Hybridization (FISH)—FISH was performed as described (41). Briefly, the cells were enriched in metaphase using colcemid (1 μg/ml, 2 h), trypsinized, and treated with 0.075 m KCl solution, fixed with methanol/acetic acid (3:1), and then dropped on slides. A peptide nucleic acid telomere probe was used to hybridize telomeres as described (2). The images were taken with Zeiss Axiovert 200M inverted microscope.
Telomeric G-overhang Measurement—The length of the telomeric G-overhang was measured by telomere overhang protection assay as described (43). Briefly, total DNA (5 μg) in gp32 protection buffer (15 μl, 10 mm HEPES, pH 7.5, 100 mm LiCl, 2.5 mm MgCl2, 5 mm CaCl2) was treated (37 °C, 30 min) with RNase A (1 μg). The single-stranded DNA was then coated (1 h, room temperature) with single-stranded DNA binding protein T4 gene protein 32 (10 pmol/μl; Roche Applied Science), followed by cross-linking (15 min, room temperature) with 0.025% glutaraldehyde. Cross-linking was stopped by adding 1 m Tris (pH 7.5) to 30 mm. The unprotected double-stranded DNA was digested (37 °C, 30 min) by DNase I (0.04 unit; Roche Applied Science), which was then inactivated (80 °C, 30 min). Protease K (1 μg/μl) and SDS (0.5%) were added, and the samples were incubated (55 °C, overnight) to digest protein and reverse cross-linking. A high specific activity 18-mer C-rich probe (5 fmol) was annealed (room temperature, overnight) to the liberated G-rich overhang. The samples were analyzed the next day on a 6% polyacrylamide gel at 4 °C with cold 0.5× TBE buffer. The gels were dried on Hybond N+ membrane (GE Healthcare), exposed to a phosphorimaging screen, and quantitated by ImageQuant software. To measure the size of the 3′ G-rich overhang, an ExoI-treated sample was used as background, and its signal was subtracted from that of the untreated sample at each measured size interval. The weighted mean size was calculated using the formula: Σ(ODi)/Σ(ODi/Li), where ODi is the PhosphorImager output (signal intensity), and Li is the length in nucleotides of the DNA at position i, using the range of the molecular weight marker standards.
RESULTS
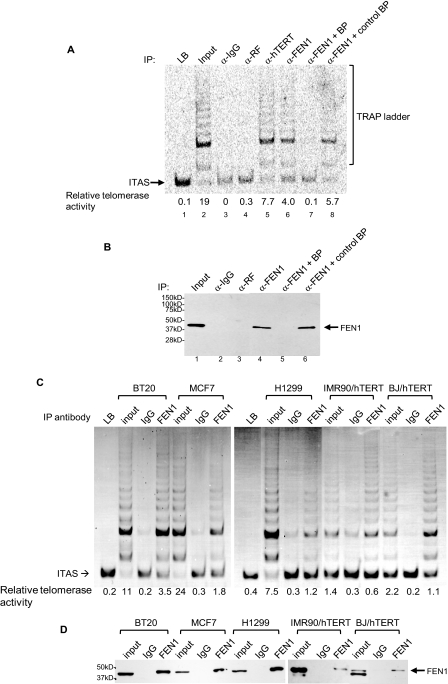
FEN1 and Telomerase Co-exist as a Complex in Both Telomerase-positive Tumor Cells and Telomerase-immortalized Human Somatic Cells—To test whether FEN1 is in complex with telomerase in vivo, we first used a polyclonal anti-FEN1 antibody to pull down telomerase from HeLa cell extracts. Following IP, the precipitates were subjected to TRAP analysis for detection of telomerase activity. As shown in Fig. 1A, FEN1 antibody precipitated telomerase activity, measured as TRAP activity, similar to an hTERT antibody (lanes 5 and 6), indicating that FEN1 was in complex with telomerase in vivo. Neither the rabbit IgG nor a control antibody to ribosomal release factor precipitated telomerase (Fig. 1A, lanes 3 and 4). Western blot analysis of the immunoprecipitates with the polyclonal anti-FEN1 antibody confirmed that the antibody was specific for FEN1 protein (Fig. 1B). Moreover, the addition of a peptide that blocks the anti-FEN1 antibody completely inhibited IP of FEN1, as shown by Western blot analysis of precipitates (Fig. 1B, lane 5), whereas a control peptide (blocking peptide for anti-GAPDH antibody) did not have any effect on anti-FEN1 recognition. Accordingly, minimal TRAP activity was detected in the precipitate when FEN1 blocking peptide was present (Fig. 1A, lane 7), whereas the control blocking peptide had no effect on the amount of telomerase precipitated by FEN1 antibody (Fig. 1A, lane 8). Taken together, these results strongly suggested that co-immunoprecipitation of FEN1 with telomerase was specific.
FIGURE 1.
FEN1 and telomerase co-exist as a complex in vivo. A, the indicated antibodies were used to immunoprecipitate proteins from HeLa cell lysate. Immunoprecipitates were subjected to nonradioactive TRAP assays for detection of telomerase activity. Aliquots equivalent to 25,000 cells were used in TRAP assays. A blocking peptide (BP) was added to the cell lysate during IP to block the interaction of FEN1 with anti-FEN1 antibody. An irrelevant peptide, which blocks anti-GAPDH recognition of GAPDH, but has no effect on FEN1 antibody recognition, was used as a control blocking peptide (control BP). The input lanes correspond to telomerase activity presented in 1,000 cells. LB, lysis buffer only; ITAS, 36-bp internal TRAP assay standards. B, specificity of the polyclonal anti-FEN1 antibody used in IP. After IP, the precipitates were subjected to FEN1 Western blotting, which demonstrates polyclonal anti-FEN1 specifically recognized FEN1. C, FEN1 is in complex with telomerase in telomerase-positive tumor cells and telomerase-immortalized human somatic cells. The anti-FEN1 antibody was used for IP of proteins from lysates of various cells expressing telomerase: telomerase-positive cancer cells BT20, MCF7, H1299 cells, IMR90 fibroblasts ectopically overexpressing hTERT (IMR90/hTERT), and BJ fibroblasts ectopically overexpressing hTERT (BJ/hTERT). Precipitates were assayed for telomerase activity by nonradioactive TRAP. D, Western blot showing that polyclonal anti-FEN1 specifically recognized FEN1 but not other proteins. After IP, precipitates were subjected to Western blotting to determine the specificity of anti-FEN1 antibody.
To test whether FEN1 was in complex with telomerase in other cell lines, we carried out the co-IP-TRAP using telomerase-positive cancer cell lines BT20, MCF7, and H1299. Anti-FEN1 antibody also precipitated telomerase activity in these cells (Fig. 1C). Normal human somatic cells such as BJ foreskin fibroblasts and IMR90 lung fibroblasts contain undetectable telomerase activity because of the low expression of the catalytic subunit of telomerase, hTERT. Ectopically overexpressing hTERT in BJ and IMR90 make them telomerase-positive. As shown in Fig. 1C, anti-hFEN1 antibody precipitated telomerase activity in BJ/hTERT and IMR90/hTERT as well. Therefore, human FEN1 forms a complex with telomerase in both telomerase-positive cancer cells and hTERT-expressing normal somatic cells.
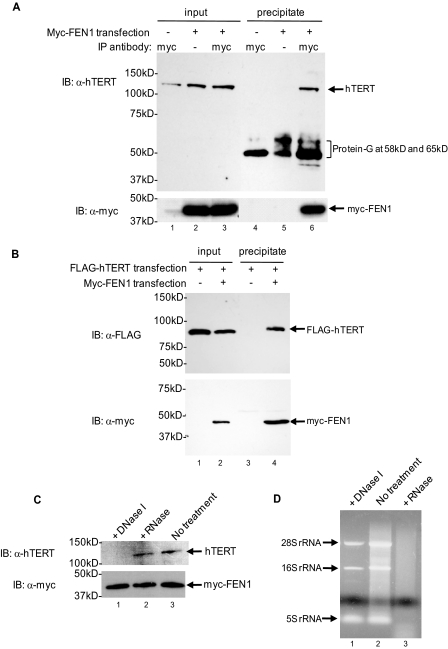
FEN1 Is in Complex with hTERT Independent of hTR but Dependent on DNA—Telomerase is a ribonucleoprotein containing an RNA subunit (human telomerase RNA) and a catalytic protein subunit (hTERT). We next investigated the potential association of FEN1 with the hTERT subunit. We first co-transfected Myc-tagged full-length FEN1 with untagged full-length hTERT into HeLa cells. Immunoprecipitation of Myc-FEN1 with anti-Myc coupled agarose beads also pulled down hTERT (Fig. 2A, lane 6), whereas hTERT was not precipitated from cells transfected with hTERT and the empty vector (Fig. 2A, lane 4). To further test the specificity of the FEN1/hTERT association, we co-expressed Myc-FEN1 and FLAG-tagged hTERT. Immunoprecipitation of Myc-FEN1 with anti-Myc-coupled agarose beads also pulled down FLAG-hTERT (Fig. 2B, lane 4), whereas FLAG-hTERT was not precipitated from cells transfected with the empty vector (Fig. 2B, lane 3). Together, these results strongly supported our observation that FEN1 was specifically in complex with telomerase (Fig. 1).
FIGURE 2.
FEN1 specifically co-immunoprecipitates with telomerase catalytic subunit hTERT. A, FEN1 and hTERT were co-immunoprecipitated from HeLa extracts. HeLa cells were transiently transfected either with the combination of hTERT and Myc-FEN1 (lanes 2, 3, 5, and 6) or with the combination of hTERT and pCl-neo empty vector (lanes 1 and 4). All of the samples therefore expressed hTERT. After IP with an anti-Myc antibody, the precipitates were subjected to immunoblotting by anti-hTERT or anti-Myc. Input represents 2.5% of the total protein used in the anti-Myc immunoprecipitations. IP, immunoprecipitating antibody; IB, immunoblotting antibody. Anti-rabbit IgG secondary antibody also detected protein G released from beads. B, exogenously expressed Myc-FEN1 and FLAG-hTERT were co-immunoprecipitated from HeLa extracts. HeLa cells were transiently transfected either with the combination of FLAG-hTERT and pCl-neo empty vector (lanes 1 and 3) or with the combination of FLAG-hTERT and Myc-FEN1 (lanes 2 and 4). IP with an anti-Myc antibody was followed by anti-FLAG Western blotting for detecting FLAG-hTERT. Input represents 2.5% of the total protein used in the anti-Myc immunoprecipitations. C, the association of Myc-FEN1 with hTERT is independent of RNA but requires DNA. HeLa cells were co-transfected with hTERT and Myc-FEN1, and cell lysates were treated with DNase I or RNase A prior to IP. Precipitates from the DNase I- or RNase A-treated lysates were immunoblotted with anti-hTERT or anti-Myc antibody. D, confirmation of degradation of DNA or cellular RNA by agarose gel electrophoresis. After DNase I or RNase A treatment, total DNA and RNA were extracted from cell lysates by phenol/chloroform (pH 7.5), precipitated by ethanol, and then loaded on 1% agarose gel. Treatment with DNase I removed genomic DNA as shown by the disappearance of the DNA smear (as the cell extracts were sonicated during lysis, the genomic DNA was sheared to small sizes). Treatment with RNase A removed RNA as shown by the disappearance of 28, 16, and 5 S ribosomal RNA bands.
Because the telomerase complex is composed of two subunits, hTERT and human telomerase RNA, we tested the possibility that the physical interaction between FEN1 and hTERT might be mediated by the telomerase RNA subunit. Treatment of cell extracts with RNase A prior to IP removed cellular RNA, as shown by the disappearance of 28, 16, and 5 S ribosomal RNA bands analyzed by agarose gel electrophoresis (Fig. 2D, lane 3). No significant reduction in the Myc-FEN1/hTERT interaction was detected (Fig. 2C, lane 2), suggesting that that the RNA component of telomerase was dispensable for the FEN1 telomerase complex.
FEN1 associates with telomere DNA during the S and G2 phases of the cell cycle in telomerase-negative human fibroblasts (37). To test whether FEN1 telomerase complex might be bridged by telomere DNA, cell extracts were treated with DNase I to remove DNA prior to IP, as shown by the disappearance of the genomic DNA smear (as cell extracts were sonicated during lysis, the genomic DNA was sheared to small sizes) (Fig. 2D, lane 1). Removal of DNA abolished the ability of Myc-FEN1 to precipitate hTERT (Fig. 2C, lane 1), suggesting that FEN1 and hTERT might associate indirectly through telomere DNA.
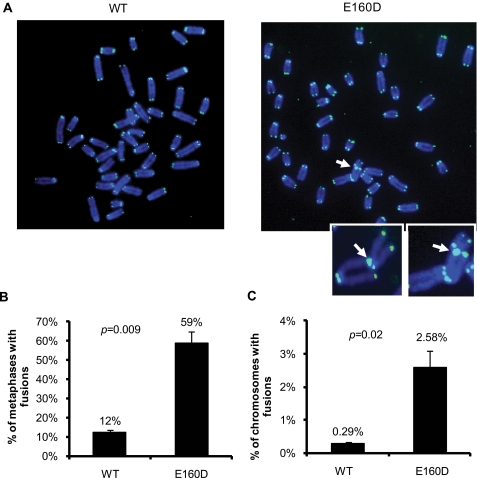
FEN1 Deficiency Leads to Telomere Instability in MEFs—Telomeres become dysfunctional when they cannot be maintained properly. Dysfunctional telomeres are recognized as damaged DNA, triggering a DNA damage response, which then leads to DNA repair at telomeres via a nonhomologous end joining pathway. We hypothesized that functional FEN1 might be essential for telomerase-mediated telomere maintenance, and deficiency in FEN1 might encumber telomerase elongation at short telomeres, leading to telomere dysfunction. To test our hypothesis, we analyzed telomere stability in MEFs carrying a FEN1 point mutation (E160D) that eliminated FEN1 exonuclease and gap-dependent endonuclease activities (33). E160D knock-in mice show a dramatic increase in spontaneous mutation rates and cancer incidences (33). When we analyzed telomere stability using FISH analysis, we found that the E160D MEFs displayed a significant increase of telomere end-to-end fusions compared with wild-type MEFs (Fig. 3), suggesting that the exonuclease and gap-dependent endonuclease activities of FEN1 were important for maintaining telomere stability.
FIGURE 3.
Telomere instability in MEFs harboring nuclease-deficient FEN1. (A) Representative images of metaphases with telomeric DNA detected by FISH (green) from wild-type (WT) and E160D knock-in MEFs (E160D). The arrow indicates telomere end-to-end fusions. The insets show enlarged images of representative telomere end-to-end fusions in E160D MEF. B and C, quantification of telomere abnormalities observed in E160D MEFs, presented as percentage of metaphases containing telomere end-to-end fusions in B and percentage of chromosomes containing end-to-end fusions in C. A minimum of 50 metaphases from two independent experiments was analyzed. The error bars represent S.D.
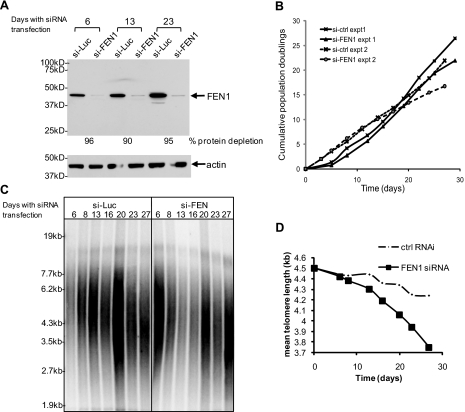
FEN1 Deficiency Induces Telomere Shortening in Telomerase-positive Cancer Cells—To test our hypothesis that functional FEN1 may be needed for telomerase-mediated telomere maintenance in human telomerase-positive cells, we determined whether telomere maintenance was affected by the reduction of FEN1 expression. One time RNA interference knockdown of FEN1 in telomerase-positive cells did not induce telomere dysfunction, as determined by telomere length measurement, FISH analysis, and γ-H2AX foci staining at dysfunctional telomeres (data not shown). This could have been due to transient protein reduction induced by short term gene knockdown. We then repeatedly treated HeLa cells with FEN1 siRNA at 3-day intervals for 4 weeks. The same method has been used to study the role of replication factor origin recognition complex in telomere maintenance (44). Quantitation of Western blot revealed that about 90–95% endogenous FEN1 protein was depleted during the course of repetitive RNA interference treatment (Fig. 4A). About 2 weeks after the initial RNA interference treatment, cells treated with FEN1 siRNA showed retarded cell growth compared with control siRNA-treated cells (Fig. 4B). We repeated the FEN1 knockdown experiment and found similar effects of siFEN1 on cell growth (Fig. 4B). Measurement of telomere length by TRF analysis revealed gradually shortened telomeres over this long term treatment (Fig. 4, C and D), suggesting that telomerase elongation of telomeres was hampered by FEN1 deficiency.
FIGURE 4.
FEN1 deficiency affects telomere maintenance in telomerase-positive cancer cells. A, HeLa cells were repeatedly transfected with FEN1 siRNA-1 or a luciferase siRNA for 4 weeks. Western blot showing FEN1 depletion in samples taken at days 6, 13, and 23 are shown. Actin was used as loading control. B, growth retardation in HeLa cells depleted of FEN1. C, telomere length measurement. Genomic DNA was isolated from collected cells over the duration of RNA interference treatment and then subjected to telomere length measurement by TRF analysis. D, mean telomere length was quantified as described (24).
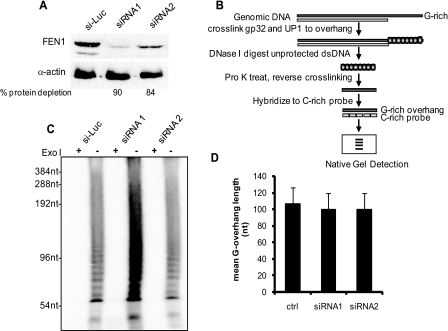
FEN1 Deficiency Did Not Alter Telomere Structure—Telomeres can form a special nucleo-protein structure called t-loop to protect chromosome stability. This structure is formed by inserting the single-stranded G-overhang at the 3′ end of the telomere back into the double-stranded telomere DNA region. Failure to maintain the protective t-loop structure leads to telomere instability (45, 46). In budding yeast, deficiency in FEN1/Rad27 leads to accumulation of G-overhangs (35, 36), which is thought to deregulate telomerase action and cause abnormal telomere shortening and lengthening (35). To test whether telomere instability caused by FEN1 deficiency in cancer cells might be due to the alteration in telomere structure, we determined whether the G-overhang is altered by FEN1 deficiency. Western blotting showed that two siRNAs targeting different coding regions of FEN1 reduced protein expression by 90 and 84%, respectively (Fig. 5A). Genomic DNA was then isolated from FEN1-depleted HeLa cells and subjected to telomere overhang protection assay for measurement of G-overhang length (Fig. 5B) (43, 47). We failed to observe any alteration in telomeric G-overhangs upon FEN1 depletion (Fig. 5, C and D). Use of a nondenaturing in-gel hybridization assay (48) to estimate the relative abundance of G-overhang also showed no change in G-overhang abundance (data not shown). Taken together, we concluded that FEN1 deficiency was unlikely to alter telomere structure.
FIGURE 5.
FEN1 deficiency does not affect telomeric G-overhang. A, extracts of HeLa cells treated with control siRNA (targeting luciferase) and FEN1 siRNAs were analyzed by Western blotting with anti-FEN1 antibody. The cells were collected 72 h after one-time siRNA transfection, and proteins were extracted for Western blotting to confirm knockdown. B, outline for measuring the size of telomeric G-overhang using telomere overhang protection assay (43). Total genomic DNA was incubated with T4 gp32 and UP1 protein, which bind to single-stranded G-overhangs. After cross-linking proteins to DNA, unprotected double-stranded DNA was removed by DNase I digestion. After heat inactivation of the DNase I, cross-linking was reversed, and proteins were digested to make the protected overhangs available for hybridization to 32P-labeled C-rich probe. Overhangs were finally size fractionated on a native polyacrylamide gel. C, telomere overhang protection assay using HeLa cells with reduced FEN1 expression. The smears represent the heterogeneity of overhang sizes. Treatment of genomic DNA with Escherichia coli ExoI (a 3′ → 5′ exonuclease) prior to the overhang protection assay specifically digests 3′ overhangs, and thus the ExoI plus (+) lanes show the background. D, FEN1 deficiency does not alter G-overhang length. Gels from overhang protection assays were analyzed by ImageQuant as described (43). The weighted mean overhang sizes were measured from three independent experiments. The error bars represent one standard deviation.
DISCUSSION
FEN1 is a multifunctional nuclease that plays a role in DNA replication and DNA repair. It associates with telomeres and interacts with telomere-binding proteins TRF2 and the Werner protein (WRN) (38, 49, 50). Here, we analyzed its role in maintaining telomere stability in telomerase-positive cancer cells. Using co-immunoprecipitation assays, we have demonstrated that FEN1 is in complex with telomerase (Figs. 1 and 2). Further analysis shows that this physical connection between FEN1 and telomerase is dependent on the presence of telomere DNA but does not require the RNA component of telomerase (Fig. 2), indicating that the FEN1 may indirectly interact with telomerase through telomere DNA. This finding is consistent with previous reports that FEN1 is localized at telomeres (37, 38). We further show that a loss of FEN1 nuclease activity in MEFs causes an increase in telomere end-to-end fusions (Fig. 3), suggesting that FEN1 nuclease activity is critical for telomere stability. Furthermore, knocking down FEN1 in telomerase-positive cancer cells induces gradual telomere shortening and retarded cell growth (Fig. 4). These data strongly suggest that FEN1 may regulate telomerase activity at telomeres and that dysfunction of FEN1 may cause failure of proper telomere maintenance by telomerase. Our results further emphasize the important roles of DNA replication machinery in regulating telomerase action at telomeres.
Telomerase elongation of telomeres is tightly regulated by various telomere-binding proteins and proteins involved in DNA repair, replication, and recombination. Deficiency in these proteins causes deregulation of telomerase action, leading to abnormal telomere shortening or lengthening. The first step during telomerase extension of telomeres is the extension of G-strands. The complementary C-strands are then filled in by the lagging strand replication machinery. Analyses in yeast and ciliates have suggested that extension of G-strand by telomerase is regulated by C-strand synthesis. When C-strand synthesis is partially inhibited by a specific inhibitor of polα and polδ, new synthesis on both C- and G-strands becomes deregulated, leading to heterogeneous length of both C- and G-strands (26). This suggests that there is a highly coordinated action of C-strand synthesis mediated by the lagging strand synthesis machinery and G-strand synthesis mediated by telomerase. Although the precise mechanism for this coordinative regulation is unclear, several studies have revealed close physical interaction between the replication machinery and telomerase, suggesting that this interaction may regulate telomerase elongation of telomeres. For example, fission yeast polα and telomerase catalytic subunit Trt1 co-exist in a complex in vivo (24). Similarly, in ciliate E. crassus, it has been demonstrated that primase and proliferating cell nuclear antigen form a complex with telomerase (25). In budding yeast, two-hybrid analysis has shown that DNA replication factor polα interacts with Cdc13p, a protein that directly interacts with Est1p and regulates telomerase activity at yeast telomeres (23). We now provide evidence that FEN1 and telomerase co-exist in a complex in human cells, suggesting that the physical connection between the lagging strand replication complexes with telomerase may be evolutionarily conserved from yeast to humans. We propose that this physical connection is perhaps one of the mechanisms regulating telomerase extension of telomeres. Disruption of the physical link between DNA replication proteins with telomerase may uncouple C-strand synthesis from telomerase-guided G-strand extension, leading to defects in telomere maintenance.
The single-stranded G-overhang at the 3′ end of the telomere is essential for the formation of the special t-loop structure. Deficiency in G-overhang generation is expected to disrupt the t-loop structure and further lead to telomere instability. Because the G-overhang is the binding site for telomerase, deficiency in G-overhang generation is also expected to hamper telomerase elongation of telomeres. Several studies have suggested that G-overhangs are generated by extensive but tightly regulated enzymatic processing events at the C-strand following telomere replication (45, 47, 51), even though the nuclease(s) has not been identified. FEN1 is a multifunctional nuclease containing endonuclease activity and 5′ → 3′ exonuclease activity and is therefore among the candidate exonucleases responsible for processing the blunt-ended telomeres left after leading strand synthesis (42, 52, 53). Increasing evidence suggests that FEN1 is unlikely to be the nuclease-processing telomere ends but rather in Okazaki fragment processing at lagging daughter telomeres. If FEN1 were the nuclease-processing telomere ends, FEN1 deficiency would leave leading strand telomeres blunt-ended after replication. In this case, we would expect that these blunt-ended DNA would be recognized as DNA double-strand breaks and be repaired by DNA repair machinery. Because half of telomeres are replicated by leading strand synthesis, we would also expect telomere instabilities on half of the chromosomes. However, the nuclease-deficient FEN1 revealed telomere end-to-end fusions at only a subset of chromosomes (Fig. 3). In support of our data, it has been reported that about 8% of chromosomes lose their telomeres when FEN1 is depleted (38). All of the affected telomeres are replicated by lagging strand synthesis, whereas telomeres replicated by leading stranded synthesis are unaffected (38). In addition, deficiency in budding yeast FEN1/Rad27p actually leads to accumulation of G-overhangs at the lagging daughter telomeres, whereas leading daughter telomere ends are normally processed (35, 36). It was interpreted that in the absence of FEN1/Rad27p, inappropriate Okazaki fragment processing might cause excessive dissociation or degradation of the last primer (perhaps by other nucleases), leading to an increase of single-stranded DNA of the template strand, i.e. G-strand (35, 36). Taken together, these data suggest that FEN1 is unlikely to be the nuclease responsible for processing telomere ends, although we cannot rule out the possibility that there may be other nuclease(s) with functions redundant to those of FEN1.
In summary, we provide the first evidence that human FEN1 protein is in complex with telomerase and regulates telomerase activity at telomeres. Together with the demonstration by others that FEN1 is localized at telomeres and affects lagging strand telomere replication, our results suggest that, similar to what has been noted in yeast and ciliates, telomerase in human cells is likely to coordinate with the lagging strand replication machinery to ensure proper telomere length control. Understanding the regulatory mechanism for this coordination will aid us in understanding the mechanisms of telomere maintenance and facilitate telomerase-based cancer therapy.
Acknowledgments
We thank L. Harrington at the Wellcome Trust Center for Cell Biology at the University of Edinburgh for FLAG-hTERT construct, S. Wilson at the National Institute of Environmental Health Sciences, and R. Bambara at University of Rochester for FEN1 cDNA, N. Popescu at NCI for breast cancer cell lines, W. E. Wright and J. W. Shay at University of Texas Southwestern Medical Center for BJ/hTERT and IMR90/hTERT, Z. Lou for help in sequencing, and K. Roberts and S. Da Costa for comments on manuscript.
This work was supported, in whole or in part, by National Institutes of Health Grants R15CA132090 (to W. C.) and R01CA073764 (to B. H. S.). This work was also supported by Texas Woman's University Research Enhancement Program (to W. C.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
The abbreviations used are: pol, polymerase; FEN1, flap endonuclease I; MEF, mouse embryonic fibroblast; TRAP, telomere repeat amplification protocol; IP, immunoprecipitation; GAPDH, glyceraldehyde-3-phosphate dehydrogenase; siRNA, small interfering RNA; TRF, terminal restriction fragment; FISH, fluorescence in situ hybridization; hTERT, human telomerase reverse transcriptase.
References
- 1.Harley, C. B., Futcher, A. B., and Greider, C. W. (1990) Nature 345 458-460 [DOI] [PubMed] [Google Scholar]
- 2.Zou, Y., Sfeir, A., Shay, J. W., and Wright, W. E. (2004) Mol. Biol. Cell 15 3709-3718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wright, W. E., Pereira-Smith, O. M., and Shay, J. W. (1989) Mol. Cell. Biol. 9 3088-3092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Campisi, J., Kim, S. H., Lim, C. S., and Rubio, M. (2001) Exp. Gerontol. 36 1619-1637 [DOI] [PubMed] [Google Scholar]
- 5.Shay, J. W., and Wright, W. E. (2001) Radiat Res. 155 188-193 [DOI] [PubMed] [Google Scholar]
- 6.d'Adda di Fagagna, F., Reaper, P. M., Clay-Farrace, L., Fiegler, H., Carr, P., Von Zglinicki, T., Saretzki, G., Carter, N. P., and Jackson, S. P. (2003) Nature 426 194-198 [DOI] [PubMed] [Google Scholar]
- 7.d'Adda di Fagagna, F., Teo, S. H., and Jackson, S. P. (2004) Genes Dev. 18 1781-1799 [DOI] [PubMed] [Google Scholar]
- 8.Longhese, M. P. (2008) Genes Dev. 22 125-140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Blasco, M. A. (2007) Nat. Chem. Biol. 3 640-649 [DOI] [PubMed] [Google Scholar]
- 10.Blackburn, E. H., Greider, C. W., Henderson, E., Lee, M. S., Shampay, J., and Shippen-Lentz, D. (1989) Genome 31 553-560 [DOI] [PubMed] [Google Scholar]
- 11.Greider, C. W., and Blackburn, E. H. (1985) Cell 43 405-413 [DOI] [PubMed] [Google Scholar]
- 12.Hiyama, E., and Hiyama, K. (2007) Br J. Cancer 96 1020-1024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Flores, I., Benetti, R., and Blasco, M. A. (2006) Curr. Opin. Cell Biol. 18 254-260 [DOI] [PubMed] [Google Scholar]
- 14.Shay, J. W., and Wright, W. E. (1996) Curr. Opin. Oncol. 8 66-71 [DOI] [PubMed] [Google Scholar]
- 15.Yago, M., Ohki, R., Hatakeyama, S., Fujita, T., and Ishikawa, F. (2002) FEBS Lett. 520 40-46 [DOI] [PubMed] [Google Scholar]
- 16.Shay, J. W., and Wright, W. E. (2006) Nat. Rev. Drug Discov. 5 577-584 [DOI] [PubMed] [Google Scholar]
- 17.Hug, N., and Lingner, J. (2006) Chromosoma 115 413-425 [DOI] [PubMed] [Google Scholar]
- 18.de Lange, T. (2006) in Telomeres (de Lange, T., Lundblad, V., and Blackburn, E. H., eds) 2nd Ed., pp. 387-431, Cold Spring Harbor Laboratory, Cold Spring Harbor, NY
- 19.Diede, S. J., and Gottschling, D. E. (1999) Cell 99 723-733 [DOI] [PubMed] [Google Scholar]
- 20.Adams Martin, A., Dionne, I., Wellinger, R. J., and Holm, C. (2000) Mol. Cell. Biol. 20 786-796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Adams, A. K., and Holm, C. (1996) Mol. Cell. Biol. 16 4614-4620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Carson, M. J., and Hartwell, L. (1985) Cell 42 249-257 [DOI] [PubMed] [Google Scholar]
- 23.Qi, H., and Zakian, V. A. (2000) Genes Dev. 14 1777-1788 [PMC free article] [PubMed] [Google Scholar]
- 24.Dahlen, M., Sunnerhagen, P., and Wang, T. S. (2003) Mol. Cell. Biol. 23 3031-3042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ray, S., Karamysheva, Z., Wang, L., Shippen, D. E., and Price, C. M. (2002) Mol. Cell. Biol. 22 5859-5868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fan, X., and Price, C. M. (1997) Mol. Biol. Cell 8 2145-2155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shen, B., Singh, P., Liu, R., Qiu, J., Zheng, L., Finger, L. D., and Alas, S. (2005) Bioessays 27 717-729 [DOI] [PubMed] [Google Scholar]
- 28.Liu, Y., Kao, H. I., and Bambara, R. A. (2004) Annu. Rev. Biochem. 73 589-615 [DOI] [PubMed] [Google Scholar]
- 29.Harrington, J. J., and Lieber, M. R. (1994) EMBO J. 13 1235-1246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kikuchi, K., Taniguchi, Y., Hatanaka, A., Sonoda, E., Hochegger, H., Adachi, N., Matsuzaki, Y., Koyama, H., van Gent, D. C., Jasin, M., and Takeda, S. (2005) Mol. Cell. Biol. 25 6948-6955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zheng, L., Zhou, M., Chai, Q., Parrish, J., Xue, D., Patrick, S. M., Turchi, J. J., Yannone, S. M., Chen, D., and Shen, B. (2005) EMBO Rep. 6 83-89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Parrish, J. Z., Yang, C., Shen, B., and Xue, D. (2003) EMBO J. 22 3451-3460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zheng, L., Dai, H., Zhou, M., Li, M., Singh, P., Qiu, J., Tsark, W., Huang, Q., Kernstine, K., Zhang, X., Lin, D., and Shen, B. (2007) Nat. Med. 13 812-819 [DOI] [PubMed] [Google Scholar]
- 34.Kucherlapati, M., Yang, K., Kuraguchi, M., Zhao, J., Lia, M., Heyer, J., Kane, M. F., Fan, K., Russell, R., Brown, A. M., Kneitz, B., Edelmann, W., Kolodner, R. D., Lipkin, M., and Kucherlapati, R. (2002) Proc. Natl. Acad. Sci. U. S. A. 99 9924-9929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Parenteau, J., and Wellinger, R. J. (1999) Mol. Cell. Biol. 19 4143-4152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Parenteau, J., and Wellinger, R. J. (2002) Genetics 162 1583-1594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Verdun, R. E., and Karlseder, J. (2006) Cell 127 709-720 [DOI] [PubMed] [Google Scholar]
- 38.Saharia, A., Guittat, L., Crocker, S., Lim, A., Steffen, M., Kulkarni, S., and Stewart, S. A. (2008) Curr. Biol. 18 496-500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chai, W., Ford, L. P., Lenertz, L., Wright, W. E., and Shay, J. W. (2002) J. Biol. Chem. 277 47242-47247 [DOI] [PubMed] [Google Scholar]
- 40.Herbert, B., Shay, J. W., and Wright, W. E. (2003) Analysis of Telomeres and Telomerase, pp. 18.6.1-18.6.6, John Wiley & Sons, Inc. [DOI] [PubMed]
- 41.Zou, Y., Gryaznov, S. M., Shay, J. W., Wright, W. E., and Cornforth, M. N. (2004) Proc. Natl. Acad. Sci. U. S. A. 101 12928-12933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ohki, R., Tsurimoto, T., and Ishikawa, F. (2001) Mol. Cell. Biol. 21 5753-5766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chai, W., Shay, J. W., and Wright, W. E. (2005) Mol. Cell. Biol. 25 2158-2168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Deng, Z., Dheekollu, J., Broccoli, D., Dutta, A., and Lieberman, P. M. (2007) Curr. Biol. 17 1989-1995 [DOI] [PubMed] [Google Scholar]
- 45.Hockemeyer, D., Sfeir, A. J., Shay, J. W., Wright, W. E., and de Lange, T. (2005) EMBO J. 24 2667-2678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Karlseder, J., Smogorzewska, A., and de Lange, T. (2002) Science 295 2446-2449 [DOI] [PubMed] [Google Scholar]
- 47.Chai, W., Du, Q., Shay, J. W., and Wright, W. E. (2006) Mol. Cell 21 427-435 [DOI] [PubMed] [Google Scholar]
- 48.Dionne, I., and Wellinger, R. J. (1996) Proc. Natl. Acad. Sci. U. S. A. 93 13902-13907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Muftuoglu, M., Wong, H. K., Imam, S. Z., Wilson, D. M., III, Bohr, V. A., and Opresko, P. L. (2006) Cancer Res. 66 113-124 [DOI] [PubMed] [Google Scholar]
- 50.Sharma, S., Sommers, J. A., Wu, L., Bohr, V. A., Hickson, I. D., and Brosh, R. M., Jr. (2004) J. Biol. Chem. 279 9847-9856 [DOI] [PubMed] [Google Scholar]
- 51.Sfeir, A. J., Chai, W., Shay, J. W., and Wright, W. E. (2005) Mol. Cell 18 131-138 [DOI] [PubMed] [Google Scholar]
- 52.Olovnikov, A. M. (1973) J. Theor. Biol. 41 181-190 [DOI] [PubMed] [Google Scholar]
- 53.Lingner, J., Cooper, J. P., and Cech, T. R. (1995) Science 269 1533-1534 [DOI] [PubMed] [Google Scholar]