Abstract
JIPs (c-Jun N-terminal kinase interacting proteins), which scaffold JNK/p38 MAP kinase signaling modules, also bind conventional kinesins and are implicated in microtubule-based membrane trafficking in neuronal cells. Here we have identified a novel splice variant of the Jip4 gene product JLPL (JNK-interacting leucine zipper protein) in yeast-two hybrid screens with the phosphoinositide kinase PIKfyve. The interaction was confirmed by pulldown and coimmunoprecipitation assays in native cells. It engages the PIKfyve cpn60_TCP1 consensus sequence and the last 75 residues of the JLP C terminus. Subpopulations of both proteins cofractionated and populated similar structures at the cell perinuclear region. Because PIKfyve is essential in endosome-to-trans-Golgi network (TGN) cargo transport, we tested whether JLP is a PIKfyve functional partner in this trafficking pathway. Short interfering RNA (siRNA)-mediated depletion of endogenous JLP or PIKfyve profoundly delayed the microtubule-based transport of chimeric furin (Tac-furin) from endosomes to the TGN in a CHO cell line, which was rescued upon ectopic expression of siRNA-resistant JLP or PIKfyve constructs. Peptides from the contact sites in PIKfyve and JLP, or a dominant-negative PIKfyve mutant introduced into cells by ectopic expression or microinjection, induced a similar defect. Because Tac-TGN38 delivery from endosomes to the TGN, unlike that of Tac-furin, does not require intact microtubules, we monitored the effect of JLP and PIKfyve depletion or the interacting peptides administration on Tac-TGN38 trafficking. Remarkably, neither maneuver altered the Tac-TGN38 delivery to the TGN. Our data indicate that JLP interacts with PIKfyve and that both proteins and their association are required in microtubule-based, but not in microtubule-independent, endosome-to-TGN cargo transport.
In mammalian cells, the endosomal/endocytic system comprises an interconnected and morphologically complex network of membrane organelles that supports fundamental functions such as nutrient entry and delivery for degradation, removal and degradation of plasma membrane or Golgi proteins, regulation and integration of signaling pathways, and protein recycling to the cell surface or the TGN2 (1–4). From the plasma membrane, the endocytosed cargo is first delivered to early endosomes/sorting endosomes. Cargoes destined for recycling to the cell surface then enter the endocytic recycling compartment, whereas others, intended for degradation, remain in early endosomes. Early endosomes undergo a series of changes, known as maturation, to give rise to maturing transport intermediates (herein ECV/MVBs; also Ref. 5) and to late endosomes that fuse with lysosomes to deliver cargo for degradation. Recycling or degradation is not the only outcome of the cell surface-originated cargoes. A set of internalized transmembrane proteins, including intracellular sorting receptors, enzymes, and toxins, are retrieved from the endosomal system and transported to the TGN. The endosome-to-TGN trafficking of the acid-hydrolase-sorting receptor, CI-MPR, the endopeptidase furin, and the putative cargo receptor TGN38 are the best studied examples. These cargoes are highly enriched in the TGN at steady state but arrive there from different compartments, utilizing distinct mechanisms. Thus, TGN38 enters the TGN from the endocytic recycling compartment by an iterative removal from the latter compartment, furin reaches the TGN by exiting the early/late endosomal system, and CI-MPR implements features of both pathways (4, 6–9).
Whereas the detailed molecular and cellular mechanisms underlying the membrane progression in the course of cargo transport through the endosomal system or retrieval from early/late endosomes to the TGN is still elusive, experimental evidence has been accumulating to implicate PIKfyve, the sole enzyme for PtdIns(3,5)P2 synthesis (10). Thus, PIKfyve has been found to interact with the late endosome-to-TGN transport factor Rab9 effector p40 (11). Furthermore, disruption of the PtdIns(3,5)P2 homeostatic mechanism by means of expression of dominant-negative kinase-deficient point mutants of PIKfyve, protein depletion, or pharmacological inhibition of PIKfyve activity was found to impair the exit of a subset of cargoes from early endosomes to the TGN and late endosomes or from the late endosomes (12–16). Phenotypically, these defects are manifested by progressive endosome swelling and cytoplasmic vacuolation, first seen by expression of dominant-negative PIKfyveK1831E in a number of mammalian cell types (17) and confirmed thereafter by other maneuvers inhibiting PIKfyve protein expression or activity (14, 16). In vitro reconstitution assays indicate that PIKfyve enzymatic activity is required in endosome processing in two ways. It triggers the formation/fission (or maturation) of ECV/MVBs from early endosomes and arrests the rate of fusion events in the endosomal system (18, 19). It is thus conceivable that impaired PIKfyve and PtdIns(3,5)P2 functioning in the fission and fusion events mechanistically underlies the constraints in the trafficking pathways traversing endosomes.
Microtubules aided by the microtubule-associated motor protein families of kinesin and dynein are required for proper performance of the endosomal/endocytic membrane system. Although their role is rather complex and not completely understood, in vivo and in vitro studies implicate microtubule-based dynamics in multiple aspects of the endocytic trafficking, including sorting of endocytic contents, fission/fusion events at early or late endosomes, early endosome maturation, and efficient motility of the transport vesicles to their destination (20–27). Accumulating evidence indicates that the binding of motor proteins to organelles or carrier vesicles is regulated by motor protein adapters. Intriguingly, this newly emerging adapter function has been found to be executed by proteins known as scaffolds of stress signaling enzymes. One such adapter for conventional kinesins is the group of JIPs that scaffold the JNK/p38 MAP kinase signaling modules (28–31). A mutation that causes mislocalization of synaptic vesicles and aberrant axonal transport in Drosophila and Caenorhabditis elegans affects the JIP3 homologs Sunday driver (dSYD) and Unc16, respectively (32, 33). In mammalian cells, JIPs are represented by four proteins (JIP1–4) derived from separate genes and several alternatively spliced variants. JIP1, the founding member, is structurally related to JIP2 (34, 35). JIP3 (also known as Unc16/JSAP1/dSYD) is structurally unrelated to JIP1 or JIP2, but as those two, it is abundant in neuronal cells (30, 32, 36). The latest addition to the group is JIP4 that occurs in three splice variants known thus far: JLP and JIP4 in mouse and SPAG9 in humans (31, 37, 38). JIP4, JLP, and SPAG9 (gene symbol, SPAG9) are structurally homologous to JIP3 but display broader distribution (37–39). Remarkably, all four members of the JIP group interact with the kinesin1 light chain, and potential cargoes for microtubule-based vesicle transport have been proposed for JIP1–JIP3 (32, 33, 38, 40–43). The role of JLP/JIP4 in the context of cargo transport or membrane trafficking events, however, has never been investigated. In the present study we report that JLP is a PIKfyve physical and functional partner in microtubule-based endosome-to-TGN trafficking. The interaction is identified by a yeast two-hybrid screen with the PIKfyve cpn60_TCP1 consensus sequence and mapped to the 75-aa peptide fragment of the extreme JLP C terminus. By monitoring divergent routes of cargo delivery to the TGN, differing by the requirement of microtubule-dependent early endosome maturation, we have determined that JLP assists PIKfyve selective functionality in microtubule-based endosome-to-TGN trafficking.
EXPERIMENTAL PROCEDURES
Yeast Two-hybrid Screening and Cloning of Full-length Mouse JLPL cDNA—The interaction mating version of two-hybrid screening of the human HeLa cell cDNA library with the PIKfyve 252-aa peptide fragment of the conserved cpn60_TCP1 domain (pNLexNLS-PIKfyve-(616–868)) was described previously (11). This screening resulted in 27 clones, confirmed as true positives based on their selective interaction with the PIKfyve-(616–868) bait and growth dependence on galactose, but not glucose, in the absence of leucine. Nucleotide sequence information revealed that seven separate clones encoded the C-terminal part of JLP/JIP4 (37, 38) spanning the last 92 to 152 aa plus a significant portion of the 3′ untranslated region (500–800 nucleotides). To obtain the full-length JLP sequence, we screened a mouse 3T3-F442A adipocyte cDNA library with the longest prey (clone YA-4) following published protocols (43). One positive clone that was the largest in size (∼7 kbp; clone 5) was sequenced in full following common strategies (43). Clone 5 was found to be identical to the reported mouse JLP (37), except for the insertion of a 13-aa fragment at position 1162, and is therefore referred to herein as JLPL, for a longer splice variant. Clone 5, however, lacked the first seven amino acids from the translational initiator. To complete the JLPL cDNA sequence, the EcoRI-XbaI fragment of the pGEM3Z-M2 clone from the mouse JLP N terminus (provided by Dr. Reddy) was ligated into the EcoRI-SalI digest of pBluescript II SK(+) along with the XbaI-SalI insert of clone 5.
JLPL and Other Constructs—The eGFP cDNA construct of mouse JLPLWT was generated by subcloning the EcoRI fragment of pBluescript II SK(+)-JLPL into the EcoRI digest of pEGFP-C2 (Clontech) in-frame with GFP. Epitope-tagged Myc-JLPL was generated in pCMV5 vector by a triple ligation using the XhoI-BamHI fragment of pEGFP-C2-JLPL and an EcoRI-XhoI-flanked double-stranded oligonucleotide encoding the 12-amino acid epitope (EQKLISEEDLLR) of the human c-Myc proto-oncogene product. For the C-terminal eGFP-JLPL-(963–1320) construct, the SacI-SalI fragment (1.9 kbp) from pEGFP-C2-JLPL was subcloned into the Sac1-Sal1 digest of pEGFP-C1. GST-based peptide fragments of JLPL were generated as follows: JLPL-(1–458), by releasing the EcoRI-SalI fragment from the pGEM4Z-JLPL-(1–458) construct and ligating into a EcoRI-SalI digest of pGEX5X-1 (BD Biosciences); JLPL-(436–794), by first ligating the PstI fragment of JLPL (nucleotides 1361–2439) into pBluescript II SK(+) and then releasing the BamHI-SalI fragment from the latter construct for subcloning into the BamHI-SalI digest of pGEX4T-2; JLPL-(963–1320), by subcloning the SacI-SalI fragment (1.9 kbp) from pBluescript II SK(+)-JLPL into the SacI-SalI digest of pGEM4Z, isolating the EcoRI-SalI fragment from this construct, and subcloning it into the EcoRI-SalI digest of pGEX4T-2; JLPL-(1246–1320), by releasing the BglII-EcoRI fragment (1 kbp) from pEGFP-C2-JLPLWT and subsequently subcloning it into the BamHI-EcoRI cohesive ends of pGEX1. Human JLPWT-S tagged sequence in pcDNA3 was kindly provided by Dr. Reddy and Dr. Lee. Construction of pEGFP-PIKfyveWT and pEGFP-PIKfyveK1831E has been described elsewhere (17, 43). pEGFP-C2-PIKfyve-(616–868) was generated by subcloning the BamHI insert of PIKfyveWT (nucleotides 1983–2742) into the BamHI digest of pEGFP-C2. Generation of a PIKfyve chimera (PIKfyve-cFab1) in which the last C-terminal 240 amino acids of the 2052-aa sequence of mouse PIKfyve are substituted with the corresponding 238-aa fragment of the yeast ortholog Fab1 is detailed in the supplemental “Experimental Procedures.” All new constructs were verified by sequencing, restriction endonuclease mapping, and Western blotting.
siRNAs and Antibodies—SmartPool™ siRNA duplexes targeting the mouse JLPL sequence were designed and generated by Dharmacon (catalog No. M-048194-00). SmartPool™ siRNA duplexes directed to mouse PIKfyve (catalog No. M-040127-01) or control mouse cyclophilin B (D-001136-01), characterized previously with mouse cells (15), were found to be similarly effective in CHO cells and were used herein. Rabbit polyclonal anti-JLP antiserum (Covance) was directed against the purified GST-JLPL-(1246–1320) peptide fragment. Unless otherwise stated, the crude antiserum (WS051) was used for immunoprecipitation and Western blotting. Rabbit polyclonal antibodies against PIKfyve (R7069) were characterized previously (44). Polyclonal anti-GFP (Ab290), anti-S, and monoclonal anti-α-tubulin antibodies were from AbCam, Santa Cruz Biotechnology, and Sigma, respectively. Anti-IRAP and anti-HA polyclonal antibodies were gifts from Drs. Paul Pilch and Mike Czech. Anti-Myc monoclonal antibody was produced by 9E10.2 hybridoma cells (ATCC). Anti-Tac monoclonal antibody was purified from 2A3A1H hybridoma cell line (ATCC) by affinity chromatography on protein G (Amersham Biosciences) following standard procedures (45). Purified anti-Tac IgG was conjugated to Alexa555 using the monoclonal antibody labeling kit and manufacturer's protocols (Invitrogen).
Cell Cultures and Transfections—CHO cell lines stably expressing human transferrin receptor and the Tac-furin (TRVb1/TTF cells) or Tac-TGN38 chimeric protein (TRVb1/Tac-TGN38 cells) under the control of a cytomegalovirus promoter were a kind gift from Dr. Fred Maxfield. Cells were maintained according to published protocols (6, 7). HEK293, COS7, and PC12 cells were cultured under conditions described in our previous studies (19, 46). Culturing and differentiation of 3T3L1 fibroblasts to adipocyte phenotype were described elsewhere (12, 15). The CHO cell lines and mouse 3T3L1 adipocytes were transfected with the mouse-specific siRNA duplexes (100 nm) by Oligofectamine (Invitrogen) and electroporation (0.2 nmol/5 × 106 cells), respectively. Seventy-two h post-transfection, cells were analyzed for the Tac chimera trafficking or lysed for immunoblotting. In the rescue experiments, 48 h following transfection with the siRNA duplexes, the TRVb1/TTF cells were transfected (Lipofectamine, Invitrogen) with the cDNAs of human JLP-S or mouse/yeast HA-PIKfyve-cFab1 chimera. Tac-furin trafficking and immunoblotting assays were performed 24 h later. COS7 cells were transfected with the indicated cDNAs by Lipofectamine. Twelve or 24 h post-transfection, cells were subjected to immunofluorescence microscopy or immunoblotting analyses.
Microinjections and Microscopy—Microinjections in single TRVb1/TTF or TRVb1/Tac-TGN38 cells, seeded on glass coverslips placed into 35-mm dishes (Corning), were performed as described previously (47). Briefly, prior to microinjection, the dish was filled with prewarmed F-12 medium supplemented with 20 mm Hepes, pH 7.4, and placed in a temperature-controlled dish heater (Warner Instruments) to maintain a constant temperature (36 °C). Purified GST fusion peptides, concentrated in the injection buffer (100 mm KCl, 4 mm sodium phosphate, pH 7.4) to 2.5 mg/ml protein, were mixed with goat IgG to aid visualization of injected cells. Reagents were micro-injected into the cytosol within a period of 15 min (>40 micro-injected cells). Cells were then returned in the incubator to recover for 45 min followed by media replacement and a Tac chimera trafficking assay with Alexa555-anti-Tac IgG (see below). Cells were fixed in 4% formaldehyde, permeabilized (0.5% Triton X-100 in phosphate-buffered saline), and stained with fluorescein isothiocyanate-conjugated donkey anti-goat IgG (Molecular Probes). Coverslips were mounted on slides using the SlowFade Antifade Kit (Molecular Probes). Fluorescence analysis was performed in a Nikon Eclipse TE 200 inverted fluorescence microscope using a ×40 lens and standard green and red fluorescence filters. Images were captured with a SPOT RT Slider charge-coupled device camera (Diagnostic Instruments) mounted on the microscope. In some experiments (Fig. 4C) the fluorescence analysis was performed with a motorized inverted confocal microscope (Olympus IX81) using a ×60 UplanApo lens. Stacks of images were taken in 1-μm steps and captured by a Retiga 1300 cooled mono 12-bit digital charge-coupled device camera (QImaging). The stack of images was subjected to deconvolution analysis, and resulting images at one and the same Z-level were used for colocalization analysis. Images were obtained using Image-Pro-Plus version 4.5.1 (Media Cybernetics), VolumScan, and Deconvolve software (VayTek, Inc.).
FIGURE 4.
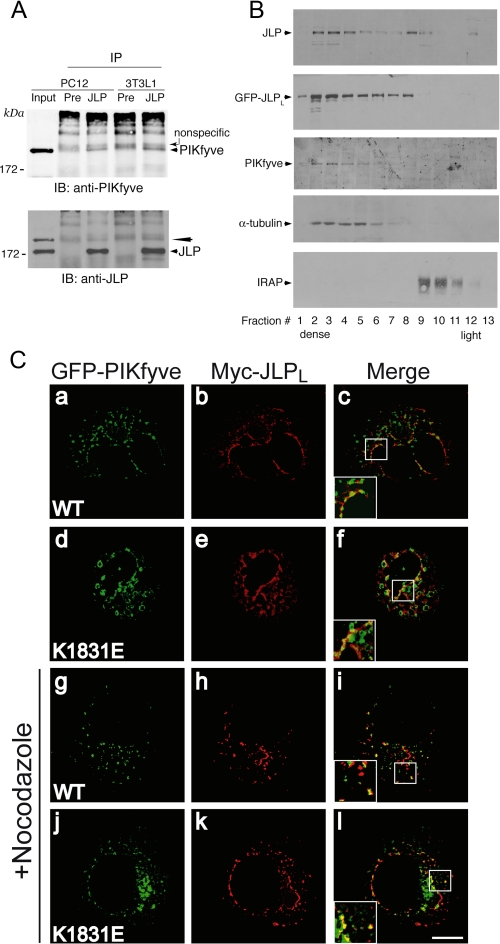
JLP interacts, cofractionates, and colocalizes with PIKfyve. A, equal protein amounts of fresh 0.1% Triton X-100 cell extracts derived from PC12 cells or 3T3L1 fibroblasts were immunoprecipitated with the preimmune serum from the JLP antibody production or anti-JLP antiserum as indicated. Washed immunoprecipitates (IP) along with the input of PC12 lysates (200 μg, representing 4% of cell protein subjected to immunoprecipitation) were resolved by SDS-PAGE and immunoblotting (IB) with the indicated antibodies, with a stripping step in between. PIKfyve coimmunoprecipitation is apparent only in the anti-JLP immunoprecipitates but not in that with preimmune serum (arrowhead). The denoted broad bands seen above the coimmunoprecipitated PIKfyve band are nonspecific, originating from the IgGs. The PIKfyve band is not fully stripped and is still seen in the JLP blot (long arrowhead). Shown are chemiluminescence detections of immunoblots from a representative experiment of three independent experiments for each cell line with similar results. B, the HEK293 cell line stably expressing HA-PIKfyveWT with or without transient coexpression of eGFP-JLPLWT was fractionated into total membranes and cytosol. Membrane fractions were subjected to equilibrium sedimentation in 30% iodixanol. Fractions were collected and analyzed by SDS-PAGE and immunoblotting with antibodies against the indicated proteins. Shown are chemiluminescence detections of blots from representative fractionations in eGFP-JLPLWT-expressing or non-expressing cells representative of four independent fractionations with similar results. C, COS7 cells were cotransfected with pCMV5-Myc-JLPLWT and either pEGFP-HA-PIKfyveWT or pEGFP-HA-PIKfyveK1831E. Twelve hours post-transfection cells were treated with nocodazole (30 min, 10 μm, 37 °C), or were left untreated as indicated, and then fixed and permeabilized. Expressed Myc-JLPLWT was detected with anti-Myc monoclonal antibody followed by Alexa568-conjugated anti-mouse IgG (panels b, e, h, and k). Expression of eGFP-PIKfyve was visualized by the GFP fluorescence (panels a, d, g, and j). Cells were viewed by confocal microscope (Olympus 1X81), and images were subjected to deconvolution analyses as described under “Experimental Procedures.” Panels on the right (c, f, i, and l) are the overlay of the deconvolved leftward images of the same Z-level through the middle of the cell. Insets represent computer-enlarged images of the boxed areas depicting a colocalization (yellow) of JLPLWT with PIKfyveWT or PIKfyveK1831E. Shown are typical images from two to three independent experiments. Bar, 10 μm.
Anti-Tac Uptake and Tac Chimera Steady-state Localization—The uptake of Alexa555-labeled anti-Tac IgG in TRVb1/TTF or TRVb1/Tac-TGN38 cells was performed as described previously (6–8). Briefly, subsequent to microinjections or transfections, cells were incubated for 10 min at 37 °C with Alexa555-anti-Tac IgG (4 μg/ml) in McCoy's 5A modified medium (Invitrogen) supplemented with 0.2% bovine serum albumin. Cells were then washed and chased with the same medium for 40 min. To reveal whether steady-state localization of the Tacchimeric protein was affected by the treatments indicated in Table 1, fixed cells were permeabilized (0.5% Triton X-100 in phosphate-buffered saline) and then incubated with anti-Tac IgG, which was detected by Alexa568-conjugated anti-mouse IgG.
TABLE 1.
Tac chimera trafficking to and steady-state localization at the TGN in TRVb1/TTF and TRVb1/Tac-TGN38 cells Cells scored for “perinuclear cluster” displayed the typical high focal concentration of Tac-associated immunofluorescence at the perinuclear TGN. Under conditions indicated by an asterisk Tac-furin was distributed predominantly in peripheral cytoplasmic punctae, but not at the perinuclear cluster, i.e. the TGN, as illustrated in Figs. 5 and 6. Quantification is based on counting at least 100 cells/condition in two to four coverslips (mean ± S.E.; *, p < 0.001 vs. respective controls).
|
Tac-Furin
|
Tac-TGN38
|
|||||
|---|---|---|---|---|---|---|
| Cell treatment | Trafficking perinuclear clustera | Steady-state localization perinuclear clusterb | Trafficking perinuclear clustera | Steady-state localization perinuclear clusterb | ||
| % cells | % cells | |||||
| siRNA-mediated protein knockdown | ||||||
| Cyclophilin (control) | 93 ± 4 | 94 ± 6 | 95 ± 6 | 96 ± 6 | ||
| PIKfyve | 12 ± 3* | 92 ± 5 | 90 ± 5 | 92 ± 5 | ||
| JLP | 15 ± 2* | 91 ± 6 | 94 ± 6 | 93 ± 2 | ||
| Expression of GFP-based constructs | ||||||
| GFP (control) | 90 ± 2 | 95 ± 4 | 94 ± 2 | 95 ± 4 | ||
| PIKfyve-(616–868) | 23 ± 3* | 93 ± 3 | 96 ± 3 | 93 ± 3 | ||
| JLPL-(963–1320) | 27 ± 4* | 94 ± 3 | 93 ± 2 | 94 ± 3 | ||
| PIKfyveK1831E | 22 ± 2* | 94 ± 4 | 94 ± 4 | 94 ± 4 | ||
| PIKfyveWT | 92 ± 2 | 96 ± 4 | 98 ± 5 | 94 ± 4 | ||
| JLPLWT | 90 ± 4 | 92 ± 4 | 92 ± 4 | 92 ± 4 | ||
| Microinjection | ||||||
| GST (control) | 92 ± 2 | 96 ± 3 | 95 ± 3 | 93 ± 4 | ||
| GST-JLPL-(1246–1320) | 15 ± 4* | 92 ± 4 | 91 ± 4 | 96 ± 4 | ||
Non-permeabilized TRVb1/TTF and TRVb1/Tac-TGN38 cells were analyzed for Tac-furin and Tac-TGN38 trafficking, respectively, as described in the legends for Figs 5 and 6
Cells were permeabilized, and Tac chimera steady-state localization was determined with anti-Tac as described under “Experimental Procedures”
Equilibrium Centrifugation—Total membrane fractions prepared from HEK293 cell lines stably expressing PIKfyveWT and resuspended in HES2+ buffer (20 mm Hepes, pH 7.5, 1 mm EDTA, supplemented with 1× protease inhibitor mixture (1 mm phenylmethylsulfonyl fluoride, 5 μg/ml leupeptin, 5 μg/ml aprotinin, 1 μg/ml pepstatin and 1 mm benzamidine) and 1× phosphatase inhibitor mixture (25 mm β-glycerophosphate, 10 mm sodium pyrophosphate, 50 mm NaF, and 2 mm NaVO3)) were mixed with iodixanol (OptiPrep; Sigma) in a polyallomer Quick-Seal centrifuge tube to 30% iodixanol and 128 mm sucrose concentrations as described previously (11). A self-generating gradient was performed by centrifugation to equilibrium for 4 h as specified previously (11). Fractions collected from the bottom of the tube were analyzed for protein content and by immunoblotting.
GST Protein Purification and Pulldown Assays—All GST fusion proteins were produced in transformed XA-90 Escherichia coli strain. Bacteria were grown for 1.5 h and then stimulated with 0.1 mm isopropyl-1-thio-β-d-galactopyranoside for an additional 6 h. Cells were lysed with 1 mg/ml lysozyme in a buffer containing 50 mm Hepes, pH 7.4, 150 mm NaCl, 5 mm EDTA, 1% Triton X-100, and 1 mm phenylmethylsulfonyl fluoride. Lysates were treated with DNase (0.1 mg/ml) in the presence of 10 mm MgCl2. Purification of the GST fusion proteins was achieved on GSH-agarose beads (Sigma) as described previously (11, 47). Cytosols, derived from HEK293 cells infected with adenoviruses encoding PIKfyveWT or empty virus prepared as previously described (11), were added to the immobilized GST fusions (2 μg/tube) and incubated for 18 h at 4 °C. Beads were washed three times in a cytosol-wash buffer (20 mm Hepes, pH 7.4, 5 mm EDTA, 150 mm NaCl, 5 mm MgCl2, 0.1% Triton X-100 and 1× protease inhibitor mixture) and then boiled in sample buffer. Captured proteins were analyzed by SDS-PAGE and immunoblotting. In some experiments, the GST proteins were eluted from the beads, dialyzed against 50 mm Hepes, pH 7.4, and used in cell microinjections. The concentration and quality of the purified proteins bound to or eluted from beads were determined electrophoretically by the intensity of the Coomassie-stained protein bands versus bovine serum albumin standard (Pierce).
Immunoprecipitation and Immunoblotting—For the immunoprecipitation analyses, cells were homogenized (10 strokes, 800 rpm) by a motor-driven homogenizer in the “homogenization” buffer (50 mm Hepes, pH 7.4, 2 mm EDTA, 150 mm NaCl, 0.1% Triton X-100, and 1× protease and phosphatase inhibitor mixtures). Cell lysates, precleared by centrifugation (20,000 × g for 15 min and then 200,000 × g for 30 min at 4 °C), were immunoprecipitated immediately after harvesting. Control immunoprecipitates with nonimmune rabbit or mouse IgG were run in parallel. Immunoprecipitations were carried out for 16 h at 4 °C with protein A-Sepharose CL-4B added in the final 1.5 h of incubation. Immunoprecipitates were washed five times with homogenization buffer. Immunoblotting with the antibodies indicated in the figure legends (Figs. 2, 3, 4) was performed subsequent to protein separation by SDS-PAGE (typically on 6% gels) and electrotransfer onto nitrocellulose membranes as described previously (11, 19). A chemiluminescence kit (Pierce) was used to detect the horse-radish peroxidase-bound secondary antibodies.
FIGURE 2.
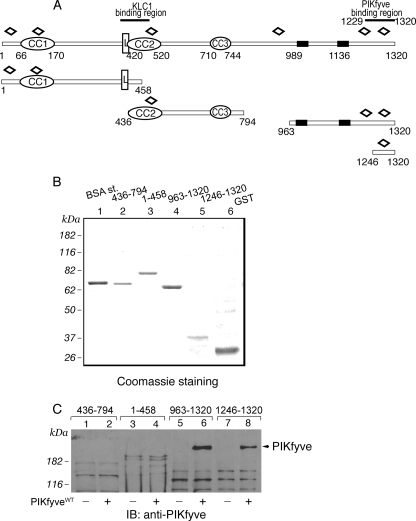
Expression and interaction of truncated peptide fragments of GST-JLPL with PIKfyveWT by a pulldown assay. A, schematic representation of the GST-JLPL fragments used in this study. CC, coiled-coil sequences; L, leucine zipper pattern; black rectangles, putative transmembrane domains; open diamonds, tetratricopeptide repeats. B, constructs indicated in A, expressed and purified from E. coli as detailed under “Experimental Procedures,” were resolved by SDS-PAGE for qualitative and quantitative estimation versus a bovine serum albumin protein standard (BSA st;1 μg). Shown is a Coomassie Blue staining from a typical GST fusion protein production. C, indicated GST fusions (2 μg), purified and immobilized on GSH-agarose beads, were incubated (18 h at 4 °C) in the presence of cytosol isolated from HEK293 cells infected with GFP-PIKfyveWT (+) or only GFP adenovirus (-). Beads were washed with cytosol wash buffer, and captured proteins were analyzed by SDS-PAGE (6% gel) followed by immunoblotting as indicated. Shown is a chemiluminescence detection of a representative immunoblot (IB) of two to four independent pulldown experiments for the individual truncated forms with similar results. Some nonspecific protein absorption on the beads is seen in all lanes, but only the C-terminal JLP fragments captured specifically PIKfyve (arrowhead).
FIGURE 3.
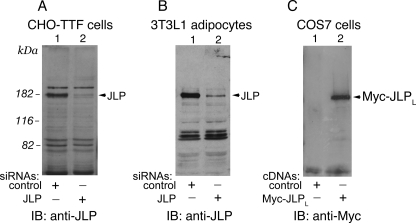
The ubiquitous 180-kDa JLP is specifically depleted in cells by siRNA. A and B, a CHO-based TRVb1/TTF cell line (A) or mouse 3T3L1 adipocytes (B) were transfected with siRNA duplexes targeting mouse JLP or control cyclophilin B as detailed under “Experimental Procedures.” Seventy-two h post-transfection, cell lysates (140 μg of protein) were analyzed by SDS-PAGE and immunoblotting (IB) with anti-JLP antiserum. Shown is a relatively high exposure blot, so that the selective ablation of endogenous JLP (lane 1 versus 2 in A and B, arrowheads) and absence of an off-target effect are apparent. C, COS7 cells were transiently transfected with pCMV5-Myc-mJLPLWT cDNA or the empty vector as indicated. Lysates were collected 24 h post-transfection. Equal protein amounts (60 μg) were analyzed by SDS-PAGE and immunoblotting with anti-Myc antibodies. Depicted is the 180-kDa Myc-JLPLWT. A–C, shown are chemiluminescence detections of representative immunoblots from two to five independent experiments with similar results.
Common Assays and Quantitation—Protein concentration was determined by bicinchoninic acid protein assay kit (Pierce). Protein levels were quantified from the intensity of the immunoblot bands by a laser scanner (Microteck) and UN-SCAN-IT software (Silk Scientific). Several films of different exposure times were quantified to assure the signals were within the linear range. Data are expressed as means ± S.E. Statistical analysis was performed by Student's t test, with p < 0.05 considered as significant.
RESULTS
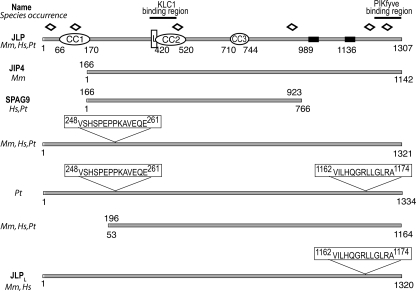
PIKfyve Interacts with the C-terminal Region of JLP/JIP4 Proteins—Our two-hybrid screens of HeLa cell cDNA library to identify proteins interacting with the PIKfyve chaperonin-like consensus sequence, thought to be engaged in various PIKfyve functional interactions (10, 17), resulted in 27 positive clones. One of them, reported previously (11), is the endosome-to-TGN transport factor Rab9 effector p40, the functionality of which has been linked to PIKfyve activity. Seven of the remaining 23 clones overlapped the extreme C-terminal 92–153 amino acids of JLP/JIP4. JIP4, JLP, and SPAG9 are well characterized splice variants of Jip4 (31). Because the former two, but not the latter, display the C-terminal sequences (Fig. 1), candidate PIKfyve interactors are therefore JLP, JIP4, both, or yet uncharacterized splice products of Jip4 displaying intact C termini.
FIGURE 1.
Schematic diagram of the JLP domain structure and splice variants of Jip4. Sequence analysis of the JLP domain structure and sequence alignment was performed using Pfam, REP, TMpred, and BLASTp. CC, coiled-coil sequences; L, leucine zipper pattern; black rectangles, putative transmembrane domains; open diamonds, tetratricopeptide repeats; Hs, Homo sapiens; Mm, Mus musculus; Pt, non-human primates; JLPL, JLP long form reported here (GenBank™ accession number FJ042496)
To obtain full-length JLP/JIP4 cDNA for further analyses, we screened a mouse adipocyte cDNA library. One of the isolated positive clones encoded the full-length JLP protein minus the initial seven amino acids. We completed the full-length sequence with a fragment from the mouse JLP N terminus (provided by Dr. Reddy). BLASTp analysis revealed that our clone represents a yet unknown alternative splicing of mouse Jip4, which incorporates 13 residues at position 1162 of the JLP sequence yielding a longer form of 1320 amino acids (Fig. 1). The work in this study was performed with this 1320-aa recombinant protein, referred to herein as JLPL (for long form) to distinguish it from the 1307-aa JLP form (Fig. 1). Data base searches revealed a transcript in Homo sapiens that is spliced identically to our 1320-aa mouse JLPL (Fig. 1). Our searches also revealed additional uncharacterized JLP/JIP4 splice variants. Thus, a 1321-aa form found in both primates and rodents differs from JLP by the inclusion of 14 residues at amino acid 247 (Fig. 1). Splice variants incorporating both the 14-aa N-terminal and the 13-aa C-terminal inclusions were also found in monkeys, but not in mice or humans, to yield the largest form among the splice variants of Jip4, comprising 1334 amino acids (Fig. 1). Intriguingly, alternative splicing of Jip4 to exclude the N-terminal region has been seen not only in JIP4 and SPAG9 (31, 38) but also in yet uncharacterized 1164-aa spliced forms found in primates and rodents, which lack the first coiled-coil/leucine zipper conserved domain shown to interact with transcription factors (37). Together, the protein sequence analyses demonstrate that Jip4 is transcribed in several splice variants, likely reflecting the need for JLPs/JIP4 to accommodate multiple proteins of different pathways and perform distinct functions.
To confirm the interaction of JLP/JIP4 C terminus with PIK-fyve by an alternative approach, several GST-conjugated JLPL peptide fragments were generated (Fig. 2A) and, after purification (Fig. 2B), tested for their ability to pull down adenovirally produced recombinant PIKfyve. As illustrated in Fig. 2C, only the fragments incorporating the C-terminal region, but not those of the N-terminal or middle region of JLPL, pulled down PIKfyveWT. Importantly, a 75-aa C-terminal fragment of JLPL (residues 1246–1320), which is 17 amino acids shorter than the shortest prey sequence obtained from the two-hybrid screens (residues 1229–1320), still displayed the ability to pull down PIKfyveWT. The extent of the interaction was slightly lower (by 30%, normalized per protein amounts) versus the C-terminal JLPL-(963–1320) fragment, as quantified by the intensity of the pulled down immunoreactive PIKfyve band (Fig. 2C). Thus, combined data from yeast two-hybrid and pull-down assays demonstrate that the C-terminal region of JLP/JIP4 interacts with PIKfyve and that as short a peptide fragment as 75 aa of the JLP/JIP4 extreme C terminus is sufficient to support this interaction.
JLP-PIKfyve Interaction in Mammalian Cells—To assess whether endogenous JLP/JIP4 and PIKfyve interact in mammalian cells, we generated anti-JLP antibodies directed to a region at the JLPL C terminus (residues 1246–1320), which is one of the most divergent regions versus the corresponding region in the JIP3 isoforms (39% similarity). On Western blots, our C-terminal antibodies detected a band of 180 kDa, for which authenticity as JLP was verified by siRNA-mediated protein knockdown (Fig. 3, A and B). We observed highly selective ablation of the 180-kDa band in mouse 3T3L1 adipocytes and CHO-based TRVb1/TTF cells transfected with the siRNA pool targeting JLPL (Fig. 3, A and B). Somewhat slower mobility in SDS-polyacrylamide gels versus the calculated molecular weight of 146,120 of the deduced JLPL sequence was also seen with ectopically expressed Myc-tagged JLPL in COS cells (Fig. 3C), further underscoring that the 180-kDa band immunoreactive with our antibodies was JLP. Concordantly, a higher apparent molecular mass of 180 kDa in SDS-gels has also been observed in other studies that utilized antibodies directed to different regions of the JLP molecule (37, 39). Consistent with previous reports characterizing JLP and JIP4 as ubiquitous proteins (37–39), the immunoprecipitation/immunoblotting analyses with our JLP antibodies found the 180-kDa JLP band to be present at variable degrees in all mouse tissues examined, such as brain, fat, liver, skeletal muscle, and mammary gland (data not shown).
Coimmunoprecipitation analysis with the generated JLP antibodies was then exploited to reveal whether endogenous JLP interacts with PIKfyve in mammalian cells. As illustrated in Fig. 4A, in accordance with our data from the two-hybrid screens and pulldown assays, we observed that PIKfyve was coimmunoprecipitated with anti-JLP in PC12 cells and 3T3L1 fibroblasts. Consistent with these observations, in equilibrium density gradients of membranes isolated from a HEK293-PIKfyveWT stable cell line characterized previously (11, 19, 46), endogenous JLP or expressed GFP-JLPLWT largely cofractionated with PIKfyve in the denser part of the gradient, where protein markers of cellular cytoskeleton (α-tubulin) but not the endosomal system (IRAP, transferrin receptors, and Rab4) were predominantly detected (Fig. 4B, and not shown).
Whereas JLP and PIKfyve antibodies, produced by us or others, readily detected endogenous levels of JLP and PIKfyve by immunoblotting, they failed to convincingly detect the endogenous proteins by immunofluorescence microscopy in most mammalian cell types tested thus far (this study, data not shown, and Refs. 10, 12, 14, 38). To overcome this limitation and learn more about the JLP intracellular distribution relative to PIKfyve, we transiently transfected COS7 cells with Myc-JLPLWT and eGFP-PIKfyveWT. Confocal microscopy was then performed in cells expressing relatively low levels of both proteins, because such conditions might more faithfully resemble the localization pattern of the endogenous proteins as observed for PIKfyve (12, 43). Consistent with previous studies (37, 38, 41), in the confocal images we observed that overexpressed JLPLWT attained primarily cytoplasmic and perinuclear distribution. Intriguingly, closer inspection of the deconvoluted confocal images from individual sections taken through the middle of the cell revealed that the JLPL-positive perinuclear structures displayed tubular profiles (Fig. 4C, panels b and e). A subpopulation of the JLPLWT-positive elongated structures was also positive for PIKfyveWT, whereas the majority of the PIKfyveWT-positive small vesicles in the cell periphery were JLPL-negative (Fig. 4C, panels a–c). This conclusion was further corroborated by inspecting the colocalization between Myc-JLPLWT and dominant-negative eGFP-PIKfyveK1831E at early expression times (∼12 h), under which conditions early endosome membrane enlargement was apparent but other phenotypic changes, such as vacuolation, were not (17, 18). Thus, predominant colocalization of eGFP-PIKfyveK1831E-positive signals with the Myc-JLPL-associated immunofluorescence was seen on a subpopulation of perinuclear vesicles with tubular profiles, whereas the dilated eGFP-PIKfyveK1831E-positive early endosomes in the cell periphery remained largely Myc-JLPLWT-negative (Fig. 4C, panels d–f). Disruption of microtubules with nocodazole caused a more fragmented staining pattern for Myc-JLPLWT, although the colocalization between the two proteins persisted (Fig. 4C, panels h–l). Intriguingly, a relative increase in the colocalization of Myc-JLPLWT with PIKfyveWT or PIKfyveK1831E on vesicles at the cell periphery was noted (Fig. 4C, panels i and l), suggesting that the colocalization in tubule-like perinuclear structures requires, in part, an intact microtubular network. Together, the data presented in Fig. 4 indicate that a subpopulation of PIKfyve and JLPL associate and colocalize in tubule-like perinuclear structures.
Several PIKfyve-interacting proteins have been identified recently, such as the p85 phosphatidylinositol 3-kinase (48), the PtdIns(3,5)P2-specific Sac3 phosphatase (19), the PIKfyve activator ArPIKfyve (46), and the p40 endosome-to-TGN transport factor (11). None of those, however, appeared to associate with JLP in a cell context, as judged by the lack of immunoreactive coimmunoprecipitated bands corresponding to the electrophoretic mobility of these proteins upon blotting of JLP immunoprecipitates with the respective antibodies (data not shown). Thus, although anticipated to do so, JLP is unlikely to function as a scaffold of the enzyme module responsible for intracellular PtdIns(3,5)P2 production (19). Importantly, unlike PIKfyve associations with Sac3 and ArPIKfyve, which persist under stringent detergent conditions of incubation during the coimmunoprecipitation reaction (i.e. NP-40 and deoxycholate (19)), PIKfyve association with JLP, as with p40 (11), requires milder conditions (0.1% Triton X-100). These data indicate that PIKfyve association with JLP is of low affinity.
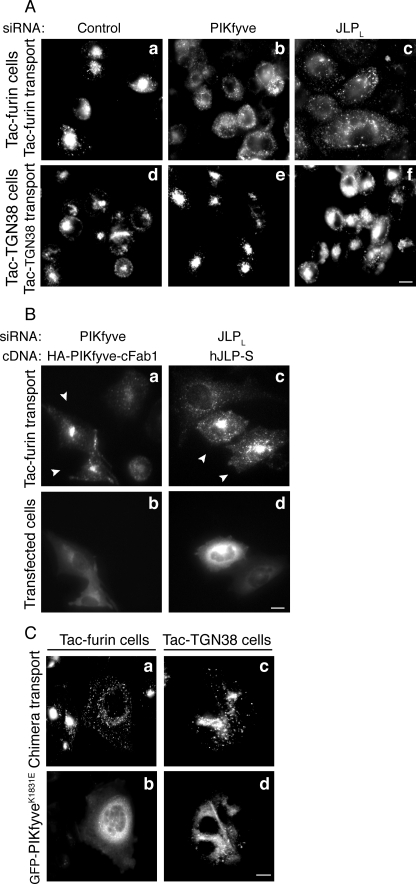
Depletion of PIKfyve or JLP Inhibits Tac-Furin but Not Tac-TGN38 Trafficking—JLP and JIP4 have been studied exclusively as scaffolds of Myc/Mac transcription factors and JNK/p38 MAP kinase signaling modules (31). Based on the well characterized abilities of JLP and JIP4 to associate with the light chain of the kinesin1 motor protein (38, 41) and functioning of the homologous isoform JIP3 in microtubule-dependent trafficking (32, 33), a JLP role in microtubule-based membrane trafficking can be anticipated. However, to date no direct information is available to implicate JLP or JIP4 in any membrane transport event. Reasoning that JLP interacts with PIKfyve to aid PIKfyve functioning in endosomal processing in the course of cargo transport, we tested the effect of the two proteins on endosome-to-TGN trafficking of furin, known to be dependent on both an intact microtubule network and the presence of PIKfyve (7, 8, 14). We utilized TRVb1/TTF cells, a CHO cell line stably expressing Tac-furin chimeric protein. This chimera consists of the extracellular/transmembrane domain of the interleukin-2 receptor α-chain (Tac) and the cytoplasmic domain of furin, which directs the chimera from the cell surface to the TGN via EE/SE without entering the recycling pathway (7, 49). From EE/SE, Tac-furin is delivered to the TGN via a single pass through late endosomes and/or the maturing ECV/MVB structures that require intact microtubules in the process of maturation (7, 20). We monitored the Tac-furin trafficking and TGN delivery by the Alexa555-labeled anti-Tac antibody, which was allowed to internalize along with Tac-furin for 10 min followed by a 40-min chase. Consistent with previous studies (6–8), we found that under these conditions Tac-furin reaches steady-state distribution at the TGN in control cells, which is manifested by the accumulation of the anti-Tac immunofluorescence signals as a cluster at the perinuclear region (Fig. 5A, panel a). siRNA-mediated PIKfyve protein depletion (>85%; Ref. 15 and this study, not shown) in the TRVb1/TTF cells profoundly inhibited the endosome-to-TGN delivery of Tac-furin. As illustrated in Fig. 5A (panel b) Tac-furin remained dispersed into peripheral cytosolic punctae in the PIKfyve-depleted cells within the time course of investigation. These data are in agreement with previous observations for an arrest of CD8-furin chimera trafficking upon PIKfyve depletion in HeLa cells (14), extending the evidence for the Tac-furin chimera transport in TRVb1/TTF cells. Tac-furin delivery to the TGN was also arrested by expression of dominant-negative kinase-deficient PIKfyveK1831E (Fig. 5C), consistent with the notion that PIKfyve-catalyzed PtdIns(3,5)P2 synthesis plays a key role in this trafficking event. Remarkably, siRNA-directed JLP knockdown (>85%, see Fig. 3A), like PIKfyve ablation or PIKfyveK1831E expression, profoundly inhibited the endosome-to-TGN transport of Tac-furin. As illustrated in Fig. 5A (panel c) the chimera was seen redistributed to peripheral punctae in cells with JLP depletion, which was observed in ∼85% of the cells (Table 1). Importantly, neither treatment disrupted the predominant steady-state distribution of Tac-furin at the perinuclear TGN, as revealed by anti-Tac IgG staining in permeabilized cells (Table 1). Collectively, these data demonstrate similar trafficking defects induced by JLP and PIKfyve depletion, indicating that both proteins are required for proper endosome-to-TGN trafficking of Tac-furin.
FIGURE 5.
Tac-furin but not Tac-TGN38 trafficking to the TGN is arrested by PIKfyve or JLP protein depletion or by PIKfyveK1831E expression. TRVb1/TTF or TRVb1/Tac-TGN38 cell lines stably expressing Tac-furin or Tac-TGN38 chimera were seeded on coverslips placed onto 35-mm plates. Cells were transfected with siRNA duplexes (100 nm) targeting specific regions in mouse JLP, PIKfyve, or cyclophilin B (Control; A and B) and after 48 h were cotransfected with pcDNA3-hJLPWT-S or the pCMV5-HA-PIKfyve-cFab1 chimera as indicated (B). Cells were transfected with pEGFP-HA-PIKfyveK1831E cDNA (C). At 72 h (A and B) or 12 h post-transfection (C), cells were incubated with Alexa555-anti-Tac monoclonal antibody (10 min, 37 °C), chased (40 min), fixed, and then processed for fluorescence microscopy as detailed under “Experimental Procedures.” Tac chimera trafficking was monitored by Alexa555 fluorescence (A, panels a–f; B, panels a and c; C, panels a and c). Expression of HA-PIKfyve-cFab1 (B, panel b), hJLP-S (B, panel d), and eGFP-PIKfyveK1831E (C, panels b and d) was detected by polyclonal anti-HA antibodies, anti-JLP antibodies (the dilution detects only the overexpressed protein), and GFP fluorescence signals, respectively. Cells were observed in a Nikon Eclipse TE 200 inverted fluorescence microscope. Note the Tac-furin distribution to cytoplasmic punctae in cells with JLP or PIKfyve depletion (A, panels b and c) or PIKfyveK1831E expression (C, panel a) and no apparent perinuclear TGN clusters. Expression of HA-PIKfyve-cFab1 and hJLPWT-S restores the steady-state TGN localization of Tac-furin in the PIKfyve- and JLP-depleted cells, respectively (B, arrowheads in panels a and c) in >85% of the expressing cells. Bar, 10 μm.
To confirm that the impaired Tac-furin trafficking was due to depletion of JLP and PIKfyve, attempts were made to restore protein levels following the siRNA treatment. For this purpose, 48 h after protein depletion, the TRVb1/TTF cells were ectopically transfected with hJLPWT-S or the HA-PIKfyve-cFab1 chimera. Both constructs displayed mismatches (6 for JLP and 26 for PIKfyve, out of 76) versus the respective sequences of mouse JLP or PIKfyve siRNA duplexes, suggesting that expression of these constructs might be relatively resistant to the siRNAs. Indeed, Western blotting (supplemental Fig. S1) and immunofluorescence microscopy analysis (Fig. 5B) demonstrated significant expression of both hJLPWT-S and the HA-PIKfyve-cFab1 chimera in the TRVb1/TTF cells subsequent to depletion of endogenous levels of the respective protein. Importantly, as illustrated in Fig. 5B, Tac-furin delivery to the TGN was rescued selectively in cells expressing hJLPWT or the PIKfyve-cFab1 chimera, indicating that the impaired trafficking was due to depletion of JLP and PIKfyve endogenous levels.
To examine whether this arrest of endosome-to-TGN transport is confined to the microtubule-dependent endocytic transport of Tac-furin, we next monitored the effect of JLP and PIKfyve protein depletion on the endocytic transport of the Tac-TGN38 chimera. Unlike Tac-furin, the Tac-TGN38 endosome-to-TGN trafficking is reportedly independent of microtubules (7). From the EE/SE it arrives at the TGN by the recycling pathway and thus bypasses the microtubule-dependent step of early endosome maturation (7). Intriguingly, neither PIKfyve nor JLP protein loss affected Tac-TGN38 arrival at the TGN. As seen from images presented in Fig. 5A (panels d–f), Tac-TGN38 acquired the control steady-state localization at the perinuclear TGN in nearly all JLP- or PIKfyve-depleted cells (Table 1). Likewise, Tac-TGN38 delivery to the TGN was unaffected upon expression of the dominant-negative kinase-deficient PIKfyve mutant (Fig. 5C, panels c and d; Table 1). This result, taken together with the selective defects on Tac-furin transport by the PIKfyve loss (Fig. 5A and Table 1), demonstrates a previously unrecognized specificity of PIKfyve functionality in microtubule-dependent endocytic trafficking of cargoes that reach the TGN from EE/SE via ECV/MVB or late endosomes but not in the microtubule-independent cargo delivery to the TGN via the endocytic recycling compartment. Collectively, these data are consistent with the notion that PIKfyve and JLP are specifically required in the microtubule-based endosome-to-TGN transport of Tac-furin but not in the microtubule-insensitive transport of Tac-TGN38.
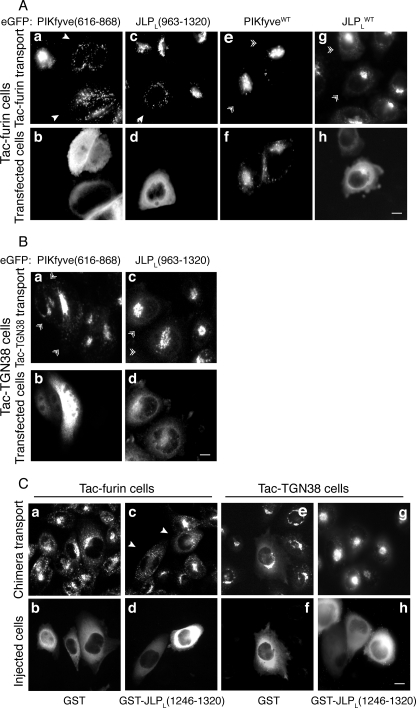
Peptide Fragments of the PIKfyve-JLP Contact Sites Arrest Tac-Furin but Not Tac-TGN38 Trafficking to the TGN—To reveal whether the association between JLP and PIKfyve is required in the microtubule-dependent Tac-furin transport to the TGN, we next examined the effect of the domains in the two proteins, found to interact with each other by the two-hybrid and pulldown assays. We reasoned that these peptide fragments, if present at high concentrations, will antagonize the functional interaction between the endogenous PIKfyve and JLP and, ultimately, inhibit the Tac-furin trafficking from endosomes to the TGN. We first examined the outcome of ectopic expression of eGFP-based fragments from the JLPL C-terminal region (residues 963–1320) or the PIKfyve cpn60_TCP1 domain (residues 616–868) by monitoring the internalized Alexa555-labeled anti-Tac antibody (10 min pulse-40 min chase) in transiently transfected TRVb1/TTF cells. As illustrated in Fig. 6A (panels a and c) expression of both peptide fragments resulted in a profound redistribution of Tac-furin into peripherally dispersed cytoplasmic punctae seen in the majority of transfected cells (Table 1). However, the Tac-furin steady-state localization at the perinuclear TGN, determined by anti-Tac staining in permeabilized cells was unaffected (Table 1). By contrast, under these conditions expressed eGFP-based wild-type forms of PIKfyve or JLPL did not alter the normal Tac-furin trafficking to, and steady-state distribution at the TGN (Fig. 6A, panels e and g; Table 1). The similar trafficking defects induced by the interacting peptide fragments of the JLPL-PIKfyve contact sites are consistent with the notion that disrupted interaction of JLP with PIKfyve underlies the retardation of the endosome-to-TGN transport of Tac-furin. By contrast, neither the interacting fragments nor the wild-type forms induced any changes in Tac-TGN38 trafficking in the TRVb1/Tac-TGN38 cells transfected with the eGFP constructs (Fig. 6B and Table 1). Together, these data implicate the interaction between PIKfyve and JLP acting selectively in the microtubule-based trafficking of Tac-furin to the TGN.
FIGURE 6.
Peptide fragments of the PIKfyve - JLPL contact sites arrest Tac-furin but not Tac-TGN38 trafficking to the TGN. A and B, TRVb1/TTF (A) and TRVb1/Tac-TGN38 cell lines (B) stably expressing Tac-furin and Tac-TGN38 chimera, respectively, seeded on coverslips were transfected with the indicated pEGFP-based constructs of PIKfyve-(616–868), JLPL-(963–1320), PIKfyveWT, or JLPLWT as indicated. Twenty-four h post-transfection the cells were incubated with Alexa555-anti-Tac monoclonal antibody (10 min, 37 °C), chased (40 min), and then fixed and processed for fluorescence microscopy as detailed under “Experimental Procedures.” Tac chimera trafficking was evaluated by Alexa555 fluorescence (A, panels a, c, e, and g; B, panels a and c), whereas expressed eGFP proteins were detected by the GFP fluorescence signals (A, panels b, d, f, and h; B, panels b and d). Denoted are the arrest of Tac-furin delivery to the TGN and distribution in cytoplasmic punctae by expression of JLPL-(963–1320) or PIKfyve-(616–868) (A, arrowheads) but not by expression of JLPLWT or PIKfyveWT (A, double arrowheads). Tac-TGN38 delivery to the TGN is unaltered by JLPL-(963–1320) or PIKfyve-(616–868) expression (B, double arrowheads). C, the cell lines were microinjected with the indicated purified GST-JLP-(1246–1320) peptide fragment or GST alone (2.5 mg/ml), mixed with goat IgG (to visualize injected cells) as described under “Experimental Procedures.” Washed cells were left to recover (45 min) and then incubated (10 min, 37 °C) with Alexa555-anti-Tac monoclonal antibody, chased (40 min), fixed, and stained with fluorescein isothiocyanate-conjugated donkey anti-goat IgG. Tac chimera trafficking was evaluated by Alexa555 fluorescence (panels a, c, e, and g), whereas the injected cells were evaluated by fluorescein isothiocyanate (panels b, d, f, and h). Arrest of Tacfurin delivery (arrowheads in panel c) but not Tac-TGN38, to the TGN and distribution in cytoplasmic punctae is apparent in cells injected with GST-JLP-(1246–1320). In A–C, cells were observed in a Nikon Eclipse TE 200 inverted fluorescence microscope. Bar, 10 μm.
The two-hybrid screen demonstrated that as few as 92 residues of the C terminus of JLP/JIP4 interact with PIKfyve. The pulldown assay further narrowed down the interacting region to a 75-aa peptide fragment of the extreme C terminus spanning amino acids 1246–1320 (see Fig. 2C). To reveal whether this short JLP C-terminal peptide still displays the ability to specifically reduce the endosome-to-TGN transport of Tacfurin as found with the JLPL-(963–1320) C-terminal peptide, the TRVb1/TTF and TRVb1/Tac-TGN38 cells were microinjected with bacterially produced and purified GST-JLPL-(1246–1320) fusion peptide or with GST alone. The kinetics of Tac-chimera trafficking was monitored with the Alexa555-labeled Tac-antibody under the protocol of a 10/40-min pulse/chase. Data illustrated in Fig. 6C demonstrate that whereas Tac-TGN38 reached the TGN over this time course (panel g), Tac-furin did not achieve the steady-state TGN distribution. Instead, it was found in an exclusively punctate distribution throughout the cytoplasm in almost all cells injected with GST-JLPL-(1246–1320) (Fig. 6C, panel c; Table 1). By contrast, when the TRVb1/TTF cells were injected with a control GST fragment or remained noninjected, the characteristic steady-state accumulation of Tac-furin at the TGN was apparent (Fig. 6C, panels a and c; Table 1). Collectively, these data demonstrate that Tac-furin trafficking, but not that of Tac-TGN38, is dependent on the 75-aa peptide fragment from the JLP C terminus that associates with PIKfyve.
DISCUSSION
In mammalian cells PIKfyve-catalyzed PtdIns(3)P-to-PtdIns(3,5)P2 conversion has been proposed to regulate endosome processing in the course of cargo transport (reviewed in Ref. 10). Trafficking pathways, both constitutive and regulated, which emanate from or traverse early endosomes en route to the TGN or to later endosomal compartments have been found to be sensitive to PIKfyve dysfunction (10). The complex functionality of PIKfyve is mediated by a number of physically associated or functionally related upstream regulators and downstream protein effectors, the identity and precise roles of which are under intensive investigation (10, 50). The data presented herein identify a novel association between PIKfyve and the JNK/p38 MAP kinase scaffolding protein JLP/JIP4, also known for the ability to interact with the light chain of the microtubule-based kinesin1 motor protein (38, 41). The association engages a region of the PIKfyve cpn60_TCP1 consensus sequence and the last 75 residues of the JLP C terminus. We further report that depletion of JLP and PIKfyve, or introduction of the interacting peptide fragments, induces defects in Tac-furin but not in Tac-TGN38 endosome-to-TGN transport. These two cargoes use divergent routes for reaching the TGN from EE/SE, with the former, but not the latter, requiring endosome maturation coordinated by the microtubule network (7). Therefore, our data mechanistically implicate JLP, PIKfyve, and their association in the microtubule-dependent endocytic trafficking to the TGN, which proceeds through maturing endosomes and late endosomes. We suggest that JLP is a PIKfyve accessory factor in endosome cargo trafficking to the TGN, which functions as a link in coupling PIKfyve to microtubule tracks to selectively facilitate the microtubule-dependent traffic from endosomes to the TGN. Our study thus unravels the previously unrecognized specificity of PIKfyve functionality in microtubule-dependent cargo trafficking from endosomes to the TGN. To the best of our knowledge, our data demonstrate for the first time that JLP is required in endosome-based trafficking to assist PIKfyve and that the PIKfyve function is limited strictly to the endosome-to-TGN trafficking of cargoes with transport requirements for an intact microtubule network.
An important point of the current study is the identification of the C-terminal region of JLP spanning amino acids 1246–1320 as the critical domain of interaction with the PIKfyve cpn60_TCP1 domain. The association is apparent only under mild detergent conditions (Figs. 2C and 3A), suggesting it is of low affinity and, therefore, transient in nature. Such an interaction will allow dynamic formation-disassembly of functional complexes on a different time scale in the course of endosome-to-TGN cargo transport. For example, the recruitment of the Rab9 effector p40, which is also required in endosome-to-TGN traffic of CI-MPR in association with the PIKfyve cpn60_TCP1 domain (11), could be coordinated with that of JLP. On the other hand, JLP interacts with KLC1 via a region distinct from that required in the PIKfyve interaction (Fig. 1 and Ref. 41). Although ternary complexes of JLP with PIKfyve and KLC subunits of kinesin1 were not addressed in our study, the fact that these two interactions proceed via non-overlapping regions is consistent with the notion that JLP associations with PIKfyve and KLC1 could occur simultaneously. Such a binding mechanism would allow JLP to couple PIKfyve-dependent endosomal functions to kinesin-driven motility on microtubules.
Although our data provide compelling evidence that JLP promotes PIKfyve-selective functioning in endosome-to-TGN cargo transport that requires an intact microtubule network, the step(s) sensitive to the JLP-PIKfyve interaction is unclear. Recent in vivo and in vitro studies have revealed mechanistic links integrating the function of motor proteins and microtubules in both vesicle transport and membrane trafficking events such as budding, fission, and fusion (3–5, 20–29). All of those appear to occur in association with microtubules, typically via complex bidirectional movements that require both the plus-end conventional kinesins and the minus-end motors (24). Because PIKfyve and PtdIns(3,5)P2 have been implicated in fission and fusion events in the endosomal system (17, 19), which are mechanistically coupled with the process of early endosome maturation in the course of cargo transport to late endosomes or the TGN (51), we suggest that JLP links PIKfyve to a motor-driven endosome processing associated with microtubules. This conclusion is corroborated in the current study by documenting selective defects in endosome-to-TGN transport of Tac-furin that, in contrast to Tac-TGN38, requires microtubule-based early endosome maturation to ECV/MVBs and late endosomes for subsequent transport progression to the TGN (Figs. 5, A and B, and 6). Furthermore, our observation of a selective retardation in Tac-furin delivery to the TGN, but not in that of Tac-TGN38, by dominant-negative kinase-deficient PIKfyveK1831E (Fig. 5C) is consistent with the notion that the microtubule-dependent endosome-to-TGN trafficking, in addition to the PIKfyve interaction with JLP, requires PIKfyve-catalyzed PtdIns(3,5)P2 synthesis. This idea is further underscored by the findings for a potential downstream effector of PtdIns(3,5P)2, the mammalian WIPI49 protein that functions in this pathway (52). Whether PIKfyve-dependent PtdIns(3,5)P2 production is related to the PIKfyve association with JLP or both events occur independently within the same segment of the endosome processing step requires further investigation.
In addition to the kinesin motor protein (38, 41) and PIK-fyve (this study), JLP and JIP4 reportedly interact with a number of proteins such as the JNK/p38 MAP kinase signaling modules, the transcription factors c-Myc and Max (37, 38), the receptor-like transmembrane protein Cdo (53), and the α-subunit of heterotrimeric G13 (54). Except for the latter, these interactions have been mapped to the N-terminal or middle region of JLP/JIP4 proteins (31). Reportedly, JLP association with Gα13 engages the C-terminal domain, spanning the last 142 residues at the C terminus (residues 1178–1320 in JLPL), but shorter fragments have not been exploited (54). Therefore, it is unclear at present whether the JLP binding regions for Gα13 and PIKfyve overlap or occupy distinct segments of the JLP C terminus and whether these interactions are related. A significant challenge for future investigation is whether in a cell context the different JLP/JIP4 splice variants displaying an intact C terminus (Fig. 1) are selectively engaged in interactions with PIKfyve, Gα13, or yet-to-be-identified partners. It is likely that spatially and temporally regulated cellular events may favor interactions with a particular JLP splice form, resulting in a distinct functional outcome. Furthermore, we have considered the possibility that JLP may also link PIKfyve to the stress kinase signaling pathways. However, our inability to detect coimmunoprecipitated p38 MAP kinase or JNK with PIKfyve and vice versa, even at highly overexposed blots (data not shown), makes this hypothesis unlikely. It should be emphasized that the relative contribution of JLP/JIP4 in the activation of JNK signaling pathways is presently unclear. Thus, whereas JLP binds to JNK, enhancing its activation (37, 41, 54, 55), JIP4 does not seem to associate with the JNK signaling module, nor does it alter JNK activation (38). Concordant with the latter observations, disruption of Jip4 does not attenuate JNK activation under various stress cues in cells and tissues of JLP-/--deficient mice (39). It is therefore conceivable that JLP/JIP4 may support specific functions outside of JNK or p38 MAP kinase signaling. Its role as a kinesin adapter assisting PIKfyve functioning in the microtubule-associated endosome events in the course of endosome-to-TGN cargo transport, suggested herein, supports this notion.
In summary, the data presented in the current study indicate an association of PIKfyve with JLP and that proper endosome operations in the course of furin, but not of TGN38, transport from endosome to the TGN require PIKfyve, JLP, and their association.
Supplementary Material
Acknowledgments
We thank Drs. Fred Maxfield and Florencia Shapiro for the stable cell lines; Dr. Tim McGraw for instructions in anti-Tac antibody purification/labeling; Drs. Reddy and Lee for the constructs of human JLP-S and the N-terminal fragment of mouse JLP; Dr. Scott Emr for the pRS416(Fab1) clone; and Drs. Mike Czech and Paul Pilch for the anti-HA and anti-IRAP antibodies, respectively. We also thank Linda McCraw for the outstanding secretarial assistance. The senior author expresses gratitude to Violeta Shisheva for the many years of support.
The nucleotide sequence(s) reported in this paper has been submitted to the GenBank™/EBI Data Bank with accession number(s) FJ042496.
This work was supported, in whole or in part, by National Institutes of Health Research Grant DK58058. This work was also supported by a grant from the American Diabetes Association (to A. S.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains supplemental “Experimental Procedures” and Fig. S1.
Footnotes
The abbreviations used are: TGN, trans-Golgi network; EE/SE, early endosome/sorting endosome; ECV/MVB, endosome carrier vesicle/multivesicular body; PIKfyve, phosphoinositide kinase for position 5 containing a fyve finger domain; PtdIns, phosphatidylinositol; CHO, Chinese hamster ovary; HEK, human embryonic kidney; CI-MPR, cation-independent mannose 6-phosphate receptor; IRAP, insulin-responsive aminopeptidase; JIP, c-Jun N-terminal kinase interacting protein; JNK, c-Jun NH2-terminal kinase; JLP, JNK-interacting leucine zipper protein; MAP, mitogen-activated protein; KLC, kinesin light chain; GFP, green fluorescent protein; eGFP, enhanced green fluorescent protein; GST, glutathione S-transferase; siRNA, short interfering RNA; SPAG9, sperm-associated antigen 9; aa, amino acid(s); HA, hemagglutinin; WT, wild type.
References
- 1.Mukherjee, S., Ghosh, R. N., and Maxfield, F. E. (1997) Physiol. Rev. 77 759-803 [DOI] [PubMed] [Google Scholar]
- 2.Miaczynska, M., and Zerial, M. (2002) Exp. Cell Res. 272 8-14 [DOI] [PubMed] [Google Scholar]
- 3.Conner, S. D., and Schmid, S. L. (2003) Nature 422 37-44 [DOI] [PubMed] [Google Scholar]
- 4.Maxfield, F. R., and McGraw, T. E. (2004) Nat. Rev. Mol. Cell Biol. 5 121-132 [DOI] [PubMed] [Google Scholar]
- 5.Gruenberg, J., and Stenmark, H. (2004) Nat. Rev. Mol. Cell Biol. 5 317-323 [DOI] [PubMed] [Google Scholar]
- 6.Ghosh, R.N., Mallet, W. G., Soe, T. T., McGraw, T. E., and Maxfield, F. R. (1998) J. Cell Biol. 142 923-936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mallet, W. G., and Maxfield, F. R. (1999) J. Cell Biol. 146 345-359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schapiro, F. B., Soe, T. T., Mallet, W. G., and Maxfield, F. R. (2004) Mol. Biol. Cell 15 2884-2894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lin, S. X., Mallet, W. G., Huang, A. Y., and Maxfield, F. R. (2004) Mol. Biol. Cell 15 721-733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shisheva, A. (2008) Cell Biol. Int. 32 541-604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ikonomov, O. C., Sbrissa, D., Mlak, K., Deeb, R., Fligger, J., Soans, A., Finley, R. L., Jr., and Shisheva, A. (2003) J. Biol. Chem. 278 50863-50871 [DOI] [PubMed] [Google Scholar]
- 12.Ikonomov, O. C., Sbrissa, D., Mlak, K., and Shisheva, A. (2002) Endocrinology 143 4742-4754 [DOI] [PubMed] [Google Scholar]
- 13.Ikonomov, O. C., Sbrissa, D., Foti, M., Carpentier, J.-L., and Shisheva, A. (2003) Mol. Biol. Cell 14 4581-4591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rutherford, A. C., Traer, C., Wassmer, T., Pattni, K., Bujny, M. V., Carlton, J. G., Stenmark, H., and Cullen, P. J. (2006) J. Cell Sci. 119 3944-3957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ikonomov, O. C., Sbrissa, D., Dondapati, R., and Shisheva, A. (2007) Exp. Cell Res. 313 2404-2416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jefferies, H. B., Cooke, F. T., Jat, P., Boucheron, C., Koizumi, T., Hayakawa, M., Kaizawa, H., Ohishi, T., Workman, P., Waterfield, M. D., and Parker, P. J. (2008) EMBO Rep. 9 164-170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ikonomov, O. C., Sbrissa, D., and Shisheva, A. (2001) J. Biol. Chem. 276 26141-26147 [DOI] [PubMed] [Google Scholar]
- 18.Ikonomov, O. C., Sbrissa, D., and Shisheva, A. (2006) Am. J. Physiol. 291 C393-C404 [DOI] [PubMed] [Google Scholar]
- 19.Sbrissa, D., Ikonomov, O. C., Fu, Z., Ijuin, T., Gruenberg, J., Takenawa, T., and Shisheva, A. (2007) J. Biol. Chem. 282 23878-238391 [DOI] [PubMed] [Google Scholar]
- 20.Bomsel, M., Parton R., Kuznetsov, S. A., Schroer, T., and Gruenberg, J. (1990) Cell 62 719-731 [DOI] [PubMed] [Google Scholar]
- 21.Aniento, F., Emans, N., Griffiths, G., and Gruenberg, J. (1993) J. Cell Biol. 123 1373-1387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nielsen, E., Severin, F., Backer, J. M., Hyman, A. A., and Zerial, M. (1999) Nat. Cell Biol. 1 376-382 [DOI] [PubMed] [Google Scholar]
- 23.Lin, S. X., Gundersen, G. G., and Maxfield, F. R. (2002) Mol. Biol. Cell 13 96-109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Murray, J. W., and Wolkoff, A. W. (2003) Adv. Drug Delivery Rev. 55 1385-1403 [DOI] [PubMed] [Google Scholar]
- 25.Hoepfner, S., Severin F., Cabezas, A., Habermann, B., Runge, A., Gillooly, D., Stenmark, H., and Zerial, M. (2005) Cell 121 437-450 [DOI] [PubMed] [Google Scholar]
- 26.Hirokawa, N., and Takemura, R. (2004) Exp. Cell Res. 301 50-59 [DOI] [PubMed] [Google Scholar]
- 27.Caviston, J. P., and Holzbaur, E. L. (2006) Trends Cell Biol. 16 530-536 [DOI] [PubMed] [Google Scholar]
- 28.Schnapp, B. J. (2003) J. Cell Sci. 116 2125-2135 [DOI] [PubMed] [Google Scholar]
- 29.Guzik, B. W., and Goldstein, L. S. (2004) Curr. Opin. Cell Biol. 16 443-450 [DOI] [PubMed] [Google Scholar]
- 30.Weston, C. R., and Davis, R. J. (2002) Curr. Opin. Genet. Dev. 12 14-21 [DOI] [PubMed] [Google Scholar]
- 31.Dhanasekaran, D. N., Kashef, K., Lee, C. M., Xu, H., and Reddy, E. P. (2007) Oncogene 26 3185-3202 [DOI] [PubMed] [Google Scholar]
- 32.Bowman, A. B., Kamal, A., Ritchings, B. W., Philp, A. V., McGrail, M., Gindhart, J. G., and Goldstein, L. S. (2000) Cell 103 583-594 [DOI] [PubMed] [Google Scholar]
- 33.Byrd, D. T., Kawasaki, M., Walcoff, M., Hisamoto, N., Matsumoto, K., and Jin, Y. (2001) Neuron 32 787-800 [DOI] [PubMed] [Google Scholar]
- 34.Whitmarsh, A. J., Cavanagh, J., Tournier, C., Yasuda, J., and Davis, R. J. (1998) Science 281 1671-1674 [DOI] [PubMed] [Google Scholar]
- 35.Yasuda, J., Whitmarsh, A. J., Cavanagh, J., Sharma, M., and Davis, R. J. (1999) Mol. Cell Biol. 19 7245-7254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kelkar, N., Gupta, S., Dickens, M., and Davis, R. J. (2000) Mol. Cell Biol. 20 1030-1043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lee, C. M., Onésime, D., Reddy, C. D., Dhanasekaran, N., and Reddy, E. P. (2002) Proc. Natl. Acad. Sci. U. S. A. 99 14189-14194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kelkar, N., Standen, C. L., and Davis, R. J. (2005) Mol. Cell Biol. 25 2733-2743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Iwanaga, A., Wang, G., Gantulga, D., Sato, T., Baljinnyam, T., Shimizu, K., Takumi, K., Hayashi, M., Akashi, T., Fuse, H., Sugihara, K., Asano, M., and Yoshioka, K. (2008) Transgenic Res. 17 1045-1058 [DOI] [PubMed] [Google Scholar]
- 40.Verhey, K. J., Meyer, D., Deeham, R., Blenis, J., Schnapp, B. J., Rapoport, T. A., and Margolis, B. (2001) J. Cell Biol. 152 959-970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nguyen, Q., Lee, C. M., Le, A., and Reddy, E. P. (2005) J. Biol. Chem. 280 30185-30191 [DOI] [PubMed] [Google Scholar]
- 42.Horiuchi, D., Barkus, R. V., Pilling, A. D., Gassman, A., and Saxton, W. M. (2005) Curr. Biol. 15 2137-2141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shisheva, A., Sbrissa, D., and Ikonomov, O. (1999) Mol. Cell Biol. 19 623-634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sbrissa, D., Ikonomov, O. C., and Shisheva, A. (1999) J. Biol. Chem. 274 21589-21597 [DOI] [PubMed] [Google Scholar]
- 45.Harlow, E., and Lane, D. (1988) Antibodies: A Laboratory Manual, pp. 271-276, Cold Spring Harbor Laboratory, Cold Spring Harbor, NY
- 46.Sbrissa, D., Ikonomov, O. C., Strakova, J., Dondapati, R., Mlak, K., Deeb, R., Silver, R., and Shisheva, A. (2004) Mol. Cell Biol. 24 10437-10447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sbrissa, D., Ikonomov, O. C., Strakova, J., and Shisheva, A. (2004) Endocrinology 145 4853-4865 [DOI] [PubMed] [Google Scholar]
- 48.Sbrissa, D., Ikonomov, O., and Shisheva, A. (2001) Mol. Cell Endocrinol. 181 35-46 [DOI] [PubMed] [Google Scholar]
- 49.Wolins, N., Bosshart, H., Küster, H., and Bonifacino, J. S. (1997) J. Cell Biol. 139 1735-1745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Michell, R. H., Heath, V. L., Lemmon, M. A., and Dove, S. K. (2006) Trends Biochem. Sci. 31 52-63 [DOI] [PubMed] [Google Scholar]
- 51.Rink, K., E. Ghigo, Kalaidzidis, Y., and Zerial, M. (2005) Cell 122 735-747 [DOI] [PubMed] [Google Scholar]
- 52.Jeffries, T. R., Dove, S. K., Michell, R. H., and Parker, P. J. (2004) Mol. Biol. Cell 15 2652-2663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Takaesu, G., Kang, J.-S., Bae, G.-U., Yi, M.-J., Lee, C. M., Reddy, E. P., and Krauss, R. S. (2006) J. Cell Biol. 175 383-388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kashef, K., Lee, C. M., Ha, J. H., Reddy, E. P., and Dhanasekaran, D. N. (2005) Biochemistry 44 14090-14096 [DOI] [PubMed] [Google Scholar]
- 55.Kashef, K., Xu, H., Reddy, E. P., and Dhanasekaran, D. N. (2006) J. Cell Biochem. 98 715-722 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.