Abstract
Leukocyte β2-integrin CD11b/CD18 mediates the firm adhesion and subsequent transepithelial migration of polymorphonuclear leukocytes, but the identity of its counter-receptor(s) on epithelia remains elusive. Here we identified a monoclonal antibody, clone C3H7, which strongly bound to the basolateral membranes of epithelial cells and inhibited both the adhesion of epithelial cells to immobilized CD11b/CD8 and the transepithelial migration of PMNs in a physiologically relevant basolateral-to-apical direction. C3H7 antigen expression in epithelial monolayers was significantly increased by treatment with proinflammatory cytokine interferon-γ or a combination of interferon-γ and tumor necrosis factor-α. Up-regulation of C3H7 antigen was also observed in the epithelium of inflamed human colon tissues. Microsequencing and Western blotting of the purified antigen showed it to be CD44 variant 3 (CD44v3), a ∼160-kDa membrane glycoprotein. Further studies demonstrated that this epithelial CD44v3 specifically binds to CD11b/CD18 through its heparan sulfate moieties. In summary, our study demonstrates for the first time that the heparan sulfate proteoglycan form of epithelial CD44v3 plays a critical role in facilitating PMN recruitment during inflammatory episodes via directly binding to CD11b/CD18.
A major component of many inflammatory diseases is the migration of large numbers of neutrophils (polymorphonuclear leukocytes, PMNs)2 across the epithelium and their accumulation within a lumen. Examples include inflammatory bowel disease (IBD), cholangitis, cholecystitis, bronchial pneumonia, bronchitis, pyelonephritis, and cystitis. Under these pathophysiological conditions, epithelial injury and disease symptoms parallel PMN infiltration of the mucosa (1, 2). The current paradigm for migration of PMN across epithelial monolayers envisions a process consisting of sequential molecularly defined events such as CD11b/CD18-mediated firm adhesion of PMN with epithelia (3) followed by CD47-SIRPα interactions at the post-adhesion stage (4). However, although PMN transepithelial migration (TEM) has been widely demonstrated to be CD11b/CD18-dependent, the epithelial counter-receptor(s) for CD11b/CD18 in mediating PMN-epithelia adhesion has not been identified.
Function mapping studies using domain-specific antibodies have demonstrated that the inserted domain (I-domain), a stretch of 200 amino acids of the CD11b subunit, is a major binding domain for CD11b/CD18 ligands (5). The I-domain of CD11b is promiscuous in ligand binding and has many known receptors including ICAM-1 (6, 7), fibrinogen (8), collagen (9), Cyr61 (CCN1), and connective tissue growth factor (CCN2) (10), heparin/heparan sulfate (11, 12), elastase (13), iC3b (14), and platelet glycoprotein Ibα (15). However, none of these ligands appear to mediate the firm adhesion of PMNs to the basolateral surfaces of epithelial monolayers at early stages of transmigration. Thus far, no epithelial basolaterally expressed CD11b/CD18 counter-receptor has been identified. ICAM-1, the best characterized cellular ligand for CD11b/CD18, cannot be the intestinal epithelial CD11b/CD18 ligand that mediates PMN firm adhesion because: (a) ICAM-1 is normally not expressed on intestinal epithelia except under inflammatory conditions (16) and (b) when ICAM-1 expression is induced it is up-regulated on the apical rather than basolateral surface of intestine epithelia. In an effort to understand the mechanisms that govern CD11b/CD18-mediated PMN TEM, previous studies by us and others have found that epithelial surface-sulfated proteoglycans (17) and junction adhesion molecule C (JAM-C) play a significant role in regulating PMN transmigration via interaction with leukocyte CD11b/CD18 (18, 19). However, compared with functional inhibitory anti-CD11b antibodies that completely block PMN TEM, soluble carbohydrates or antibodies against JAM-C create only partial inhibition. These results clearly suggest the existence of unknown epithelial adhesion molecule(s) that bind to leukocyte CD11b/CD18 and regulate PMN TEM. Heparin and heparan sulfate have also been shown to block the adhesion and PMN TEM via binding to CD11b/CD18 (11, 12); thus it is reasonable to suggest that a basolateral membrane glycoprotein decorated with heparan sulfate moieties may serve as a counter-receptor for CD11b/CD18. However, the nature of this epithelial heparan sulfate proteoglycan has not been identified.
Here we sought to identify novel epithelial adhesive ligand(s) important in PMN transmigration, in particular, a ligand that can bind to CD11b/CD18 on migrating PMNs and mediate the firm adhesion of PMNs to the epithelial basolateral surfaces. To do this, we screened a panel of monoclonal antibodies generated against epithelial plasma membranes. This screening identified one mAb, termed C3H7, that recognized a basolateral membrane protein and inhibited PMN TEM in a physiologically relevant basolateral-to-apical direction. Further study of these results identified the C3H7 antigen as a v3-type human epithelial CD44 variant, a ∼160-kDa glycoprotein that is decorated with heparan sulfate moieties. Subsequent studies revealed that the C3H7 antigen appears to function as a cellular ligand for CD11b/CD18 in regulating the firm adhesion of PMNs to the epithelium during their transmigration process.
EXPERIMENTAL PROCEDURES
Cells, Tissue Samples, and Reagents—Human intestinal epithelial T84 cells were grown in Dulbecco's modified Eagle's medium/F-12 supplemented with 6% fetal bovine serum. Two other lines of human intestinal epithelial cells, HT29 cells and Caco2-BBE (Caco2) cells (passages 30–50), were grown in high-glucose Dulbecco's modified Eagle's medium (Invitrogen) supplemented with 14 mmol/liter NaHCO3 and 10% newborn calf serum (20). Subculturing of epithelial cells was performed every 6–8 days by treatment with 0.1% trypsin and 1.0 mm EDTA in Ca2+- and Mg2+-free phosphate-buffered saline. For transmigration experiments, epithelial cells were grown on collagen-coated, permeable polycarbonate filters as described previously (21, 22). In particular, for apical-to-basolateral transmigration experiments, epithelial monolayers were grown on permeable collagen-coated, polycarbonate supports (inserts) with 5-μm pore size and a surface area of 0.33 cm2 (Costar Inc., Cambridge, MA) as described previously (23, 24). For physiologically directed, basolateral-to-apical transmigration, T84 cells were plated on the underside of permeable filters to produce inverted monolayers (23, 24). PMNs were isolated from the whole blood of normal human volunteers by dextran and Ficoll sedimentation as described previously in detail (22). All of the protocols were approved by the Georgia State University institutional review board committee. Isolated PMNs were resuspended in HBSS without calcium and magnesium (HBSS-) and were used within 4 h. Human large intestine/colon tissue specimens from normal volunteer donors and active IBD patients were obtained from the National Disease Research Interchange of the National Resource Center (Philadelphia, PA). These tissue specimens, which were freshly frozen or snap frozen shortly after surgical removal, were sectioned at 5–10-μm thickness using a Leica CM3050S cryostat (Leica Microsystems) and mounted on coverslides followed by air drying before immunostaining was performed. Normal human colon sections were purchased from Biochain (Hayward, CA). Recombinant human TNF-α and interferon-γ were purchased from Upstate Biotechnology and used at 20 and 10 ng/ml, respectively. Heparitinase I, II, and III (from Flavobacterium heparinum), neuraminidase, and chondroitinase ABC were from Calbiochem (La Jolla, CA). Bovine serum albumin (BSA), Triton X-100, n-octyl-β-d-glucopyranoside (OG), human fibrinogen (FBG), heparan sulfate (HS), and protein A-agarose were purchased from Sigma.
Antibodies—T84 cell plasma membranes were used as an antigen to generate a panel of antibodies as described previously (25). Briefly, female Balb/c mice were injected intraperitoneally with a 1:1 mix of complete Freund's adjuvant and T84 membranes (100 μg) (25). The mice were inoculated every 2 weeks for 6 weeks with 50 μg of T84 cell membranes in a mixture with incomplete Freund's adjuvant. Splenocytes were harvested, washed in phosphate-buffered saline, and mixed 2:1 with hybridoma cells in the presence of 50% polyethyleneglycol. The cells were plated in Dulbecco's modified Eagle's medium supplemented with 1% hypoxanthine/aminopterin/thymidine medium for selection, and single clones were obtained by limiting dilution. Cultures of T84 cells in 96-well plates were used to select for clones producing antibodies against epithelial surfaces. Supernatants from these clones were used to assay PMN transmigration and T84 cell-CD11b/CD18 adhesion. Of these, one mAb (clone C3H7, IgG1) was further characterized. Monoclonal anti-CD44H, anti-CD44v3, anti-CD44v4, anti-CD44v6, anti-CD44v7, and anti-CD44v9 were obtained from R & D Systems (Minneapolis, MN). Rabbit polyclonal antibodies to CD44v3 variant (AB2081) was from Chemicon International (Temecula, CA). The hybridoma of the inhibitory anti-CD11b mAb 44a (IgG2a) was obtained from ATCC, and the produced antibody was purified using protein A-conjugated Sepharose. The noninhibitory monoclonal anti-CD11b antibody, LM2/1, and anti-CD11c antibody CBR-p150/4G1 were obtained from Abcam and Cell Sciences (Canton, MA), respectively. Mouse isotype-matched mAb (clone C1F3) was used as a control. HRP-conjugated or Alexa Fluor 488/568-conjugated secondary antibodies were obtained from Molecular Probes.
PMN Transmigration—PMN transmigration was performed using confluent T84 cell monolayers cultured on Transwells or collagen-coated Transwell filters as described previously (22, 23, 25). All PMN TEM experiments were performed in a basolateral-to-apical (b-to-a) direction, unless otherwise indicated. Briefly, confluent T84 monolayers were washed free of media followed by the addition of 50 μl of HBSS containing antibodies and incubation for 20 min (20 °C). After a 20-min preincubation, HBSS was added (100 μ1) followed by 1 × 106 PMN in 25 μl of HBSS-. Transmigration was initiated by transfer monolayers to 24-well tissue culture plates containing 1 ml of HBSS with n-formyl-methionyl-leucyl-phenylalanine (1 μm for epithelial monolayers and 0.1 μm for Transwell filters, respectively). After incubation for indicated time at 37 °C, neutrophil migration across monolayers into the chemoattractant-containing lower chambers was quantitated by myeloperoxidase assay (22, 25, 26).
Cytokine Treatment and C3H7 Antigen Measurement—T84 monolayers were treated with TNF-α (20 ng/ml), interferon-γ (10 ng/ml), and a combination of the two for 48 and 72 h, respectively (27). After washing, the monolayers were fixed, blocked with 1% BSA, and labeled with mAb C3H7. Bound mAb was detected by a HRP-conjugated secondary antibody followed by color development and A405 nm measurement. The A405 nm reading from isotope-matched mAb C1F3 labeling served as the background and was subtracted from the values of experimental groups.
Immunofluorescence Labeling and Confocal Microscopy—Labeling of epithelial monolayers cultured on Transwell filters was performed as described previously (27). Briefly, monolayers were fixed and permeabilized using cold ethanol (-20 °C, 20 min) and subsequently blocked with 5% normal goat serum. Monolayers were then incubated with primary antibodies followed by Alexa Fluor-conjugated secondary antibodies. After three washes, the monolayers were mounted in ProLong anti-fading embedding solution (Molecular Probes) and analyzed using a laser scanning confocal microscope (FV1000; Olympus). The images shown are representative of at least three experiments with multiple images taken per slide. For Z-series, optical sections were imaged at 0.5-μm intervals. Frozen human intestine/colon tissue sections were briefly fixed with 3.7% paraformaldehyde and permeabilized with 0.03% Triton X-100 (5 min, 4 °C), followed by immunofluorescence staining and image analysis. For background labeling, the samples were incubated with C1F3.
Differential Biotinylation of Apical and Basolateral Surface Proteins—T84 cells grown on Transwell inserts were washed twice with HBSS. 1 mm sulfo-NHS-biotin (Pierce) was added apically or basolaterally to monolayers and then incubated for 10 min at 4 °C. The monolayers were washed three times with 150 mm NH4Cl to quench the residual biotin. The monolayers were lysed in lysis buffer. 10 μg of mAb C3H7 was added to the cell lysate to immunoprecipitate C3H7 antigen. Biotinylated cell surface antigen was detected with avidin-horseradish peroxidase (Pierce).
Tryptic Digestion and Identification of C3H7 Antigen—Bulk antigen was purified from ∼500 cm2 of confluent T84 cell plasma membranes using mAb C3H7-coupled affinity column (CnBr-activated Sepharose 4B; Pierce). Antigen was eluted by 1% OG-containing elute solution (pH10.5). The eluant was pH-neutralized and resolved by SDS-PAGE, and protein bands were localized by Gelcode 250 (Pierce). A protein band near 160 kDa was extracted for trypsin digestion and microsequence analysis.
In Vitro Protein Binding Assay—Human CD11b/CD18 and CD11c/CD18 were purified as described previously (28, 29). Briefly, ∼1010 PMNs were solubilized in lysis buffer, and precleared lysate was passed over a mAb LM2/1 immunoaffinity column, followed by washing and detergent exchange to 1% OG. Bound CD11b/CD18 was eluted with 50 mm triethylamine, pH 10.5, 2 mm MgCl2, and 1% OG followed by neutralization. For CD11c/CD18 purification, 30 g of human spleen tissue was solubilized in lysis buffer, and the precleared lysate was passed over a mAb 4G1 immunoaffinity column. After detergent exchange, bound CD11c/CD18 was eluted and neutralized as described above. Purified CD11b/CD18 and CD11c/CD18 (100 μg/ml) in 150 mm NaCl, 2 mm MgCl2, 2 mm CaCl2, 100 mm Tris, and 1% OG, pH 7.4, were diluted 15-fold with HBSS and immediately added to the 96-well microtiter plate (flat-bottomed; ICN Biomedical) (50 μl/well) followed by incubation overnight at 4 °C. After blocking the nonspecific binding with 1% BSA, the wells coated with β2-integrins were then incubated with purified C3H7 antigen (5 μg/ml each) in binding buffer (HBSS plus 0.1% Triton X-100) with or without inhibitory reagents for 1 h at 37 °C. After washing, bound C3H7 antigen was detected by an anti-CD44H antibody, followed by HRP-conjugated secondary antibodies and color development using 2,2′-azino-bis(3-ethylbenzthiazoline-6-sulfonic acid). In separate experiments, the wells were also coated with purified C3H7 antigen, and the wells were preincubated with different deglycosidases, including a combination of heparitinase I/II/III, chondroitinase ABC, and neuraminidase at 37 °C for 30 min (17). Purified CD11b/CD18 in HBSS containing 0.1% Triton X-100 was then added into the wells and incubated at 37 °C for 1 h. After washing, bound CD11b/CD18 was detected by mAb LM2/1 and HRP-conjugated secondary antibodies. BSA-coated wells served as blank controls.
Cell Adhesion Assays—Adhesion of epithelial Caco2 cells to immobilized CD11b/CD18 was performed as described previously (17). Briefly, 96-well plates were coated with isolated functionally active CD11b/CD18 and blocked by 1% BSA in HBSS. Caco2 cells elicited by trypsin and EDTA were loaded with 5 μg/ml BCECF-AM (Molecular Probes). After removing free dye, fluorescence-loaded Caco2 cells were added into CD11b/CD18-coated wells (∼2 × 105 cells/well in 150 μl) in the presence or absence of antibodies/reagents. The wells were incubated at 37 °C for 1 h. To quantify cell adhesion, the fluorescent intensity of each well before and after three gentle washes with HBSS was determined using a fluorescent plate reader (Molecular Device) at excitation/emission wavelengths of 485/535 nm. Cells adhesion to BSA-coated wells served as blank controls. In separate experiments, Caco2 cells were transfected with (si)RNA duplexes directed CD44v3 (Ambion) using Lipofectin (Invitrogen) in serum-free medium (20). A nontargeting siRNA (Ambion) was used as the control for nonsequence-specific effects of siRNAs. The expression levels of CD44v3 in transfected cells were determined by Western blot analysis and flow cytometry. The cells were then used in CD11b/CD18 adhesion assay.
Data Analysis—All of the data are shown as the means ± S.D. Paired Student's t test was used to determine the significance of differences between population means (*, p < 0.05; **, p < 0.01).
RESULTS
Fusion of splenocytes from mice immunized with T84 cell plasma membranes yielded nearly 1200 antibody-producing clones. We screened each clone for reactivity to intact T84 cells, inhibition of T84 cells adhesion to immobilized CD11b/CD18, and blockade of PMN transmigration across T84 cell monolayers. One subclone, IgG1, termed clone C3H7, was selected because it satisfied all three criteria of our antibody screening.
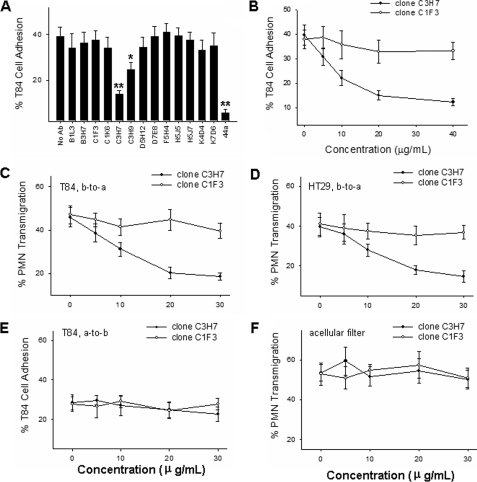
mAb C3H7 Inhibits T84-CD11b/CD18 Adhesion and PMN TEM—Monoclonal antibodies were tested using three epithelial cell lines (T84, HT29, and Caco2 cells) to determine their influence on epithelial cell adhesion to immobilized CD11b/CD18 and PMN TEM in response to n-formyl-methionyl-leucyl-phenylalanine, respectively. As shown in Fig. 1A, among a panel of epithelial-specific monoclonal antibodies, only mAb C3H7 strongly inhibited the adhesion of T84 cells to immobilized CD11b/CD18. The inhibitory effect of mAb C3H7 showed a dose-dependent manner (Fig. 1B). mAb C3H7 also dose-dependently inhibited PMN migration across T84 (Fig. 1C) and HT29 (Fig. 1D) epithelial monolayers in a physiologically relevant b-to-a direction. At 5 μg/ml, mAb C3H7 reduced PMN TEM by over 50%. At the same concentration, mAb C1F3, an isotype-matched antibody that also binds to epithelial monolayers, had no effect on PMN TEM. Interestingly, mAb C3H7 had significant less inhibition on migration of PMNs in apical-to-basolateral direction (Fig. 1E), suggesting that mAb C3H7 influences PMN TEM in a polarized fashion. Furthermore, mAb C3H7 also showed no effect on PMN transfilter migration (Fig. 1F), implying that the inhibitory role of mAb C3H7 on PMN TEM is due exclusively to interactions between mAb C3H7 and epithelial monolayers.
FIGURE 1.
Functional influence of mAb C3H7 on adhesion of T84 cells to immobilized CD11b/CD18 (A and B) and PMN transmigration across epithelial monolayers (C–E) and acellular Transwell filters (F), respectively. The antibody concentrations are the same (20 μg/ml IgG). PMN migration across T84 monolayers in b-to-a direction (C), HT29 monolayers in b-to-a direction (D), T84 monolayers in apical-to-basolateral direction (E), and acellular Transwell filters (F) in response to n-formyl-methionyl-leucyl-phenylalanine were performed in the presence or absence of indicated concentrations of mAb C3H7 and control mAb C1F3, respectively. The data are presented as the means ± S.D. (n = 3).
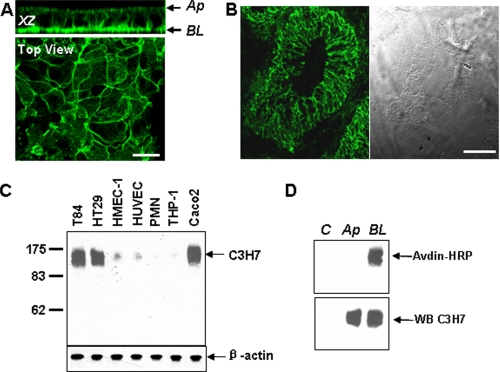
Basolateral Localization of C3H7 Antigen in Epithelial Cells—The polarized inhibitory effect of mAb C3H7 on PMN TEM prompts us to believe that the C3H7 antigen is a critical adhesion molecule located at the basolateral surfaces of epithelial monolayers. Immunofluorescence labeling and Western blotting were therefore performed in both cultured T84 monolayers and normal human colon tissue sections to define the localization of the C3H7 antigen. As shown by the X-Z image and en face image in Fig. 2A, C3H7 antigen was mainly localized at the basolateral and lateral membranes of T84 cell monolayers cultured on permeable supports. A similar localization pattern of C3H7 antigen was confirmed in normal colon tissue sections (Fig. 2B). Western blotting using mAb C3H7 revealed a heavily glycosylated ∼160-kDa protein expressed in intestinal T84, HT29, and Caco2 epithelial cells (Fig. 2C). Compared with epithelial cells, endothelial cells (human umbilical vein endothelial cells and human microvascular endothelial cells) expressed C3H7 antigen at a much lower level (Fig. 2C). The relative small difference in apparent molecular weight between epithelial and endothelial cell lines likely represents cell tissue-specific differences in glycosylation. By contrast, C3H7 antigen was not detected in the lysate of PMN and THP-1, a neoplastic monocytic cell line (Fig. 2C). Immunoprecipitation of differential surface biotinylated T84 cells confirmed localization of C3H7 antigen illustrated by confocal microscopy, with the nearly exclusive expression in the basolateral, but not apical, cell surface (Fig. 2D). Together, these results suggest that the C3H7 antigen is an epithelial-specific basolateral membrane protein, which is agreement with the observation that mAb C3H7 blocked PMN TEM in a polarized fashion and did not affect PMN migration across collagen-coated acellular membranes.
FIGURE 2.
Localization of C3H7 antigen to the basolateral and lateral membranes of epithelium. A, immunofluorescence labeling and confocal microscopy was used to define the pattern of C3H7 antigen expression. Shown here are an X-Z orientation series and an en face image demonstrating nearly complete localization to the basolateral and subapical lateral membranes. B, immunofluorescence labeling of mAb C3H7 in normal human colon tissue sections. Scale bar, 20 μm. C, Western blot analysis of equal amounts of lysates from indicated cell types with mAb C3H7. D, confluent T84 monolayers were biotinylated on the apical (Ap) or basolateral (BL) surface. Lysates were immunoprecipitated with either C3H7 or isotype-matched control mAb C1F3. Proteins were visualized with streptavidin-HRP to identify only surface biotinylated C3H7 antigen (upper panel) or with mAb C3H7 to demonstrate total C3H7 antigen (lower panel). The control lane (lane C) represents mAb C1F3 immunoprecipitation.
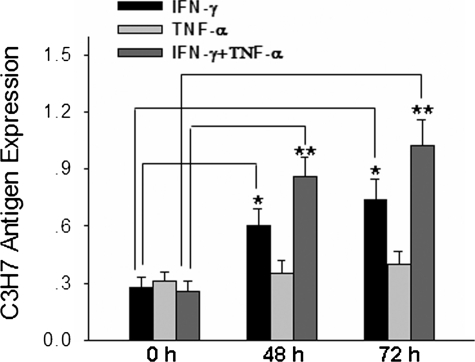
Up-regulation of C3H7 Antigen by Pro-inflammatory Cytokines—Because PMN transmigration across mucosal epithelia is a hallmark of inflammatory conditions including ulcerative colitis and Crohn disease, we determined the expression of C3H7 antigen in T84 epithelia under the stimulation of several pro-inflammatory cytokines to illustrate the physiological role of C3H7 antigen during inflammatory episodes. As shown in Fig. 3, T84 monolayer surface expression of C3H7 antigen was significantly increased by stimulation of INF-γ and particularly by a combination of INF-γ and TNF-α. The enhancement of C3H7 antigen expression by INF-γ or combination of INF-γ and TNF-α was time-dependent. Interestingly, treatment with TNF-α alone did not lead to enhanced expression of C3H7 antigen until 72 h of incubation. Up-regulation of the C3H7 antigen expression on T84 cell surfaces by pro-inflammatory cytokines was confirmed by flow cytometry, and these flow cytometric analyses also failed to demonstrate binding of mAb C3H7 to PMN (data not shown).
FIGURE 3.
Up-regulation of C3H7 antigen on epithelial cells by pro-inflammatory cytokines. T84 monolayers cultured on Transwell inserts were treated with cytokines or left untreated. Surface binding of mAb C3H7 to control and cytokine-stimulated T84 monolayers was assessed by enzyme-linked immunosorbent assay after incubation for 48 and 72 h as detailed under “Experimental Procedures.” The data are presented as the means ± S.D. (n = 4). IFN, interferon.
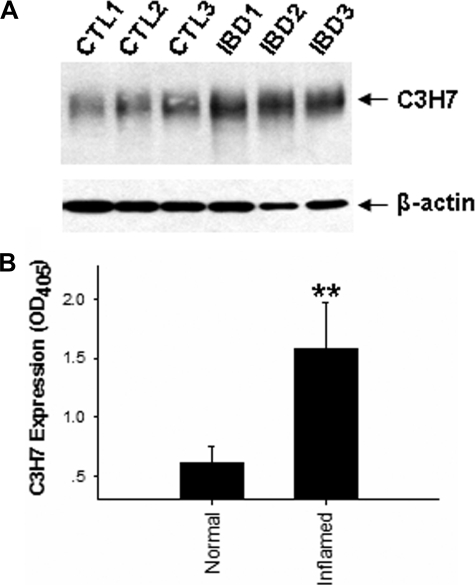
To exclude the possibility that enhanced expression of C3H7 antigen by cytokines was due to cell culture, the expression and localization of C3H7 antigen in intestinal epithelia was further examined by studying normal human colon tissue samples from three healthy donors and inflammatory colon tissue samples from three IBD patients, including two patients with active ulcerative colitis and one patient with Crohn disease. As shown in Fig. 4A, Western blot analysis clearly showed that inflammatory colon tissues had a significant increase in C3H7 antigen expression, which is in agreement with the observation that the C3H7 antigen is up-regulated in pro-inflammatory cytokine-treated epithelial monolayers (Fig. 3). Fig. 4B showed a quantitative analysis of mAb C3H7 labeling in three pairs of normal and inflamed tissue blocks.
FIGURE 4.
Up-regulation of C3H7 antigen in the epithelia of human inflamed colon tissues. A, Western blot analysis of C3H7 antigen in colon tissues. Note that expression levels of C3H7 in inflamed colon tissues (IBD1–3) are increased compared with those of normal colon tissues (CTL1–3). B, quantitative analysis of C3H7 antigen expression levels in human colon tissues. The data are presented as the means ± S.D. (n = 3).
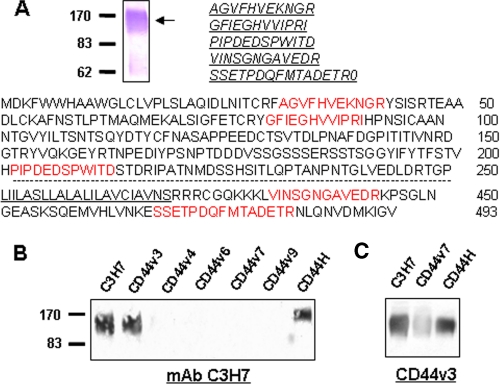
Identification of the C3H7 Antigen as v3-type Epithelial CD44 Variant—Further experiments were performed to identify the antigen recognized by mAb C3H7. Approximately 900 cm2 of confluent HT29 cells were used to obtain sufficient C3H7 antigen for microsequence analysis. Purified mAb C3H7 was conjugated to Sepharose 4B to construct an affinity column. After immunoaffinity purification, the antigen was digested with trypsin, and four tryptic peptides resulting from mass spectroscopy showed direct sequence homology (Fig. 5A) with human CD44, a multistructural and multifunctional cell surface adhesion molecule involved in cell-cell and cell-matrix interactions (30). Because the genomic organization of CD44 involves 20 exons, and the first 5 and the last 5 exons are constant, whereas the 10 exons between those regions are subjected to alternative splicing, the tryptic peptides identified could not determine which CD44 variant the C3H7 antigen represents. To determine the CD44 splice variant of the C3H7 antigen, we blotted C3H7 antigen purified from HT29 cell lysates with a panel of antibodies specifically against different variants of CD44. As shown in Fig. 5B, the C3H7 antigen was recognized by an antibody against CD44H, the smallest common form shared by all CD44 variants, confirming that the C3H7 antigen is indeed a member of the human CD44 family. Furthermore, purified C3H7 antigen was also recognized by an anti-CD44v3 antibody but not by antibody specifically against CD44v4, CD44v6, CD44v7, and CD44v9. In agreement with this, immunoprecipitated protein from HT29 cell lysate by anti-CD44v3 monoclonal antibody was also recognized by mAb C3H7. The results suggest that the C3H7 antigen may represent an epithelial CD44 variant containing the v3 domain.
FIGURE 5.
Biochemical identification of C3H7 antigen as an epithelial CD44 variant containing v3 domain (v3-type variant). A, purified C3H7 antigen from HT29 cells and five sequenced tryptic peptides (red) aligned with human CD44. The transmembrane region is underlined. B, immunoprecipitated product by mAb C3H7 was blotted with the antibodies against various CD44 variants. Note that the C3H7 antigen is recognized by both anti-CD44H and anti-CD44v3 antibodies but not by other anti-CD44 antibodies. C, immunoprecipitated product by anti-CD44v3 mAb was blotted with mAb C3H7, CD44v7, and anti-CD44H, respectively.
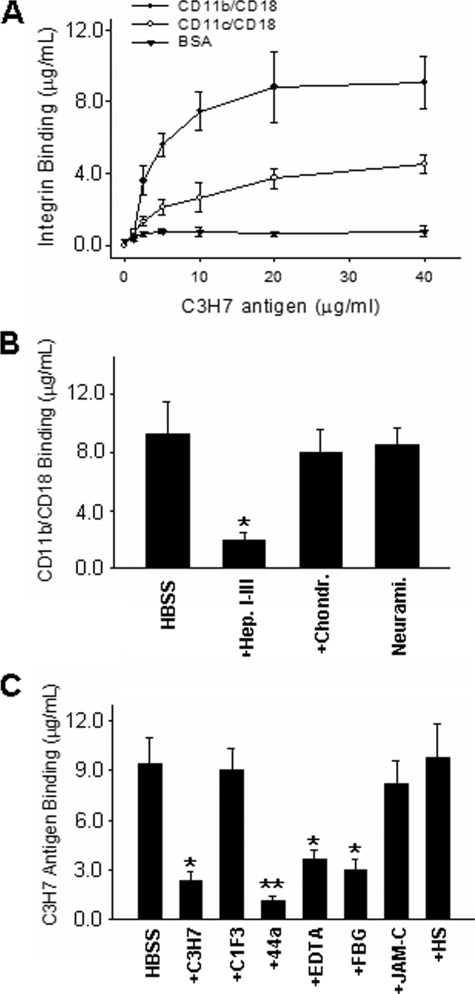
CD44v3 Modulates PMN-Epithelial Adhesion via Binding to CD11b/CD18—Based on the findings that mAb C3H7 inhibits both the adhesion of T84 cells to immobilized CD11b/CD18 and the TEM of PMNs in a b-to-a direction, we next tested whether the CD44v3 (C3H7 antigen) serves as an adhesive ligand for CD11b/CD18. In this experiment, purified functionally active CD11b/CD18 and CD11c/CD18 were immobilized on 96-well plates and blocked with 1% BSA. Isolated CD44v3 was then added into the wells and incubated at 37 °C for 1 h. Bound CD44v3 was detected by anti-CD44H followed by HRP-conjugated secondary antibody detection and color development. As shown in Fig. 6A, CD44v3 bound strongly to CD11b/CD18 but only weakly to CD11c/CD18. No binding of CD44v3 to immobilized BSA was observed. Because previous studies suggested that epithelial CD44v3 is a heparan sulfate intrinsic membrane proteoglycan (31, 32) and that heparin or heparan sulfate is able to bind CD11b/CD18 (11, 12), we further treated the immobilized C3H7 antigen with a combination of three heparinases (I, II, and III) or other deglycosidases, including chondroitinase ABC and neuraminidase, prior to the binding assay of CD11b/CD18 (Fig. 6B). Heparinase treatment almost completely inhibited the binding of epithelial CD44v3 with CD11b/CD18, whereas treatment with chondroitinase ABC or neuraminidase had no significant effect. Together, these results suggest that CD44v3 specifically binds to CD11b through its heparan sulfate moieties and that negative charges associated with carbohydrates contribute minimally to CD44v3-CD11b/CD18 binding. As shown in Fig. 6C, the binding of epithelial CD44v3 with CD11b/CD18 was comparable with FBG-CD11b/CD18 binding. The binding was largely abolished by anti-CD11b antibody 44a, a mAb against the I-domain of CD11b, and by EDTA, an effective Mg2+ chelator. These results suggest that binding of CD44v3 to CD11b/CD18 is likely through the I-domain of CD11b. Binding of CD44v3 with CD11b/CD18 was also reduced by FBG but not by soluble JAM-C, suggesting that epithelial CD44v3 and FBG may share the same binding region on CD11b. Furthermore, epithelial CD44v3 binding to CD11b/CD18 was not affected by purified heparan sulfate, suggesting that the binding may depend not only on heparan sulfate moieties but also on the structural configuration of proteoglycan.
FIGURE 6.
Specific binding of the heparan sulfate proteoglycan form of epithelial CD44v3 to CD11b/CD18 via its decorating heparan sulfate moieties. A, binding curve of purified CD44v3 to β2-integrins and BSA. B, binding of CD11b/CD18 to immobilized CD44v3 before and after treatment with various deglycosidases, including heparinase I, II, and III (Hep. I-III), chondroitinase ABC (Chondr.), and neuraminidase (Neurami.). C, binding of CD44v3 to CD11b/CD18 in the presence or absence of various inhibitors, including antibody C3H7, C1F3, 44a, and anti-JAM-C at 20 μg/ml each, FBG (50 μg/ml), heparin sulfate (HS) (150 μg/ml), and 3 mm EDTA. The data are presented as the means ± S.D. (n = 3).
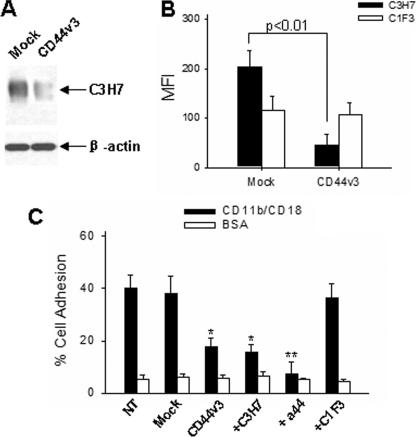
Because CD11b/CD18-mediated adhesion of PMN to epithelial is a critical step for PMN transmigration process, next we determine the role of CD44v3 in regulating epithelial cell-specific binding to CD11b/CD18. In this experiment, Caco2 cells were transfected with CD44v3-specific siRNA or control siRNA before cell adhesion assays. As shown in Fig. 7A, Western blot and flow cytometry showed that both total CD44v3 in Caco2 cells and Caco2 cell surface expression of CD44v3 were significantly reduced by CD44v3 siRNA transfection, whereas no such alteration of CD44v3 was observed in Caco2 cell with Mock transfection. Caco2 cells were further labeled with BCECF-AM and added into 96-well microtiter plates coated with functional active CD11b/CD18 for cell adhesion assay (17). As can be seen in Fig. 7B, Caco2 cells, as well as mock-transfected Caco2 cells, strongly adhered to immobilized CD11b/CD18; the adhesion rates were 40.5 ± 6.1 and 37.9 ± 4.1% of total applied cells, respectively. In contrast, Caco2 cells transfected with CD44v3 siRNA showed a significant reduction of adhesion to immobilized CD11b/CD18, and the adhesion rate was decreased to 17.1 ± 3.7% of total applied cells. The reduction of cell adhesion to CD11b/CD18 by CD44v3 siRNA transfection was comparable with that by mAb C3H7.
FIGURE 7.
Inhibition of Caco2 cell adhesion to CD11b/CD18 by reduction of CD44v3 via CD44v3 siRNA and by anti-CD44v3 antibody. A and B, reduction of CD44v3 in Caco2 cells transfected with CD44v3 siRNA shown by Western blot analysis and flow cytometry, respectively. MFI, mean fluorescence intensity. C, adhesion of Caco2 cells to immobilized CD11b/CD18. Antibodies were used at 25 μg/ml. The data are presented as the means ± S.D. (n = 3).
DISCUSSION
The present report demonstrates for the first time that epithelial CD44v3 heparan sulfate proteoglycan serves as a novel counter-receptor for the leukocyte β2-inetgrin CD11b/CD18 and is directly involved in PMN adhesion to and migration across intestinal epithelial monolayers in a physiologically relevant direction. Originally identified as a homing receptor for lymphocyte, CD44 has been widely characterized (33). Structurally, the genomic organization of CD44 involves 20 exons, with the first five and the last five exons constant and the 10 exons (v1–v10) located between these regions subjected to alternative splicing. The standard CD44, CD44H, is the smallest variant, containing only 10 common exons (first five and last five) without any exon insertion in between. Various CD44 isoforms show strong tissue specificity. As indicated by our results, the CD44v3 recognized by mAb C3H7 is predominantly expressed in intestinal epithelial basolateral surfaces but not in PMNs and monocytes. This result of CD44v3 expression in colonic epithelium is in agreement with numerous previous reports (34–38). CD44 and its numerous variants are thought to mediate a variety of functions, including cell-extracellular matrix binding, leukocyte transmigration across endothelial and epithelial monolayers, and lymphocyte activation (39). For example, epithelial CD44v3 has been reported to be involved in inflammation such as that seen in IBD (35, 40). The expression of CD44v3 is significantly enhanced under inflammatory conditions (41). CD44v3, as well as other CD44 variants such as CD44v6, is also involved in regulating growth and metastasis of tumors of an epithelial origin (42). The mechanism by which CD44 variants govern leukocyte recruitment, however, is not fully understood, although several possibilities have been suggested, such as binding of CD44 variants with hyaluronan (33, 43–46) and sequestering chemoattractant via their decorated heparan sulfate moieties (15, 47, 48). Besides binding to hyaluronan, CD44 is also involved in interactions with other proteins including leukocyte integrins. For example, stimulation of CD44 can induce inside out activation of CD11b/CD18 through the GTPase Rap1 (49). Epithelial CD44 variant is also found to be up-regulated by cytokine interleukin-4 and mediate the adhesion between lymphocytes and colon epithelial cells (34). By showing that epithelial CD44v3 serves as a binding partner for the β2-inetgrin CD11b/CD18, our findings help to clarify the functional role of epithelial CD44v3. Unlike ICAM-1, whose exclusive expression on the apical membranes of inflammatory epithelium could not account for the phenomenon of leukocyte adhesion and transmigration across epithelial monolayers, epithelial CD44v3 is selectively expressed at basolateral surfaces of epithelium (Fig. 2), thus making it an ideal counter-receptor for leukocyte integrins that also contributes to the adhesion and TEM of leukocytes, particularly in infection or under inflammatory conditions such as in IBD.
The following features are consistent with a specific interaction of epithelial CD44v3 with the leukocyte β2-integrin CD11b/CD18: (a) Adhesion of epithelial cells to immobilized CD11b/CD18 is blocked by mAb C3H7, and mAb C3H7 specifically labels epithelial basolateral membranes, indicating that the C3H7 antigen (epithelial CD44v3) is an epithelial basolateral membrane protein that mediates T84 cell-CD11b/CD18 binding. (b) In vitro protein binding assays directly show that epithelial CD44v3 binds to leukocyte β2-integrin CD11b/CD18, possibly the I-domain of CD11b subunit (Fig. 6A). This binding can be abolished by mAb C3H7, anti-CD11b antibody 44a, CD11b/CD18 counter-receptor FBG, and EDTA, a chelator of Mg2+ and Ca2+, which are required for integrin stability and binding activity (Fig. 6C). (c) Binding assays also show that epithelial CD44v3 binds to CD11b/CD18 through its decorated heparan sulfate moieties (Fig. 6B). Previous studies have reported that heparin and heparan sulfates are binding partners of CD11b/CD18 (11, 12), and soluble carbohydrates with similar structures can inhibit leukocyte adhesion to epithelial monolayers and subsequent transmigration (21). Our data suggest that epithelial CD44v3 may be a heparan sulfate proteoglycan that serves as a counter-receptor for β2-integrin CD11b/CD18 during leukocyte TEM. In support of this, a previous study suggests that among all CD44 variants, CD44v3 is the only one decorated with heparan sulfate moieties (32). (d) Epithelial CD44v3 expression along the basolateral and lateral membranes of cultured T84 monolayers (Fig. 3B) is significantly enhanced after treatment with either interferon-γ or combination of interferon-γ and TNF-α. The up-regulation of epithelial CD44v3 by pro-inflammatory cytokines, which is also seen in inflamed colon tissues, indirectly supports the role of epithelial CD44v3 as a CD11b/CD18 counter-receptor and also provides a novel explanation of increased PMN adhesion and transmigration during inflammatory episodes.
The up-regulation of expression of epithelial C3H7 antigen under pro-inflammatory cytokine treatment is also in agreement with previous reports that showed the overexpression of CD44v3 in inflammatory diseases and epithelium damage (35, 41, 50). It has been well known that CD44 undergoes differential glycosylation to yield both N- and O-glycan modified forms of CD44 in addition to proteoglycan-like variants containing heparin sulfate and chondroitin sulfate moieties (32). Our data suggest that epithelial CD44v3 recognized by mAb C3H7 is a membrane proteoglycan decorated with heparan sulfate moieties, and these moieties mediate its binding interactions with β2-integrin CD11b/CD18.
In summary, we demonstrate here that the heparan sulfate proteoglycan form of epithelial CD44v3, recognized by mAb C3H7, represents a basolateral expressed counter-receptor for leukocyte β2-integrin CD11b/CD18 during PMN TEM. The epithelial CD44v3-CD11b/CD18 interaction defines a novel pathway of leukocyte recruitment relevant in inflammatory disorders associated with increased CD44v3 expression, such as IBD, and could provide the basis for the development of novel therapeutic applications.
Acknowledgments
We thank Zhen Bian and Rui Bai for cell isolation and culture and other technical assistance.
This work was supported, in whole or in part, by National Institutes of Health Grant R21 AI073622. This work was also supported by Scientist Development Grant 0535263N from the American Heart Association. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
The abbreviations used are: PMN, polymorphonuclear leukocyte; ICAM-1, intercellular adhesion molecule 1; TEM, transepithelial migration; IBD, inflammatory bowel disease; mAb, monoclonal antibody; TNF, tumor necrosis factor; I-domain, inserted domain; JAM-C, junction adhesion molecule C; BSA, bovine serum albumin; OG, n-octyl-β-d-glucopyranoside; FBG, fibrinogen; HRP, horseradish peroxidase; b-to-a, basolateral-to-apical; HBSS, Hanks' balance salt solution; BCECF-AM, 2′,7′-bis-(2-carboxyethyl)-carboxyfluorescein; siRNA, small interfering RNA.
References
- 1.Hawker, P. C., McKay, J. S., and Turnberg, L. A. (1980) Gastroenterology 79 508-511 [PubMed] [Google Scholar]
- 2.Weiland, J. E., Davis, W. B., Holter, J. F., Mohammed, J. R., Dorinsky, P. M., and Gadek, J. E. (1986) Am. Rev. Respir. Dis. 133 218-225 [DOI] [PubMed] [Google Scholar]
- 3.Colgan, S. P., Comerford, K. M., and Lawrence, D. W. (2002) Sci. World J. 2 76-88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Liu, Y., Buhring, H. J., Zen, K., Burst, S. L., Schnell, F. J., Williams, I. R., and Parkos, C. A. (2002) J. Biol. Chem. 277 10028-10036 [DOI] [PubMed] [Google Scholar]
- 5.Diamond, M. S., Garcia-Aguilar, J., Bickford, J. K., Corbi, A. L., and Springer, T. A. (1993) J. Cell Biol. 120 1031-1043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Smith, C. W., Rothlein, R., Hughes, B. J., Mariscalco, M. M., Rudloff, H. E., Schmalstieg, F. C., and Anderson, D. C. (1988) J. Clin. Investig. 82 1746-1756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Diamond, M. S., Staunton, D. E., de Fougerolles, A. R., Stacker, S. A., Garcia-Aguilar, J., Hibbs, M. L., and Springer, T. A. (1990) J. Cell Biol. 111 3129-3139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Altieri, D. C., Bader, R., Mannucci, P. M., and Edgington, T. S. (1988) J. Cell Biol. 107 1893-1900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Walzog, B., Schuppan, D., Heimpel, C., Hafezi-Moghadam, A., Gaehtgens, P., and Ley, K. (1995) Exp. Cell Res. 218 28-38 [DOI] [PubMed] [Google Scholar]
- 10.Schober, J. M., Chen, N., Grzeszkiewicz, T. M., Jovanovic, I., Emeson, E. E., Ugarova, T. P., Ye, R. D., Lau, L. F., and Lam, S. C. (2002) Blood 99 4457-4465 [DOI] [PubMed] [Google Scholar]
- 11.Diamond, M. S., Alon, R., Parkos, C. A., Quinn, M. T., and Springer, T. A. (1995) J. Cell Biol. 130 1473-1482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Peter, K., Schwarz, M., Conradt, C., Nordt, T., Moser, M., Kubler, W., and Bode, C. (1999) Circulation 100 1533-1539 [DOI] [PubMed] [Google Scholar]
- 13.Cai, T. Q., and Wright, S. D. (1996) J. Exp. Med. 184 1213-1223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dana, N., Fathallah, D. M., and Arnaout, M. A. (1991) Proc. Natl. Acad. Sci. U. S. A. 88 3106-3110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang, L., Fuster, M., Sriramarao, P., and Esko, J. D. (2005) Nat. Immunol. 6 902-910 [DOI] [PubMed] [Google Scholar]
- 16.Huang, G. T., Eckmann, L., Savidge, T. C., and Kagnoff, M. F. (1996) J. Clin. Investig. 98 572-583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zen, K., Liu, Y., Cairo, D., and Parkos, C. A. (2002) J. Immunol. 169 5270-5278 [DOI] [PubMed] [Google Scholar]
- 18.Santoso, S., Sachs, U. J., Kroll, H., Linder, M., Ruf, A., Preissner, K. T., and Chavakis, T. (2002) J. Exp. Med. 196 679-691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zen, K., Babbin, B. A., Liu, Y., Whelan, J. B., Nusrat, A., and Parkos, C. A. (2004) Mol. Biol. Cell 15 3926-3937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yan, Y., Dalmasso, G., Sitaraman, S., and Merlin, D. (2007) Am. J. Physiol. 292 G535-G545 [DOI] [PubMed] [Google Scholar]
- 21.Colgan, S. P., Parkos, C. A., McGuirk, D., Brady, H. R., Papayianni, A. A., Frendl, G., and Madara, J. L. (1995) J. Biol. Chem. 270 10531-10539 [DOI] [PubMed] [Google Scholar]
- 22.Liu, Y., Merlin, D., Burst, S. L., Pochet, M., Madara, J. L., and Parkos, C. A. (2001) J. Biol. Chem. 276 40156-40166 [DOI] [PubMed] [Google Scholar]
- 23.Parkos, C. A., Colgan, S. P., Liang, T. W., Nusrat, A., Bacarra, A. E., Carnes, D. K., and Madara, J. L. (1996) J. Cell Biol. 132 437-450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Parkos, C. A., Delp, C., Arnaout, M. A., and Madara, J. L. (1991) J. Clin. Investig. 88 1605-1612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lawrence, D. W., Bruyninckx, W. J., Louis, N. A., Lublin, D. M., Stahl, G. L., Parkos, C. A., and Colgan, S. P. (2003) J. Exp. Med. 198 999-1010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Colgan, S. P., Parkos, C. A., Matthews, J. B., D'Andrea, L., Awtrey, C. S., Lichtman, A. H., Delp-Archer, C., and Madara, J. L. (1994) Am. J. Physiol. 267 C402-C410 [DOI] [PubMed] [Google Scholar]
- 27.Reaves, T. A., Colgan, S. P., Selvaraj, P., Pochet, M. M., Walsh, S., Nusrat, A., Liang, T. W., Madara, J. L., and Parkos, C. A. (2001) Am. J. Physiol. Gastrointest. 280 G746-G754 [DOI] [PubMed] [Google Scholar]
- 28.Frick, C., Odermatt, A., Zen, K., Mandell, K. J., Edens, H., Portmann, R., Mazzucchelli, L., Jaye, D. L., and Parkos, C. A. (2005) Eur. J. Immunol. 35 3610-3621 [DOI] [PubMed] [Google Scholar]
- 29.Staunton, D. E., Dustin, M. L., Erickson, H. P., and Springer, T. A. (1990) Cell 61 243-254 [DOI] [PubMed] [Google Scholar]
- 30.Naor, D., Sionov, R. V., and Ish-Shalom, D. (1997) Adv. Cancer Res. 71 241-319 [DOI] [PubMed] [Google Scholar]
- 31.Brown, T. A., Bouchard, T., St John, T., Wayner, E., and Carter, W. G. (1991) J. Cell Biol. 113 207-221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jackson, D. G., Bell, J. I., Dickinson, R., Timans, J., Shields, J., and Whittle, N. (1995) J. Cell Biol. 128 673-685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Khan, A. I., Kerfoot, S. M., Heit, B., Liu, L., Andonegui, G., Ruffell, B., Johnson, P., and Kubes, P. (2004) J. Immunol. 173 7594-7601 [DOI] [PubMed] [Google Scholar]
- 34.Macdonald, D. C., Leir, S. H., Brooks, C., Sanders, E., Lackie, P., and Rosenberg, W. (2003) Eur. J. Gastroenterol. Hepatol. 15 1101-1110 [DOI] [PubMed] [Google Scholar]
- 35.Camacho, F. I., Munoz, C., Sanchez-Verde, L., Saez, A. I., Alcantara, M., and Rodriguez, R. (1999) Histopathology 35 144-149 [DOI] [PubMed] [Google Scholar]
- 36.Pituch-Noworolska, A., Wieckiewicz, J., Krzeszowiak, A., Stachura, J., Ruggiero, I., Gawlicka, M., Szczepanik, A., Karcz, D., Popiela, T., and Zembala, M. (1998) Anticancer. Res. 18 3747-3752 [PubMed] [Google Scholar]
- 37.Reinisch, W., Heider, K. H., Oberhuber, G., Dejaco, C., Mullner, M., Adolf, G. R., and Gasche, C. (1998) Gut 43 375-382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Trejdosiewicz, L. K., Morton, R., Yang, Y., Banks, R. E., Selby, P. J., and Southgate, J. (1998) Cytokine 10 756-765 [DOI] [PubMed] [Google Scholar]
- 39.Lesley, J., Hyman, R., and Kincade, P. W. (1993) Adv. Immunol. 54 271-335 [DOI] [PubMed] [Google Scholar]
- 40.Wittig, B., Seiter, S., Schmidt, D. S., Zuber, M., Neurath, M., and Zoller, M. (1999) Lab. Investig. 79 747-759 [PubMed] [Google Scholar]
- 41.Rosenberg, W. M., Prince, C., Kaklamanis, L., Fox, S. B., Jackson, D. G., Simmons, D. L., Chapman, R. W., Trowell, J. M., Jewell, D. P., and Bell, J. I. (1995) Lancet 345 1205-1209 [DOI] [PubMed] [Google Scholar]
- 42.Weg-Remers, S., Hildebrandt, U., Feifel, G., Moser, C., Zeitz, M., and Stallmach, A. (1998) Am. J. Gastroenterol. 93 790-794 [DOI] [PubMed] [Google Scholar]
- 43.Cichy, J., and Pure, E. (2003) J. Cell Biol. 161 839-843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Siegelman, M. H., DeGrendele, H. C., and Estess, P. (1999) J. Leukocyte Biol. 66 315-321 [DOI] [PubMed] [Google Scholar]
- 45.DeGrendele, H. C., Estess, P., Picker, L. J., and Siegelman, M. H. (1996) J. Exp. Med. 183 1119-1130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Underhill, C. (1992) J. Cell Sci. 103 293-298 [DOI] [PubMed] [Google Scholar]
- 47.Parish, C. R. (2006) Nat. Rev. 6 633-643 [DOI] [PubMed] [Google Scholar]
- 48.Schenauer, M. R., Yu, Y., Sweeney, M. D., and Leary, J. A. (2007) J. Biol. Chem. 282 25182-25188 [DOI] [PubMed] [Google Scholar]
- 49.Vachon, E., Martin, R., Kwok, V., Cherepanov, V., Chow, C. W., Doerschuk, C. M., Plumb, J., Grinstein, S., and Downey, G. P. (2007) Blood 110 4492-4502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Leir, S. H., Baker, J. E., Holgate, S. T., and Lackie, P. M. (2000) Am. J. Physiol. 278 L1129-L1137 [DOI] [PubMed] [Google Scholar]