Abstract
Pulmonary function after birth is dependent upon surfactant lipids that reduce surface tension in the alveoli. The sterol-responsive element-binding proteins (SREBPs) are transcription factors regulating expression of genes controlling lipid homeostasis in many tissues. To identify the role of SREBPs in the lung, we conditionally deleted the SREBP cleavage-activating protein gene, Scap, in respiratory epithelial cells (ScapΔ/Δ) in vivo. Prior to birth (E18.5), deletion of Scap decreased the expression of both SREBPs and a number of genes regulating fatty acid and cholesterol metabolism. Nevertheless, ScapΔ/Δ mice survived postnatally, surfactant and lung tissue lipids being substantially normalized in adult ScapΔ/Δ mice. Although phospholipid synthesis was decreased in type II cells from adult ScapΔ/Δ mice, lipid storage, synthesis, and transfer by lung lipofibroblasts were increased. mRNA microarray data indicated that SCAP influenced two major gene networks, one regulating lipid metabolism and the other stress-related responses. Deletion of the SCAP/SREBP pathway in respiratory epithelial cells altered lung lipid homeostasis and induced compensatory lipid accumulation and synthesis in lung lipofibroblasts.
Pulmonary surfactant consists of lipids and associated proteins that reduce surface tension at the air-liquid interface. Surfactant is required for adaptation to air breathing after birth. Lack of surfactant in preterm infants causes infantile respiratory distress syndrome and acute respiratory distress syndrome in older individuals. Likewise, mutations in genes regulating surfactant homeostasis, including SFTPB, SFTPC, and ABCA3, disrupt surfactant homeostasis causing fatal respiratory distress or chronic lung disease (1, 2). Although various lipids play a critical role in surfactant function, transcriptional mechanisms regulating surfactant homeostasis in the respiratory epithelium remain poorly understood.
In other tissues, transcriptional mechanisms regulating lipid synthesis are known to be dependent on a number of transcription factors, including CCAAT/enhancer-binding protein (C/EBP)2 isoforms, liver X receptor, peroxisome proliferator-activated receptors (PPARs), and sterol regulatory element-binding proteins (SREBPs) (3-6). Although transcriptional networks regulating lipid homeostasis have been extensively studied in other cell types, including hepatocytes and adipocytes, less is known regarding transcriptional control of lipid homeostasis in the respiratory epithelium. SREBP-1c, C/EBPα, and C/EBPδ regulate lipogenic enzymes and transport proteins in the lung (7-11). C/EBP isoforms and SREBP-1c mRNAs are increased in the fetal rat lung during late gestation in association with increased expression of surfactant proteins (A, B, C, and D) (1). Induction of proteins regulating lipid synthesis and surfactant proteins occurs during perinatal lung maturation and is required for respiratory function at birth.
Three SREBP isoforms, SREBP-1a, SREBP-1c, and SREBP-2, are synthesized as inactive precursors that are inserted into the membranes of the endoplasmic reticulum (ER), where they bind to SREBP cleavage-activating protein (SCAP). In response to cholesterol depletion, SCAP transports the SREBPs from the endoplasmic reticulum to the Golgi, where the NH2-terminal domain of SREBP, the active form of the transcription factor, is released by proteolytic cleavage by two proteases, S2P and S2P, allowing the active SREBP to enter the nucleus where it binds and activates transcription of target genes. SREBPs regulate many aspects of lipid biosynthesis; SREBP-1a and, particularly, SREBP-1c are relatively selective for the regulation of fatty acid synthesis, whereas SREBP-2 is a more potent activator of cholesterol synthesis (3, 12).
Although SREBP-1c regulates a number of genes that are known to influence surfactant homeostasis, lung pathology was not detected in adult Srebf-1 gene-targeted mice (13), indicating that compensatory or redundant pathways maintain surfactant lipid synthesis for respiratory function after birth (14). To determine the role of the SREBP-regulating pathways in lung lipid homeostasis, we conditionally deleted the Scap gene in respiratory epithelial cells in the developing lung, thereby inactivating all three SREBP isoforms. In this study, we show that SCAP regulates both SREBPs and a number of genes controlling lipid homeostasis in the lung. Deletion of Scap altered lipid content and synthesis. Maintenance of surfactant function in Scap-deleted mice after birth was associated with compensatory lipid synthesis, accumulation, and transfer by lung lipofibroblasts.
EXPERIMENTAL PROCEDURES
Transgenic Mice—Scapflox/flox mice bearing a loxP-flanked neo cassette located 3 kb 5′ of Scap exon 1 and a third loxP site located in intron 1 (15) were purchased from The Jackson Laboratory (Bar Harbor, ME). Homologous recombination between loxP sites was accomplished by expression of Cre recombinase using (tetO)7CMV-Cretg/tg mice. The SP-C-rtTA--/tg transgenic mouse line (16, 17) was used for respiratory epithelium-specific expression of reverse tetracycline transactivation (rtTA) to cause permanent recombination of the floxed allele after exposure of the dam to doxycycline (16, 17). Triple transgenic mice, herein termed ScapΔ/Δ mice, were generated by mating (tetO)7CMV-Cre-/tg/Scapflox/flox to SP-C-rtTA-/tg/Scapflox/flox mice. Scapflox/flox littermates lacking either rtTA or Cre genes served as controls. Triple transgenic mice SP-C-rtTAtg/-, TetO-Cretg/-, Scapflox/flox are maintained on a mixed background issued from the FVB/N strain (SP-C-rtTAtg/-, TetO-Cretg/-) and the B6;129S6 background (Scapflox/flox). Genotypes were identified by PCR with genomic DNA from the tails of mice using the forward primer 5′-GCT CTG CGC ATC CTA TCC AAT TCC C-3′ and the reverse primer 5′-CAG CCG GCA AGT AAC AAG GGA TCC G-3′ for Scapflox/flox. Genotyping for SP-C-rtTA and (tetO)7CMV-Cre DNA was performed by PCR as described previously (16).
Animal Husbandry and Doxycycline Administration—Mice were maintained in a pathogen-free environment in accordance with protocols approved by the Institutional Animal Care and Use Committee of the Cincinnati Children's Hospital Research Foundation. All animals were housed in humidity- and temperature-controlled rooms on a 12-12-h light-dark cycle. Mice were allowed food and water ad libitum. There was no serological evidence of pulmonary pathogens or bacterial infections in sentinel mice maintained within the colony. Gestation was dated by detection of the vaginal plug (as E0.5) and correlated with weight of each pup at the time of sacrifice. Dams bearing control and ScapΔ/Δ mice were maintained on doxycycline in food (625 mg/kg; Harlan Teklad, Madison, WI) from E6.5 to E12.5 to cause widespread deletion of Scap in progenitor cells that form the peripheral lung (18).
Tissue Preparation—Four- to eight-week-old mice were anesthetized by an injection of 0.25 ml of anesthetic (ketamine, xylazine, and acepromazine) and then exsanguinated by severing the inferior vena cava. For fetal lung tissue analyses, dams were killed by exsanguination, and fetal lungs were dissected for analysis. Fetuses were removed from the uterine sac, weighed, and crown-to-rump lengths determined to assess developmental age (19). Lung weights were measured after the heart, trachea, and bronchi were removed.
In Situ Hybridization—In situ hybridization was performed using 35S-UTP-labeled riboprobe for Scap (a 2175-bp mouse cDNA (GenBank™ accession number GI 47564087), subcloned into pCRII, Invitrogen). In situ hybridizations were performed on lung sections at E12.5, E14.5, E16.5, and E18.5 as described previously (23). The exposure period was 5 weeks for the material presented in Fig. 1. The material was photographed after counterstaining the sections with toluidine blue.
FIGURE 1.
SCAP deletion in the respiratory epithelium regulates SREBP gene expression in vivo. A-F, in situ hybridization for Scap mRNA was performed on embryonic sections from E14.5. Scap mRNA was readily detected in the respiratory epithelium (arrow, inset), as well as in the surrounding mesenchyme, in the littermate controls (A and D). In contrast, Scap mRNA was absent in the respiratory epithelium (arrow, inset) of the ScapΔ/Δ mice (B and E). No specific ISH signals were observed in sections from the ScapΔ/Δ mice, which were hybridized with the sense (control) probe (C and F). Insets in D and E show a higher magnification of the respiratory tubules indicated with the red asterisk in both the bright field (A and B) and corresponding dark field images (D and E). Sections were hybridized with radiolabeled riboprobes as described previously (23), coated with photographic emulsion, exposed for 5 weeks, and then counter-stained with toluidine blue prior to imaging. V, vessel. Scale bar, 100 μm for A-F, and 50 μm for the insets in D and E. G, quantitative RT-PCR was performed to estimate Scap, Srebp-1c, Srebp-1a, and Srebp-2 mRNAs in E18.5 lungs, E18.5, and adult isolated alveolar type II cells from ScapΔ/Δ (white bar) and control littermates (black bar) and normalized to β-actin mRNA. Dams were treated with doxycycline from E6.5 to E12.5 to delete the Scap gene from respiratory epithelial cells. Results are expressed as the means ± S.E. of five animals per group. *, p < 0.01 versus control littermates.
Isolation of Lung Fibroblasts and Alveolar Type II Epithelial Cells—Fibroblasts and alveolar type II cells were isolated from 6-week-old control and ScapΔ/Δ mouse lung, using collagenase and differential plating as described previously (22, 24). Fibroblasts and type II cells were used 2 h after isolation for RNA analysis or cultured for study of lipid synthesis. Cell purity for the alveolar type II cell and fibroblast fractions was determined by immunochemistry using rabbit polyclonal pro-SP-C (1:2000, Seven Hills Bioreagents, Cincinnati, OH) and vimentin (1:600, mouse IgM, Sigma) as described below.
RNA Isolation and Analysis—RNA was isolated from whole lung, isolated lung fibroblasts, or alveolar type II cells from 8-week-old mice using TRIzol (Invitrogen). RNA was treated with DNase at room temperature for 15 min before cDNA synthesis. RNA (4 μg) was reverse-transcribed and then analyzed by quantitative RT-PCR for using Smart Cycler (Cepheid, Sunnyvale, CA). A list of genes and associated primers are described in supplemental Table 1.
Cell Transfection and Luciferase Assay—Mouse lung epithelial cells, MLE-15, an immortalized mouse lung epithelial cell line that maintains some morphological and functional characteristics of type II epithelial cells, were cultured in HITES medium (20, 21). The surfactant protein reporter constructs were described previously (20) and cotransfected with either empty vector pcDNA3 or pCMV-nSREBP-1c plasmid, encoding amino acids 1-436 of human nuclear (n) SREBP-1c purchased from the American Type Culture Collection (ATCC, Manassas, VA) or pcDNA3-nSREBP-1a and pcDNA3-nSREBP-2 described previously (21). Forty eight hours after transfection, luciferase activity was assessed and normalized for cotransfection efficiency to β-galactosidase activity. All transfections were performed in triplicate. pcDNA3 (Invitrogen) and pCMV-β-galactosidase (Clontech) vectors were used to normalize DNA and transfection efficiency, respectively.
Lung Histology, Histochemistry, and Immunostaining—Embryonic lungs were immersed in 4% paraformaldehyde in phosphate-buffered saline (20 mm Tris·HCl, pH 7.6, 137 mm NaCl). Four- to eight-week-old mice lungs were inflation fixed with the same fixative at 25 cm of H2O before immersion. Lungs were fixed overnight, washed with phosphate-buffered saline, dehydrated in a series of alcohols, and embedded in paraffin. Immunohistochemistry was performed on tissue sections using a microwave antigen-retrieval technique for detection of transcription factors (Ki-67). Sections were pretreated with 3% H2O2 in methanol for inactivation of endogenous peroxidase and then blocked in 4% normal serum for 2 h before incubation with the primary antibody overnight at 4 °C. Antibodies used were generated to Ki-67 (1:500, M7249, rat monoclonal, DAKO, Carpinteria, CA), cleaved caspase-3 (1:1000, rabbit polyclonal; R&D Systems, Inc., Minneapolis, MN), and adipose differentiation related protein (ADFP) (1:1000, rabbit polyclonal) (22). After being rinsed, tissue sections were incubated with biotinylated goat anti-rabbit or goat anti-rat IgGs (7.5 μg/ml; Vector Laboratories, Burlingame, CA) for 30 min and detected with an avidin-biotin peroxidase complex detection kit (Vectastain Elite ABC kit; Vector Laboratories, Burlingame, CA) using nickel-diaminobenzidine as a substrate. The precipitation reaction was enhanced with Tris-cobalt, and the sections were counterstained with 0.1% of nuclear fast red. All experiments shown are representative of findings from at least four independent dams, each generating at least one or two ScapΔ/Δ mice that were compared with control littermates.
Oil Red O Staining—Lung sections were stained with Oil Red O to detect neutral lipids according to the manufacturer's protocol (Poly Scientific, Bayshore, NY). The number of Oil Red O-stained cells was determined by a point-counting method on 6-week-old mice (30, 31). Five mice of both genotypes were studied. Measurements were performed on two sections taken at intervals throughout the left, right upper, or right lower lobes. Sections were viewed and photograph using a ×20 objective, and the digital images (fields) were analyzed using Meta-Morph imaging software (Molecular Devices, Downingtown, PA). A computer-generated, 121-point lattice grid was superimposed on each field, and the number of Oil Red O-stained cells was counted manually. The density of Oil Red O stained cells was calculated by dividing the number of Oil Red O-stained cells by alveolar area (mm2). Fifteen fields per section were analyzed to gather the data. The x and y coordinates for each field measured were selected by a random number generator. Bronchioles, large vessels, and smaller arterioles and venules were excluded from the study.
Saturated Phosphatidylcholine (Sat PC) Measurement—Eight-week-old mice (n = 5/group) were anesthetized and killed by exsanguination. Tracheas were cannulated, and five 1-ml aliquots of 0.9% NaCl were flushed into the lungs and withdrawn by syringe three times for each aliquot. The lavaged lung tissue was removed and homogenized in 2 ml of 0.9% NaCl. Sat PC in lipid extracts of bronchoalveolar lavage fluid (BALF) and lung tissue were isolated with osmium tetroxide (25) followed by phosphorus measurement, as described previously (26). The volumes of recovered BALF from all the groups were similar.
Electrospray Ionization-Mass Spectrometry of Phospholipid Molecular Species—BALF and lung tissue phospholipids were extracted with chloroform and methanol according to Bligh and Dyer (27) after adding the following internal standards (nmol/107 cells): PC14:0/14:0 (15 nmol), PE14:0/14:0 (4 nmol), PG14:0/14:0 (2 nmol), and PS14:0/14:0 (2 nmol). Electrospray ionization mass spectrometry (ESI-MS) of phospholipids was performed as described previously (28). The volumes of recovered BALF from all the groups were similar.
Precursor Incorporation into Phosphatidylcholine (PC) and Secretion—After cell isolation and differential plating, fibroblasts were grown for 24 h in Dulbecco's modified Eagle's medium containing 20 mm HEPES, 50 units of penicillin/ml, 50 μg of streptomycin/ml, and 5% fetal calf serum and supplemented with 1 μCi/ml [3H]choline chloride (PerkinElmer Life Sciences). Alveolar type II cells were cultured on Matrigel matrix-rat tail collagen (70:30 v/v) in bronchial epithelial cell growth medium minus hydrocortisone plus 5% charcoal-stripped fetal bovine serum and 10 ng/ml keratinocyte growth factor for 5 days (24) and then cultured for 48 h with the same medium supplemented with 1 μCi/ml [3H]choline chloride. After culture in the presence of the isotope, cells were washed with saline solution and replaced in culture medium without the isotope for 3 h. After 3 h of secretion, conditioned media and adherent cells were harvested separately. Lipids were extracted with chloroform and methanol (27), and radioactivity in PC was counted. Percent secretion at 3 h was estimated from the amount of radioactive PC in the conditioned media divided by the sum of the radioactive PC in the conditioned media and cell extract. Cell numbers and volumes of recovered conditioned media from all the groups were similar.
Lipid Uptake—After cell isolation and differential plating, fibroblasts were cultured for 24 h in medium, as described previously, supplemented with 1 μCi/ml [3H]choline chloride or 1 μCi/ml [14C]acetate (PerkinElmer Life Sciences). After culture in the presence of the isotope, cells were washed with saline solution and replaced in culture medium without the isotope for 4 h. After 4 h of secretion, fibroblast-conditioned media were harvested, and radioactivity was counted and stored at -20 °C. Alveolar type II cells were cultured on Matrigel as described previously for 3 days (24), washed with saline solution, and then cultured for 4 h with fibroblast-conditioned media. After 4 h in fibroblast-conditioned media, adherent cells were washed and harvested. An aliquot of the cell suspension was taken to estimate protein concentration determined by the Bradford assay, and the remaining cell suspension was extracted for lipid content with chloroform and methanol, and radioactivity in PC was counted. Percentage of uptake at 4 h was estimated from the amount of radioactive PC in the type II cells divided by the sum of the radioactive PC in the conditioned media and cell extract.
Triglyceride and Cholesteryl Ester Measurements—Lipid extracts of BALF, lung tissue, alveolar type II cells, and lung fibroblasts recovered from 8-week-old mice (n = 5/group) were assayed for triglyceride and cholesteryl ester content according to the manufacturer's protocol using the triglyceride quantification kit (BioVision, Mountain View, CA) and the cholesterol/cholesteryl ester quantification kit (EMD Chemicals, Inc., San Diego, CA), respectively. The volumes of recovered BALF from all the groups were similar. The protein concentration in each fraction was measured by the Bradford assay (Bio-Rad) and was used to normalize triglyceride and cholesteryl ester contents in each fraction.
Ultrastructure Studies—Lung tissue from 8-week-old ScapΔ/Δ mice and littermate controls was fixed in modified Karnovsky's fixative consisting of 2% glutaraldehyde and 2% paraformaldehyde in 0.1 m sodium cacodylate buffer (SCB) and 0.1% calcium chloride, pH 7.3. The tissue was postfixed in 1% osmium tetroxide in 0.1 m SCB, dehydrated, and embedded in epoxy resin (EMbed 812; Electron Microscopy Sciences, Fort Washington, PA), as described previously (29). Ultrathin sections were viewed in a Morgagni 268 transmission electron microscope, and digitized images were collected with an AMT Advantage Plus 2K × 2K digital camera (Advanced Microscopy Techniques, Danvers, MA).
RNA Microarray Analysis—For Affymetrix MicroArray, E18.5 lung RNAs from ScapΔ/Δ mice and littermate controls were prepared using RNeasy Protect mini kit (Qiagen, Valencia, CA) according to the manufacturer's protocol. RNA was treated with DNase at room temperature for 15 min. Lung cRNA was hybridized to the murine genome 430 2.0 array (consisting of 45,000 probe sets representing over 34,000 mouse genes; Affymetrix Inc.) using the manufacturer's protocol. Affymetrix MicroArray Suite version 5.0 was used to scan and quantitate the gene chips using default scan settings. Normalization was performed using the Robust Multichip Average model that consists of three steps, namely background adjustment, quartile normalization, and summarization (32, 33). Microarray analysis was performed with the software package BRB Array Tools, developed by the Biometric Research Branch of the NCI, National Institutes of Health (linus.nci.nih.gov). Differentially expressed genes were identified using a univariate F test and permutation test (n = 100) with a significance level of 0.05. Data were prefiltered by excluding probe sets as follows: 1) whose expression differed ≤1.2 from the median in 80% of the samples, 2) whose expression data were missing in more than 50% of the samples, or 3) with more than 70% of absent calls by the Affymetrix algorithm in six samples. Gene ontology and KEGG pathway analysis were performed using the public available web-based tool David (Data base for annotation, visualization, and integrated discovery) (34). Gene frequency and probability in each functional category were calculated by Fisher Exact Test using Mouse Genome 430 2.0 as background control. Biological association networks were built using Ingenuity Pathway Analysis (Ingenuity Systems, Inc., Redwood City, CA).
Statistical Analysis—Either Mann-Whitney U test or Student's t test were used to determine the levels of difference between groups. Values for all measurements were expressed as the mean ± S.E., and p values for significance are indicated for each experiment.
RESULTS
Conditional Deletion of the Scap Gene in the Lung Alveolar Epithelium—Srebp mRNAs were previously detected in alveolar type II cells in the murine lung, supporting their potential role in the regulation of surfactant homeostasis (10). To identify the role of SREBPs in alveolar type II cells, triple transgenic mice SP-C-rtTAtg/-, TetO-Cretg/tg, and Scapflox/flox were produced in which Scap was selectively and permanently deleted in the respiratory epithelium following administration of doxycycline to the dam from E6.5 to E12.5 (21). To assess the efficiency of Cre-mediated Scap gene deletion in the ScapΔ/Δ mice, in situ hybridization was performed on lung sections prepared from fetal mouse lung at E14.5 (Fig. 1, A-F). Although readily detected in control littermates (Fig. 1D), Scap mRNA was decreased or absent in the epithelial cells of developing lung tubules of the ScapΔ/Δ mice (Fig. 1E). Likewise, Scap gene deletion was assessed by quantitative RT-PCR on E18.5 fetal lung, E18.5, and adult alveolar type II cells mRNAs, demonstrating a marked decrease in Scap mRNA in ScapΔ/Δ mice (Fig. 1G). As observed in other tissues (15, 21), deletion of Scap reduced Srebp-1c and Srebp-1a mRNAs.
Expression of Genes Encoding Lipogenic and Surfactant-associated Proteins in ScapΔ/Δ Mice—Previous studies demonstrated that SREBPs regulate genes playing important roles in fatty acid and cholesterol metabolism. To identify genes influenced by the SCAP/SREBP pathway in the lung, the expression of mRNA encoding a number of known transcriptional targets of the SREBPs were estimated by quantitative RT-PCR from E18.5 lung, adult whole lung, and alveolar type II cells isolated from ScapΔ/Δ and control littermates (Fig. 2). A marked decrease in mRNAs influencing fatty acid metabolism (Scd1, Scd2, Fasn, Fdps1, Fad1, Fabp5, and Thrsp) and cholesterol metabolism (Hmgcs1, Hmgcs2, Ldlr, Abca1, and Gpam) was observed at E18.5 in the lungs of ScapΔ/Δ mice (Fig. 2, B and C). Similar Scap-dependent alterations were observed in alveolar type II cells isolated from the adult ScapΔ/Δ mice (Fig. 2, H and I). Differences in expression of these mRNAs were not observed in mRNA isolated from total adult lung, indicating the potential for compensatory regulation of gene expression in other lung cell types in the adult. Expression of Abca3, a phospholipid transport protein critical for surfactant production (35, 36), was significantly decreased at E18.5, consistent with previous findings demonstrating that SREBP-1c directly induced the Abca3 gene promoter (21). Because the promoter regions of lung surfactant-associated genes (Sftpa, Sftpb, Sftpc, and Sftpd) contain potential DNA-binding sites for SREBPs, we sought to determine whether SREBPs activate the Sftpa, Sftpb, Sftpc, and Sftpd promoters in vitro. Promoter constructs for these genes were cotransfected with expression vectors encoding a nuclear form of SREBP-1a (nSREBP-1a, amino acids 1-460), SREBP-1c (nSREBP-1c, amino acids 1-436), or SREBP-2 (nSREBP2, amino acids 1-431) in MLE-15 cells. nSREBP-1a and nSREBP-1c significantly increased the transcriptional activity of the Sftpa, Sftpb, and Sftpd constructs (Fig. 2J). In contrast, none of the SREBPs significantly changed Sftpc promoter activity. In vivo, the mRNA encoding SP-A (Sftpa) was decreased at E18.5 in ScapΔ/Δ mice (Fig. 2, A, D and G), whereas expression of other Sftp genes was not significantly altered by deletion of Scap.
FIGURE 2.
The SCAP/SREBP pathway regulates lipid metabolism genes in the lung. Quantitative RT-PCR was performed to estimate lung surfactant-associated (A, D, and G), fatty acid metabolism (B, E, and H), or cholesterol metabolism (C, F and I) gene mRNA levels in fetal lung from ScapΔ/Δ (white bar) and control littermates (black bar) at E18.5 (A-C), as well as whole lung (D-F) and alveolar type II cells (G-I) isolated from adults. Results were normalized to β-actin mRNA. Dams were treated with doxycycline from E6.5 to E12.5 to delete the Scap gene from respiratory epithelial cells. Results are expressed as the means ± S.E. of 3-5 animals per group. *, p < 0.01 versus control littermates. J, effect of SREBPs on surfactant protein promoter activity was assessed after cotransfection of either rat Sftpa-Luc, murine (m) SftpB-Luc, mSftpc-Luc, or mSftpd-Luc with either pcDNA3, pcDNA3-nSREBP-1a, pCMV-nSREBP-1c, or pcDNA3-SREBP-2 into MLE15 cells. Results are expressed as the mean ± S.E. of three separate experiments performed in triplicate, *, p < 0.05 versus pcDNA3 condition.
Modification of the Lipid Content in ScapΔ/Δ Mice Lungs—To evaluate the effects of Scap gene deletion on lung lipid content, surfactant Sat PC pool size in BALF and total lung (alveolar lavage plus lung tissue after lavage) was measured in adult mice. Although total lung Sat PC was significantly decreased (Fig. 3B), BALF Sat PC was similar in ScapΔ/Δ mice and control littermates (Fig. 3A). To determine the effects of Scap gene deletion on lung lipid composition, phospholipids from both BALF and lung tissues were assessed by ESI-MS (Fig. 3, C and D). The concentrations of individual phospholipid classes in BALF and lung tissue extracts were calculated as the sums of their individual molecular species determined by ESI-MS. There was a 30% reduction in total PG in both surfactant and lung tissue in the ScapΔ/Δ mice; however, significant differences in the fractional concentrations of PC, PI, PE, and PS were not observed in either BALF or lung tissue. Similarly, there were no differences in the individual molecular species of PG or PI in BALF or in PG, PI, PE, or PS in lung tissue from ScapΔ/Δ mice. In contrast, the relative concentration of PC16:0/16:1 in surfactant and lung tissue PC, expressed relative to total PC, was significantly decreased in adult ScapΔ/Δ mice. The decrease in PC16:0/16:1 content occurred without changes in the content of the disaturated species PC16:0/14:0, PC16:0/16:0, and PC16:0/18:0, but with an increase in longer chain PC species (data not shown).
FIGURE 3.
Phospholipid composition and metabolism are altered in lungs of ScapΔ/Δ mice. Sat PC pool size in BALF was similar in adult ScapΔ/Δ and control littermates (A). In contrast, Sat PC pool size in lung tissue after BALF was significantly decreased in adult ScapΔ/Δ (B). Results are expressed as the means ± S.E. of five animals per group. The relative distributions of the phospholipid classes, phosphatidylcholine (PC), phosphatidylethanolamine (PE), phosphatidylinositol (PI), phosphatidylserine (PS), phosphatidylglycerol (PG), in BALF (C) or lung tissue (D) from ScapΔ/Δ (white bar) and control littermates (black bar) were determined by ESI-MS analysis. The sums of individual molecular species from each phospholipid class are expressed as means ± S.E. of five animals per group.
To assess whether the changes in expression of genes influencing phospholipid metabolism altered phospholipid synthesis and secretion, incorporation of [3H]choline into PC in cultured alveolar type II cells was assessed (Fig. 4, A and B). Although PC synthesis was significantly decreased by 43% in type II cells from adult ScapΔ/Δ mice (Fig. 4A), there were no differences in PC secretion (Fig. 4B). To evaluate the effects of Scap gene deletion on lung neutral lipid content, triglyceride and cholesteryl ester content was measured in adult mice. Surprisingly, total lung triglyceride and cholesteryl ester content was significantly increased in ScapΔ/Δ mice (Fig. 4, C and D). Although BALF triglyceride content was undetectable under our experimental conditions, BALF cholesteryl ester content was significantly decreased in ScapΔ/Δ mice (data not shown). In isolated alveolar type II cells, cholesteryl ester content was similar in ScapΔ/Δ mice and control littermates, whereas triglyceride content was slightly reduced in the type II cells from ScapΔ/Δ mice. Taken together, these data indicate that deletion of Scap in the respiratory epithelium may be compensated by the accumulation and synthesis of lipids in non-epithelial compartments of the lung.
FIGURE 4.
Phospholipid synthesis and neutral lipid content are altered in lungs of ScapΔ/Δ mice. Incorporation of [3H]choline into PC was measured in primary cultures of lung alveolar type II cells (A and B) from adult ScapΔ/Δ (white bar) and control littermates (black bar). Radiolabeled PC was recovered from cells (A) or after 3 h of secretion (B). Results are expressed as the means ± S.E. of five animals per group. Neutral lipids from whole lung tissue, isolated alveolar type II cells (TIIC), and fibroblasts were assessed for triglycerides (C) and cholesteryl esters (D) content in adult ScapΔ/Δ (white bar) and control littermates (black bar). Results are expressed as the means ± S.E. of five animals per group.
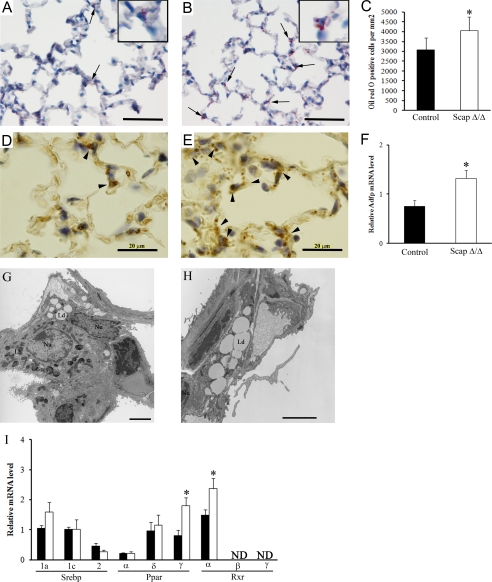
Identification of a Compensatory Pathway in the Lipofibroblasts of ScapΔ/Δ Mice—Lipofibroblasts present numerous lipid droplets containing triglycerides and other lipids that can be transferred to the majority of alveolar type II cells and used in the synthesis of pulmonary surfactant (37-39). In general, lung morphology in adult ScapΔ/Δ mice was normal; however, focal lobar and panacinar emphysema was noted in ∼30% of the mice (data not shown). Deletion of Scap in alveolar type II cells increased the number and size of lipid droplets present in the lipofibroblasts as assessed by Oil Red O staining, a marker for neutral lipid accumulation (Fig. 5, A and B). Quantitative analysis demonstrated a significant increase in the number of cells stained for Oil Red O in the alveolar walls (Fig. 5C). Measurement of triglyceride and cholesteryl ester content demonstrated an increase of neutral lipids in fibroblasts from ScapΔ/Δ mice (Fig. 4, C and D), suggesting that the increase in triglyceride and cholesteryl ester content observed in whole tissue was related to the increase in the content in the fibroblasts. In addition, mRNA expression of ADFP, a marker of cells storing neutral lipids, was markedly increased in lungs from ScapΔ/Δ mice (Fig. 5F), where it was localized to interstitial fibroblasts in the alveolar walls (Fig. 5, D and E). Electron microscopy demonstrated that the increase in lipid droplets occurred in lung lipofibroblasts and not in other lung cell types (Fig. 5, G and H), suggesting that lipid accumulation in lipofibroblasts may compensate for decreased lipid synthesis after Scap deletion in type II epithelial cells. Consistent with this concept, expression of Pparg and Rxra mRNAs (both transcriptional regulators of lipogenesis) was significantly increased in lung lipofibroblasts from adult ScapΔ/Δ mice (Fig. 5I).
FIGURE 5.
CompensatorylipidaccumulationinlipofibroblastsafterdeletionofScap.NeutrallipidswereassessedbyOilRedOstaininginadultScapΔ/Δ (whitebar) (A) and control littermates (black bar)(B). Extensive lipid inclusions were noted in the lipofibroblasts within the alveoli in ScapΔ/Δ mice (arrowhead). Density of lipid droplet-containing cells (C) was estimated in adult ScapΔ/Δ and control littermates. Data are expressed as the number of lipid stained cells per lung area and represent mean ± S.E. (*, p < 0.001 for ScapΔ/Δ versus control littermates). Extensive immunostaining for adipose differentiation-related protein (ADFP)(arrowhead) was observed in adult ScapΔ/Δ (E) compared with control littermates (D). F, quantitative RT-PCR was performed to estimate Adfp mRNA levels in isolated fibroblasts from adult ScapΔ/Δ and control littermates. Results were normalized toβ-actin mRNA. Results are expressed as the means ± S.E. of five animals per groups. *, p < 0.01 versus control littermates. Electron microscopy showed that type II cells from ScapΔ/Δ (G) were similar to these in control mice (data not shown), and higher magnification confirmed the presence of the lipid droplets in the fibroblasts adjacent to the cuboidal type II cells in ScapΔ/Δ mice (H). Scale bar, 20 μm. I, Srebp, Ppar, and Rxr isoform mRNAs were assessed by quantitative RT-PCR in fibroblasts isolated from ScapΔ/Δ mice (white bar) and control littermates (black bar). Results were normalized to β-actin mRNA. Results are expressed as the mean ± S.E. of five animals per group, *, p < 0.01 versus control littermates. ND, nondetectable.
Enhanced Transfer of Labeled Lipids from Lipofibroblasts to Type II Cells after Deletion of Scap—To determine whether the increase in lipid content in the lipofibroblasts of the ScapΔ/Δ mice was because of increased lipid synthesis, incorporation of phospholipid precursor was analyzed in isolated lung fibroblasts. Incorporation of [3H]choline into PC was increased by 25% in lung fibroblasts isolated from ScapΔ/Δ mice compared with control mice (Fig. 6A), whereas the percent of PC secretion was not significantly altered (Fig. 6B). To evaluate whether alveolar type II cells import lipids secreted by the lipofibroblasts, conditioned media from isolated fibroblasts were prelabeled with [3H]choline or [14C]acetate, and conditioned media were added to cultured type II cells for 4 h. After 4 h of incubation with fibroblast-conditioned media, the percentage of lipid uptake was increased by 50% in alveolar type II cells isolated from ScapΔ/Δ mice compared with control mice (Fig. 6C). Uptake of [14C]acetate-labeled lipids by type II cells from ScapΔ/Δ mice was increased to a lesser extent (Fig. 6D).
FIGURE 6.
Enhanced synthesis and transfer of lipids from lipofibroblasts to type II cells after deletion of Scap. Incorporation of [3H]choline into PC was measured in primary cultures of lung fibroblasts (A and B) from adult ScapΔ/Δ (white bar) and control littermates (black bar). Radiolabeled PC was recovered from cells (A) or after 3 h of secretion (B). Results are expressed as the means ± S.E. of five animals per group. Percentage of uptake by type II cells of lipids secreted by fibroblasts was determined by radiolabeling and expressed relative to the total label added to the culture wells as described under “Experimental Procedures.” Transfer of [3H]choline and [14C]acetate labeled lipids was measured from lipofibroblasts to alveolar type II cells of adult ScapΔ/Δ (white bar) and control littermates (black bar). Uptake percentage of [3H]choline-PC (C) and [14C]acetate-lipids (D) by adult alveolar type II cells from ScapΔ/Δ mice (white bar) was significantly increased compared with control mice (black bar). Results are expressed as the means ± S.E. of five animals per group.
RNA Microarray Analysis of SCAP/SREBP-regulated Genes—Microarray analysis of lung RNA from E18.5 ScapΔ/Δ and control mice identified 578 genes that were significantly altered (p value ≤ 0.05); 340 mRNAs were increased, and 238 mRNAs were decreased. Among these, 118 genes exhibited more than a 1.5-fold change in expression (45 were up-regulated and 73 were down-regulated; see supplemental Table 2). Differentially expressed genes were classified according to Gene Ontology classification. Gene Ontology analysis indicated that lipid/sterol metabolism was the most overly represented biological process down-regulated by Scap deletion (33%, 1.7E-12). In contrast, genes involved in amino acid biosynthesis (7.3%, 3.68 E-04) and cell death (34%, 1.38 E-03) were induced in response to the deletion of Scap. Many of the induced genes encode proteins located in the endoplasmic reticulum, making the ER the most enriched cellular compartment identified after deletion of Scap when compared with the distribution of cellular sites in the mouse genome (11%, 5.9 E-04). Gene set enrichment analysis, using the differentially expressed genes overlapping with KEGG pathways, suggested the enrichment of down-regulated genes in “biosynthesis of steroids” (1.33E-09), “PPAR signaling” (9.77E-04), and “polyunsaturated fatty acid biosynthesis” (1.09E-04) pathways. Genes involved in lipid metabolism, including sterol, isoprenoid, fatty acid, and carboxylic acid metabolism and biosynthesis, were markedly decreased in lungs of ScapΔ/Δ mice, demonstrating that Scap plays an important role in the regulation of genes influencing lipid homeostasis in the lung. Changes in gene expression represented known direct transcriptional targets of the active forms of SREBPs (40) and were likely to include many indirect effects on gene expression related to change in lipid homeostasis. Expression of mRNAs of genes regulating lipid synthesis were decreased in ScapΔ/Δ mice as assessed by real time PCR, consistent with the RNA microarray data (Fig. 2). Biological association networks in which the functions of the proteins encoded by genes whose expression was changed by more than 1.5-fold revealed only two major functional networks influenced by deletion of Scap as follows: 1) those associated with lipid/sterol metabolism, and 2) ER stress-mediated cell death (Fig. 7).
FIGURE 7.
Network analysis of mRNAs influenced by the deletion of Scap in the respiratory epithelium. Differentially expressed genes identified from the mRNA microarray study of lung tissue at E18.5 were used to build a gene regulation interactome as described under “Experimental Procedures.” Genes connected to SCAP/SREBP were visualized by building a SCAP/SREBP centric network. Biological association networks of differentially expressed genes revealed two major functional networks lipid/sterol metabolism and ER-stress mediated cell death. Genes/proteins are represented as nodes, and the biological relationship between two nodes is represented as an edge (line). Uncolored gene nodes were not identified as differentially expressed in our microarray and were integrated into the networks on the basis of literature evidence indicating the likelihood of their relevance to the network. The node shapes denote different types of molecule as indicated in the figure legend.
To determine whether the deletion of Scap altered cell proliferation or cell death in the respiratory epithelium, cell proliferation and apoptosis were assessed by immunohistochemistry using Ki-67 (supplemental Fig. 1, A-D) and cleaved caspase 3 (supplemental Fig. 1, E-H) as markers. No differences in Ki-67 and cleaved caspase-3 staining were observed in lungs of the ScapΔ/Δ mice at E18.5 or in adults.
DISCUSSION
SCAP and the SREBP transcription factors are expressed in diverse tissues, including liver, adipose tissue, kidney, lung, intestine, and brain, where they regulate lipid homeostasis (12, 41-43). After conditional deletion of Scap in the respiratory epithelium, expression of genes encoding SREBP 1 and -2 (Srebf1 and Srebf2) and multiple SREBP target genes involved in lipid metabolism were decreased at E18.5 as well as in adult mice. As expected, genes dedicated to the biosynthesis of fatty acids, phospholipids, triglycerides, cholesterol, and sterols were decreased by deletion of Scap. A number of these genes are known to be directly regulated by SREBPs in vitro and in vivo (40). Decreased expression of several genes regulating fatty acid, triglyceride, and phospholipid synthesis, including Scd1, Scd2, Fabp5, Fdps, Fasn, Acsl4, and Aytl2, as well as cholesterol synthesis, Acox1, Pon1, Idi1, Hmgcs1, Sqle, and Lss, was observed, demonstrating that deletion of Scap was likely to influence lipid biosynthesis in type II epithelial cells. In addition to genes regulating lipid biosynthesis, expression of a number of lipid transporters, including the low density lipoprotein receptor mediating cholesterol uptake, and lipid transport like ATP-binding cassette a3 (Abca3) and Abca1, were decreased in the ScapΔ/Δ mice. Besnard et al. (21) demonstrated the direct transcriptional regulation of Abca3 by SREBPs. Several members of the ABC family of lipid transport proteins (ABCA1, ABCA7, ABCB4, ABCD2, ABCG3, ABCG5, and ABCA3) are regulated by SREBPs either directly or indirectly (21, 40, 44-47). Both surfactant lipids and proteins are required for surfactant function at birth. In this study, we found that SREBPs directly regulated the transcription of Sftpa, Sftpb, and Sftpd promoter constructs in vitro. However, only the expression of Sftpa mRNA was significantly decreased in adult ScapΔ/Δ mice. Lack of alteration in the expression of the other surfactant protein genes suggests that other transcription factors compensate for the SREBP decrease to maintain surfactant protein activity.
Consistent with the reduction of SREBPs caused by deletion of Scap in alveolar type II cells, expression of genes regulating lipid biosynthesis was reduced in fetal and adult mice. Consistent with this finding, PC synthesis from [3H]choline was significantly decreased in type II alveolar cells isolated from adult ScapΔ/Δ mice. Mice did not develop respiratory distress or death following birth, indicating that alveolar surfactant lipid content was sufficient to reduce surface tension in the alveoli. This observation suggested that there must be compensatory pathways that are capable of maintaining surfactant lipid homeostasis in vivo. This conclusion was supported by analysis of surfactant PC in BALF from adult ScapΔ/Δ mice, which demonstrated a normal concentration of Sat PC and unaltered fractional concentration of disaturated PC molecular species compared with control mice. The enhanced accumulation of neutral lipid in lung lipofibroblasts of the ScapΔ/Δ mice supports the presence of a compensatory pathway supplying lipid precursors to type II alveolar cells from adjacent pulmonary lipofibroblasts. This observation was supported by both increased incorporation of [3H]choline into PC in isolated lung fibroblasts from adult ScapΔ/Δ mice and increased uptake of [3H]choline-labeled lipids from the lipofibroblasts into the type II cells. The numbers of lipid droplets, the concentration of neutral lipids, and staining for ADFP were increased in lungs from ScapΔ/Δ mice, indicating an increase in lipid storage in the non-epithelial compartment in ScapΔ/Δ mice. Magra et al. (48) have proposed that ADFP may be involved in the transfer of lipid from lipofibroblasts to lung alveolar type II epithelial cells for the production of surfactant phospholipid. This study provides further support for a model in which lipid fibroblasts interact closely with type II epithelial cells to modulate lipid substrate supply needed for surfactant synthesis and support the role of this interaction in perinatal lung development as proposed by Torday et al. (37-39). One additional implication of these observations is that the decreased PC synthesis in isolated adult type II alveolar cells must be a consequence of their inadequate ability to synthesize fatty acids de novo, rather than being due to fundamental abnormalities in either the CDP:choline pathway for PC synthesis or in mechanisms regulating surfactant packaging in lamellar bodies. The decreased PG in both BALF and lung tissue may be related to the decreased expression of ABCA3 in ScapΔ/Δ mice, a result consistent with the reported virtual absence of PG in the ABCA3-/- mouse (49). Although mechanisms controlling compensatory changes in lipid metabolism in lipofibroblasts are unknown, we showed that the expression of transcription factors PPARγ and retinoid X receptor-α, known to regulate lipid synthesis in other tissues, was increased in lipofibroblasts isolated from ScapΔ/Δ mice. Both immunostaining for ADFP and Adfp mRNA levels were increased in the lungs of ScapΔ/Δ mice. The promoter region of the Adfp gene contains binding sites for SREBP, PPAR, and retinoid X receptor (50, 51) providing a potential mechanism influencing expression of lipid synthesis pathways in lung fibroblasts. The potential for local compensatory regulation of phospholipid synthesis in pulmonary cells is consistent with previous findings in the liver wherein the decrease in fatty acid synthesis observed after hepatic deletion of Scap was balanced by an equal increase in lipogenesis in nonhepatic tissues, occurring primarily in adipose tissue of the L-SCAP-deleted mice. These findings support the likelihood that lipid accumulation by lipofibroblasts, perhaps mediated by the activation of transcriptional programs regulating lipogenesis, compensates foe the lack of SCAP in alveolar type II cells. mRNA microarray analysis indicated an increased expression of PlagL2, Stx3, Plscr1, Acox2, Mthfd2, and Ptgs2 that may serve to maintain cholesterol and phospholipid levels after deletion of Scap (supplemental Table 2). PlagL2 is a transcription factor known to enhance the activity of the Sftpc gene in the lung (52). Deletion of PlagL2 is associated with lipid uptake deficiency in the intestine and peripheral tissues due to reduction of genes linked to intracellular transport of lipids, membranes, and secretory products (53). Increased PlagL2 expression may represent the activation of a compensatory pathway that facilitates lipid uptake from extracellular sites. In support of this concept, the expression of several genes known to be regulated by PlagL2 (Tmem9, Snx5, and Slc5a1) were also increased after deletion of Scap.
In the mRNA microarray analysis, expression of genes mediating intracellular transport was altered in ScapΔ/Δ mice, including decreased expression of genes associated with microtubule formation (Dynlt and Dctn5) and release of secretory granules from the trans-Golgi network (Rab27a and Rab27b). In contrast, mRNAs encoding proteins influencing endocytic membrane trafficking, including Rab17, Rin1, Rab3c, and Anxa1, were increased, suggesting the induction of endocytic processes that may be involved in the provision of lipids and other nutrients. Expression of genes encoding proteins modulating inflammatory responses also was increased after deletion of Scap, including multiple eicosanoid pathway members (Pla2g4c, Ptgs2, Ptger4, Ptger3, Ptger1, and Alox5ap).
Of the differentially expressed mRNAs identified after deletion of Scap, those encoding proteins resident in the ER were selectively enriched. Because the ER is highly sensitive to various cell stresses, this response may indicate a generalized response to changes in phospholipid metabolism. A number of oxidative stress response genes were altered by deletion of Scap, including Pon1, Scd1 and -2, Mme, MT1 and -2, Alb, Cyp2b6, and Ptgs2. The expression of genes involved in the amino acid biosynthesis and transport were significantly induced (Asns, Mthfd2, Phgdh, Pycr1, Slc1a1, Slc7a1, and Slc7a3) in ScapΔ/Δ mice, perhaps indicating an adaptive response to nutrient deprivation. Of interest, asparagine synthase (Asns) catalyzes the synthesis of asparagine and glutamate from aspartate and serves as a nutrient deprivation sensor. The transcriptional response to nutrient deprivation is mediated via ER stress response cis-elements in the Asns promoter (54, 55). Both Asns and Slc7a1, the arginine/lysine transporter, are induced during amino acid starvation by ATF3/ATF4 (56, 57). Expression of several ER stress-responsive transcription factors was significantly induced by deletion of Scap, including Atf3, Atf5, Ddit3, and Trib3. Activating transcription factors (ATF) 3 and 5 are members of the ATF/CREB subfamily of basic region leucine zipper proteins; both are induced in response to oxidative stress or amino acid starvation (58, 59). ATF3 and DDIT3 (alias CHOP or GADD153) are transcription factors induced in response to ER overload, unfolded protein response, and ER stress-induced cell death. Although ATF3 is required for the induction of DDIT3 expression during amino acid deprivation, DDIT3 mediates the feedback inhibition of ATF3 (58, 60). TRIB3 is a transcriptional target of DDIT3/ATF4 and represses DDIT3/ATF4 function. TRIB3 regulates DDIT3-dependent cell death during ER stress (61, 62). Interestingly, all of these transcription factors are influenced by phosphorylation of eukaryotic initiation factor 2 by eukaryotic initiation factor 2 kinases (59, 63). The levels of the transcription factor ATF4 are rapidly increased in response to the phosphorylation of eukaryotic initiation factor 2 during ER stress (58, 64). ATF4 directly or indirectly influences the transcription of ATF3/ATF5, DDIT3, and TRIB3 (58, 63). In ScapΔ/Δ mice, the induction of ATF4, ATF3, DDIT3, and TRIB3 suggests these transcription factors may function in a network to coordinate gene expression in response to the cellular stress related to the lack of SCAP/SREBP and its influence on lipid homeostasis in the lung.
The balance among pro- and anti-apoptosis regulators and effectors determines ER stress-mediated cell survival versus cell death. In this study, 15 of 44 genes induced by Scap deletion have been functionally related to cell death. Among these, proapoptotic factors include several ER-responsive transcription factors Atf3, Ddit3, and Trib3 and many apoptosis-related genes (Emp1, Derl3, Hmga2, Lgals3, Olr1, and Pla2g4c). Atf3, Ddit3, and Trib3 induce cell cycle arrest and apoptosis in many cell types. Derlin-3 (Derl3) expression is induced by the unfolded protein response and is required for ER-associated degradation of misfolded glycoprotein and cell death (65). Anti-apoptotic factors, including Atf5, Asns, Cyp2b6, Mt1, Naip5, Ptger4, Ptgs2, Rb11, and Sprr1a, were also induced. Both Atf5 and its target, Cyp2b6, are induced under ER stress (66). The increased expression of this group of genes in ScapΔ/Δ mice suggests the activation of a response protecting the cell from stress-induced apoptosis. Neither immunostaining for cleaved caspase 3 nor cell proliferation was affected by deletion of Scap, supporting the concept that pro- and anti-apoptosis regulators are relatively balanced in ScapΔ/Δ mice.
Recent studies demonstrated a direct relationship between altered cellular lipid composition and ER stress. Increase or depletion of the fatty acid or cholesterol content activated ER stress responses indicating the requirement for precise regulation of cellular lipid composition to maintain normal cell function (67-71). This study and other models in which lipid homeostasis is perturbed share an increase in expression of ER stress-induced transcription factors (ATF3, ATF5, and CHOP), as well as elevated expression of oxidative stress response genes and those influencing amino acid biosynthesis and transport pathways, suggesting a common response to alterations in cellular lipid composition. ER stress can modulate the activation of SREBP (69, 72, 73), indicating potential reciprocal interactions between ER stress and the SCAP/SREBP pathway.
This study demonstrates that although the SCAP/SREBP pathway was not required for respiration at birth, deletion of Scap influenced lung phospholipid homeostasis, regulating lipid biosynthesis and cellular stress responses. These findings identify a complex lipid sensing pathway mediating lung lipid homeostasis that depends on interactions between the alveolar epithelium and lung lipofibroblasts. Maintenance of respiratory function despite alterations in lipid synthesis by type II epithelial cells indicates a mechanism by which lipofibroblasts compensate for changes in lipid homeostasis in the alveolar epithelium. Because SREBPs regulate a number of genes critical for pulmonary homeostasis, perturbation of the SCAP/SREBP pathway may render individuals susceptible to pulmonary disease. Likewise, the SCAP/SREBP pathway may influence the susceptibility of newborns, infants, and older individuals to acute and chronic lung diseases.
Supplementary Material
Acknowledgments
We acknowledge the technical contributions from Sharon Dingle, Paula Blair, Gail Macke, and Sandra Olson.
This work was supported, in whole or in part, by National Institutes of Health Grants HL-85610 and HL-90156 (to J. A. W.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Fig. 1 and Tables 1 and 2.
Footnotes
The abbreviations used are: C/EBP, CCAAT/enhancer-binding protein; Ldlr, low density lipoprotein receptor; SREBP, sterol regulatory element-binding protein; nSREBP, nuclear form of SREBP; ADFP, adipose differentiation related protein; SCAP, SREBP cleavage-activating protein; Sftp, surfactant protein; Sat PC, saturated phosphatidylcholine; PPAR, peroxisome proliferator-activated receptor; ER, endoplasmic reticulum; ADFP, adipose differentiation-related protein; RT, reverse transcription; BALF, bronchoalveolar lavage fluid; ESI-MS, electrospray ionization mass spectrometry; PC, phosphatidylcholine; PE, phosphatidylethanolamine; PI, phosphatidylinositol; PS, phosphatidylserine; rtTA, reverse tetracycline transactivation; ATF, activating transcription factor.
References
- 1.Whitsett, J. A., and Weaver, T. E. (2002) N. Engl. J. Med. 347 2141-2148 [DOI] [PubMed] [Google Scholar]
- 2.Whitsett, J. A. (2006) Paediatr. Respir. Rev. 7 Suppl. 1, 240-242 [DOI] [PubMed] [Google Scholar]
- 3.Eberle, D., Hegarty, B., Bossard, P., Ferre, P., and Foufelle, F. (2004) Biochimie (Paris) 86 839-848 [DOI] [PubMed] [Google Scholar]
- 4.Flodby, P., Barlow, C., Kylefjord, H., Ahrlund-Richter, L., and Xanthopoulos, K. G. (1996) J. Biol. Chem. 271 24753-24760 [DOI] [PubMed] [Google Scholar]
- 5.Rosen, E. D., and Spiegelman, B. M. (2000) Annu. Rev. Cell Dev. Biol. 16 145-171 [DOI] [PubMed] [Google Scholar]
- 6.Rosen, E. D., Walkey, C. J., Puigserver, P., and Spiegelman, B. M. (2000) Genes Dev. 14 1293-1307 [PubMed] [Google Scholar]
- 7.Chang, Y., Edeen, K., Lu, X., De Leon, M., and Mason, R. J. (2006) Am. J. Respir. Cell Mol. Biol. 35 268-274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chang, Y., Wang, J., Lu, X., Thewke, D. P., and Mason, R. J. (2005) J. Lipid Res. 46 2624-2635 [DOI] [PubMed] [Google Scholar]
- 9.Martis, P. C., Whitsett, J. A., Xu, Y., Perl, A. K., Wan, H., and Ikegami, M. (2006) Development (Camb.) 133 1155-1164 [DOI] [PubMed] [Google Scholar]
- 10.Mason, R. J., Pan, T., Edeen, K. E., Nielsen, L. D., Zhang, F., Longphre, M., Eckart, M. R., and Neben, S. (2003) J. Clin. Investig. 112 244-255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang, F., Pan, T., Nielsen, L. D., and Mason, R. J. (2004) Am. J. Respir. Cell Mol. Biol. 30 174-183 [DOI] [PubMed] [Google Scholar]
- 12.Horton, J. D., Goldstein, J. L., and Brown, M. S. (2002) J. Clin. Investig. 109 1125-1131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shimano, H., Shimomura, I., Hammer, R. E., Herz, J., Goldstein, J. L., Brown, M. S., and Horton, J. D. (1997) J. Clin. Investig. 100 2115-2124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Horton, J. D., Shimomura, I., Brown, M. S., Hammer, R. E., Goldstein, J. L., and Shimano, H. (1998) J. Clin. Investig. 101 2331-2339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Matsuda, M., Korn, B. S., Hammer, R. E., Moon, Y. A., Komuro, R., Horton, J. D., Goldstein, J. L., Brown, M. S., and Shimomura, I. (2001) Genes Dev. 15 1206-1216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Perl, A. K., Tichelaar, J. W., and Whitsett, J. A. (2002) Transgenic Res. 11 21-29 [DOI] [PubMed] [Google Scholar]
- 17.Perl, A. K., Wert, S. E., Loudy, D. E., Shan, Z., Blair, P. A., and Whitsett, J. A. (2005) Am. J. Respir. Cell Mol. Biol. 33 455-462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Perl, A. K., Wert, S. E., Nagy, A., Lobe, C. G., and Whitsett, J. A. (2002) Proc. Natl. Acad. Sci. U. S. A. 99 10482-10487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rugh, R. (1993) The Mouse: Its Reproduction and Development, p. 299, Oxford University Press, New York
- 20.Besnard, V., Wert, S. E., Kaestner, K. H., and Whitsett, J. A. (2005) Am. J. Physiol. 289 L750-L759 [DOI] [PubMed] [Google Scholar]
- 21.Besnard, V., Xu, Y., and Whitsett, J. A. (2007) Am. J. Physiol. 293 L1395-L1405 [DOI] [PubMed] [Google Scholar]
- 22.Schultz, C. J., Torres, E., Londos, C., and Torday, J. S. (2002) Am. J. Physiol. 283 L288-L296 [DOI] [PubMed] [Google Scholar]
- 23.Wert, S. E., Glasser, S. W., Korfhagen, T. R., and Whitsett, J. A. (1993) Dev. Biol. 156 426-443 [DOI] [PubMed] [Google Scholar]
- 24.Rice, W. R., Conkright, J. J., Na, C. L., Ikegami, M., Shannon, J. M., and Weaver, T. E. (2002) Am. J. Physiol. 283 L256-264 [DOI] [PubMed] [Google Scholar]
- 25.Glasser, S. W., Burhans, M. S., Korfhagen, T. R., Na, C. L., Sly, P. D., Ross, G. F., Ikegami, M., and Whitsett, J. A. (2001) Proc. Natl. Acad. Sci. U. S. A. 98 6366-6371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bartlett, G. R. (1959) J. Biol. Chem. 234 466-468 [PubMed] [Google Scholar]
- 27.Bligh, E. G., and Dyer, W. J. (1959) Can. J. Biochem. Physiol. 37 911-917 [DOI] [PubMed] [Google Scholar]
- 28.Postle, A. D., Gonzales, L. W., Bernhard, W., Clark, G. T., Godinez, M. H., Godinez, R. I., and Ballard, P. L. (2006) J. Lipid Res. 47 1322-1331 [DOI] [PubMed] [Google Scholar]
- 29.Zhou, L., Dey, C. R., Wert, S. E., Yan, C., Costa, R. H., and Whitsett, J. A. (1997) Dev. Dyn. 210 305-314 [DOI] [PubMed] [Google Scholar]
- 30.Bolender, R. P., Hyde, D. M., and Dehoff, R. T. (1993) Am. J. Physiol. 265 L521-L548 [DOI] [PubMed] [Google Scholar]
- 31.Wert, S. E., Yoshida, M., LeVine, A. M., Ikegami, M., Jones, T., Ross, G. F., Fisher, J. H., Korfhagen, T. R., and Whitsett, J. A. (2000) Proc. Natl. Acad. Sci. U. S. A. 97 5972-5977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Irizarry, R. A., Bolstad, B. M., Collin, F., Cope, L. M., Hobbs, B., and Speed, T. P. (2003) Nucleic Acids Res. 31 e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Irizarry, R. A., Hobbs, B., Collin, F., Beazer-Barclay, Y. D., Antonellis, K. J., Scherf, U., and Speed, T. P. (2003) Biostatistics 4 249-264 [DOI] [PubMed] [Google Scholar]
- 34.Dennis, G., Jr., Sherman, B. T., Hosack, D. A., Yang, J., Gao, W., Lane, H. C., and Lempicki, R. A. (2003) Genome Biol. 4 P3. [PubMed] [Google Scholar]
- 35.Garmany, T. H., Moxley, M. A., White, F. V., Dean, M., Hull, W. M., Whitsett, J. A., Nogee, L. M., and Hamvas, A. (2006) Pediatr. Res. 59 801-805 [DOI] [PubMed] [Google Scholar]
- 36.Shulenin, S., Nogee, L. M., Annilo, T., Wert, S. E., Whitsett, J. A., and Dean, M. (2004) N. Engl. J. Med. 350 1296-1303 [DOI] [PubMed] [Google Scholar]
- 37.Torday, J., Hua, J., and Slavin, R. (1995) Biochim. Biophys. Acta 1254 198-206 [DOI] [PubMed] [Google Scholar]
- 38.Torday, J. S., and Rehan, V. K. (2002) Am. J. Physiol. 283 L130-L135 [DOI] [PubMed] [Google Scholar]
- 39.Torday, J. S., Sun, H., Wang, L., Torres, E., Sunday, M. E., and Rubin, L. P. (2002) Am. J. Physiol. 282 L405-L410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Horton, J. D., Shah, N. A., Warrington, J. A., Anderson, N. N., Park, S. W., Brown, M. S., and Goldstein, J. L. (2003) Proc. Natl. Acad. Sci. U. S. A. 100 12027-12032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Felder, T. K., Klein, K., Patsch, W., and Oberkofler, H. (2005) Biochim. Biophys. Acta 1731 41-47 [DOI] [PubMed] [Google Scholar]
- 42.Okamoto, K., Kakuma, T., Fukuchi, S., Masaki, T., Sakata, T., and Yoshimatsu, H. (2006) Brain Res. 1081 19-27 [DOI] [PubMed] [Google Scholar]
- 43.Wang, Z., Jiang, T., Li, J., Proctor, G., McManaman, J. L., Lucia, S., Chua, S., and Levi, M. (2005) Diabetes 54 2328-2335 [DOI] [PubMed] [Google Scholar]
- 44.Iwamoto, N., Abe-Dohmae, S., Sato, R., and Yokoyama, S. (2006) J. Lipid Res. 47 1915-1927 [DOI] [PubMed] [Google Scholar]
- 45.Weinhofer, I., Kunze, M., Rampler, H., Bookout, A. L., Forss-Petter, S., and Berger, J. (2005) J. Biol. Chem. 280 41243-41251 [DOI] [PubMed] [Google Scholar]
- 46.Wong, J., Quinn, C. M., and Brown, A. J. (2006) Biochem. J. 400 485-491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zeng, L., Liao, H., Liu, Y., Lee, T. S., Zhu, M., Wang, X., Stemerman, M. B., Zhu, Y., and Shyy, J. Y. (2004) J. Biol. Chem. 279 48801-48807 [DOI] [PubMed] [Google Scholar]
- 48.Magra, A. L., Mertz, P. S., Torday, J. S., and Londos, C. (2006) J. Lipid Res. 47 2367-2373 [DOI] [PubMed] [Google Scholar]
- 49.Fitzgerald, M. L., Xavier, R., Haley, K. J., Welti, R., Goss, J. L., Brown, C. E., Zhuang, D. Z., Bell, S. A., Lu, N., McKee, M., Seed, B., and Freeman, M. W. (2007) J. Lipid Res. 48 621-632 [DOI] [PubMed] [Google Scholar]
- 50.Dalen, K. T., Ulven, S. M., Arntsen, B. M., Solaas, K., and Nebb, H. I. (2006) J. Lipid Res. 47 931-943 [DOI] [PubMed] [Google Scholar]
- 51.Edvardsson, U., Ljungberg, A., Linden, D., William-Olsson, L., Peilot-Sjogren, H., Ahnmark, A., and Oscarsson, J. (2006) J. Lipid Res. 47 329-340 [DOI] [PubMed] [Google Scholar]
- 52.Yang, M. C., Weissler, J. C., Terada, L. S., Deng, F., and Yang, Y. S. (2005) Am. J. Respir. Cell Mol. Biol. 32 35-43 [DOI] [PubMed] [Google Scholar]
- 53.Van Dyck, F., Braem, C. V., Chen, Z., Declercq, J., Deckers, R., Kim, B. M., Ito, S., Wu, M. K., Cohen, D. E., Dewerchin, M., Derua, R., Waelkens, E., Fiette, L., Roebroek, A., Schuit, F., van de Ven, W. J., and Shivdasani, R. A. (2007) Cell Metab. 6 406-413 [DOI] [PubMed] [Google Scholar]
- 54.Barbosa-Tessmann, I. P., Chen, C., Zhong, C., Schuster, S. M., Nick, H. S., and Kilberg, M. S. (1999) J. Biol. Chem. 274 31139-31144 [DOI] [PubMed] [Google Scholar]
- 55.Kilberg, M. S., and Barbosa-Tessmann, I. P. (2002) J. Nutr. 132 1801-1804 [DOI] [PubMed] [Google Scholar]
- 56.Lopez, A. B., Wang, C., Huang, C. C., Yaman, I., Li, Y., Chakravarty, K., Johnson, P. F., Chiang, C. M., Snider, M. D., Wek, R. C., and Hatzoglou, M. (2007) Biochem. J. 402 163-173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Pan, Y., Chen, H., Siu, F., and Kilberg, M. S. (2003) J. Biol. Chem. 278 38402-38412 [DOI] [PubMed] [Google Scholar]
- 58.Jiang, H. Y., Wek, S. A., McGrath, B. C., Lu, D., Hai, T., Harding, H. P., Wang, X., Ron, D., Cavener, D. R., and Wek, R. C. (2004) Mol. Cell. Biol. 24 1365-1377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Watatani, Y., Ichikawa, K., Nakanishi, N., Fujimoto, M., Takeda, H., Kimura, N., Hirose, H., Takahashi, S., and Takahashi, Y. (2008) J. Biol. Chem. 283 2543-2553 [DOI] [PubMed] [Google Scholar]
- 60.Wolfgang, C. D., Chen, B. P., Martindale, J. L., Holbrook, N. J., and Hai, T. (1997) Mol. Cell. Biol. 17 6700-6707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Jousse, C., Deval, C., Maurin, A. C., Parry, L., Cherasse, Y., Chaveroux, C., Lefloch, R., Lenormand, P., Bruhat, A., and Fafournoux, P. (2007) J. Biol. Chem. 282 15851-15861 [DOI] [PubMed] [Google Scholar]
- 62.Ohoka, N., Yoshii, S., Hattori, T., Onozaki, K., and Hayashi, H. (2005) EMBO J. 24 1243-1255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zhou, D., Palam, L. R., Jiang, L., Narasimhan, J., Staschke, K. A., and Wek, R. C. (2008) J. Biol. Chem. 283 7064-7073 [DOI] [PubMed] [Google Scholar]
- 64.Harding, H. P., Zhang, Y., Zeng, H., Novoa, I., Lu, P. D., Calfon, M., Sadri, N., Yun, C., Popko, B., Paules, R., Stojdl, D. F., Bell, J. C., Hettmann, T., Leiden, J. M., and Ron, D. (2003) Mol. Cell 11 619-633 [DOI] [PubMed] [Google Scholar]
- 65.Oda, Y., Okada, T., Yoshida, H., Kaufman, R. J., Nagata, K., and Mori, K. (2006) J. Cell Biol. 172 383-393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Pascual, M., Gomez-Lechon, M. J., Castell, J. V., and Jover, R. (2008) Drug Metab. Dispos. 36 1063-1072 [DOI] [PubMed] [Google Scholar]
- 67.Feng, B., Yao, P. M., Li, Y., Devlin, C. M., Zhang, D., Harding, H. P., Sweeney, M., Rong, J. X., Kuriakose, G., Fisher, E. A., Marks, A. R., Ron, D., and Tabas, I. (2003) Nat. Cell Biol. 5 781-792 [DOI] [PubMed] [Google Scholar]
- 68.Flowers, M. T., Keller, M. P., Choi, Y., Lan, H., Kendziorski, C., Ntambi, J. M., and Attie, A. D. (2008) Physiol. Genomics 33 361-372 [DOI] [PubMed] [Google Scholar]
- 69.Harding, H. P., Zhang, Y., Khersonsky, S., Marciniak, S., Scheuner, D., Kaufman, R. J., Javitt, N., Chang, Y. T., and Ron, D. (2005) Cell Metab. 2 361-371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Little, J. L., Wheeler, F. B., Fels, D. R., Koumenis, C., and Kridel, S. J. (2007) Cancer Res. 67 1262-1269 [DOI] [PubMed] [Google Scholar]
- 71.Wei, Y., Wang, D., Topczewski, F., and Pagliassotti, M. J. (2006) Am. J. Physiol. 291 E275-E281 [DOI] [PubMed] [Google Scholar]
- 72.Lee, J. N., and Ye, J. (2004) J. Biol. Chem. 279 45257-45265 [DOI] [PubMed] [Google Scholar]
- 73.Wang, H., Kouri, G., and Wollheim, C. B. (2005) J. Cell Sci. 118 3905-3915 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.