Abstract
Using recombinant bead-conjugated emerin we affinity-purified seven proteins from HeLa cell nuclear lysates that bind emerin either directly or indirectly. These proteins were identified by mass spectrometry as nuclear αII-spectrin, non-muscle myosin heavy chain alpha, Lmo7 (a predicted transcription regulator; reported separately), nuclear myosin I, β-actin (reported separately), calponin 3 and SIKE. We now report emerin binds nuclear myosin I (NMI, a molecular motor) directly in vitro. Furthermore, bead-conjugated emerin bound nuclear αII-spectrin and NMI equally well with or without ATP (which stimulates motor activity), whereas ATP decreased actin binding by 65%. Thus αII-spectrin and NMI interact stably with emerin. To investigate the physiological relevance of these interactions, we used antibodies against emerin to affinity-purify emerin-associated protein complexes from HeLa cells, then further purified by ion exchange chromatography to resolve by net charge, and size exclusion chromatography yielding six distinct emerin-containing fractions (0.5 to 1.6 MDa). Western blotting suggested each complex had distinct components involved in nuclear architecture (e.g., NMI, αII-spectrin, lamins) or gene or chromatin regulation (BAF, transcription regulators, HDACs). Additional constituents were identified by mass spectrometry. One putative gene-regulatory complex (complex 32) included core components of the Nuclear Co-Repressor (NCoR) Complex, which mediates gene regulation by thyroid hormone and other nuclear receptors. When expressed in HeLa cells, FLAG-tagged NCoR subunits Gps2, HDAC3, TBLR1 and NCoR each co-immunoprecipitated emerin, validating one putative complex. These findings support the hypothesis that emerin scaffolds a variety of functionally distinct multi-protein complexes at the nuclear envelope in vivo. Notably included are nuclear myosin I-containing complexes that might sense and regulate mechanical tension at the nuclear envelope.
Keywords: laminopathy, emerin, LEM-domain, nuclear envelope, lamin, Emery-Dreifuss muscular dystrophy, DNA repair, 14-3-3 proteins, Lmo7, retinoblastoma, nuclear myosin I, Sun2
INTRODUCTION
Emery-Dreifuss muscular dystrophy (EDMD) is inherited through mutations in either of two genes: LMNA, encoding A-type lamins, and EMD, which encodes a nuclear membrane protein named emerin (1–3). Emerin belongs to the ‘LEM-domain’ family, defined by a ~40-residue folded domain (the ‘LEM domain’) that binds a chromatin protein named Barrier-to-Autointegration Factor (BAF; 4, 5). BAF is a small, essential metazoan protein with direct roles in higher-order chromatin structure (6–8), nuclear assembly (6, 8, 9), and gene regulation (10, 11). Emerin also binds lamin filaments, which anchor emerin at the nuclear inner membrane (12, 13). Together, emerin and lamin A form stable ternary complexes with other binding partners in vitro (10), with the unproven potential to form a variety of oligomeric protein complexes in vivo (2, 14). Emerin is expressed in all cells tested (15), but EDMD affects only three tissues, with progressive skeletal muscle weakening, contractures of major tendons and potentially fatal cardiac conduction system defects (1, 16). To explain this selectivity, emerin was proposed to have tissue-specific binding partners that might regulate gene expression, including transcription factors and signaling molecules (16–19). Growing evidence supports such models, including the identification of three gene-regulatory proteins that bind emerin: germ cell-less (GCL; 10), Btf (20) and Lmo7 (21). Emerin also binds β-catenin directly and appears to attenuate β-catenin-mediated gene expression (22), and is involved in alternative mRNA splicing in vivo, through direct binding to YT521-B (23). Binding of four aforementioned gene-regulatory partners is disrupted by mutations in the same regions of emerin, termed repressor binding domains-1 (RBD-1) and RBD-2, suggesting emerin scaffolds a variety of gene-regulatory partners at the inner nuclear membrane (21).
Emerin also maintains the structural integrity of the nucleus (24), potentially by anchoring a proposed actin-spectrin filament network at the inner nuclear membrane (25). Architectural models predict that loss of emerin might selectively disrupt tissues under high mechanical stress, including skeletal muscle and tendons (18, 19). Although architectural models fail to explain the cardiac conduction pathology in EDMD, they are consistent with structural defects (aberrant shape and nuclear envelope herniations) seen in a subset of nuclei from EDMD patients (26) and patients with other diseases linked to mutations in LMNA (‘laminopathies’; 18, 27).
Here we report the purification of novel emerin-associated proteins from cultured human (HeLa) cell nuclei, including the actin-binding proteins NMI and αII-spectrin. We also independently purified six distinct emerin-containing multiprotein complexes from HeLa nuclei. One putative complex includes actin, NMI, αII-spectrin, A- and B-type lamins and other candidate constituents including Sun2, suggesting molecular mechanisms by which emerin influences nuclear architecture. Interestingly, other complexes had components involved in gene regulation, chromatin structure, RNA processing and other activities including DNA repair. The presence of core components of the Nuclear Co-Repressor (NCoR) complex in one putative gene-regulatory complex was independently validated in vivo, providing proof-of-principle that these putative complexes may be physiologically relevant. The implications of each emerin-containing complex in terms of emerin function and EDMD disease mechanisms are discussed.
EXPERIMENTAL PROCEDURES
Plasmids and antibodies
Recombinant emerin protein comprising the entire nucleoplasmic domain of emerin (residues 1-222) and lacking the transmembrane domain was expressed in bacteria from pET11c-emerin and purified as described (10). Plasmids encoding FLAG-HDAC3, FLAG-TBL1, FLAG-TBLR1 and FLAG-NCoR were generous gifts from Robert Roeder (The Rockefeller University). The plasmid encoding FLAG-GPS was a generous gift from Saul Silverstein (Columbia University). Antibodies against each named protein, and the dilution used for immunoblotting, were: actin (A-5060, Sigma-Aldrich, Inc., 1:2500), β-actin (A-2228, Sigma-Aldrich, Inc., 1:5000), NMI (generous gift from Primal de Lanerolle; 0.2 μg/ml), αII-spectrin (SC-7465, Santa Cruz Biotechnology, Inc, 1:200), emerin (serum 2999, 1:10000; 28) and NCL-emerin, Novocastra, Ltd, 1:1000), BAF (serum 3273, 1:1000; 29), H1 (SC-8030, Santa Cruz Biotechnology, Inc., 1:500), lamin A (NCL-Lamin A, Novocastra Ltd, 1:500), lamin B (NA-12, EMD Biosciences, Inc, 1:5000), H3 (ab7834, Abcam, Inc, 1:500), p107 (SC-318, Santa Cruz Biotechnology, Inc, 1:1000), Lmo7 (serum 4990, 1:500; 21), BAF-L (serum 4176, 1:1000; 30), MAN1 (serum 4279, 1:500; 31), Rb (SC-102, Santa Cruz Biotechnology, Inc, 1:500), p130 (SC-317, Santa Cruz Biotechnology, 1:500), and FLAG (F-1804, Sigma-Aldrich, Inc, 1:500 dilution).
Affinity purification using emerin-conjugated beads and mass spectrometry
Wildtype emerin residues 1-222 or BSA (negative control) were coupled to Affigel-15 beads (Bio-Rad Laboratories) per manufacturer instructions. Nuclear extracts were prepared from 1010 HeLa-S3 cells by hypotonic lysis (32). HeLa cells were obtained from the National Cell Culture Center. For affinity purification, 50 mg of nuclear extract were incubated (4 h at 4°C) with 0.5 mg/ml (0.5 mg) bead-coupled emerin or BSA in Binding Buffer (BB: 50 mM HEPES, 250 mM NaCl, 0.1% Triton X-100) containing protease inhibitors and 2 mM DTT. Beads were collected by centrifugation at 500g, washed five times with BB and bound proteins were eluted with SDS-PAGE sample buffer. Proteins were identified using a QSTAR/Pulsar mass spectrometer at the Mass Spectrometry/Proteomics Facility at The Johns Hopkins University School of Medicine (www.hopkinsmedicine.org/msf/).
Binding Assays
Co-immunoprecipitation assays were done using equal masses (5 μg) of purified NMI (a generous gift from Primal deLanerolle) and wildtype emerin (residues 1-222), essentially as described (28). Proteins were incubated 2 h at 22°C in PBS with 0.1% Triton X-100 (PBS-T), 2 mM DTT, 10 μg/ml aprotinin, 10 μg/ml leupeptin, 10 μg/ml pepstatin A, 1 mM PMSF, and then incubated 4 h at 22°C with emerin monoclonal antibodies (NCL-Emerin; Novocastra Laboratories, Ltd.) coupled to Protein-A Sepharose. The beads were washed five times with PBS-T, and bound proteins were eluted with SDS-sample buffer, resolved by SDS-PAGE and Western blotted with antibodies against either emerin (rabbit serum 2999; 28) or NMI (33).
For the in vivo co-immunoprecipitation assays, HeLa cells were transfected using Mirus LT-1 transfection reagent per manufacturer specifications, using 12 μg of plasmid per 100 mm dish. After 36–48 h incubation, cells were lysed with 400 μl of modified NEHN buffer (500 mM NaCl, 1% NP40, 20 mM HEPES, pH 8, 1 mM EDTA, 2 mM DTT, 20% glycerol, 1 mM PMSF, 5 μg/ml each of aprotinin, leupeptin, pepstatin A). The lysate was diluted to 1 ml with NEHN dilution buffer (20 mM HEPES, pH 8, 1 mM EDTA, 20% glycerol, 2 mM DTT, 1 mM PMSF, 5 μg/ml each of aprotinin, leupeptin, pepstatin A) and incubated with 2 μl of M2-agarose (Sigma Corp) for 16 h at 4°C. The beads were washed five times with modified wash buffer (150 mM NaCl, 20 mM HEPES, pH 8, 0.2 mM EDTA, 0.5 mM DTT, 20% glycerol, 0.2% NP-40, 1 mM PMSF, 5 μg/ml each of aprotinin, leupeptin, pepstatin A) and eluted with 40 μl SDS-PAGE buffer. Samples were resolved by SDS-PAGE, transferred to nitrocellulose, and immunoblotted with antibodies against emerin (serum 2999; 1:10,000 dilution) and FLAG (M2, Sigma F-1804; 1:500 dilution).
Emerin complex purification
Nuclear extracts were prepared as described (32), with the following changes. Purified nuclei were resuspended in 1 M NaCl, 1% Triton X-100, 20 mM HEPES (pH 8.0) to extract nuclear lamina components and integral inner nuclear membrane proteins and centrifuged for 30 min at 40,000g. The supernatant fraction was diluted tenfold with 20 mM HEPES (pH 8.0), incubated 10 min at 4°C to allow reformation of complexes that may have dissociated during cell lysis, and then centrifuged 30 min at 40,000g. The resulting supernatant was incubated 16 h at 4°C with 10 mg of serum 2999 covalently coupled to a CarboLink (Pierce) column, per manufacturer instructions. Emerin-containing complexes were eluted four times each with 400 μg recombinant emerin (400 μg/ml). Eluted fractions were combined and loaded onto a 1 ml Mono Q column (GE Healthcare). The Mono Q column was washed with 20 ml of PBS, 0.05% Triton X-100 and complexes were eluted with a 20 ml linear gradient (0–1 M NaCl) at 0.5 ml/min. Aliquots of each fraction were resolved by SDS-PAGE and Western blotted to identify those containing emerin. Emerin-containing peak fractions were each pooled (three total) and resolved by size exclusion chromatography using a S300 preparative column (GE Healthcare) at 0.5 ml/min in PBS, 0.05% Triton X-100; emerin-containing fractions were then identified by Western blotting. Each resolved emerin-containing complex was then immunoblotted with antibodies against H1, BAF, lamin B, lamin A, NMI, αII-spectrin, actin, emerin, H3, p107 and Lmo7. Complexes that purified under similar conditions at least twice were selected for protein identification by LCMS/MS at the Mass Spectrometry/Proteomics Facility (www.hopkinsmedicine.org/msf/). Both ion exchange and size exclusion chromatography were performed using an LCC-501 plus FPLC (GE Healthcare).
RESULTS
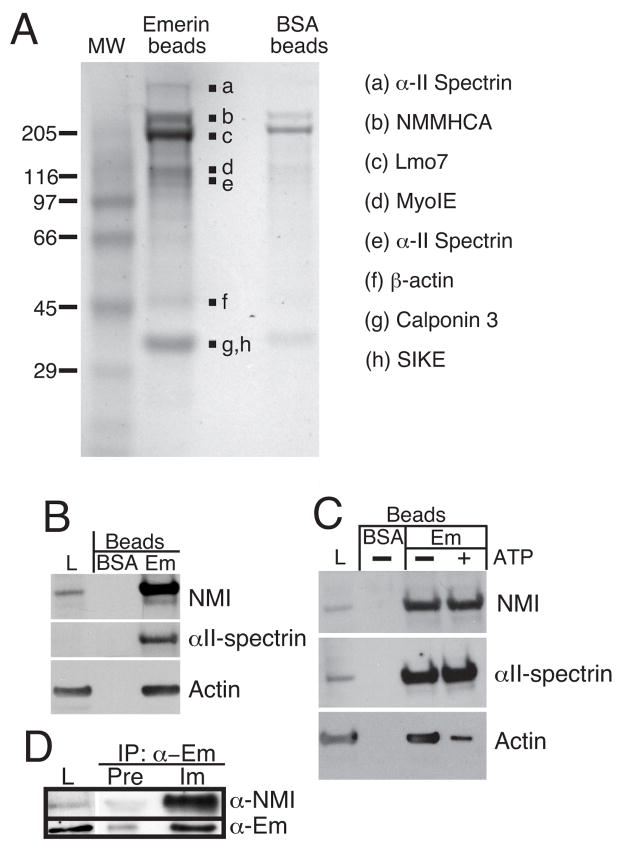
We first used affinity chromatography to purify emerin-binding proteins from human (HeLa) cells. Purified recombinant emerin protein (residues 1-222), which lacks the transmembrane domain, was covalently attached to Affi-gel beads as described previously (10). BSA-conjugated beads served as the negative control. We then incubated the emerin-beads and BSA-beads each with 50 mg protein from nuclear extracts of HeLa cells (see Methods). Beads were washed, and bound proteins were eluted with SDS-sample buffer, resolved by SDS-PAGE and stained with Coomassie Blue. Seven bands associated specifically with emerin beads (Figure 1A, solid squares), and not BSA beads. All seven bands were identified unambiguously by MALDI-TOF mass spectrometry, with significant coverage throughout the length of each protein (data not shown). One band contained two different proteins, Calponin 3 and SIKE (Figure 1A, bands g, h). Five of the purified proteins were structural (myosin IE, αII-spectrin, actin, calponin, and non-muscle myosin heavy chain A ‘NMMHCA’), one was a suppressor of IKKε- and TBK1-activation of interferon response elements (SIKE; 34) and one was a transcription regulator (Lmo7). Lmo7 activates transcription of many muscle-relevant and heart-relevant genes, including the emerin gene itself (21) and Lmo7 activity is proposed to be inhibited by direct binding to emerin (21). Characterization of the direct functional interaction between emerin and SIKE will be reported elsewhere (Holaska and Wilson, unpublished observations).
FIGURE 1.
Affinity purification of emerin-associated proteins. A, Coomassie-stained gel showing proteins that bound to emerin-beads or BSA-beads. Bands a-h were identified by mass spectrometry, and are named at right. B, Immunoblots of HeLa nuclear lysate proteins (L) or proteins affinity-purified using either BSA-beads or emerin-beads in the absence of ATP. Blots were probed with antibodies to NMI, actin (all isoforms) or αII-spectrin. C, Effects of ATP treatment. Nuclear extracts were left untreated, as in (B), or treated with 10 mM ATP prior to affinity purification on BSA-beads or emerin-beads in the continuous presence of ATP. Bound proteins were resolved by SDS-PAGE and immunoblotted as in (B). D, Co-immunoprecipitation of recombinant purified emerin and NMI proteins. Recombinant NMI and emerin (residues 1-222) proteins were incubated together (L), immunoprecipitated using either immune (IM) or pre-immune (Pre) serum 2999 against emerin, and resolved by SDS-PAGE and western blotted using antibodies against NMI (α-NMI) or emerin (α-Em).
To focus further study, we considered the structural proteins that purified with emerin. NMMHCA is a myosin II heavy chain that is expressed in most tissues tested (35), and localizes primarily in the cytoplasm. Thus its physiological relevance to emerin was uncertain. Also uncertain was the physiological relevance of calponin 3 (Cnn3), which associates with the cytoskeleton, but is not required for muscle contraction (36). Calponins bind both actin and myosin, and inhibit the actin-activated ATPase activity of myosin (37–40). In contrast, the remaining proteins (Myosin IE, β-actin and αII-spectrin) have known relevance to nuclear structure and function. β-actin and nuclear-specific isoforms of both myosin I and αII-spectrin have diverse roles in mRNA transcription (33, 41) (42), DNA repair (43–45), nuclear export (46) and directed chromosome movement within nuclei (47). However, the peptides identified by MALDI-TOF did not reveal whether we had purified the nuclear-specific isoforms of myosin I or αII-spectrin. To answer this question and explore the possible significance of these results, we first focused on β-actin, NMI and αII-spectrin.
Emerin binds NMI directly and associates stably with αII-spectrin
To distinguish between cytoplasmic myosin and NMI we immunoblotted the HeLa nuclear lysates and our emerin affinity-purified proteins using an antibody that specifically recognizes the 16 N-terminal residues unique to NMI (33). NMI was detected in the nuclear extracts (Figure 1B, load ‘L’) and was highly and specifically enriched in the emerin affinity-purified fraction (Figure 1B, Em-beads), showing that emerin associates (directly or indirectly) with NMI. Parallel blots probed with antibodies specific for either αII-spectrin or actin confirmed the mass spectrometry results: both actin and αII-spectrin were present in the affinity-purified fraction, with significant enrichment for αII-spectrin relative to the starting lysate (Figure 1B). Interestingly, a different antibody (S-1515, Sigma-Aldrich, Inc) that recognizes most isoforms of spectrin except αII-spectrin, gave no signal in either the nuclear extract or emerin-bound fraction (data not shown). We concluded that αII-spectrin is enriched in the nucleus and associates with emerin either directly or indirectly (e.g., via actin).
The above experiments were done without any added ATP. To distinguish between direct versus indirect association with emerin, we first tested the hypothesis that ATP hydrolysis by NMI, a molecular motor, might release actin plus any actin-associated proteins from the emerin-beads. The BSA-beads and emerin-beads were incubated with HeLa nuclear lysates in the continuous presence or absence of 10 mM ATP, and bound proteins were resolved by SDS-PAGE and immunoblotted (Figure 1C). Remarkably, ATP had no effect on the binding of NMI or αII-spectrin, but decreased the amount of bound actin by ~65% (n=3). We concluded that most actin associated with emerin-beads indirectly, probably via direct binding to NMI. However, a significant fraction of actin was insensitive to ATP, consistent with either (a) direct binding to emerin, as reported previously (25, 48), or (b) direct binding to a different co-purified protein. The ATP experiment further suggested that nuclear myosin might bind directly to emerin. To test this possibility, recombinant purified emerin and NMI proteins were incubated and then immunoprecipitated using either immune or pre-immune antibodies against human emerin (Figure 1D). NMI was efficiently co-immunoprecipitated with emerin by immune but not pre-immune antibodies (Figure 1D, Im and Pre, respectively), demonstrating direct binding of NMI to emerin in vitro. These results showed that NMI binds directly to emerin in an ATP-insensitive manner in vitro (see Discussion).
Purification of emerin-containing complexes
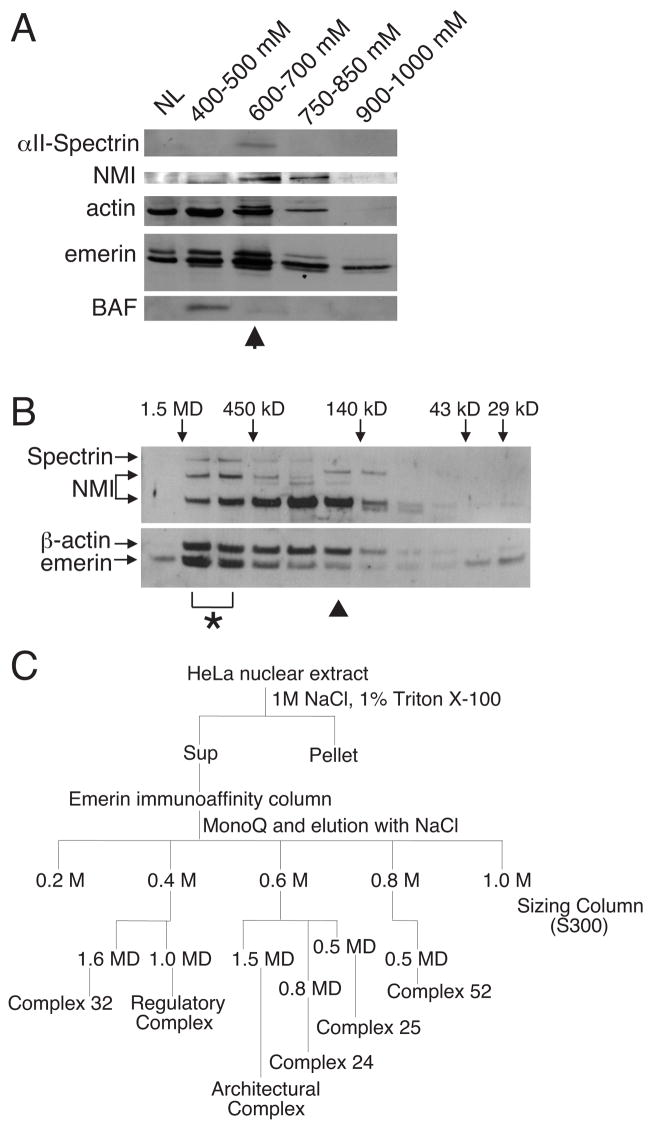
To determine if emerin, actin, NMI and αII-spectrin formed a bona fide complex in vivo, we purified emerin-containing complexes from HeLa nuclei in three steps: affinity purification using antibodies against emerin, ion exchange chromatography and size-exclusion chromatography (Superose 12). Supporting a possible emerin-actin-NMI-αII-spectrin complex, these proteins were co-eluted by 600–700 mM NaCl (Figure 2A) during ion exchange chromatography, and subsequently co-resolved at a molecular mass between 0.8 and 1.4 MDa (Figure 2B, *), suggesting that actin, NMI and αII-spectrin co-fractionated. Assuming one-to-one stoichiometry, the predicted mass of this putative complex was 476 kD, suggesting the 0.8–1.4 MDa complex had multiple copies of one or more components, or additional unidentified components. Interestingly, a smaller putative complex that lacked αII-spectrin, but included emerin, actin and NMI eluted at a mass of ~ 200 kD (Figure 2B, σ), consistent with a single copy of each component (191 kD). These results suggested that emerin, actin and NMI might comprise a sub-complex within a larger architectural complex(es) in vivo.
FIGURE 2.
Purification of a putative emerin-NMI-actin-αII-spectrin-containing complex, and five other complexes, from HeLa cells. A, Results from a two-step purification of emerin-associated protein complexes (emerin affinity purification, ion-exchange chromatography eluted with a linear NaCl gradient). Shown is a representative Western blot of the MonoQ elution profile probed with antibodies to each indicated protein. The NaCl concentration that eluted each fraction is indicated. NL, nuclear lysate. B, The Mono-Q fraction that eluted with 0.6–0.7 M NaCl (arrow, Figure 2A) was further resolved by size exclusion (Superose 12) chromatography and fractions were Western blotted with antibodies against each indicated protein. This analysis (n=3) suggested the presence of at least two complexes: a putative 0.8–1.4 MD complex that included emerin, NMI, actin and αII-spectrin (*), and a separate complex containing emerin, NMI and actin (σ). C, Protocol used for subsequent independent purifications of emerin-containing complexes from HeLa cell nuclear lysates (n=3).
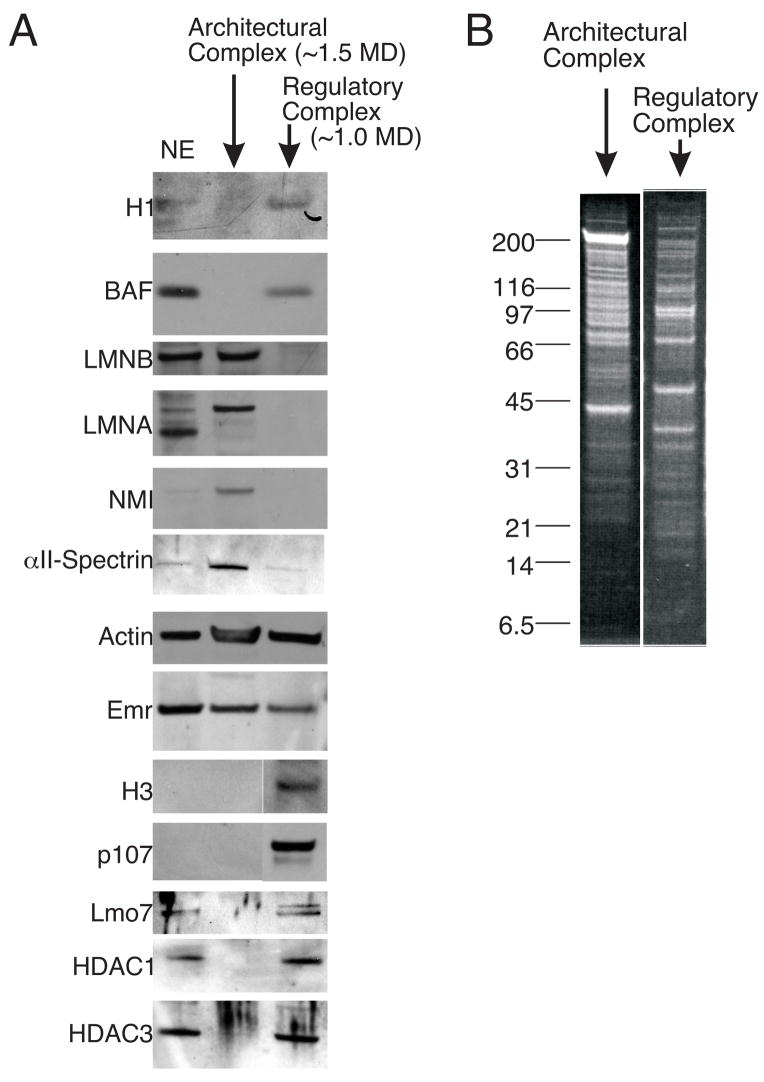
To further characterize the putative emerin-actin-NMI-containing complex, we repeated the purification using a size exclusion column (S300) with greater resolving power (Figure 2C). The proposed spectrin-containing architectural complex eluted with a molecular mass of 1.5 MD (n=3; data not shown). To identify the components of this putative complex, we first resolved the constituent proteins by SDS-PAGE and Western blotted for candidate components using antibodies against linker histones (H1), BAF, lamin B, lamins A/C, NMI, αII-spectrin, actin, emerin, H3, the Rb-related protein p107, and Lmo7 (Figure 3A, architectural complex). Consistent with potential roles in nuclear architecture, we detected lamin B, lamin A, NMI, αII-spectrin, actin and emerin, whereas chromatin and gene-regulatory proteins (H1, BAF, H3, p107, Lmo7) were not detected. The putative architectural complex also had a distinct profile of at least 20 polypeptide bands (Figure 2B, Architectural complex) compared to other potential complexes (see below). To identify additional components, proteins were identified by LCMS/MS. This analysis revealed other known structural proteins, including three nuclear pore complex components (gp210, Tpr, Nup155), NuMA, beta-filamin b, Sun2, myoferlin, AHNAK-like and other putative structural proteins (see Discussion).
FIGURE 3.
Purification of distinct emerin-containing architectural and regulatory complexes. A, Immunoblots of the proposed Architectural and Regulatory complexes show distinct protein compositions. Among candidate proteins tested, the architectural complex had detectable emerin, lamin A, lamin B, NMI and actin, whereas the regulatory complex had detectable BAF, H1, H3, p107, Lmo7, HDAC1, HDAC3 and actin. B, Analytical SDS-PAGE protein profiles of the Architectural and Regulatory complexes stained with SYPRO-Ruby to assess the number and sizes of constituent polypeptides. Proteins in each sample were identified by LCMS/MS. Selected results are described in the text and Table 1; full datasets are available upon request.
Our original ion exchange chromatography further suggested the presence of other emerin-containing complex(es), since the fraction eluted by 400–500 mM NaCl lacked NMI and αII-spectrin, but contained BAF (Figure 2A). To characterize this complex(es), the 400 mM-eluted fraction was resolved by size exclusion (S300 column) chromatography, and each eluted fraction was Western-blotted for candidate proteins (Figure 3A). One fraction that eluted at 1.0 MD included linker histones, BAF, actin, emerin, H3, p107, Lmo7, HDAC1 and HDAC3, but had no detectable lamins, NMI or αII-spectrin (Figure 3A, Regulatory complex). This fraction, known as the ‘Regulatory complex’, had many polypeptide bands that differed from the putative Architectural complex in analytical SDS-PAGE (Figure 3B). Interestingly, LCMS/MS analysis of the putative Regulatory complex revealed additional components that regulate gene expression or chromatin structure, including Far Upstream element-Binding Protein (FUBP1), EBNA2 co-activator p100/SND1, nuclear transcription factor Y-beta (NF-YB), nuclear transcription factor Y-gamma (NF-YC), KRAB-associated protein-1 (KAP1), DNA methyltransferase 1 (dnmt1), TATA box-binding protein interacting-protein 49b (Tip49b), mammalian sterile 20-like 3 (MST-3) and XAP-5 (see Discussion).
Purification of four additional emerin-containing complexes
This purification procedure yielded four other emerin-containing complexes (n=3), which were named based on their S300 elution fraction number as complexes 24, 25, 32 and 52 (Figure 2C). These putative complexes also had distinct polypeptide compositions in analytical SDS-PAGE (Figure 4A) and were Western-blotted for candidate proteins using antibodies against emerin, lamins A/C, actin, BAF, BAF-Like (BAF-L; 30), αII-spectrin, lamin B, MAN1, NMI, H3, p107, Rb, p130 and linker histones (Figure 4B). Complex 24 had prominent emerin, lamin A, H3 and linker histone bands, and weaker bands for BAF-L, lamin B, NMI, Rb and p130 (Figure 4B). Complex 25 had detectable emerin and low levels of actin, lamin B (arrow) and BAF (Figure 4B). Complex 32 was strongly positive for emerin, actin, BAF-L, αII-spectrin, lamin B (arrow), NMI, H3, p107, and p130 (Figure 4B). The emerin antibody recognized two bands, suggesting Complex 32 might also contain a post-translationally modified form of emerin. Complex 52 had detectable emerin, actin, lamin B (arrow), MAN1, NMI and H1, plus a weak p130 signal (Figure 4B). LCMS/MS analysis of each putative complex revealed additional components, many of which are listed in Table 1. The full list of identified proteins for each complex is available upon request. Complex 24 included RNA processing proteins (hnRNPU, SF2p32, SAP130/SF3b3) and proteins involved in DNA replication (MCM2, MCM4, MCM6). Complex 25 contained predominantly RNA processing proteins (SmD1-16K, ZNF265, SAP130/SF3b3, hnRNPK, Lsm8, Lsm2, La), but also included a number of signaling proteins (KCIP1/14-3-3-epsilon, 14-3-3 beta, BCCIP-beta, G3BP1, CDC37 homolog; Table 1). Complex 52 was rich in both RNA processing proteins (SmD1-16K, SF2p32, hnRNPU, hnRNPK, Lsm8, hnRNPL, NFAR-1/NF-90, BAT1) and signaling components (KCIP1/14-3-3-epsilon, 14-3-3 theta, creatine kinase b, Api5/Aac11, IQGAP, PDCD4, HDGF, ERBA1; Table 1). Interestingly, complex 32 was unique in having many subunits of chromatin remodeling complexes (HDAC3, TBL1, TBLR1, KAP1/TIF1β, BAF155, BAF170, Snf5/Ini, DRIP36/TRAP36, CoREST, dpy-30-like, Mi-2β, Rpd3, SAP18; Table 1). Of these, four (TBL1, TBLR1, HDAC3, KAP1) were core components of the previously characterized NCoR complex, which also contains NCoR and GPS2 (49, 50). The NCoR complex silences chromatin by deacetylating the Lys-5 residue of histone 4 (H4K5; 51). The potential purification of NCoR with emerin was exciting, since a large fraction of chromatin associated with the nuclear envelope is silenced (52).
FIGURE 4.
Purification of four additional emerin-containing complexes. A, Analytical SDS-PAGE analysis of complexes 24, 25, 32 and 52 stained with SYPRO-Ruby to assess the number and sizes of constituent polypeptides. Proteins in each sample were identified by LCMS/MS. B, Western blots probed for candidate components suggest complexes 24, 25, 32 and 52 have distinct protein compositions. A subset of identified components is described in the text and Table 1; full datasets are available upon request.
TABLE 1.
Components of putative emerin-containing complexes
| Complex # | 24 | 25 | 52 | 32 |
|---|---|---|---|---|
| RNA Processing Proteins | hnRNPU
SF2p32 SAP130 |
SmD1-16K
ZNF265 SAP130 hnRNPK Lsm8 Lsm2 La |
SmD1-16K
SF2p32 hnRNPU hnRNPK Lsm8 hnRNPL NFAR1/NF90 BAT1 |
|
| Signaling Proteins | KCIP/14-3-3 epsilon
14-3-3 beta BCCIP beta G3BP CDC37 homolog |
KCIP/14-3-3 epsilon
14-3-3 theta Creatine kinase b Api5/Aac11 IQGAP PDCD4 HDGF ERBA1 |
||
| DNA Replication Factors | MCM2
MCM4 MCM6 |
|||
| Chromatin Modifying Proteins | HDAC3
TBL1 TBLR1 KAP-1 BAF155 BAF170 Snf5/Ini DRIP36/TRAP36 CoREST Dpy30-like Mi-23 Rpd3 SAP18 |
Emerin interacts with NCoR complex components in vivo
To independently test whether emerin interacted with the NCoR complex, we transiently transfected FLAG-HDAC3, FLAG-TBL1, FLAG-TBLR1, or vector alone into HeLa cells, lysed the cells 36 h later, diluted the lysates 2.5-fold and incubated them with M2-agarose to immunoprecipitate each FLAG-tagged protein. Emerin co-immunoprecipitated with FLAG-HDAC3 and FLAG-TBLR1 well above the vector-only background signal; in contrast the emerin signal with FLAG-TBL1 was only slightly above background (Figure 5A, n=5). These results confirmed the emerin interaction with at least two components of the NCoR complex in vivo. To test other components of this complex, the experiment was repeated for FLAG-HDAC3, FLAG-GPS and FLAG-NCoR. Emerin co-immunoprecipitated above background with all three proteins, and particularly well with FLAG-GPS and FLAG-NCoR (Figure 5B), implicating GPS and/or NCoR as potential direct binding partners for emerin in complex 32. These results validated a subset of the LCMS/MS results for complex 32 by confirming the association of major components of the NCoR complex with emerin in vivo. We therefore concluded that emerin interacts with the NCoR complex in vivo. These findings provided proof-of-principle that our purification yielded at least one valid novel emerin-associated protein complex.
FIGURE 5.
Emerin interacts with FLAG-tagged NCoR complex components in vivo. A,B, HeLa cells were mock transfected (−) or transfected to transiently express either FLAG-HDAC3, FLAG-TBL1, FLAG-TBLR1, FLAG-GPS or FLAG-NCoR, lysed in modified NEHN buffer and incubated with M2-agarose to immunoprecipitate FLAG-tagged proteins. Load (L, 10%), supernatant (S, 10%) and pellet (P, 50%) were immunoblotted with antibodies against the FLAG-tag (α-FLAG) or emerin (α-emerin).
DISCUSSION
We discovered that emerin interacts directly with the nuclear isoform of myosin I and also associates with αII-spectrin. αII-spectrin was highly enriched on emerin beads, and may interact with emerin either directly, or indirectly through binding to NMI or another component of the proposed architectural complex. NMI, a molecular motor, is a new direct binding partner for emerin. Our other findings strongly support the hypothesis that emerin forms a variety of functionally distinct multi-protein complexes in a single cell. These putative complexes are hypothesized to have roles in nuclear actin-myosin-spectrin-based architecture, gene regulation or signaling, which are discussed further below, and potentially other novel functions. Complex 32, in particular, included key components of the NCoR complex, which were independently validated as emerin-associated in vivo, supporting a direct role for emerin in transcription repression or in tethering repressed chromatin at the nuclear envelope. We hypothesize that emerin binding to the NCoR complex might contribute to the maintenance of repressed chromatin at the nuclear envelope. Considerable work is needed to characterize other putative components of complex 32, and to independently validate the compositions of the other five complexes, since some putative components may have co-purified artifactually. This analysis of the emerin proteome in HeLa cells can be viewed as a ‘high throughput’ study: it provides a molecular starting point to propose and test specific roles for emerin in tissues affected by EDMD disease.
Affinity purification of new direct emerin-binding proteins
The emerin-conjugated beads were remarkably effective in identifying new direct binding partners for emerin. Four of the seven affinity-purified proteins bind emerin directly: actin (25), Lmo7 (21), NMI (reported here) and SIKE (to be reported separately). The three remaining proteins, NMMHCA, calponin 3 and αII-spectrin are actin-binding proteins and therefore might associate with emerin non-specifically, although direct binding to emerin has not been tested. Previously identified emerin-binding proteins, including lamins, BAF, GCL, YT521-B, nesprin-1α, nesprin-2β or β-catenin (10, 23, 28, 53) (22, 54) were not recovered by affinity for emerin-beads. This was no surprise for lamins, which were inefficiently solubilized by the modified hypotonic lysis method used, which probably favored recovery of readily-solubilized, abundant higher affinity partners for bead-bound emerin (Figure 1). Lamins were solubilized by the more disruptive conditions (sonication) used for the three-step chromatographic purification of emerin-associated complexes (Figure 3).
Emerin directly binds and caps the pointed-end of F-actin in vitro (25) and also binds directly to nuclear myosin I. Thus, other actin-binding proteins recovered in our emerin pull-down study (NMMHCA, calponin 3, αII-spectrin; Figure 1) are either plausible components of emerin-containing complexes, or artifacts with no nuclear relevance. However several actin-binding proteins previously thought to localize exclusively in the cytoplasm are now known to shuttle into the nucleus, including tropomodulin and profilin (55–57), so NMMHCA and calponin 3 cannot be ruled out as nuclear-relevant solely based on their cytoplasmic localization. Interestingly, nuclear-specific isoforms of αII-spectrin are involved in DNA repair (43–45). The presence of αII-spectrin in two of the putative emerin-containing complexes (Architectural complex and complex 32) independently supports the hypothesis that emerin stabilizes an actin cortical network at the nuclear inner membrane (25), and further suggests emerin might influence DNA repair.
Implications of emerin binding directly to nuclear myosin I
NMI is an alternatively translated isoform of myosin I-β (also known as myosin IC, ‘MYOIC’, in humans). The nuclear and cytosolic isoforms are identical except for 16 extra residues at the N-terminus of the nuclear isoform (33, 58). NMI is necessary for mRNA transcription in vitro, binds directly to the large subunit of RNA polymerase II, and is proposed to facilitate movement of the transcription machinery (33, 59) or chromatin itself during transcription (47). Why would NMI bind emerin? Emerin and NMI might influence nuclear architecture or transcription activity, or both. Emerin might sequester inactive NMI at the nuclear inner membrane, or facilitate its motor activity by anchoring NMI at the nuclear membrane. Alternatively, since NMI is a barbed end-directed motor (60), a third possibility is that emerin ‘loads’ NMI onto the pointed end of actin filaments and thereby promotes the movement of myosin and unknown cargo into the nucleoplasm. Supporting this idea, Chuang and colleagues showed that an active reporter gene locus moved, in an actin- and NMI-dependent manner, from the nuclear envelope to the nuclear interior (47). A fourth possibility is that emerin-and-lamin A-anchored NMI might pull actin filaments towards, or along, the nuclear envelope. This action, coupled to emerin-promoted actin polymerization at the inner membrane (22), has the potential to generate outward force, and/or tension along, the nuclear envelope. Interestingly the cytoplasmic counterpart of NMI is a tension-regulated motor (Mike Ostap, personal communication); thus emerin-bound NMI has the potential to both sense and regulate the mechanical stiffness of the peripheral nuclear lamina network, a potentially fundamental contribution to nuclear shape and architecture. These proposed roles are consistent with, and suggest molecular mechanisms for, emerin’s roles in mechano-sensitive gene expression and nuclear shape (24). Emerin-associated myosin motors might also function during mitosis to ‘shrink’ the nuclear envelope, similar to myosin action during cytokinesis (61).
The proposed emerin-containing architectural complex
The core elements of the cytoplasmic actin cortical network, defined in erythrocytes, include integral membrane proteins (e.g., Band 3), anchoring proteins (ankyrin), spectrin filaments and ‘junctional complexes’ consisting of short actin filaments, Band 4.1, adducin, tropomodulin, and tropomyosin (62). Spectrin filaments, composed of α/β-spectrin heterodimers, are anchored to the plasma membrane by the Band 3-ankyrin complex, and bind junctional complexes through direct interactions with three components (Band 4.1, adducin, and actin). Tropomodulin and tropomyosin stabilize the junctional complex. Actin cortical networks in nonerythroid cells can include α-actinin, vinculin, paxillin and talin (63); talin binds directly to integrins, thereby anchoring the actin cortical network to focal adhesions. The composition of our putative Architectural Complex suggests emerin might provide similar ‘cortical-networking’ functions at the nuclear inner membrane by interlinking actin-NMI-spectrin complexes with A- and B-type lamins, and potentially also with AHNAK or nuclear isoforms of titin (64, 65), or myoferlin (66, 67). Titin binds directly to both A- and B-type lamins, and overexpression of the lamin-binding region of titin profoundly disrupts nuclear shape and nuclear envelope integrity (65). The peptides identified in our study did not distinguish between titin and AHNAK in the putative Architectural Complex. Interestingly, myoferlin is present at the nuclear envelope, is required for myotube formation and is linked to muscular dystrophy (66, 67). Further work is clearly needed to validate the existence and understand the composition of the putative emerin-containing Architectural complex(es) in vivo.
Purification of multiple emerin-containing regulatory complexes
The other five potential complexes included many proteins that function in gene expression, RNA processing, cell signaling and/or chromatin modification. The putative Regulatory Complex included known gene-regulatory proteins. For example, NF-YB and NF-YC are essential subunits of the NF-Y complex, which binds the CCAAT box, and helps activate many different genes (68, 69). The NF-YB/C heterodimer recruits the DNA-binding subunit, NF-YA, allowing formation of the NF-Y complex (70). Since only NF-YB and NF-YC were detected in this complex, we speculate that emerin might repress transcription by sequestering NF-YB/C heterodimers at the nuclear envelope. Three other transcription activators (SND-1, FUBP, XAP-5) were also detected in this complex, suggesting that emerin might sequester transcription activators at the nuclear envelope. Other potential components, including KAP-1 and DNMT1, are direct repressors of gene expression. KAP-1 scaffolds Kruppel-associated-box (KRAB)-zinc finger proteins and coordinates both SETDB1-mediated methylation of H3K9 and deposition of heterochromatin-binding protein 1 (HP1) to silence gene expression (71). DNMT1 methylates CpG dinucleotides, organizes large heterochromatic regions and is proposed to transmit methylation patterns during replication (72). Interestingly, DNMT1 is also required to maintain H3K9 methylation in vivo (72), functionally linking DNMT-1 to KAP-1 in the proposed regulatory complex. Heterochromatin is often associated with the nuclear envelope, and the association of emerin with gene silencing machinery might begin to explain why. Two additional putative components (Tip49b, Mst-3) of the putative Regulatory Complex regulate chromatin dynamics. Tip49b is a component of the human INO80 complex (73), an ATP-dependent chromatin-remodeling complex. Mst-3 is a kinase that can phosphorylate H3, but not other histones (74), and might therefore regulate chromatin structure at the nuclear envelope.
Three putative complexes included RNA processing proteins (Table 1), consistent with a previous report that emerin binds splicing factor YT521-B and influences splice-site selection (23). Complex 24 also contained replication components MCM2, MCM4, MCM6, which in S. pombe form stable complexes with a fourth protein, MCM7, not detected in complex 24 (75). Yeast MCM2 inhibits formation of MCM 4/6/7 dimers and hence blocks MCM 4/6/7 helicase activity (75).
Complex 25 contained a mixture of RNA processing and signaling molecules, suggesting the presence of two distinct complexes that were not resolved during purification. Lsm2, Lsm8 and SmD1-16K form a bona fide RNA processing complex with a predicted mass of 35.5 kD in vivo (76). The signaling proteins 14-3-3-beta and KCIP/14-3-3-epsilon bind each other in vitro and in vivo (77, 78), and mediate TNFα/NF-κB signaling in vivo (79). The 14-3-3 family of proteins has over 200 known interactors, including kinases, and has many conserved roles in cell proliferation, signaling and apoptosis (80), hence their potential association with emerin has broad implications for signal regulation at the nuclear envelope.
Complex 52 included several RNA processing proteins known to mutually interact: hnRNPU, hnRNPK, hnRNPL, and NFAR interact (79), as do Lsm8 and SmD1-16K (76). Bat1 and SmD1-16K can interact with each other (81), or Bat1 and SF2p32 can interact with 14-3-3 proteins (82). Thus complex 52 might include one or more previously characterized complexes involved in RNA processing or signaling. We did not assay for nucleic acids.
Validation of emerin-associated NCoR complex components in vivo
Our findings suggest emerin influences gene silencing, via the NCoR complex. HDAC3, TBL1 and TBLR1, which co-immunoprecipitated emerin from cells, are core components of all three previously-reported NCoR deacetylase complexes (49, 83), the largest of which is ~ 1.5 MDa (49). Other putative components of NCoR complexes include BAF155, BAF170, Snf5, KAP1, NCoR, SAP130, SAP120 and SRCAP (50). Interestingly, all but SRCAP also co-purified with emerin in Complex 32, which had a mass of ~1.5 MD (range 1.2–1.8 MD), suggesting emerin might tether the large NCoR complex. It remains unknown which if any candidates identified by western blotting (e.g., actin, BAF-L, αII-spectrin, lamin B, NMI, p107, p130) co-associate with the NCoR complex in vivo. This will be important to determine in the future, since Complex 32 might include independent complexes that failed to resolve from emerin in this purification protocol. For example this fraction also included components of the human SWI/SNF complex (BAF155, BAF170, Ini/Snf5; 84–86), or other chromatin silencing machines: Dpy-30-like is probably involved in H3K4 methylation, since it is homologous to yeast Saf19, a component of the SET1 complex that methylates H3K4 (87), CoREST (co-repressor for REST; 88) is an integral component of the CoREST histone deacetylase complex (89), and Rpd3 is a component of several HDAC complexes, including NuRD and Sin3 (90–92). Interestingly, SAP18 (a component of the Sin3 complex; 93) and Mi-2β (a component of the NuRD complex; 94) were also found in Complex 32. We therefore speculate that emerin might tether a variety of silencing complexes to the inner nuclear membrane, as proposed for the related LEM-domain protein LAP2β (95).
The NCoR complex regulates genes important for muscle differentiation, including MyoD, Myf5 and myogenin (96), suggesting a specific pathway by which emerin might influence muscle differentiation. We therefore propose that emerin may regulate chromatin dynamics, at least in part, by associating with the NCoR complex in vivo, and that loss of this interaction may contribute to EDMD disease. Other nuclear envelope proteins are proposed to tether heterochromatin at the nuclear periphery (52). For example, lamin B receptor (LBR) binds heterochromatin protein 1 (HP1; 97), which binds silenced chromatin via H3K9me (98, 99). LAP2β, an inner nuclear membrane protein that is related to emerin outside the LEM-domain, binds HDAC3 and regulates its activity (95).
“Adaptor/Scaffold” hypothesis
Our findings support the hypothesis that emerin scaffolds a variety of multi-protein complexes at the nuclear inner membrane. How might emerin, a relatively small protein (254 residues), interact purposefully with so many partners? Not all scaffolding proteins are large; for example Grb2 is a comparably sized (217-residue) adaptor/scaffolding protein that binds many receptor tyrosine kinases at the cell surface and recruits signaling complex components (100). Current knowledge of emerin’s structure is compatible with scaffolding activity: emerin has only one demonstrated folded domain (the N-terminal ~40-residue LEM-domain; 101–103), and is predicted by SCRATCH software (data not shown) to be highly (~60%) unstructured and extended between the LEM-domain and the C-terminal transmembrane domain. Within this extended region is a proposed APC homology domain, which is sufficient to bind β-catenin (22) and overlaps one of two functional domains in which mutations disrupt binding to transcription regulators, and certain other partners (10, 20, 21, 23). Emerin’s interactions are likely to be regulated posttranslationally. Emerin is phosphorylated at multiple sites (104–107), and phosphorylation regulates binding to at least one known partner: BAF (107); K. Tifft and K.L.W., unpublished observations). Biochemical studies showed that emerin’s partners themselves might influence the composition of a complex, since certain partners (e.g., BAF and lamin A, or GCL and lamin A) can co-bind emerin in vitro, whereas others (e.g., BAF and transcription factor GCL) compete with each other for binding to emerin (10).
Given so many proposed partners and functions for emerin, why is the emerin-null phenotype (EDMD) not more severe? There are at least three plausible reasons. First, partial functional redundancy between emerin and other LEM-domain proteins (e.g., LAP2β, or Lem2/NET25; 108–110) might compensate for loss of emerin in some, but not all, tissues (14). Second, emerin may ‘fine-tune’ appropriate responses to specific stimuli, such as mechanical stress, critical only for certain tissues. Third, emerin may be regulated by, or participate in, tissue-specific signaling pathways. Based on these considerations, we predict no single mechanism will account for disease in all three EDMD-affected tissues. However further study of the emerin-containing complexes (emerin ‘proteome’) may be a feasible way to understand the disease mechanism for each specific tissue. For skeletal muscle, Complex 32 (discussed above) and the putative Regulatory and Architectural complexes are of particular interest. The Regulatory complex is proposed to include Rb family members and HDAC subunits (Rb-dependent gene regulation is impaired in EDMD muscle; 111, 112) plus Lmo7, which regulates emerin expression and is required to maintain muscle (21, 113). The Architectural complex, which may include Sun2 (114) has the potential to interlink the nucleoskeleton and cytoskeleton, and mediate mechano-sensing functions of emerin, potentially via nuclear myosin I activity. Validated emerin-containing complexes can also be examined in the future in cells bearing EDMD-causing mutations in A-type lamins, to identify shared molecular mechanisms for EDMD disease.
Acknowledgments
We are grateful to Dr. Robert N. Cole at the AB Mass Spectrometry/Proteomics Facility at Johns Hopkins School of Medicine (http://www.hopkinsmedicine.org/msf), for protein identification. This facility is supported by the National Center for Research Resources shared instrumentation grant 1S10-RR14702, the Johns Hopkins Fund for Medical Discovery, and the Institute for Cell Engineering. We are grateful to Dr. Primal de Lanerolle (University of Illinois at Chicago) for his generous gifts of NMI protein and antibodies. We thank Dr. Robert Roeder (The Rockefeller University) for the FLAG-GPS, FLAG-TBL1, FLAG-TBLR1, FLAG-HDAC3 and FLAG-NCoR constructs. We thank the National Cell Culture Center for HeLa cell pellets. Many thanks to Dr. Doug Robinson and members of the Wilson lab for discussions, advice and comments on this manuscript. These studies were supported by the National Institutes of Health (R01GM48646 to KLW and F32GM067397 to JMH), the American Heart Association (Scott B. Deutschman Memorial Research Award to KLW, and 2004 Scholarship in Cardiovascular Disease & Stroke Research to SRB), and the Ray Mills Fund (to K.L.W.).
Footnotes
The abbreviations used are: Api1, Apoptosis inhibitor 1; BAF, Barrier-to-autointegration factor; BAF155/170, Brg-associated factor 155/170; BAT, HLA-B-associated transcript 1; BCCIP, BRCA2 and CDKN1A-interacting protein; BSA, Bovine serum albumin; Btf, Bcl-2-associated transcription factor; Dnmt1, DNA methyltransferase 1; DRIP, Vitamin D receptor-interacting protein; EDMD, Emery-Dreifuss muscular dystrophy; ERBA1, Thyroid hormone receptor alpha; FUBP1, far upstream element binding protein 1; G3BP1, Ras-GTPase-activating protein-binding protein; GCL, Germ-cell less; H1, histone 1; H3, histone 3; H3K9me, H3-lysine-9-methyl; HDAC, Histone deacetylase; Gps2, G protein pathway suppressor 2; HDGF, hepatoma-derived growth factor; hnRNP, heterogeneous nuclear ribonucleoprotein; HP1, heterochromatin protein 1; IQGAP1, IQ motif-containing GTPase-activating protein 1; KAP1, KRAB-associated protein 1; KCIP-1, Protein kinase C inhibitor protein 1; LBR, lamin B receptor; Lmo7, Lim domain only 7; Lsm, Sm-like; MCM, minichromosome maintenance; Mst-3, mammalian sterile 20 kinase-like 3; NCoR, Nuclear Co-repressor; NFAR, nuclear factor associated with double-stranded RNA; NF-Y, nuclear transcription factor Y; NMI, nuclear myosin I; NMMHCA, non-muscle myosin heavy chain A; NuMA, nuclear mitotic apparatus protein; NURD, nucleosome remodeling and histone deacetylation; PDCD4, programmed cell death 4; Rb, Retinoblastoma protein; RBD, repressor-binding domain; Rpd3, reduced potassium dependency 3; SAP18, Sin3-associated polypeptide 18 kD; SAP130, spliceosome-associated polypeptide 130 kD; SETDB1, SET-domain protein bifurcated 1; SF2, Splicing factor 2; SIKE, suppressor of IKKε- and TBK1-activation of interferon response elements; SmD1-16k, small nuclear ribonucleoprotein D1 polypeptide 16 kD; SND1, Staphylococcal nuclease domain-containing protein 1; Snf, sucrose non-fermenting; SRCAP, Snf2-related CBP-activator protein; TBL1, transducin-beta-like 1; TBLR1, TBL1-related protein 1; Tip49b, TBP-interacting protein 48kD; ZNF265, zinc finger protein 265.
References
- 1.Emery AE. Emery-Dreifuss muscular dystrophy-a 40 year retrospective. Neuromuscul Disord. 2000;10:228–232. doi: 10.1016/s0960-8966(00)00105-x. [DOI] [PubMed] [Google Scholar]
- 2.Bengtsson L, Wilson KL. Multiple and surprising new functions for emerin, a nuclear membrane protein. Curr Opin Cell Biol. 2004;16:73–79. doi: 10.1016/j.ceb.2003.11.012. [DOI] [PubMed] [Google Scholar]
- 3.Nagano A, Koga R, Ogawa M, Kurano Y, Kawada J, Okada R, Hayashi YK, Tsukahara T, Arahata K. Emerin deficiency at the nuclear membrane in patients with Emery- Dreifuss muscular dystrophy. Nat Genet. 1996;12:254–259. doi: 10.1038/ng0396-254. [DOI] [PubMed] [Google Scholar]
- 4.Segura-Totten M, Wilson K. BAF: roles in chromatin, nuclear structure and retrovirus integration. Trends Cell Biol. 2004;14:261–266. doi: 10.1016/j.tcb.2004.03.004. [DOI] [PubMed] [Google Scholar]
- 5.Margalit A, Brachner A, Gotzmann J, Foisner R, Gruenbaum Y. Barrier-to-autointegration factor--a BAFfling little protein. Trends Cell Biol. 2007;17:202–208. doi: 10.1016/j.tcb.2007.02.004. [DOI] [PubMed] [Google Scholar]
- 6.Segura-Totten M, Kowalski AM, Craigie R, Wilson KL. Barrier-to-autointegration factor: major roles in chromatin decondensation and nuclear assembly. J Cell Biol. 2002;158:475–485. doi: 10.1083/jcb.200202019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Montes de Oca R, Lee KK, Wilson KL. Binding of barrier to autointegration factor (BAF) to histone H3 and selected linker histones including H1.1. J Biol Chem. 2005;280:42252–42262. doi: 10.1074/jbc.M509917200. [DOI] [PubMed] [Google Scholar]
- 8.Margalit A, Segura-Totten M, Gruenbaum Y, Wilson KL. Barrier-to-autointegration factor is required to segregate and enclose chromosomes within the nuclear envelope and assemble the nuclear lamina. Proc Natl Acad Sci U S A. 2005;102:3290–3295. doi: 10.1073/pnas.0408364102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Haraguchi T, Koujin T, Segura-Totten M, Lee KK, Matsuoka Y, Yoneda Y, Wilson KL, Hiraoka Y. BAF is required for emerin assembly into the reforming nuclear envelope. J Cell Sci. 2001;114:4575–4585. doi: 10.1242/jcs.114.24.4575. [DOI] [PubMed] [Google Scholar]
- 10.Holaska J, Lee K, Kowalski A, Wilson K. Transcriptional repressor germ cell-less (GCL) and barrier-to-autointegration factor (BAF) compete for binding to emerin in vitro. J Biol Chem. 2003;278:6969–6975. doi: 10.1074/jbc.M208811200. [DOI] [PubMed] [Google Scholar]
- 11.Wang X, Xu S, Rivolta C, Li LY, Peng G, Swain PK, Sung C, Swaroop A, Berson EL, Dryja TP, Chen S. Barrier to autointegration factor interacts with the cone-rod homeobox and represses its transactivation function. J Biol Chem. 2002;277:43288–43300. doi: 10.1074/jbc.M207952200. [DOI] [PubMed] [Google Scholar]
- 12.Burke B, Stewart CL. Life at the edge: the nuclear envelope and human disease. Nat Rev Mol Cell Biol. 2002;3:575–585. doi: 10.1038/nrm879. [DOI] [PubMed] [Google Scholar]
- 13.Holaska JM, Wilson KL, Mansharamani M. The nuclear envelope, lamins and nuclear assembly. Curr Opin Cell Biol. 2002;14:357–364. doi: 10.1016/s0955-0674(02)00329-0. [DOI] [PubMed] [Google Scholar]
- 14.Gruenbaum Y, Margalit A, Goldman RD, Shumaker DK, Wilson KL. The nuclear lamina comes of age. Nat Rev Mol Cell Biol. 2005;6:21–31. doi: 10.1038/nrm1550. [DOI] [PubMed] [Google Scholar]
- 15.Tunnah D, Sewry CA, Vaux D, Schirmer EC, Morris GE. The apparent absence of lamin B1 and emerin in many tissue nuclei is due to epitope masking. J Mol Histol. 2005;36:337–344. doi: 10.1007/s10735-005-9004-7. [DOI] [PubMed] [Google Scholar]
- 16.Cohen M, Lee KK, Wilson KL, Gruenbaum Y. Transcriptional repression, apoptosis, human disease and the functional evolution of the nuclear lamina. Trends Biochem Sci. 2001;26:41–47. doi: 10.1016/s0968-0004(00)01727-8. [DOI] [PubMed] [Google Scholar]
- 17.Holaska JM, Wilson KL. Multiple roles for emerin: Implications for Emery-Dreifuss muscular dystrophy. Anat Rec A Discov Mol Cell Evol Biol. 2006;288:676–680. doi: 10.1002/ar.a.20334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ostlund C, Worman HJ. Nuclear envelope proteins and neuromuscular diseases. Muscle Nerve. 2003;27:393–406. doi: 10.1002/mus.10302. [DOI] [PubMed] [Google Scholar]
- 19.Bonne G, Yaou RB, Beroud C, Boriani G, Brown S, de Visser M, Duboc D, Ellis J, Hausmanowa-Petrusewicz I, Lattanzi G, Merlini L, Morris G, Muntoni F, Opolski G, Pinto YM, Sangiuolo F, Toniolo D, Trembath R, van Berlo JH, van der Kooi AJ, Wehnert M. 108th ENMC International Workshop, 3rd Workshop of the MYO-CLUSTER project: EUROMEN, 7th International Emery-Dreifuss Muscular Dystrophy (EDMD) Workshop, 13–15 September 2002, Naarden, The Netherlands. Neuromuscul Disord. 2003;13:508–515. doi: 10.1016/s0960-8966(03)00063-4. [DOI] [PubMed] [Google Scholar]
- 20.Haraguchi T, Holaska JM, Yamane M, Wilson KL, Hiraoka Y. Emerin binding to Btf, a death-promoting transcriptional repressor, is disrupted by a missense mutation that causes Emery-Dreifuss muscular dystrophy. Eur J Biochem. 2004;271:1035–1045. doi: 10.1111/j.1432-1033.2004.04007.x. [DOI] [PubMed] [Google Scholar]
- 21.Holaska JM, Rais-Bahrami S, Wilson KL. Lmo7 is an emerin-binding protein that regulates the transcription of emerin and many other muscle-relevant genes. Hum Mol Genet. 2006;15:3459–3472. doi: 10.1093/hmg/ddl423. [DOI] [PubMed] [Google Scholar]
- 22.Markiewicz E, Tilgner K, Barker N, van de Wetering M, Clevers H, Dorobek M, Hausmanowa-Petrusewicz I, Ramaekers FC, Broers JL, Blankesteijn WM, Salpingidou G, Wilson RG, Ellis JA, Hutchison CJ. The inner nuclear membrane protein emerin regulates beta-catenin activity by restricting its accumulation in the nucleus. EMBO J. 2006;25:3275–3285. doi: 10.1038/sj.emboj.7601230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wilkinson FL, Holaska JM, Zhang Z, Sharma A, Manilal S, Holt I, Stamm S, Wilson KL, Morris GE. Emerin interacts in vitro with the splicing-associated factor, YT521-B. Eur J Biochem. 2003;270:2459–2466. doi: 10.1046/j.1432-1033.2003.03617.x. [DOI] [PubMed] [Google Scholar]
- 24.Lammerding J, Hsiao J, Schulze PC, Kozlov S, Stewart CL, Lee RT. Abnormal nuclear shape and impaired mechanotransduction in emerin-deficient cells. J Cell Biol. 2005;170:781–791. doi: 10.1083/jcb.200502148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Holaska JM, Kowalski AM, Wilson KL. Emerin caps the pointed end of actin filaments: evidence for an actin cortical network at the nuclear inner membrane. PLoS Biol. 2004;2:1354–1362. doi: 10.1371/journal.pbio.0020231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fidzianska A, Hausmanowa-Petrusewicz I. Architectural abnormalities in muscle nuclei. Ultrastructural differences between X-linked and autosomal dominant forms of EDMD. J Neurol Sci. 2003;210:47–51. doi: 10.1016/s0022-510x(03)00012-1. [DOI] [PubMed] [Google Scholar]
- 27.Burke B, Stewart CL. The Laminopathies: The Structure of the Nucleus and Its Involvement in Disease. Annu Rev Genomics Hum Genet. 2005 doi: 10.1146/annurev.genom.7.080505.115732. NEEDS VOL/page #. [DOI] [PubMed] [Google Scholar]
- 28.Lee KK, Haraguchi T, Lee RS, Koujin T, Hiraoka Y, Wilson KL. Distinct functional domains in emerin bind lamin A and DNA-bridging protein BAF. J Cell Sci. 2001;114:4567–4573. doi: 10.1242/jcs.114.24.4567. [DOI] [PubMed] [Google Scholar]
- 29.Bengtsson L, Wilson KL. Barrier-to-autointegration factor phosphorylation on Ser-4 regulates emerin binding to lamin A in vitro and emerin localization in vivo. Mol Biol Cell. 2006;17:1154–1163. doi: 10.1091/mbc.E05-04-0356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tifft KE, Segura-Totten M, Lee KK, Wilson KL. Barrier-to-autointegration factor-like (BAF-L): a proposed regulator of BAF. Exp Cell Res. 2006;312:478–487. doi: 10.1016/j.yexcr.2005.11.013. [DOI] [PubMed] [Google Scholar]
- 31.Mansharamani M, Wilson KL. Direct binding of nuclear membrane protein MAN1 to emerin in vitro and two modes of binding to barrier-to-autointegration factor. J Biol Chem. 2005;280:13863–13870. doi: 10.1074/jbc.M413020200. [DOI] [PubMed] [Google Scholar]
- 32.Offterdinger M, Schofer C, Weispoltshammer K, Grunt TW. c-erb-3: a nuclear protein in mammary epithelial cells. J Cell Biol. 2002;157:929–939. doi: 10.1083/jcb.200109033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pestic-Dragovich L, Stojiljkovic L, Philimonenko AA, Nowak G, Ke Y, Settlage RE, Shabanowitz J, Hunt DF, Hozak P, de Lanerolle P. A myosin I isoform in the nucleus. Science. 2000;290:337–341. doi: 10.1126/science.290.5490.337. [DOI] [PubMed] [Google Scholar]
- 34.Huang J, Liu T, Xu LG, Chen D, Zhai Z, Shu HB. SIKE is an IKK epsilon/TBK1-associated suppressor of TLR3- and virus-triggered IRF-3 activation pathways. EMBO J. 2005;24:4018–4028. doi: 10.1038/sj.emboj.7600863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Heath KE, Campos-Barros A, Toren A, Rozenfeld-Granot G, Carlsson LE, Savige J, Denison JC, Gregory MC, White JG, Barker DF, Greinacher A, Epstein CJ, Glucksman MJ, Martignetti JA. Nonmuscle myosin heavy chain IIA mutations define a spectrum of autosomal dominant macrothrombocytopenias: May-Hegglin anomaly and Fechtner, Sebastian, Epstein, and Alport-like syndromes. Am J Hum Genet. 2001;69:1033–1045. doi: 10.1086/324267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Maguchi M, Nishida W, Kohara K, Kuwano A, Kondo I, Hiwada K. Molecular cloning and gene mapping of human basic and acidic calponins. Biochem Biophys Res Commun. 1995;217:238–244. doi: 10.1006/bbrc.1995.2769. [DOI] [PubMed] [Google Scholar]
- 37.Winder SJ, Walsh MP. Smooth muscle calponin. Inhibition of actomyosin MgATPase and regulation by phosphorylation. J Biol Chem. 1990;265:10148–10155. [PubMed] [Google Scholar]
- 38.Winder S, Walsh M. Inhibition of the actomyosin MgATPase by chicken gizzard calponin. Prog Clin Biol Res. 1990;327:141–148. [PubMed] [Google Scholar]
- 39.Makuch R, Birukov K, Shirinsky V, Dabrowska R. Functional interrelationship between calponin and caldesmon. Biochem J. 1991;280(Pt 1):33–38. doi: 10.1042/bj2800033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Abe M, Takahashi K, Hiwada K. Effect of calponin on actin-activated myosin ATPase activity. J Biochem (Tokyo) 1990;108:835–838. doi: 10.1093/oxfordjournals.jbchem.a123289. [DOI] [PubMed] [Google Scholar]
- 41.Olave IA, Reck-Peterson SL, Crabtree GR. Nuclear actin and actin-related proteins in chromatin remodeling. Annu Rev Biochem. 2002;71:755–781. doi: 10.1146/annurev.biochem.71.110601.135507. [DOI] [PubMed] [Google Scholar]
- 42.Hofmann WA, Johnson T, Klapczynski M, Fan JL, de Lanerolle P. From transcription to transport: emerging roles for nuclear myosin I. Biochem Cell Biol. 2006;84:418–426. doi: 10.1139/o06-069. [DOI] [PubMed] [Google Scholar]
- 43.McMahon LW, Walsh CE, Lambert MW. Human alpha spectrin II and the Fanconi anemia proteins FANCA and FANCC interact to form a nuclear complex. J Biol Chem. 1999;274:32904–32908. doi: 10.1074/jbc.274.46.32904. [DOI] [PubMed] [Google Scholar]
- 44.McMahon LW, Sangerman J, Goodman SR, Kumaresan K, Lambert MW. Human alpha spectrin II and the FANCA, FANCC, and FANCG proteins bind to DNA containing psoralen interstrand cross-links. Biochemistry. 2001;40:7025–7034. doi: 10.1021/bi002917g. [DOI] [PubMed] [Google Scholar]
- 45.Sridharan D, Brown M, Lambert WC, McMahon LW, Lambert MW. Nonerythroid alphaII spectrin is required for recruitment of FANCA and XPF to nuclear foci induced by DNA interstrand cross-links. J Cell Sci. 2003;116:823–835. doi: 10.1242/jcs.00294. [DOI] [PubMed] [Google Scholar]
- 46.Hofmann W, Reichart B, Ewald A, Muller E, Schmitt I, Stauber RH, Lottspeich F, Jockusch BM, Scheer U, Hauber J, Dabauvalle MC. Cofactor requirements for nuclear export of Rev response element (RRE)- and constitutive transport element (CTE)-containing retroviral RNAs. An unexpected role for actin. J Cell Biol. 2001;152:895–910. doi: 10.1083/jcb.152.5.895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chuang CH, Carpenter AE, Fuchsova B, Johnson T, de Lanerolle P, Belmont AS. Long-range directional movement of an interphase chromosome site. Curr Biol. 2006;16:825–831. doi: 10.1016/j.cub.2006.03.059. [DOI] [PubMed] [Google Scholar]
- 48.Fairley E, Kendrick-Jones J, Ellis J. The Emery-Dreifuss muscular dystrophy phenotype arises from aberrant targeting and binding of emerin at the inner nuclear membrane. J Cell Sci. 1999;112:2571–2582. doi: 10.1242/jcs.112.15.2571. [DOI] [PubMed] [Google Scholar]
- 49.Zhang J, Kalkum M, Chait BT, Roeder RG. The N-CoR-HDAC3 nuclear receptor corepressor complex inhibits the JNK pathway through the integral subunit GPS2. Mol Cell. 2002;9:611–623. doi: 10.1016/s1097-2765(02)00468-9. [DOI] [PubMed] [Google Scholar]
- 50.Underhill C, Qutob MS, Yee SP, Torchia J. A novel nuclear receptor corepressor complex, N-CoR, contains components of the mammalian SWI/SNF complex and the corepressor KAP-1. J Biol Chem. 2000;275:40463–40470. doi: 10.1074/jbc.M007864200. [DOI] [PubMed] [Google Scholar]
- 51.Hartman HB, Yu J, Alenghat T, Ishizuka T, Lazar MA. The histone-binding code of nuclear receptor co-repressors matches the substrate specificity of histone deacetylase 3. EMBO Rep. 2005;6:445–451. doi: 10.1038/sj.embor.7400391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Spector DL. The dynamics of chromosome organization and gene regulation. Annu Rev Biochem. 2003;72:573–608. doi: 10.1146/annurev.biochem.72.121801.161724. [DOI] [PubMed] [Google Scholar]
- 53.Mislow JM, Holaska JM, Kim MS, Lee KK, Segura-Totten M, Wilson KL, McNally EM. Nesprin-1alpha self-associates and binds directly to emerin and lamin A in vitro. FEBS Lett. 2002;525:135–140. doi: 10.1016/s0014-5793(02)03105-8. [DOI] [PubMed] [Google Scholar]
- 54.Wheeler MA, Davies JD, Zhang Q, Emerson LJ, Hunt J, Shanahan CM, Ellis JA. Distinct functional domains in nesprin-1alpha and nesprin-2beta bind directly to emerin and both interactions are disrupted in X-linked Emery-Dreifuss muscular dystrophy. Exp Cell Res. 2007 doi: 10.1016/j.yexcr.2007.03.025. [DOI] [PubMed] [Google Scholar]
- 55.Kong KY, Kedes L. Cytoplasmic nuclear transfer of the actin-capping protein tropomodulin. J Biol Chem. 2004;279:30856–30864. doi: 10.1074/jbc.M302845200. [DOI] [PubMed] [Google Scholar]
- 56.Stuven T, Hartmann E, Gorlich D. Exportin 6: a novel nuclear export receptor that is specific for profilin. actin complexes. EMBO J. 2003;22:5928–5940. doi: 10.1093/emboj/cdg565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Pendleton A, Pope B, Weeds A, Koffer A. Latrunculin B or ATP depletion induces cofilin-dependent translocation of actin into nuclei of mast cells. J Biol Chem. 2003;278:14394–14400. doi: 10.1074/jbc.M206393200. [DOI] [PubMed] [Google Scholar]
- 58.Nowak G, Pestic-Dragovich L, Hozak P, Philimonenko A, Simerly C, Schatten G, de Lanerolle P. Evidence for the presence of myosin I in the nucleus. J Biol Chem. 1997;272:17176–17181. doi: 10.1074/jbc.272.27.17176. [DOI] [PubMed] [Google Scholar]
- 59.Philimonenko VV, Zhao J, Iben S, Dingova H, Kysela K, Kahle M, Zentgraf H, Hofmann WA, de Lanerolle P, Hozak P, Grummt I. Nuclear actin and myosin I are required for RNA polymerase I transcription. Nat Cell Biol. 2004;6:1165–1172. doi: 10.1038/ncb1190. [DOI] [PubMed] [Google Scholar]
- 60.De La Cruz EM, Ostap EM. Relating biochemistry and function in the myosin superfamily. Curr Opin Cell Biol. 2004;16:61–67. doi: 10.1016/j.ceb.2003.11.011. [DOI] [PubMed] [Google Scholar]
- 61.Piekny A, Werner M, Glotzer M. Cytokinesis: welcome to the Rho zone. Trends Cell Biol. 2005;15:651–658. doi: 10.1016/j.tcb.2005.10.006. [DOI] [PubMed] [Google Scholar]
- 62.Delaunay J. Molecular basis of red cell membrane disorders. Acta Haematol. 2002;108:210–218. doi: 10.1159/000065657. [DOI] [PubMed] [Google Scholar]
- 63.Brakebusch C, Fassler R. The integrin-actin connection, an eternal love affair. EMBO J. 2003;22:2324–2333. doi: 10.1093/emboj/cdg245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sussman J, Stokoe D, Ossina N, Shtivelman E. Protein kinase B phosphorylates AHNAK and regulates its subcellular localization. J Cell Biol. 2001;154:1019–1030. doi: 10.1083/jcb.200105121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zastrow MS, Flaherty DB, Benian GM, Wilson KL. Nuclear titin interacts with A- and B-type lamins in vitro and in vivo. J Cell Sci. 2006;119:239–249. doi: 10.1242/jcs.02728. [DOI] [PubMed] [Google Scholar]
- 66.Davis DB, Delmonte AJ, Ly CT, McNally EM. Myoferlin, a candidate gene and potential modifier of muscular dystrophy. Hum Mol Genet. 2000;9:217–226. doi: 10.1093/hmg/9.2.217. [DOI] [PubMed] [Google Scholar]
- 67.Doherty KR, Cave A, Davis DB, Delmonte AJ, Posey A, Earley JU, Hadhazy M, McNally EM. Normal myoblast fusion requires myoferlin. Development. 2005;132:5565–5575. doi: 10.1242/dev.02155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kabe Y, Yamada J, Uga H, Yamaguchi Y, Wada T, Handa H. NF-Y is essential for the recruitment of RNA polymerase II and inducible transcription of several CCAAT box-containing genes. Mol Cell Biol. 2005;25:512–522. doi: 10.1128/MCB.25.1.512-522.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Maity SN, de Crombrugghe B. Role of the CCAAT-binding protein CBF/NF-Y in transcription. Trends Biochem Sci. 1998;23:174–178. doi: 10.1016/s0968-0004(98)01201-8. [DOI] [PubMed] [Google Scholar]
- 70.Mantovani R. The molecular biology of the CCAAT-binding factor NF-Y. Gene. 1999;239:15–27. doi: 10.1016/s0378-1119(99)00368-6. [DOI] [PubMed] [Google Scholar]
- 71.Schubert CMLR, de Vries CJ, Plasterk RH, Priess JR. MEX-5 and MEX-6 function to establish soma/germline asymmetry in early C. elegans embryos. Mol Cell. 2000;5:671–682. doi: 10.1016/s1097-2765(00)80246-4. [DOI] [PubMed] [Google Scholar]
- 72.Espada J, Ballestar E, Fraga MF, Villar-Garea A, Juarranz A, Stockert JC, Robertson KD, Fuks F, Esteller M. Human DNA methyltransferase 1 is required for maintenance of the histone H3 modification pattern. J Biol Chem. 2004;279:37175–37184. doi: 10.1074/jbc.M404842200. [DOI] [PubMed] [Google Scholar]
- 73.Jin J, Cai Y, Yao T, Gottschalk AJ, Florens L, Swanson SK, Gutierrez JL, Coleman MK, Workman JL, Mushegian A, Washburn MP, Conaway RC, Conaway JW. A mammalian chromatin remodeling complex with similarities to the yeast INO80 complex. J Biol Chem. 2005;280:41207–41212. doi: 10.1074/jbc.M509128200. [DOI] [PubMed] [Google Scholar]
- 74.Schinkmann K, Blenis J. Cloning and characterization of a human STE20-like protein kinase with unusual cofactor requirements. J Biol Chem. 1997;272:28695–28703. doi: 10.1074/jbc.272.45.28695. [DOI] [PubMed] [Google Scholar]
- 75.Lee JK, Hurwitz J. Isolation and characterization of various complexes of the minichromosome maintenance proteins of Schizosaccharomyces pombe. J Biol Chem. 2000;275:18871–18878. doi: 10.1074/jbc.M001118200. [DOI] [PubMed] [Google Scholar]
- 76.Lehner B, Sanderson CM. A protein interaction framework for human mRNA degradation. Genome Res. 2004;14:1315–1323. doi: 10.1101/gr.2122004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Chaudhri M, Scarabel M, Aitken A. Mammalian and yeast 14-3-3 isoforms form distinct patterns of dimers in vivo. Biochem Biophys Res Commun. 2003;300:679–685. doi: 10.1016/s0006-291x(02)02902-9. [DOI] [PubMed] [Google Scholar]
- 78.Benton R, Palacios IM, St Johnston D. Drosophila 14-3-3/PAR-5 is an essential mediator of PAR-1 function in axis formation. Dev Cell. 2002;3:659–671. doi: 10.1016/s1534-5807(02)00320-9. [DOI] [PubMed] [Google Scholar]
- 79.Bouwmeester T, Bauch A, Ruffner H, Angrand PO, Bergamini G, Croughton K, Cruciat C, Eberhard D, Gagneur J, Ghidelli S, Hopf C, Huhse B, Mangano R, Michon AM, Schirle M, Schlegl J, Schwab M, Stein MA, Bauer A, Casari G, Drewes G, Gavin AC, Jackson DB, Joberty G, Neubauer G, Rick J, Kuster B, Superti-Furga G. A physical and functional map of the human TNF-alpha/NF-kappa B signal transduction pathway. Nat Cell Biol. 2004;6:97–105. doi: 10.1038/ncb1086. [DOI] [PubMed] [Google Scholar]
- 80.Tzivion G, Gupta VS, Kaplun L, Balan V. 14-3-3 proteins as potential oncogenes. Semin Cancer Biol. 2006;16:203–213. doi: 10.1016/j.semcancer.2006.03.004. [DOI] [PubMed] [Google Scholar]
- 81.Sato S, Tomomori-Sato C, Parmely TJ, Florens L, Zybailov B, Swanson SK, Banks CA, Jin J, Cai Y, Washburn MP, Conaway JW, Conaway RC. A set of consensus mammalian mediator subunits identified by multidimensional protein identification technology. Mol Cell. 2004;14:685–691. doi: 10.1016/j.molcel.2004.05.006. [DOI] [PubMed] [Google Scholar]
- 82.Jin J, Smith FD, Stark C, Wells CD, Fawcett JP, Kulkarni S, Metalnikov P, O’Donnell P, Taylor P, Taylor L, Zougman A, Woodgett JR, Langeberg LK, Scott JD, Pawson T. Proteomic, functional, and domain-based analysis of in vivo 14-3-3 binding proteins involved in cytoskeletal regulation and cellular organization. Curr Biol. 2004;14:1436–1450. doi: 10.1016/j.cub.2004.07.051. [DOI] [PubMed] [Google Scholar]
- 83.Yoon HG, Chan DW, Huang ZQ, Li J, Fondell JD, Qin J, Wong J. Purification and functional characterization of the human N-CoR complex: the roles of HDAC3, TBL1 and TBLR1. EMBO J. 2003;22:1336–1346. doi: 10.1093/emboj/cdg120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Rachez C, Lemon BD, Suldan Z, Bromleigh V, Gamble M, Naar AM, Erdjument-Bromage H, Tempst P, Freedman LP. Ligand-dependent transcription activation by nuclear receptors requires the DRIP complex. Nature. 1999;398:824–828. doi: 10.1038/19783. [DOI] [PubMed] [Google Scholar]
- 85.Rachez C, Freedman LP. Mediator complexes and transcription. Curr Biol. 2001;13:274–280. doi: 10.1016/s0955-0674(00)00209-x. [DOI] [PubMed] [Google Scholar]
- 86.Johnson CN, Adkins NL, Georgel P. Chromatin remodeling complexes: ATP-dependent machines in action. Biochem Cell Biol. 2005;83:405–417. doi: 10.1139/o05-115. [DOI] [PubMed] [Google Scholar]
- 87.Nagy PL, Griesenbeck J, Kornberg RD, Cleary ML. A trithorax-group complex purified from Saccharomyces cerevisiae is required for methylation of histone H3. Proc Natl Acad Sci U S A. 2002;99:90–94. doi: 10.1073/pnas.221596698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Andres ME, Burger C, Peral-Rubio MJ, Battaglioli E, Anderson ME, Grimes J, Dallman J, Ballas N, Mandel G. CoREST: a functional corepressor required for regulation of neural-specific gene expression. Proc Natl Acad Sci U S A. 1999;96:9873–9878. doi: 10.1073/pnas.96.17.9873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.You A, Tong JK, Grozinger CM, Schreiber SL. CoREST is an integral component of the CoREST- human histone deacetylase complex. Proc Natl Acad Sci U S A. 2001;98:1454–1458. doi: 10.1073/pnas.98.4.1454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Zhang P, Spardling AC. Efficient and dispersed local P element transposition from Drosophila females. Genetics. 1993;133:361–373. doi: 10.1093/genetics/133.2.361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Luo J, Su F, Chen D, Shiloh A, Gu W. Deacetylation of p53 modulates its effect on cell growth and apoptosis. Nature. 2000;408:377–381. doi: 10.1038/35042612. [DOI] [PubMed] [Google Scholar]
- 92.Harikrishnan KN, Chow MZ, Baker EK, Pal S, Bassal S, Brasacchio D, Wang L, Craig JM, Jones PL, Sif S, El-Osta A. Brahma links the SWI/SNF chromatin-remodeling complex with MeCP2-dependent transcriptional silencing. Nat Genet. 2005;37:254–264. doi: 10.1038/ng1516. [DOI] [PubMed] [Google Scholar]
- 93.Zhang Y, Iratni R, Erdjument-Bromage H, Tempst P, Reinberg D. Histone deacetylases and SAP18, a novel polypeptide, are components of a human Sin3 complex. Cell. 1997;89:357–364. doi: 10.1016/s0092-8674(00)80216-0. [DOI] [PubMed] [Google Scholar]
- 94.Xue Y, Wong J, Moreno GT, Young MK, Cote J, Wang W. NURD, a novel complex with both ATP-dependent chromatin-remodeling and histone deacetylase activities. Mol Cell. 1998;2:851–861. doi: 10.1016/s1097-2765(00)80299-3. [DOI] [PubMed] [Google Scholar]
- 95.Somech R, Shaklai S, Geller O, Amariglio N, Simon AJ, Rechavi G, Gal-Yam EN. The nuclear-envelope protein and transcriptional repressor LAP2beta interacts with HDAC3 at the nuclear periphery, and induces histone H4 deacetylation. J Cell Sci. 2005;118:4017–4025. doi: 10.1242/jcs.02521. [DOI] [PubMed] [Google Scholar]
- 96.Busson M, Carazo A, Seyer P, Grandemange S, Casas F, Pessemesse L, Rouault JP, Wrutniak-Cabello C, Cabello G. Coactivation of nuclear receptors and myogenic factors induces the major BTG1 influence on muscle differentiation. Oncogene. 2005;24:1698–1710. doi: 10.1038/sj.onc.1208373. [DOI] [PubMed] [Google Scholar]
- 97.Ye Q, Worman HJ. Interaction between an integral protein of the nuclear envelope inner membrane and human chromodomain proteins homologous to Drosophila HP1. J Biol Chem. 1996;271:14653–14656. doi: 10.1074/jbc.271.25.14653. [DOI] [PubMed] [Google Scholar]
- 98.Fischle W, Tseng BS, Dormann HL, Ueberheide BM, Garcia BA, Shabanowitz J, Hunt DF, Funabiki H, Allis CD. Regulation of HP1-chromatin binding by histone H3 methylation and phosphorylation. Nature. 2005;438:1116–1122. doi: 10.1038/nature04219. [DOI] [PubMed] [Google Scholar]
- 99.Jacobs SA, Khorasanizadeh S. Structure of HP1 chromodomain bound to a lysine 9-methylated histone H3 tail. Science. 2002;295:2080–2083. doi: 10.1126/science.1069473. [DOI] [PubMed] [Google Scholar]
- 100.Dharmawardana PG, Peruzzi B, Giubellino A, Burke TR, Jr, Bottaro DP. Molecular targeting of growth factor receptor-bound 2 (Grb2) as an anti-cancer strategy. Anticancer Drugs. 2006;17:13–20. doi: 10.1097/01.cad.0000185180.72604.ac. [DOI] [PubMed] [Google Scholar]
- 101.Cai M, Huang Y, Ghirlando R, Wilson KL, Craigie R, Clore GM. Solution structure of the constant region of nuclear envelope protein LAP2 reveals two LEM-domain structures: one binds BAF and the other binds DNA. EMBO J. 2001;20:4399–4407. doi: 10.1093/emboj/20.16.4399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Cai M, Huang Y, Suh JY, Louis JM, Ghirlando R, Craigie R, Clore GM. Solution NMR Structure of the Barrier-to-Autointegration Factor-Emerin Complex. J Biol Chem. 2007;282:14525–14535. doi: 10.1074/jbc.M700576200. [DOI] [PubMed] [Google Scholar]
- 103.Wolff N, Gilquin B, Courchay K, Callebaut I, Worman HJ, Zinn-Justin S. Structural analysis of emerin, an inner nuclear membrane protein mutated in X-linked Emery-Dreifuss muscular dystrophy. FEBS Lett. 2001;501:171–176. doi: 10.1016/s0014-5793(01)02649-7. [DOI] [PubMed] [Google Scholar]
- 104.Roberts RC, Sutherland-Smith AJ, Wheeler MA, Jensen ON, Emerson LJ, Spiliotis, Tate CG, Kendrick-Jones J, Ellis JA. The Emery-Dreifuss muscular dystrophy associated-protein emerin is phosphorylated on serine 49 by protein kinase A. FEBS J. 2006;273:4562–4575. doi: 10.1111/j.1742-4658.2006.05464.x. [DOI] [PubMed] [Google Scholar]
- 105.Amanchy R, Kalume DE, Iwahori A, Zhong J, Pandey A. Phosphoproteome analysis of HeLa cells using stable isotope labeling with amino acids in cell culture (SILAC) J Proteome Res. 2005;4:1661–1671. doi: 10.1021/pr050134h. [DOI] [PubMed] [Google Scholar]
- 106.Schlosser A, Amanchy R, Otto H. Identification of tyrosine-phosphorylation sites in the nuclear membrane protein emerin. FEBS J. 2006;273:3204–3215. doi: 10.1111/j.1742-4658.2006.05329.x. [DOI] [PubMed] [Google Scholar]
- 107.Hirano Y, Segawa M, Ouchi FS, Yamakawa Y, Furukawa K, Takeyasu K, Horigome T. Dissociation of emerin from barrier-to-autointegration factor is regulated through mitotic phosphorylation of emerin in a xenopus egg cell-free system. J Biol Chem. 2005;280:39925–39933. doi: 10.1074/jbc.M503214200. [DOI] [PubMed] [Google Scholar]
- 108.Schirmer EC, Florens L, Guan T, Yates JR, 3rd, Gerace L. Nuclear membrane proteins with potential disease links found by subtractive proteomics. Science. 2003;301:1380–1382. doi: 10.1126/science.1088176. [DOI] [PubMed] [Google Scholar]
- 109.Liu J, Lee KK, Segura-Totten M, Neufeld E, Wilson KL, Gruenbaum Y. MAN1 and emerin have overlapping function(s) essential for chromosome segregation and cell division in Caenorhabditis elegans. Proc Natl Acad Sci U S A. 2003;100:4598–4603. doi: 10.1073/pnas.0730821100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Nili E, Cojocaru GS, Kalma Y, Ginsberg D, Copeland NG, Gilbert DJ, Jenkins NA, Berger R, Shaklai S, Amariglio N, Brok-Simoni F, Simon AJ, Rechavi G. Nuclear membrane protein LAP2beta mediates transcriptional repression alone and together with its binding partner GCL (germ-cell-less) J Cell Sci. 2001;114:3297–3307. doi: 10.1242/jcs.114.18.3297. [DOI] [PubMed] [Google Scholar]
- 111.Frock RL, Kudlow BA, Evans AM, Jameson SA, Hauschka SD, Kennedy BK. Lamin A/C and emerin are critical for skeletal muscle satellite cell differentiation. Genes Dev. 2006;20:486–500. doi: 10.1101/gad.1364906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Bakay M, Wang Z, Melcon G, Schiltz L, Xuan J, Zhao P, Sartorelli V, Seo J, Pegoraro E, Angelini C, Shneiderman B, Escolar D, Chen YW, Winokur ST, Pachman LM, Fan C, Mandler R, Nevo Y, Gordon E, Zhu Y, Dong Y, Wang Y, Hoffman EP. Nuclear envelope dystrophies show a transcriptional fingerprint suggesting disruption of Rb-MyoD pathways in muscle regeneration. Brain. 2006;129:996–1013. doi: 10.1093/brain/awl023. [DOI] [PubMed] [Google Scholar]
- 113.Semenova E, Wang X, Jablonski MM, Levorse J, Tilghman SM. An engineered 800 kilobase deletion of Uchl3 and Lmo7 on mouse chromosome 14 causes defects in viability, postnatal growth and degeneration of muscle and retina. Hum Mol Genet. 2003;12:1301–1312. doi: 10.1093/hmg/ddg140. [DOI] [PubMed] [Google Scholar]
- 114.Tzur YB, Wilson KL, Gruenbaum Y. SUN-domain proteins: ‘Velcro’ that links the nucleoskeleton to the cytoskeleton. Nat Rev Mol Cell Biol. 2006;7:782–788. doi: 10.1038/nrm2003. [DOI] [PubMed] [Google Scholar]