The findings of this study indicate that CD56 is expressed on a subset of adult T-cell acute lymphoblastic leukemia cells with distinct immunophenotypic features and greater resistance to therapy. See related perspective article on page 160.
Keywords: T-cell ALL, CD56, natural killer, immunophenotype
Abstract
Background
Expression of CD56 has been associated with poor prognosis in acute myeloid leukemia and aggressive lymphoma.
Design and Methods
We analyzed the impact of CD56 expression in a cohort of 452 newly diagnosed adult T-cell acute lymphoblastic leukemia (T-ALL) patients; clinical data were available for 306 patients. Treatment was according to the GMALL study protocols 06/99 and 07/03 stipulating stratification into standard (thymic T-ALL) and high risk (pre- and mature T-ALL) groups.
Results
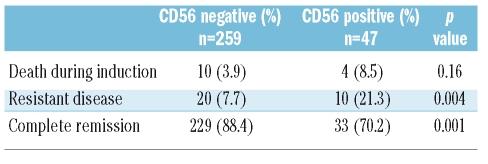
CD56 expression was detected in 63/452 (13.9%) patients. CD56+ T-ALL were predominantly of non-thymic (pre-T 35%, mature 41%) immunophenotypic subtypes, whereas 53% of the CD56− cases were thymic T-ALL (p=0.00002). CD13, CD33, CD34 and HLA-DR were significantly more frequently expressed. A clonal T-cell receptor rearrangement was detected in 22/23 CD56+ ALL. No major clinical differences were observed at presentation. Treatment of CD56+ ALL resulted in a lower rate of complete remissions (70% vs. 88%) (p=0.001) and a higher rate of resistant disease (21% vs. 8%) (p=0.004). CD56 expression had no significant influence on overall (48% vs. 59%) and disease free survival (67% vs. 57%) at three years.
Conclusions
CD56 is expressed on a subset of adult T-ALL with distinct immunophenotypical features and higher resistance to therapy. Most CD56+ ALL were treated in the high-risk arm of the GMALL study protocols owing to their non-thymic phenotype. Thus after risk adapted treatment a prognostic impact of CD56 expression was not detectable.
Introduction
CD56 (neural-cell adhesion molecule; NCAM) is a marker for natural killer (NK) cells and is also expressed on a subset of normal T cells. However, neoplastic myeloid,1,2 lymphoid, plasmacytoid dendritic3 or myeloma4 cells can also express CD56.
Expression of CD56 has been shown to impact prognosis in different hematologic malignancies. In acute myeloid leukemia with t(15;17) and t(8;21) as well as in anaplastic large cell lymphoma it was associated with poor clinical outcome.5–7 In T-cell acute lymphoblastic leukemia/lymphoma (T-ALL/LBL) CD56 expression has been reported for a small number of cases and an association with worse outcome has been suggested.8–11 The expression of other single antigens such as CD2, CD10 or myeloic antigens has been evaluated for their prognostic impact in T-ALL.12–14 However, the immunological subtype is still the leading prognostic factor, with thymic T-ALL having a considerably better prognosis than pre-T or mature T-ALL.15,16
As the central immunocytological reference laboratory for the prospective GMALL therapy trials we could take advantage of receiving a large number of leukemia samples. Therefore, we tried to answer the question whether CD56+ T-ALL forms a distinct subgroup concerning clinicopathological presentation, differences in immunophenotype and genotype as compared to other T-ALL subgroups. We also determined the impact of CD56 expression on treatment response and survival. In addition, by analyzing the immunophenotypical data, we tried to gain more information on a putative common T/NK precursor cell of origin for this ALL subtype.
Design and Methods
Patients
Between November 1999 and September 2006 pre-treatment peripheral blood or bone marrow specimens were obtained from 452 adult (≥15 years of age) patients. Diagnosis of ALL was confirmed at the central cytological laboratory according to the French-American-British criteria17 and by immunophenotyping at the central GMALL reference laboratory in Berlin. Patients fulfilling the entry criteria (amongst others: age 15–65 years, ≥25% bone marrow infiltration) were treated within the GMALL therapy trials 06/1999 and 07/2003. Details of the treatment protocols have been published elsewhere.18,19 Treatment of patients with T-cell ALL was stratified into a standard (CD1-positive, thymic T-ALL) and a high-risk group (pre-T-ALL and mature T-ALL). Until the first amendment in October 2000, hyperleukocytosis (>100×109/L) was considered an additional poor prognostic factor, leading to high-risk allocation of 10 patients with thymic T-ALL, 2 of whom were CD56-positive. A hematopoietic stem cell transplantation (SCT) (allogeneic or autologous, depending on availability of a donor) in first complete remission (CR) was recommended for patients in the high-risk group. A SCT in the standard risk arm, i.e. thymic T-ALL, was also possible in case of a level of minimal residual disease (MRD) >10−4 after induction therapy and first CR as assessed by quantitative real-time PCR of clone specific markers. Methods and frequency of MRD assessment have been reported recently.18 Five patients with CD56− thymic T-ALL were transplanted due to an unfavorable course of MRD.
The protocols have been approved by the ethics committees of the participating study clinics and all patients had given their consent to the scientific use of their data and the use of residual sample material obtained during diagnostic procedures for scientific purposes.
Immunophenotyping
Methods used for immunophenotyping and classification of ALL subtypes were applied as previously described in detail.15,20,21 Briefly, ALL samples were immunologically classified as pre-T cell (cyCD3+, CD7+, CD5+/−, CD2−/+, CD1a−, CD4−, CD8−, sCD3−, thymic T cell (cyCD3+, CD7+, CD5+/−, CD2+/−, CD1a+, CD4+/−, CD8+/−, sCD3−/+), or mature T cell (cyCD3+, CD7+, CD5+/−, CD2+, CD1a−, CD4+/−, CD8−/+, sCD3+). Furthermore, expression of the markers CD56, CD13, CD33, CD10, CD117, TCR α/β, TCR γ/δ, CD34 and HLA-DR was investigated. Expression of a marker on ≥20% of the lymphatic blasts was defined as positive.
Cytogenetic and molecular genetic analysis
Cytogenetic analysis and fluorescence in situ hybridization (FISH) were performed before start of therapy. Methods for chromosome analysis, G-banding and FISH analyses, as well as classification of chromosome abnormalities according to the International System for Human Cytogenetic Nomenclature, have been published previously.22,23 Samples were also tested for clonal T-cell receptor (TCR) gene rearrangement at the TCR β, γ and δ locus using real-time quantitative PCR as previously described.24,25 Both cytogenetic and molecular genetic analyses were performed in a central reference laboratory.
Statistical analysis
CD56 subgroups were tested for the clinical variables sex, age, WBC, hemoglobin level, platelet count; presence or absence of hepatomegaly, splenomegaly, mediastinal tumor and lymphadenopathy and infiltration of the central nervous system (CNS), for the distribution of immunological subtypes as well as for abberant antigen expression. A descriptive analysis of cytogenetics and molecular biology was performed. The characteristics of the patients and their response to treatment were compared by χ2 and Wilcoxon rank-sum tests. All tests were 2-sided, and adjusted for multiple testing with a p-value of less than 0.01 indicating a statistically significant difference.
Remission duration was calculated as time from achievement of CR to time of relapse, death in CR, SCT or last follow-up; patients with continuous complete remission, death in CR or SCT in first CR were censored. Survival was calculated from time of diagnosis to death or last follow-up. Curves for survival and remission duration were constructed using the Kaplan-Meier method, with differences compared by the log-rank test. Statistical analyses were performed using the SAS software, release 8.02 and the SPSS software package for Windows, release 14.
Results
Patients’ characteristics
Of 452 patients analyzed by flow cytometry, 344 were included in the GMALL trials 06/99 and 07/03; the remaining patients were treated in different protocols or lost to follow-up. At the time-point of our analysis 306 patients were evaluable for outcome; their characteristics are given in Table 1. Of those patients, 47 (15.4%) had CD56-positive T-ALL. One-hundred and seventy-four patients were treated in the 06/99 study and 132 in the 07/03 study with no significant differences in the proportion of CD56+ cases.
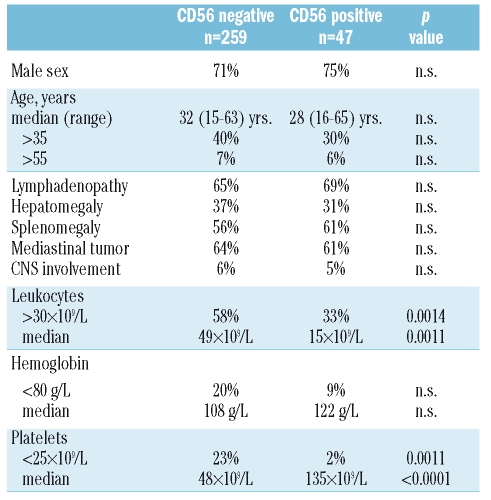
Table 1.
Pre-treatment characteristics.
There were no significant differences in both the CD56+ and CD56− subgroups concerning sex, median age, age distribution or clinical presentation in terms of lymphadenopathy, hepatomegaly, splenomegaly, mediastinal tumor or CNS spread. A high proportion of patients in both groups presented with lymphadenopathy or splenomegaly, whereas CNS infiltration at presentation was infrequent.
CD56+ T-ALL presented with lower leukocyte counts and less frequently with leukocytes >30×109/L, and also with higher platelet counts compared to the CD56− cases. These differences were statistically significant (Table 1).
Immunophenotype
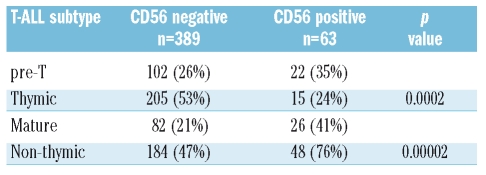
Pre-treatment immunophenotypical data were available for 452 patients. CD56 expression (≥20% of blasts) was detected in 63 of these patients (13.9%). The level of CD56 expression (i.e. the percentage of CD56+ blasts) had a steady distribution within the CD56+ group, therefore the existence of further distinct subgroups with especially high or low levels of CD56 expression was considered unlikely. The median proportion of CD56+ blasts was 62% in CD56+ T-ALL and 2% in the negative group. CD56 expression was seen in all subtypes of TALL, however, a significant difference in distribution of CD56+ T-ALL over the subgroups was detected (Table 2). Whereas CD1+ thymic T-ALL comprised the largest subgroup of CD56− T-ALL (53%), in CD56+ T-ALL the thymic subtype was rather rare (24%). Thus, non-thymic phenotypes (mature or pre-T) were the dominant subtypes in CD56+ T-ALL (p=0.00002).
Table 2.
CD56 expression in T-ALL subtypes.
A separate analysis of CD56 expression in T-ALL subgroups defined according to the European Group for the Immunological Characterization of Acute Leukemias (EGIL),26 where mature T-ALL is defined solely by its surface CD3 expression, showed again a significant difference for the thymic subgroup, but not for pro-, pre- or mature T-ALL (data not shown).
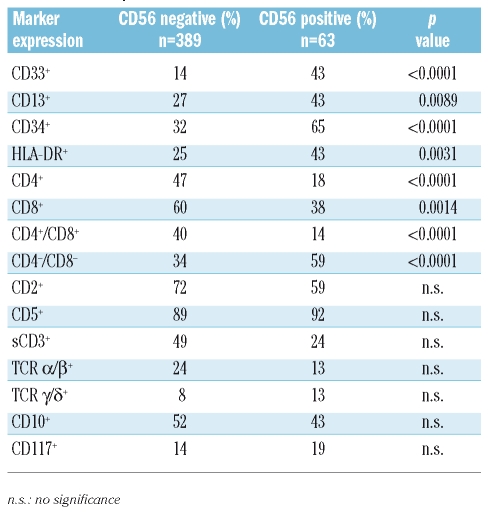
The myeloid markers CD33 and CD13, as well as CD34 and HLA-DR, were significantly more frequently expressed in the CD56+ group. CD56+ T-ALL was more often negative for CD4, CD8 or double negative. There were no significant differences in the expression of CD10, CD117, CD2, CD5, surface CD3 (sCD3) or the TCR molecules (Table 3).
Table 3.
Marker profile of T-ALL.
In 10 T-ALL cases (5 CD56+ including two pre-T, 1 mature and 2 thymic T-ALL) we also looked for the expression of further natural killer cell associated markers. Thus, expression of CD161, CD94, NKG2D, CD158a, CD158b and NKp46 was investigated by flow cytometry. All cases tested were negative for these markers (usually less than 5% of positive cells) except one case of a mature, sCD3 expressing CD56+ ALL, which exhibited partial expression (27%) of NKG2D.
Cytogenetic and molecular biological analysis
Cytogenetic data were available for six CD56+ cases. Four had a normal karyotype and two had aberrations involving the chromosomes 1, 5, 7, 12 and 13 [t(7;13), del 1(p), del 5(q), dic (7;12)] including one case with hypodiploid karyotype.
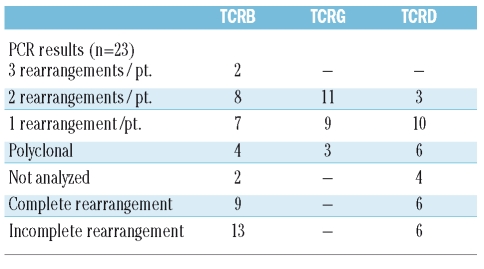
Twenty-three patients with CD56+ T-ALL could be evaluated for clonal TCR rearrangement. In some cases not all PCR reactions were performed or sequencing of the specific rearrangement was omitted due to scant material. Results of the molecular biological analysis are given in Table 4. All except one case had rearranged TCR genes: in 11 cases clonality in all three TCR loci was detected, in an additional 11 cases in one or two TCR loci. In one case a single incomplete TCRD (Dd2Dd3) rearrangement was found only. TCRB rearrangements were complete in 9 and incomplete in 13 patients, TCRD rearrangements were complete in 6 and incomplete in 6 patients. The usage of particular gene segments was heterogeneous and is not given in detail. In TCRB rearrangements Jb2.3 and Db2 were most frequently used; Vd1-Jd1 was the most frequently observed TCRD rearrangement.
Table 4.
TCR rearrangements in 23 CD56+ T-ALL.
Treatment results
Treatment results and follow-up data of 306 patients (259 CD56− and 47 CD56+ T-ALL) were available for analysis. Results after induction chemotherapy and follow-up of responders are given in Table 5. The CR rate was significantly lower (70.2%) in the CD56+ group accompanied by a higher rate of induction failure (21%) compared to CD56− cases. Of those patients who achieved a CR, 71 CD56− and 20 CD56+ patients received a hematopoietic stem cell transplantation (31% vs. 60.6%), thus the proportion of transplanted CD56+ patients was significantly higher (p=0.01). There were no significant differences in terms of death either during induction or in CR.
Table 5.
Response to induction therapy and clinical course of responders.
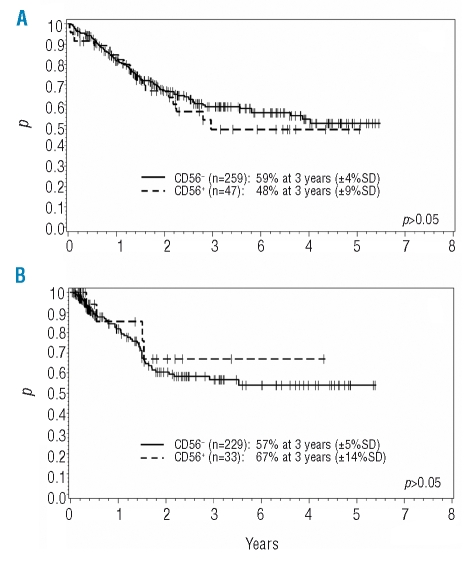
After a median follow-up of 31 months we observed 60 ALL relapses, and relapse rates were similar in both subgroups. There were 8 relapses including or confined to the CNS; all were CD56− (p=0.4). Furthermore, we did not find a significant influence of CD56 expression on overall survival (OAS) nor on remission duration. The 3-year probability of OAS for the CD56− group was 59% compared to 48% for CD56+ T-ALL; the probability of continuous CR at three years was 57% vs. 67% (Figure 1). An analysis of T-ALL subroups (pre-T, thymic, and mature) concerning OAS and remission duration was also not able to reveal a significant influence of CD56 expression.
Figure 1.
Overall survival in adult T-ALL according to CD56-expression, probability of survival at three years (A), and remission duration in adult T-ALL according to CD56-expression; probability of continuing complete remission at three years (B).
Discussion
In this study we investigated a large number of patients with adult T-ALL diagnosed in our reference immunocytological laboratory and treated in the GMALL trial concerning presence of CD56 expression. CD56+ T-ALL comprised a small subgroup in our collective. Of 452 newly diagnosed T-ALL patients analyzed, 13.9% did express CD56. Expression was detected in all maturational subgroups, but there was a significant predominance of non-thymic subtypes (76%), whereas in the CD56− group thymic T-ALL was the most common subtype (53%).
CD56 expression has been reported for only a small number of ALL cases so far. The Eastern Cooperative Oncology Group (ECOG) gave a descriptive analysis of 4 CD56+ cases out of a series of 107 T-ALL.10 Montero et al. gave an immunophenotypical characterization of 4 patients with CD56+ T-ALL, and Onishi et al. described the clinical characteristics of 5 cases of CD56+ T-ALL/LBL.8,9 Ravandi et al. reported a series of 16 patients out of 200 adult ALL cases.11
In these studies, some influence of CD56 expression on clinical presentation has been suggested, such as predominance of systemic lymphadenopathy in the absence of a mediastinal tumor or a correlation with CNS involvement.9,11
In our study CD56+ T-ALL exhibited few differences at presentation, namely a lower white blood count and higher platelet levels. For all other characteristics no difference was found. Interestingly, there was no difference concerning CNS involvement. CD56 expression has been associated with infiltration of extranodal sites including the CNS in lymphoma27 and a higher rate of CNS involvement in T-ALL.11 We could not confirm these findings in our large series. The association with CNS disease found in the study of Ravandi et al. might be explained by a generally higher risk of CNS infiltration in T-cell ALL,15,28 which were most probably accounted for by the CD56+ cases in this report. There, ALL were only distinguished according to their CD56 expression without reporting the frequency of B- and T-lineage ALL or the immunological subtypes.11
The expression of CD56 has been shown to be of prognostic relevance in certain hematologic malignancies.5–7,29 In the small series of Montero et al., an inferior survival of CD56+ T-ALL was reported.8 Therefore, we analyzed the impact of CD56 expression on outcome in our series.
In the GMALL trial, T-ALL patients have been treated either in a standard or high-risk arm, immunophenotype (thymic or non-thymic) being the main risk factor for stratification. Only very few patients were allocated to a high-risk arm due to other factors such as MRD. Therapy intensification for high-risk patients was usually by means of hematopoietic SCT after induction and consolidation therapy.
After the induction phase significantly less CD56+ patients achieved a complete remission, and significantly more CD56+ patients received a SCT. However, the overall survival of adult T-ALL was not significantly influenced by CD56 expression. After three years the probability of survival was comparable in the CD56+ and the CD56− group.
The differences in remission rates might be attributable to the strong association of CD56 expression with non-thymic subtypes. This association also explains the frequent application of SCT in CD56+ patients according to high-risk stratification for non-thymic subtypes in the GMALL protocol. These findings ultimately confirm the inferior prognosis of early and mature T-ALL which can be improved by therapy intensification according to the GMALL protocol. An additional negative prognostic impact of CD56 expression cannot be supported from our data. However, only a therapy trial without stratification according to GMALL subtypes would allow a definitive conclusion to be drawn.
In our study we also analyzed the immunophenotype of CD56+ T-ALL. We detected an accumulation of coexpression of myeloid markers and CD34 as well as a significant bypassing of the thymic subtype in conjunction with a lower rate of CD4 and CD8 (significant) and surface CD3 (not significant) expression. CD56 was differentially expressed on T-ALL subtypes and one might speculate on a common precursor with a mixed T- and NK-differentiation.
T and NK cells develop from a bipotent T/NK progenitor (CD34+/CD33+/CD13+/CD7+/CD2+/−). TCR gene rearrangement determines T differentiation in this context whereas TCR genes in NK progenitors remain in germline. CD56 expression then normally appears only on more mature NK cells.30 Almost all of our CD56+ T-ALL cases showed rearranged TCR genes. The use of TCR gene segments in rearrangement revealed no distinctive pattern. The frequency of complete and incomplete rearrangements and the number of detectable rearrangements per patient were not different to what one would expect from T-ALL.24,25 CD56+ and CD56− T-ALL were also tested for the expression of further NK-associated markers such as CD161, KIR, CD94, NKG2D or NKp46,31 whereas CD161 has been shown to be an early marker expressed on lymphoblastic lymphomas of true NK lineage.32 No expression of these markers could be detected on both T-ALL subtypes. Taken together these findings cannot support the hypothesis of CD56+ T-ALL directly arising from a common T/NK progenitor. However, the mechanisms leading to CD56 expression after TCR rearrangement remain unclear.
In conclusion, CD56 is expressed differentially on adult T-ALL with additional distinct immunophenotypical features but does not correlate fundamentally with a different clinical presentation. Response to current therapy is worse in CD56+ T-ALL, mostly due to frequent non-thymic subtype. However, outcome is similar to CD56− T-ALL after therapy within the GMALL protocols.
Footnotes
A complete list of the members of the German Multicenter Adult ALL study group appears in the Online Supplementary Appendix.
Authorship and Disclosures
LF, NG, SS, TB, HR, MB, DH, ET: conception and design, or analysis and interpretation of data; LF, NG, SS, TB, HR, MB, DH, ET: drafting the article or revising it critically for important intellectual content; LF, NG, DH, ET: final approval of the version to be published.
The authors reported no potential conflicts of interest.
References
- 1.Seymour JF, Pierce SA, Kantarjian HM, Keating MJ, Estey EH. Investigation of karyotypic, morphologic and clinical features in patients with acute myeloid leukemia blast cells expressing the neural cell adhesion molecule ( CD56) Leukemia. 1994;8:823–6. [PubMed] [Google Scholar]
- 2.Suzuki R, Yamamoto K, Seto M, Kagami Y, Ogura M, Yatabe Y, et al. CD7+ and CD56+ myeloid/natural killer cell precursor acute leukemia: a distinct hematolymphoid disease entity. Blood. 1997;90:2417–28. [PubMed] [Google Scholar]
- 3.Chaperot L, Bendriss N, Manches O, Gressin R, Maynadie M, Trimoreau F, et al. Identification of a leukemic counterpart of the plasmacytoid dendritic cells. Blood. 2001;97:3210–7. doi: 10.1182/blood.v97.10.3210. [DOI] [PubMed] [Google Scholar]
- 4.Van Camp B, Durie BG, Spier C, De Waele M, Van Riet I, Vela E, et al. Plasma cells in multiple myeloma express a natural killer cell-associated antigen: CD56 (NKH-1; Leu-19) Blood. 1990;76:377–82. [PubMed] [Google Scholar]
- 5.Baer MR, Stewart CC, Lawrence D, Arthur DC, Byrd JC, Davey FR, et al. Expression of the neural cell adhesion molecule CD56 is associated with short remission duration and survival in acute myeloid leukemia with t(8;21)(q22;q22) Blood. 1997;90:1643–8. [PubMed] [Google Scholar]
- 6.Ferrara F, Morabito F, Martino B, Specchia G, Liso V, Nobile F, et al. CD56 expression is an indicator of poor clinical outcome in patients with acute promyelocytic leukemia treated with simultaneous all-transretinoic acid and chemotherapy. J Clin Oncol. 2000;18:1295–300. doi: 10.1200/JCO.2000.18.6.1295. [DOI] [PubMed] [Google Scholar]
- 7.Suzuki R, Kagami Y, Takeuchi K, Kami M, Okamoto M, Ichinohasama R, et al. Prognostic significance of CD56 expression for ALK-positive and ALK-negative anaplastic large-cell lymphoma of T/null cell phenotype. Blood. 2000;96:2993–3000. [PubMed] [Google Scholar]
- 8.Montero I, Rios E, Parody R, Perez-Hurtado JM, Martin-Noya A, Rodriguez JM. CD56 in T-cell acute lymphoblastic leukaemia: a malignant transformation of an early myeloid-lymphoid progenitor? Haematologica. 2003;88:ELT26. [PubMed] [Google Scholar]
- 9.Onishi Y, Matsuno Y, Tateishi U, Maeshima AM, Kusumoto M, Terauchi T, et al. Two entities of precursor T-cell lymphoblastic leukemia/lymphoma based on radiologic and immunophenotypic findings. Int J Hematol. 2004;80:43–51. doi: 10.1532/ijh97.04061. [DOI] [PubMed] [Google Scholar]
- 10.Paietta E, Neuberg D, Richards S, Bennett JM, Han L, Racevskis J, et al. Rare adult acute lymphocytic leukemia with CD56 expression in the ECOG experience shows unexpected phenotypic and genotypic heterogeneity. Eastern Cooperative Oncology Group. Am J Hematol. 2001;66:189–96. doi: 10.1002/1096-8652(200103)66:3<189::aid-ajh1043>3.0.co;2-a. [DOI] [PubMed] [Google Scholar]
- 11.Ravandi F, Cortes J, Estrov Z, Thomas D, Giles FJ, Huh YO, et al. CD56 expression predicts occurrence of CNS disease in acute lymphoblastic leukemia. Leuk Res. 2002;26:643–9. doi: 10.1016/s0145-2126(01)00188-6. [DOI] [PubMed] [Google Scholar]
- 12.Uckun FM, Steinherz PG, Sather H, Trigg M, Arthur D, Tubergen D, et al. CD2 antigen expression on leukemic cells as a predictor of event-free survival after chemotherapy for T-lineage acute lymphoblastic leukemia: a Children’s Cancer Group study. Blood. 1996;88:4288–95. [PubMed] [Google Scholar]
- 13.Boucheix C, David B, Sebban C, Racadot E, Bené MC, Bernard A, et al. Immunophenotype of adult acute lymphoblastic leukemia, clinical parameters, and outcome: an analysis of a prospective trial including 562 tested patients (LALA87). French Group on Therapy for Adult Acute Lymphoblastic Leukemia. Blood. 1994;84:1603–12. [PubMed] [Google Scholar]
- 14.Vitale A, Guarini A, Ariola C, Meloni G, Perbellini O, Pizzuti M, et al. Absence of prognostic impact of CD13 and/or CD33 antigen expression in adult acute lymphoblastic leukemia. Results of the GIMEMA ALL 0496 trial. Haematologica. 2007;92:342–8. doi: 10.3324/haematol.10385. [DOI] [PubMed] [Google Scholar]
- 15.Thiel E, Kranz BR, Raghavachar A, Bartram CR, Löffler H, Messerer D, et al. Prethymic phenotype and genotype of pre-T (CD7+/ER−)-cell leukemia and its clinical significance within adult acute lymphoblastic leukemia. Blood. 1989;73:1247–58. [PubMed] [Google Scholar]
- 16.Hoelzer D, Arnold R, Freund M, et al. Characteristics, outcome and risk factors in adult T-lineage acute lymphoblastic leukemia (ALL) Blood. 1999;94:2926a. [Abstract] [Google Scholar]
- 17.Bennett JM, Catovsky D, Daniel MT, Flandrin G, Galton DA, Gralnick HR, et al. The morphological classification of acute lymphoblastic leukaemia: concordance among observers and clinical correlations. Br J Haematol. 1981;47:553–61. doi: 10.1111/j.1365-2141.1981.tb02684.x. [DOI] [PubMed] [Google Scholar]
- 18.Brüggemann M, Raff T, Flohr T, Gökbuget N, Nakao M, Droese J, et al. Clinical significance of minimal residual disease quantification in adult patients with standard-risk acute lymphoblastic leukemia. German Multicenter Study Group for Adult Acute Lymphoblastic Leukemia. Blood. 2006;107:1116–23. doi: 10.1182/blood-2005-07-2708. [DOI] [PubMed] [Google Scholar]
- 19.Raff T, Gökbuget N, Lüschen S, Reutzel R, Ritgen M, Irmer S, et al. Molecular relapse in adult standard-risk ALL patients detected by prospective MRD monitoring during and after maintenance treatment: data from the GMALL 06/99 and 07/03 trials. Blood. 2007;109:910–5. doi: 10.1182/blood-2006-07-037093. [DOI] [PubMed] [Google Scholar]
- 20.Ludwig WD, Rieder H, Bartram CR, Heinze B, Schwartz S, Gassmann W, et al. Immunophenotypic and genotypic features, clinical characteristics, and treatment outcome of adult proB acute lymphoblastic leukemia: results of the German multicenter trials GMALL 03/87 and 04/89. Blood. 1998;92:1898–909. [PubMed] [Google Scholar]
- 21.Burmeister T, Gökbuget N, Reinhardt R, Rieder H, Hoelzer D, Schwartz S. NUP214-ABL1 in adult T-ALL: the GMALL study group experience. Blood. 2006;108:3556–9. doi: 10.1182/blood-2006-04-014514. [DOI] [PubMed] [Google Scholar]
- 22.Fonatsch C, Schaadt M, Kirchner H, Diehl V. A possible correlation between the degree of karyotype aberrations and the rate of sister chromatid exchanges in lymphoma lines. Int J Cancer. 1980;26:749–56. doi: 10.1002/ijc.2910260608. [DOI] [PubMed] [Google Scholar]
- 23.Mitelmann F. ISCN An International System of Human Cytogenetic Nomenclature. Basel: Karger; 1995. [Google Scholar]
- 24.Brüggemann M, van der Velden VH, Raff T, Droese J, Ritgen M, Pott C, et al. Rearranged T-cell receptor β genes represent powerful targets for quantification of minimal residual disease in childhood and adult T-cell acute lymphoblastic leukemia. Leukemia. 2004;18:709–19. doi: 10.1038/sj.leu.2403263. [DOI] [PubMed] [Google Scholar]
- 25.Szczepaƒski T, van der Velden VH, Raff T, Jacobs DC, van Wering ER, Brüggemann M, et al. Comparative analysis of T-cell receptor gene rearrangements at diagnosis and relapse of T-cell acute lymphoblastic leukemia (T-ALL) shows high stability of clonal markers for monitoring of minimal residual disease and reveals the occurrence of second TALL. Leukemia. 2003;17:2149–56. doi: 10.1038/sj.leu.2403081. [DOI] [PubMed] [Google Scholar]
- 26.Bene MC, Castoldi G, Knapp W, Ludwig WD, Matutes E, Orfao A, et al. Proposals for the immunological classification of acute leukemias. European Group for the Immunological Characterization of Leukemias (EGIL) Leukemia. 1995;9:1783–6. [PubMed] [Google Scholar]
- 27.Kern WF, Spier CM, Hanneman EH, Miller TP, Matzner M, Grogan TM. Neural cell adhesion molecule-positive peripheral T-cell lymphoma: a rare variant with a propensity for unusual sites of involvement. Blood. 1992;79:2432–7. [PubMed] [Google Scholar]
- 28.Gokbuget N, Hoelzer D. Meningeosis leukaemica in adult acute lymphoblastic leukaemia. J Neurooncol. 1998;38:167–80. doi: 10.1023/a:1005963732481. [DOI] [PubMed] [Google Scholar]
- 29.Suzuki R, Murata M, Kami M, Ohtake S, Asou N, Kodera Y, et al. Prognostic significance of CD7+ CD56+ phenotype and chromosome 5 abnormalities for acute myeloid leukemia M0. Int J Hematol. 2003;77:482–9. doi: 10.1007/BF02986617. [DOI] [PubMed] [Google Scholar]
- 30.Sanchez MJ, Muench MO, Roncarolo MG, Lanier LL, Phillips JH. Identification of a common T/natural killer cell progenitor in human fetal thymus. J Exp Med. 1994;180:569–76. doi: 10.1084/jem.180.2.569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fischer L, Hummel M, Burmeister T, Schwartz S, Thiel E. Skewed expression of natural-killer (NK)-associated antigens on lymphoproliferations of large granular lymphocytes (LGL) Hematol Oncol. 2006;24:78–85. doi: 10.1002/hon.777. [DOI] [PubMed] [Google Scholar]
- 32.Lin CW, Liu TY, Chen SU, Wang KT, Medeiros LJ, Hsu SM. CD94 1A transcripts characterize lymphoblastic lymphoma/leukemia of immature natural killer cell origin with distinct clinical features. Blood. 2005;106:3567–74. doi: 10.1182/blood-2005-02-0519. [DOI] [PubMed] [Google Scholar]