Abstract
The assembly of β-barrel proteins into membranes is a fundamental process that is essential in Gram-negative bacteria, mitochondria and plastids. Our understanding of the mechanism of β-barrel assembly is progressing from studies carried out in Escherichia coli and Neisseria meningitidis. Comparative sequence analysis suggests that while many components mediating β-barrel protein assembly are conserved in all groups of bacteria with outer membranes, some components are notably absent. The Alphaproteobacteria in particular seem prone to gene loss and show the presence or absence of specific components mediating the assembly of β-barrels: some components of the pathway appear to be missing from whole groups of bacteria (e.g. Skp, YfgL and NlpB), other proteins are conserved but are missing characteristic domains (e.g. SurA). This comparative analysis is also revealing important structural signatures that are vague unless multiple members from a protein family are considered as a group (e.g. tetratricopeptide repeat (TPR) motifs in YfiO, β-propeller signatures in YfgL). Given that the process of the β-barrel assembly is conserved, analysis of outer membrane biogenesis in Alphaproteobacteria, the bacterial group that gave rise to mitochondria, also promises insight into the assembly of β-barrel proteins in eukaryotes.
Keywords: outer membrane assembly, membrane structure, Omp85, β-barrel proteins, Alphaproteobacteria, mitochondria
Introduction
Bacterial cells precisely organize their cellular activities within the cytoplasmic compartment, and the stability and integrity of a bacterial cell relies on the presence of a cell wall that encases the cytoplasm. The cell wall comprises the cytoplasmic membrane, peptidoglycan layer and, in Gram-negative bacteria, an outer membrane composed of integral membrane proteins, lipids, and lipopolysaccharides (Nikaido, 2003; Scheffers & Pinho, 2005; Ruiz et al., 2006; Bos et al., 2007). Understanding the properties of this structural and biochemical barrier is critical to devising strategies to inhibit growth of bacterial pathogens.
Analysing the structure of membranes and membrane proteins remains a challenging problem, given the difficulty associated with purifying membrane fractions and analysing membrane protein structures (Elofsson & von Heijne, 2007). Yet understanding how membranes function, and particularly how membranes are assembled, depends on knowledge of the structures of the functionally important membrane proteins. As detailed in this review, bacterial outer membranes are largely composed of proteins with a ‘β-barrel’ architecture. Recent research provides the means to appreciate the secretion, folding and assembly of β-barrel proteins in bacterial outer membranes. These recent studies build on classic studies of protein secretion in bacteria, of membrane protein crystallization and biophysical analyses. With the knowledge that mitochondria share an evolutionary relationship with Alphaproteobacteria, comparisons between the outer membrane assembly machinery found in mitochondria with that found in bacteria are shedding light on the fundamental aspects of β-barrel protein assembly. Analysis of genome sequences suggests the outer membrane assembly machinery might be similar in all classes of bacteria that have outer membranes.
Bacterial outer membranes: structure and function
Generally, bacterial outer membranes have been found to be asymmetric, with the inner leaflet of the outer membrane composed mainly of phospholipids and the outer leaflet of lipopolysaccharide. The lipid A moiety of lipopolysaccharide sits within the hydrophobic interior of the membrane and to this is anchored the hydrophilic core polysaccharide, followed by the highly variable O-antigen (Raetz & Whitfield, 2002). Some structural variations on this general theme exist: lipopolysaccharide is sometimes absent (e.g. in Deinococcus radiodurans, the spirochaetes Treponema and Borrelia and Alphaproteobacteria such as Sphingomonas; Work & Griffiths, 1968; Thompson & Murray, 1981; Hardy & Levin, 1983; Belisle et al., 1994; Kawasaki et al., 1994; Huang & Anderson, 1995; Wiese & Seydel, 1999; Porcella & Schwan, 2001; Raetz & Whitfield, 2002; Cullen et al., 2004), and the peptidoglycan layer can be more intimately associated with the inner, rather than the outer membrane (e.g. in spirochaetes; Holt, 1978). For all these bacteria, a common theme is the presence of a diverse array of β-barrel proteins in the outer membrane (Buchanan, 1999; Schulz, 2000, 2002; Wimley, 2003). There are two other topologies in which outer membrane proteins might be found: lipoproteins (reviewed by Narita et al., 2004; Tokuda & Matsuyama, 2004) anchored to the inner or outer leaflet of the outer membrane by covalently attached lipids, and a new class of proteins typified by Wza, that have an α-/β-structure that has not been observed previously in any type of integral membrane protein (Collins & Derrick, 2007).
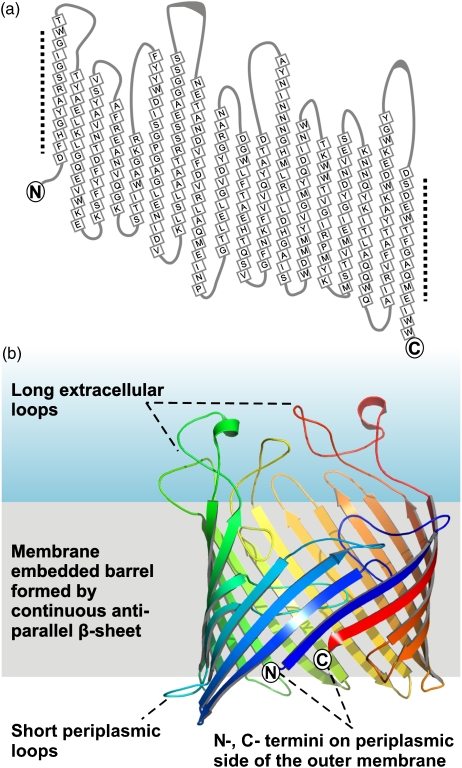
The best-characterized class of outer membrane proteins are the β-barrels. In this well-defined structure, the polypeptide chain folds into a series of antiparallel β-strands that join through hydrogen bonds between the first and last β-strand, to form a barrel (Fig. 1a). In cases such as LamB, the N-terminus is exposed to the periplasm (Fig. 1b), while in some β-barrel structures the N-terminus is folded back into the barrel lumen. The outer surface of the β-barrel is formed from amino acid residues with hydrophobic side chains, and the central space of the barrel filled by more hydrophilic (including charged) residues. If the diameter of the barrel is large enough, it will form a water-filled cavity. The cavity can form a pore through the outer membrane, but can be partially (or fully) occluded by the infolding of interstrand loops and the N-terminus (Cowan et al., 1992; Buchanan, 1999; Basle et al., 2006). This provides several possibilities to regulate the semi-permeable nature of the outer membrane, such that β-barrel membrane proteins can define the barrier characteristics of the cell wall in Gram-negative bacteria.
Fig. 1.
What is a β-barrel? (a) The polypeptide backbone of LamB from Escherichia coli (Wang et al., 1997) is traced to show the residues contributing to each β-strand (in squares) aligned to based on the hydrogen bond interactions between each strand. The unpaired hydrogen bond donors and acceptors in the first and last strand are highlighted by the dotted lines. (b) The structure of the assembled β-barrel membrane protein, represented as if in three dimensions, shows sequential β-strands (shown as colored arrows) form an antiparallel sheet that wraps into a cylinder: the final β-strand hydrogen bonds to the first strand to complete the barrel. Loops of polypeptide between the strands tend to be short on the periplasmic rim of the barrel, while longer loops are exposed to the extracellular face of the membrane. These longer loops are structured and can be folded back into the barrel lumen.
The Alpha- (and other) proteobacteria
The structures of almost all known β-barrel proteins are from species of Proteobacteria. The Proteobacteria are classified into five distinct groups: the Alpha-, Beta-, Gamma-, Delta- and Epsilonproteobacteria based on molecular phylogenetics of rRNA sequences (Woese, 1987). A number of prediction methods have been developed to detect β-barrel proteins from genome sequence (Wimley, 2002; Berven et al., 2004; Bigelow et al., 2004) and these suggest that most classes of Proteobacteria code for more than 100 β-barrel proteins. For example, analysis with the bioinformatics platform BOMP (Berven et al., 2004), suggests that 139 β-barrel membrane proteins are encoded in the genome of Escherichia coli, and a similar number in the genomes of the deltaproteobacterium Bdellovibrio and the epsilonproteobacterium Helicobacter pylori. The Alphaproteobacteria provide exceptions to this rule, with some species apparently coding for very few β-barrel proteins; only 31 are predicted from the genome of Brucella mellitensis and only 17 in the genome of Rickettsia prowazekii.
A large proportion of the bacterial species within the alphaproteobacterial group are intracellular symbionts or pathogens (Batut et al., 2004). Rickettsia live in the cytoplasm of human host cells and rely on their host for a range of nutrients that, due to redundancy and gene loss, the bacteria are no longer capable of making. As it has been discussed previously (Andersson & Kurland, 1998; Ogata et al., 2001; Sallstrom & Andersson, 2005), Alphaproteobacteria that live as symbionts and parasites tend to dispense with genes that are not essential for core functions. Studying their outer membrane protein assembly pathway offers insight into general features of the process. It is possible that, with such a narrow set of substrate proteins, some nonessential components of the assembly pathway have become superfluous. Given that mitochondria from eukaryotic cells share ancestry with Alphaproteobacteria, studies on the import and assembly of β-barrel proteins into intracellular bacteria and detailed analysis of the assembly of β-barrel proteins into mitochondrial outer membranes promise further insight (Pfanner et al., 2004; Paschen et al., 2005; Dolezal et al., 2006; Bolender et al., 2008). An intriguing recent report showed Rickettsia assembles host-derived voltage-dependent anion channel (VDAC) into its outer membrane – perhaps making up for a shortfall in its own membrane protein genes by ‘importing’β-barrel proteins from its host's cytoplasm (Emelyanov & Vyssokikh, 2006).
The export and assembly of outer membrane proteins in E. coli
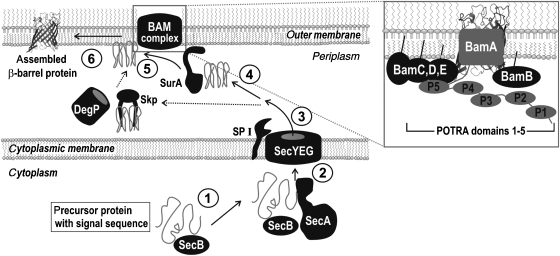
Our understanding of β-barrel protein assembly into the outer membrane of E. coli is summarized in Fig. 2. Classic studies by Beckwith and Silhavy defined many of the components of the protein secretion pathway (Bassford et al., 1991; Tamm et al., 2004; Mogensen & Otzen, 2005; Ruiz et al., 2006) and recent work by many labs, as reviewed in the following pages, is revealing the mechanisms driving protein secretion and membrane protein assembly.
Fig. 2.
In Escherichia coli, outer membrane proteins are synthesized in the cytoplasm as precursors with a signal sequence where they are recognized by the chaperones SecB and SecA (1). SecA assists translocation through the SecYEG complex in the inner membrane (2), and the signal sequence is processed by SP I (3). The substrate proteins are assisted across the periplasm by the chaperone SurA (4), and delivered to the BAM complex (5), to catalyse insertion into the outer membrane (6). Other chaperones, Skp and DegP, might cooperate to help ensure transfer to the outer membrane; DegP can also function as a protease to degrade misfolded outer membrane proteins in situations of environmental stress (Young & Hartl, 2003; Krojer et al., 2008).
The β-barrel outer membrane proteins are made as precursors with an N-terminal leader sequence, and these precursor proteins interact with the cytoplasmic factors SecB and SecA, in a process that can also be facilitated by the cytoplasmic chaperone DnaK (Qi et al., 2002; Scott & Barnett, 2006; Chen et al., 2007; Driessen & Nouwen, 2007; Papanikou et al., 2007). After translocation through the SecYEG complex in the inner membrane, the leader sequence is processed by the SP-I signal peptidase and the unfolded protein then associates with the chaperone SurA (Tamm et al., 2004; Mogensen & Otzen, 2005). A second chaperone, Skp, can bind these substrate proteins but perhaps only if they fail to productively interact with SurA (Bitto & McKay, 2003; Kleinschmidt, 2003; Walton & Sousa, 2004; Sklar et al., 2007a). SurA transfers partially folded β-barrels to an ‘insertion and assembly complex’ in the outer membrane. In E. coli, the core subunit of this complex is YaeT, a member of the Omp85 protein family, and it is associated tightly with four lipoproteins called YfgL, YfiO, NlpB and SmpA (Wu et al., 2005; Ruiz et al., 2006). This complex, recently dubbed the β-barrel assembly machinery complex (forBAM; Misra, 2007), completes the insertion and assembly of β-barrel protein substrates.
To determine whether equivalent components are found ubiquitously and particularly in the Alphaproteobacteria, we made use of hidden Markov models (HMMs) developed to describe each of the known components of the β-barrel assembly pathway. Table 1 summarizes the results for the intracellular pathogens R. prowazekii and Brucella melitensis and the free-living species Caulobacter crescentus. For more broad comparison, we included in the analysis a betaproteobacterium Neisseria meningitidis; two seminal studies on outer membrane assembly in N. meningitidis first identified Omp85 (Genevrois et al., 2003; Voulhoux et al., 2003). The process of β-barrel protein assembly is highly conserved with Omp85 family members found in all bacteria that have outer membranes (Gentle et al., 2004; Cavalier-Smith, 2006), and many of the other components of the pathway are also highly conserved. However, several distinctions are apparent, as discussed in the following sections.
Table 1.
HMMs for each protein family were used to find components of the outer membrane assembly pathway in all bacteria for which complete genome information is available
| Component* | Bacterial species† | ||||
|---|---|---|---|---|---|
| Caulobacter | Rickettsia | Brucella | Escherichia | Neisseria | |
| SecB | Q9A224 | Q9ZE76 | Q8YE23 | P0AG86 | Q9JY16 |
| SecA | P38380 | Q9ZCX7 | Q8YJG2 | P10408 | Q9JYK8 |
| SecY | Q9A8T3 | Q9ZCS5 | Q8YHM0 | P0AGA2 | Q7DDS8 |
| SecE | Q9A3J8 | P50054 | Q8YHQ3 | P0AG96 | Q9K1J4 |
| SecG | Q9A7K4 | Q9ZE68 | Q8YHF4 | P0AG99 | Q7DD68 |
| SPase I | Q9A806 | Q9ZE32 | Q8YG73 | P00803 | Q9K056 |
| SPase II | Q9AAA6 | Q9ZDC4 | Q8YES8 | P00804 | P65265 |
| Skp | Q9A712 | Q9ZDR1 | Q8YB18 | P0AEU7 | Q9K1H1 |
| SurA | Q9A7N3 | O05951 | Q8YG95 | P21202 | Q9K186 |
| Omp85 | Q9A711 | Q9ZE03 | Q8YHH0 | P0A940 | Q9K1H0 |
| YfgL | Q9A7R7 | Q9ZDU2 | None | P77774 | None |
| YfiO | Q9A6U9 | Q9ZDY1 | Q8YI58 | P0AC02 | Q9K0B1 |
| NlpB | None | None | None | P0A903 | Q9JZR5 |
| SmpA | Q9A8I8 | Q9ZCG9 | Q8YGH5 | P0A938 | Q9K1F0 |
Building of HMM models and database search were performed with the hmmer package version 2.3.2. (Eddy, 1998), and used to search the uniprot data set (Release 12.4, containing swiss-prot Release 54.4 and trembl Release 37.4) as described previously for mitochondrial protein translocase components (Dolezal et al., 2006).
The species listed in the table are Caulobacter cresentus CB15, Rickettsia prowazekii, Brucella melitensis 16M, Escherichia coli K12 and Neisseria meningitidis serogroup B.
Cytoplasmic chaperones SecA and SecB
SecB is a molecular chaperone that is involved in binding signal sequences of various outer membrane protein precursors as they emerge from the ribosome (Lecker et al., 1990; de Cock et al., 1992; Ernst et al., 1994; Behrmann et al., 1998; Baars et al., 2006; Ureta et al., 2007). It then shuttles these proteins to the SecA motor at the SecYEG translocon found in the inner membrane. The interaction between the two chaperones results in the transfer of the preproteins to SecA and the release of SecB (Fekkes et al., 1997). SecB was suggested to be present only in Proteobacteria (Driessen, 2001; Scott & Barnett, 2006), and this exclusive distribution is confirmed in the HMM search. SecB is highly conserved across the five classes of Proteobacteria, including Alphaproteobacteria. In the crystal structure of SecB the protein is a dimer of dimers, and in solution the tetrameric form is in dynamic equilibrium with the dimeric form and this equilibrium is important for substrate transfer to SecA (Topping et al., 2001; Dekker et al., 2003; Randall et al., 2005). A serine residue (Ser22) that sits at the dimer interface is phosphorylated in the SecB protein of E. coli (Macek et al., 2008). The functional consequence of phosphorylation is unknown, but the Ser22 residue is conserved in the alphaproteobacterial (and other) SecB sequences.
SecA is a multidomain ATPase, which binds to nascent outer membrane proteins after their synthesis in the cytoplasm, assisting their translocation through the SecYEG translocon in the cytoplasmic membrane (Driessen & Nouwen, 2007; Gelis et al., 2007; Papanikou et al., 2007; Rapoport, 2007). In Alphaproteobacteria, SecA has a conserved structure with all the domains known for the E. coli homologue: the core DEAD (or helicase) motor, the C-domain and preprotein-binding domain (PBD), which confer the specificity on the DEAD motor and presumably enables protein insertion into the SecYEG complex in the inner membrane.
SecYEG translocon
Most of the bacterial proteins which cross the cytoplasmic membrane are translocated through a heterotrimeric SecYEG complex (Breyton et al., 2002; van den Berg et al., 2004; Smith et al., 2005; Driessen & Nouwen, 2007; Rapoport, 2007). Biochemical and genetic experiments (Bieker et al., 1990; Driessen et al., 1991) together with structures of bacterial (E. coli; Breyton et al., 2002) and archaeal (Methanoccocus janaschii; Van den Berg et al., 2004) SecYEG complexes revealed that the membrane pore is formed solely by the 10 transmembrane segments of the SecY subunit, with the smaller SecE and SecG subunits on the periphery. SecE forms an external clamp for the helical bundle of SecY, while the proposed role of the nonessential SecG is to stabilize the complex and stimulate cytosolic SecA activity (Papanikou et al., 2007; Sugai et al., 2007). In each species of Alphaproteobacteria analysed, the SecYEG complex is predicted to be equivalent to that found in other bacteria: SecY predicts to have 10 transmembrane helices with both the N- and C-termini facing the cytoplasm, SecE has a single predicted transmembrane segment, as found in most bacteria, with the exception of the Gammaproteobacteria (e.g. E. coli) and some Betaproteobacteria. SecG would span the cytoplasmic membrane twice, as is expected to be typical for all bacteria.
The signal peptidase
Several signal peptidases have been implicated in aspects of protein secretion, and the signal sequences that dictate β-barrel protein export across the inner membrane are removed by the leader peptidase SP I (signal peptidase I). The structure and function of the leader peptidase in E. coli has been studied and shown to consist of a protease domain, oriented in the periplasm, which is anchored to the cytoplasmic membrane by two transmembrane α-helices (Dalbey et al., 1997). The same topology is predicted for the leader peptidase from the betaproteobacterium N. meningitidis and from other Proteobacteria.
Bioinformatic protein sequence analysis tools, TMpred and DAS, both suggest that the leader peptidase from Alphaproteobacteria each have a single predicted transmembrane domain, but otherwise conform to the domain structure expected for leader peptidases (Paetzel et al., 2002). Leader peptidase is considered the ancestor of the mitochondrial IMP complex that consists of two related proteases Imp1 and Imp2 (Gakh et al., 2002). While most Alphaproteobacteria have a single peptidase similar to Imp1 (Burri et al., 2005), in C. crescentus there are two genes encoding leader peptidases where one, CC1559, has the sequence signatures characteristic of the mitochondrial Imp2, rather than Imp1. It is through a similar evolutionary process of gene duplication and specialization, that the Imp2 protease of mitochondria is thought to have been derived (Burri et al., 2005).
The periplasmic chaperones Skp and SurA
Work with E. coli mutants shows SurA to be the major chaperone for delivery of proteins across the periplasm to the outer membrane (Tamm et al., 2004; Hennecke et al., 2005; Ruiz et al., 2006; Sklar et al., 2007a). At least two other periplasmic chaperones, Skp and DegP, are involved in outer membrane assembly (Tamm et al., 2004; Mogensen & Otzen, 2005), but have been suggested to serve as ‘back-up’ to assist proteins that fail to interact productively with SurA (Sklar et al., 2007a). DegP appears to assist folding for a subset of outer membrane proteins; degP-null strains show decreased levels of the major outer membrane proteins OmpA, OmpF and, to a lesser extent, OmpC. This chaperone assembles into large oligomers of 6, 12 or 24 copies, with the 12- and 24-mer sequestering folded outer membrane protein protomers, but not oligomers, prior to their insertion and assembly in the outer membrane (Krojer et al., 2008). DegP can also play a degradative role, being directly responsible for proteolysis of ‘terminally misfolded’ outer membrane proteins (Young & Hartl, 2003).
Members of the Skp protein family are present in diverse bacteria, including all proteobacterial groups and other bacteria that have outer membranes including Treponema pallidum, Bacteroides thetaiotaomicron, Chlamydophila caviae and Deinococcus radiodurans (Alcock et al., 2008). The cyanobacteria have an outer membrane and Omp85 (Bölter et al., 1998; Gentle et al., 2004; Schleiff & Soll, 2005), but their genomes do not encode a Skp homologue (Alcock et al., 2008).
SurA sequences have been identified in many groups of bacteria, such as the Chlorobiaceae and groups typified by D. radiodurans and Cytophaga hutchinsonii and are highly conserved suggesting that this chaperone, like Skp, was derived early in evolution. However, at least two genera of Alphaproteobacteria (Anaplasma and Ehrlichia) have no sequence that matches the characteristic features of SurA. It is likely then that a secondary loss of SurA has occurred in these groups, reflecting the common trend of gene loss that occurs in intracellular Alphaproteobacteria (Batut et al., 2004; Sallstrom & Andersson, 2005). In the absence of SurA, either the Skp and DegP chaperones that are present in Anaplasma and Ehrlichia perform the essential function provided by SurA in other organisms or, perhaps, a novel chaperone might work in the place of SurA.
In E. coli and other Gammaproteobacteria, SurA is composed from four domains: an N-terminal domain, two PPIase domains and a C-terminal domain (Behrens, 2002; Bitto & McKay, 2003). Curiously, the SurA sequences found in Alphaproteobacteria have either one or no PPIase domains (Alcock et al., 2008), demonstrating that these domains are not fundamental to SurA function. While the PPIase domains are characteristic of SurA from E. coli, a mutant SurA protein lacking both PPIase domains still binds β-barrel substrates effectively (Behrens et al., 2001; Hennecke et al., 2005).
Omp85
Members of the Omp85 family of proteins are found in all groups of bacteria with outer membranes, and in mitochondria and chloroplasts which were derived from intracellular bacteria in the course of evolution (Gentle et al., 2004, 2005; Schleiff & Soll, 2005; Cavalier-Smith, 2006; Bos et al., 2007). Omp85 family members had been identified as vaccine candidates in several groups of bacterial pathogens (reviewed by Gentle et al., 2005) when pioneering work in N. meningitidis showed Omp85 to be essential for outer membrane biogenesis (Genevrois et al., 2003; Voulhoux et al., 2003).
Omp85 proteins are defined by the occurrence of two domains: an N-terminal region containing multiple polypeptide-transport-associated (POTRA) domains and a C-terminal β-barrel domain. The recent structure of a related protein, FhaC, provides a reasonable model for the barrel domain of Omp85 (Clantin et al., 2007). In E. coli and in Neisseria, deletion of any one of the POTRA domains has at least partial effects on outer membrane protein assembly (Robert et al., 2006; Kim et al., 2007). Recently, the three-dimensional structure of the POTRA domains from the Omp85 protein YaeT were solved and provide the basis for a model for how Omp85 can bind diverse peptide sequences, enabling it to handle its numerous β-barrel protein substrates (Kim et al., 2007; Misra, 2007). The alphaproteobacterial homologs of Omp85 all appear to have five POTRA domains.
In E. coli, the Omp85 protein YaeT is in a complex with four lipoproteins: YfgL, NlpB, YfiO and SmpA (Stenberg et al., 2005; Wu et al., 2005; Malinverni et al., 2006; Sklar et al., 2007b). These have recently been renamed BamA(YaeT), BamB(YfgL), BamC(NlpB), BamD(YfiO) and BamE(SmpA) to reflect their functional interactions in the BAM complex (Misra, 2007). Analysis of each of the POTRA domain deletion mutants suggests that while the first POTRA domain is not required for interactions with the lipoprotein partners, POTRA domains 2, 3 and 4 are required to mediate interactions with BamB. In addition, POTRA domain 3 has been suggested to mediate augmentation of β-strand formation in substrate proteins. The fifth POTRA domain is crucial for interactions with the other partner lipoproteins (Kim et al., 2007) and, in Neisseria meningitidis, a truncated form of Omp85 lacking the first four POTRA domains is sufficient for outer membrane biogenesis (Robert et al., 2006; Bos et al., 2007).
Lipoprotein partners of Omp85
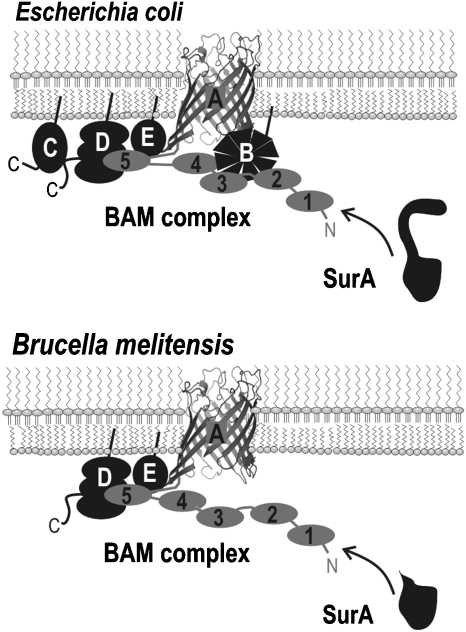
Outer membrane lipoproteins are synthesized as precursor proteins in the cytoplasm, with a characteristic N-terminal signal sequence followed by a key cysteine residue. The precursor protein is translocated into the periplasm by the Sec protein machinery in the inner membrane, and processed by signal peptidase II (Table 1) and modified by a lipoprotein diacylglyceryl transferase (Narita et al., 2004; Tokuda & Matsuyama, 2004). The lipid moiety anchors the lipoprotein in the inner or outer leaflet of the outer membrane: leaving a large proportion of the lipoprotein available for interaction with other outer membrane proteins. Genetic analysis identified the four lipoproteins BamB, BamC, BamD and BamE (YfgL, YfiO, NlpB and SmpA, respectively) that interact with Omp85 (Fig. 3), and coprecipitation and other analyses (Wu et al., 2005; Malinverni et al., 2006; Sklar et al., 2007b) suggest a basis for the overall architecture of the BAM complex: (1) there is direct interaction between BamA and BamC (Malinverni et al., 2006), (2) there is a direct interaction between BamA and BamB (Malinverni et al., 2006), (3) binding of BamD occurs through interactions it makes with the C-terminus of BamC (Malinverni et al., 2006; Vuong et al., 2008) and (4) BamE interacts directly with BamA, BamC and BamD, but not with BamB (Sklar et al., 2007b).
Fig. 3.
The BAM complexes in Escherichia coli and Brucella melitensis. The interactions between subunits of the BAM complex in E. coli are depicted. The POTRA domains are labelled numerically 1–5: POTRA domains 2, 3 and 4 are required to mediate interactions with YfgL (BamB=‘B’), while the fifth POTRA domain is crucial for interactions with YfiO (‘D’) and SmpA (‘E’), with BamD serving as the docking point for NlpB (‘C’) (Malinverni et al., 2006; Kim et al., 2007; Sklar et al., 2007b). In Brucella melitensis, the SurA chaperone is diminished lacking the PPIase domains, and BamB (YfgL) and BamC (NlpB) are lacking from the BAM complex.
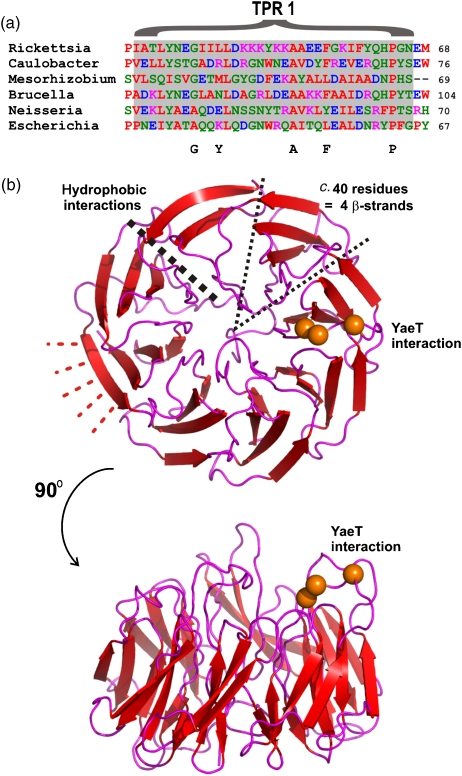
The gene encoding BamD is essential for viability of E. coli (Onufryk et al., 2005; Wu et al., 2005; Malinverni et al., 2006) and the BamD lipoprotein is found ubiquitously in Gram-negative bacteria (Malinverni et al., 2006; Table 1). This includes non-Proteobacteria such as Treponema, Chlorobium and Chlamydophila. In N. gonorrhoeae the homologous protein is a peptidoglycan-associated lipoprotein called competence lipoprotein (ComL) and, as a consequence, BamD homologs are often annotated as encoding a ‘DNA uptake lipoprotein’; a transposon insertion into the middle of ComL resulted in reduced cell size, aberrant cellular morphology and transformation deficiency (Fussenegger et al., 1996), presumably a result of the altered outer membrane properties of these mutants. The BamD protein in Rickettsia and other Alphaproteobacteria each strongly predict to have at least three tetratricopeptide repeat (TPR) motifs, but comparisons of all these sequences in multiple sequence alignments suggest each protein probably contains six TPR helix-turn-helix structures. The consensus sequence for the TPRs is not stringent, and is not well conserved in the BamD from E. coli, however, overall sequence similarities would suggest a homologous TPR-rich structure is present in all BamD proteins (Fig. 4a). TPRs are structural elements that enable protein–protein interactions and have been found operating in a number of protein transport pathways (Blatch & Lassle, 1999; D'Andrea & Regan, 2003). For example, the mitochondrial protein import receptor Tom70 is built from multiple TPR elements (Chan et al., 2006; Wu & Sha, 2006) and binds β-barrel substrate proteins en route to the mitochondrial equivalent of the BAM complex (Chan et al., 2006). A TPR-rich structure might enable BamD to bind partner proteins (like BamC) and/or substrate proteins.
Fig. 4.
Sequence analyses of the Alphaproteobacteria suggest BamD (YfiO) has at least five TPR motifs and BamB (YfgL) could have a β-propeller structure. (a) BamD (YfiO) sequences from Alphaproteobacteria were analysed with three independent TPR prediction strategies (SMART, http://smart.embl-heidelberg.de/; HHpred, http://toolkit.tuebingen.mpg.de/hhpred; and TPRpred, http://toolkit.tuebingen.mpg.de/tprpred), revealing the presence of five TPR motifs in various Alphaproteobacteria, including Rickettsia, Caulobacter and Mesorhizobium. The TPR is a degenerate motif with very few strictly conserved positions – the position of the consensus residues G–Y–A–F–P are shown. Although TPR signatures are not clear in all BamD sequences (such as the one from Escherichia coli), clustalw aligns the BamD homologs readily and at least some of the key residues found in TPR motifs are evident in all species. The alignment corresponding to the first TPR motif (‘TPR1’) is shown. (b) BamB (YfgL) sequences from Alphaproteobacteria were analysed with SMART to determine the presence of seven or eight β-propeller motifs. Homology searching using HHpred suggests BamB from E. coli is most similar to proteins with an eight-bladed β-propeller fold. Each of the blades would interact via hydrophobic contacts (heavy black line) and the outer β-strand can make additional hydrogen bond contacts (red lines). BamB from E. coli can readily be modelled using six of the β-propeller structures in the Protein Data Bank (2AD6, 1YIQ, 1KB0, 1W6S, 1FLG and 1KV9) as template structures. The model structure of BamB is shown from two views. In this structural model the three mutations in BamB (L173, L175, and R176; the Cα atoms of each are highlighted as orange spheres), that cause defects for docking to the POTRA domains of BamA (Vuong et al., 2008), come together in one of the β-propeller motifs.
Escherichia coli mutants lacking BamC have mild defects in outer membrane biogenesis (Onufryk et al., 2005; Wu et al., 2005; Malinverni et al., 2006; Sklar et al., 2007b. A BamC homolog has not been found in any of the Alphaproteobacteria (Table 1), a distinct (or highly diverged) protein might fulfil the function of BamC. Alternatively, if the function of BamC is redundant another component of the BAM complex might compensate in these organisms.
Mutants in which the yfgL gene, encoding BamB, has been deleted have reduced levels of outer membrane proteins (Onufryk et al., 2005; Ruiz et al., 2005). The BamB protein from Alphaproteobacteria has seven or eight predicted Pyrrolo-quinoline quinone (PQQ) enzyme repeats, a motif representative of β-propeller structures (Jawad & Paoli, 2002) found in some enzymes and in protein domains involved in protein-protein interactions. Homology searches suggest that both the alphaproteobacterial and the E. coli BamB fit the profile of domains with an eight-bladed β-propeller fold (Fig. 4b). The PQQ enzyme active site residues are not present in BamB, indicating this protein is unlikely to be involved in enzymatic catalysis. If BamB does have a β-propeller structure, it could interact with β-strands in the POTRA of BamA and/or assist in stabilizing nascent β-strands in substrate proteins. While BamB is found in several species of Alphaproteobacteria including Caulobacter and Rickettsia, no related sequence was found in Brucella. BamB is also absent from the Omp85 complex in Neisseria (Table 1; Bos et al., 2007). It was recently shown that cargo-bound SurA can bypass interaction with BamB and interact directly with BamA (Vuong et al., 2008). It may be that only some protein substrates interact with BamB prior to assembly.
Escherichia coli mutants lacking BamE have defects in outer membrane protein assembly (Sklar et al., 2007b). This protein is found ubiquitously in Proteobacteria, though no related proteins were found in other groups of bacteria with outer membranes. The structure of the homologue, called OmlA, from Xanthomonas has been solved (Vanini et al., 2008). As in E. coli, OmlA in Pseudomonas and Xanthomonas is required for outer membrane integrity (Ochsner et al., 1999; Sklar et al., 2007b; Fuangthong et al., 2008) omlA mutants show increased susceptibility to antibiotics (such as rifampin and chloramphenicol) and detergents. Given their high degree of sequence similarity, the structure of OmlA will be important for interpreting interactions BamE makes with other components of the BAM complex.
How do β-barrels assemble?
Understanding the folding pathway for any membrane protein is a considerable challenge. For β-barrel assembly, it is now possible to start to reconcile biophysical analyses that have looked at folding reactions undergone by purified proteins, with our growing appreciation of the components mediating the assembly pathway in vivo. A β-barrel can be considered a continuous β-sheet, wrapped to a cylinder. The assembly of a β-barrel, therefore, consists of three processes:
preventing the somewhat hydrophobic polypeptide from misfolding before its localization within a membrane environment,
folding of β-strands into the β-sheet, with the first and final strand interactions completing the cylindrical shape and
insertion of the barrel into the lipid phase of the outer membrane.
Biophysical evidence suggests the substrate proteins would be in a partially structured form in the periplasm (reviewed by Tamm et al., 2004) and that periplasmic chaperones such as SurA are needed to maintain the ‘periplasmic intermediate’ (Lazar & Kolter, 1996; Bulieris et al., 2003; Hennecke et al., 2005). In vitro models of outer membrane protein insertion predict a so called ‘molten disc’ intermediate at the bilayer interface, with β-strands sitting flat on the membrane (Kleinschmidt & Tamm, 2002; Tamm et al., 2004). SurA is, therefore, the prime candidate to assist in process (1).
The β-strands in outer membrane proteins have a sequence Φ–X–Φ (where Φ represents an aromatic residue) at least in the C-terminal strand, and mutations in this motif impair assembly of outer membrane proteins (Struyve et al., 1991; de Cock et al., 1997; Robert et al., 2006). SurA has been shown to bind directly to the C-terminal Φ–X–Φ motif (Bitto & McKay, 2003), as has Omp85 (de Cock et al., 1997; Robert et al., 2006; Bos et al., 2007). The POTRA domains of Omp85/BamA provide a means to receive substrates from chaperones like SurA. The POTRA domains have been suggested to facilitate templating of β-strands in the nascent outer membrane protein substrate and, if the lipoprotein BamB has a β-propeller structure, it too might facilitate strand formation in this way to assist in the completion of process (2). If its role were simply to assist the function of SurA and Omp85, this redundancy of function would explain why BamB is absent in some bacteria and why corresponding yfgL mutants of E. coli show only relatively minor defects in outer membrane assembly.
The third process is perhaps the one we understand least about. What mechanism is used to insert proteins into the hydrophobic core of the outer membrane? The β-barrel domain of Omp85/BamA might increase the kinetics of strand insertion by providing some local distortions in the lipid population, assisting intermediate forms of a barrel to gradually assemble and enter the membrane (3). Alternatively, the proteinaceous environment created by the components of the BAM complex in the periplasm might favour a barrel forming, to enable a more dramatic en bloc insertion of the barrel into the plane of the outer membrane.
One class of substrate protein, the autotransporters, is providing an excellent means to interrogate the mechanism of Omp85/BamA function. Autotransporters consist of an N-terminal ‘passenger domain’ and a C-terminal β-barrel domain (Henderson et al., 2004; Jacob-Dubuisson et al., 2004; Newman & Stathopoulos, 2004; Dautin & Bernstein, 2007). The secretion of the passenger domain requires the insertion of the β-barrel into the outer membrane and the translocation of the passenger domain across the outer membrane. But do these two reactions occur one after the other, as suggested by the name ‘autotransporter’? When secretion is complete, what remains of the fragment connecting the two domains can be found within the lumen of the barrel (Oomen et al., 2004; Meng et al., 2006; Barnard et al., 2007), and this ‘vapour trail’ suggests the passenger passes through the barrel pore to cross the outer membrane. However, recent work identified a novel intermediate in the periplasm whose topology resembles that of the protein after passenger domain translocation, suggesting the fragment connecting the two domains was placed inside the barrel lumen before insertion into the outer membrane (Ieva et al., 2008). It represents a thermodynamically impressive feat if the BAM complex is capable of inserting into the outer membrane an autotransporter with the huge passenger domain prepositioned on its outer surface (Jain & Goldberg, 2007; Ieva et al., 2008). It should be noted, whichever mechanism is used to translocate autotransporter passengers across the outer membrane, the β-barrel has a limited tolerance for folded elements in the passenger domain (Jong et al., 2007).
What mitochondria tell us about outer membrane assembly
Mitochondria are found in all eukaryotic cells, having been derived from intracellular bacterial symbionts (Yang et al., 1985; Gray et al., 1999). There has been debate over whether hydrogenosomes and mitosomes found, in place of ‘classic’ mitochondria, in some unicellular eukaryotes are metabolically specialized mitochondria. However it is now clear that these organelles all share a common ancestry, with a common set of protein translocases that were developed in mitochondria for the import of proteins from the cytosol (Dolezal et al., 2006). In the course of evolution most of the genes encoding mitochondrial proteins, including those coding for β-barrel outer membrane proteins, were transferred to the nucleus (Lang et al., 1999; Andersson et al., 2003). The number of β-barrel proteins in the outer mitochondrial membrane is probably small (estimates in yeast are c. 15), though the major porin VDAC accounts for around 25% of the protein mass of the membrane (Burri et al., 2006; Zahedi et al., 2006; Hoogenboom et al., 2007; Young et al., 2007). The outer membrane of mitochondria is a phospholipid bilayer, with phosphatidylcholine, phosphatidylethanolamine and phosphatidylinositol the major lipid constituents (de Kroon et al., 1997, 1999).
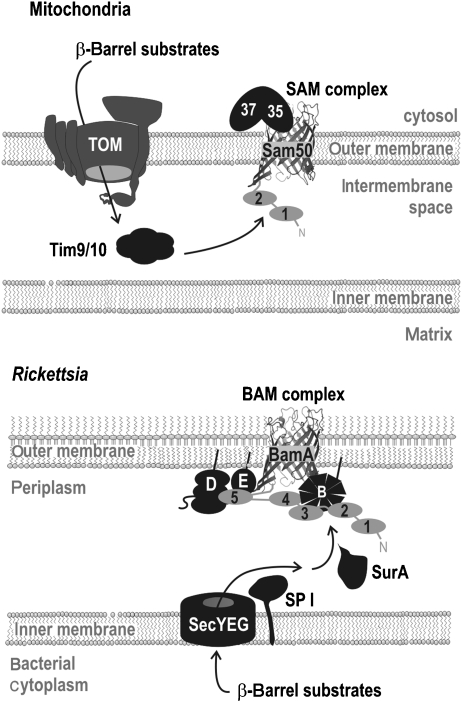
Despite being imported from the cytosol, mitochondrial β-barrel proteins are still inserted into the outer membrane from the inner surface (Fig. 5). This requires that proteins be recognized and translocated through the outer membrane by the protein translocase in the outer mitochondrial membrane (TOM complex). The TOM complex has no counterpart in bacteria. In the intermembrane space, topologically equivalent to the bacterial periplasm, β-barrel proteins are chaperoned by tiny TIM chaperones (Fig. 5). While the tiny TIM proteins show some structural similarities to Skp chaperones (Webb et al., 2006), and functional similarities to SurA (Alcock et al., 2008), there is no obvious ancestry to relate the mitochondrial and bacterial chaperones. The assembly of β-barrel proteins into the mitochondrial outer membrane is driven by the sorting and assembly machinery (SAM complex) and at the core of the SAM complex is a protein called Sam50, a member of the Omp85 protein family (Kozjak et al., 2003; Paschen et al., 2003; Gentle et al., 2004). Reflecting their ancestry, mitochondrial Sam50 proteins are most closely related to the Omp85 proteins from Alphaproteobacteria (Gentle et al., 2004).
Fig. 5.
Assembly pathways for β-barrel proteins in mitochondria and Alphaproteobacteria. In eukaryotes, β-barrel proteins are translated in the cytosol, transported across the mitochondrial outer membrane by the TOM complex and passed to the inner surface of the SAM complex for assembly. Passage through the intermembrane space depends on tiny TIM chaperones, such as the Tim9/10 complex (Pfanner et al., 2004; Paschen et al., 2005; Bolender et al., 2008). The final steps of assembly depend on the metaxins, Sam35 and Sam37 (‘35’ and ‘37’), on the outer face of the membrane (Pfanner et al., 2004; Paschen et al., 2005; Bolender et al., 2008; Chan & Lithgow, 2008). The Rickettsia are considered the closest living relatives to the progenitor of mitochondria, and use a BAM complex mechanism largely equivalent to that found in other bacteria.
The mitochondrial SAM complex receives substrate proteins from the tiny TIM chaperones, and in no eukaryotes have homologs of SurA or Skp been found (Alcock et al., 2008). This demonstrates that Omp85 proteins do not need to contact a SurA chaperone to productively receive substrate proteins. The hidden Markov model searches reported here find no evidence for BamB, BamC, BamD or BamE homologs in eukaryotes. In a sense then, mitochondria can be considered as representing an end-point to the gene loss seen in Rickettsia and Brucella in regards to components of the outer membrane assembly machinery.
The absence of the four lipoproteins and the SurA chaperone in mitochondria has an important corollary, with the mitochondrial protein Sam50 predicted to have only 1–2 POTRA domains. These POTRA domains are not essential for Sam50 to function, with the metaxin Sam35 serving as the ‘receptor’ for β-barrel substrates to bind the SAM complex and initiate assembly (Chan & Lithgow, 2008; Kutik et al., 2008). It is impossible to tell whether, in the course of evolution, the loss of the POTRA domains was causative for loss of the genes encoding SurA and the lipoproteins, or whether it happened subsequently. However, the findings in the mitochondrial system provide independent support to the proposition that the five POTRA domains in the bacterial Omp85 are important because they are needed to organize the lipoprotein partners and the SurA chaperone.
Concluding remarks
The assembly of β-barrel proteins into membranes is an essential process in Gram-negative bacteria, mitochondria and plastids. The significance of the process is underscored by observations that even relatively small perturbations to the assembly process, such as mutations in nonessential components of the BAM complex, compromises the barrier function of the outer membrane leaving mutant bacteria hypersensitive to antibiotics. Our understanding of the mechanism of β-barrel assembly is benefiting from comparative approaches, with findings on the assembly pathway in organelles informing studies on the pathway in bacteria, and vice versa.
With the availability of vast genome sequence data from so many species of bacteria it has become possible to make comparative sequence analyses, to add further value to the work carried out with model bacteria such as E. coli and N. meningitidis. Some protein features, the presence of multiple TPR motifs in BamD and of multiple β-propeller signatures in BamB, are not readily apparent in an analysis of a single polypeptide. However, these structural features become strikingly apparent when a set of homologous proteins from across bacterial species is analysed as a group.
This is an exciting time for the study of membrane proteins. An increase, slow but sure, in the number of structures solved for membrane proteins has increased our appreciation of how polypeptides can be organized in biological membranes. Improved assays for tracking the assembly of membrane proteins in distinct systems provides a means to cross-fertilize our knowledge of any given system. A prime example of this is how the discovery of Omp85 in bacterial outer membranes led to the characterization of the SAM complex in the outer membrane of mitochondria, and the further understanding of the ‘BAM complex’ in bacteria. Three clear questions now present themselves, with answers that would largely complete our understanding of β-barrel membrane protein assembly.
Firstly, where and how does folding of a polypeptide into a β-barrel occur? The barrels may fold into the lipid environment of the outer membrane, but it seems increasingly likely that folding occurs before the encounter with the bilayer. But would this be in the periplasm per se, assisted by soluble chaperones such as SurA, or in the grasp of the various periplasmically exposed components of the BAM complex?
Secondly, how do the external loops of β-barrels fold? These loops are crucial for the function of β-barrel membrane proteins often forming the occlusions that provide selectivity to the β-barrel pores. In mitochondria, there are external factors, the metaxins, bound to the SAM complex that might assist these folding reactions (Pfanner et al., 2004; Paschen et al., 2005; Dolezal et al., 2006; Chan & Lithgow, 2008), but we know of no specific factors bound to the outer surface of the bacterial BAM complex.
Thirdly, can Omp85 do more than just assemble β-barrels? While it is clear Omp85 plays a direct role in outer membrane protein insertion (Voulhoux et al., 2003; Doerrler & Raetz, 2005), other studies have shown that cells in which the expression of Omp85 has been shut down accumulate lipopolysaccharide and phospholipids in their inner membranes, likely en route to the outer membrane. This defect could be a secondary effect of improper outer membrane protein assembly, but it remains possible that Omp85 also aids lipid insertion via a similar mechanism to protein insertion: through creating a ‘disturbance’ in the outer membrane that could accelerate the transfer of substrates into the hydrophobic core.
Addressing these questions promises a more complete picture of the assembly of bacterial outer membranes.
Acknowledgments
We thank Ben Adler, Travis Barnard and Kip Gabriel for critical reading of the manuscript. Work in the authors' laboratories are supported by grants from the National Health and Medical Research Council (to A.W.P. and T.L.) and the Australian Research Council (to S.K.B. and T.L.). S.K.B. is supported by the Intramural Research Program of the NIH, National Institute of Diabetes and Digestive and Kidney Diseases.
References
- Alcock FH, Grossmann JG, Gentle IE, Likić VA, Lithgow T, Tokatlidis K. Conserved substrate binding by chaperones in the bacterial periplasm and the mitochondrial intermembrane space. Biochem J. 2008;409:377–387. doi: 10.1042/BJ20070877. [DOI] [PubMed] [Google Scholar]
- Andersson SG, Kurland CG. Reductive evolution of resident genomes. Trends Microbiol. 1998;6:263–268. doi: 10.1016/s0966-842x(98)01312-2. [DOI] [PubMed] [Google Scholar]
- Andersson SG, Karlberg O, Canback B, Kurland CG. On the origin of mitochondria: a genomics perspective. Philos Trans R Soc Lond B Biol Sci. 2003;358:165–177. doi: 10.1098/rstb.2002.1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baars L, Ytterberg AJ, Drew D, Wagner S, Thilo C, van Wijk KJ, de Gier JW. Defining the role of the Escherichia coli chaperone SecB using comparative proteomics. J Biol Chem. 2006;281:10024–10034. doi: 10.1074/jbc.M509929200. [DOI] [PubMed] [Google Scholar]
- Barnard TJ, Dautin N, Lukacik P, Bernstein HD, Buchanan SK. Autotransporter structure reveals intra-barrel cleavage followed by conformational changes. Nat Struct Mol Biol. 2007;14:1214–1220. doi: 10.1038/nsmb1322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basle A, Rummel G, Storici P, Rosenbusch JP, Schirmer T. Crystal structure of osmoporin OmpC from E. coli at 2.0 A. J Mol Biol. 2006;362:933–942. doi: 10.1016/j.jmb.2006.08.002. [DOI] [PubMed] [Google Scholar]
- Bassford P, Beckwith J, Ito K, Kumamoto C, Mizushima S, Oliver D, Randall L, Silhavy T, Tai PC, Wickner B. The primary pathway of protein export in E. coli. Cell. 1991;65:367–368. doi: 10.1016/0092-8674(91)90453-6. [DOI] [PubMed] [Google Scholar]
- Batut J, Andersson SG, O'Callaghan D. The evolution of chronic infection strategies in the Alphaproteobacteria. Nat Rev Microbiol. 2004;2:933–945. doi: 10.1038/nrmicro1044. [DOI] [PubMed] [Google Scholar]
- Behrens S. Periplasmic chaperones – new structural and functional insights. Structure. 2002;10:1469–1471. doi: 10.1016/s0969-2126(02)00893-6. [DOI] [PubMed] [Google Scholar]
- Behrens S, Maier R, de Cock H, Schmid FX, Gross CA. The SurA periplasmic PPIase lacking its parvulin domains functions in vivo and has chaperone activity. EMBO J. 2001;20:285–294. doi: 10.1093/emboj/20.1.285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behrmann M, Koch HG, Hengelage T, Wieseler B, Hoffschulte HK, Müller M. Requirements for the translocation of elongation-arrested, ribosome-associated OmpA across the plasma membrane of Escherichia coli. J Biol Chem. 1998;273:13898–13904. doi: 10.1074/jbc.273.22.13898. [DOI] [PubMed] [Google Scholar]
- Belisle JT, Brandt ME, Radolf JD, Norgard MV. Fatty acids of Treponema pallidum and Borrelia burgdorferi lipoproteins. J Bacteriol. 1994;176:2151–2157. doi: 10.1128/jb.176.8.2151-2157.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berven FS, Flikka K, Jensen HB, Eidhammer I. BOMP: a program to predict integral beta-barrel outer membrane proteins encoded within genomes of Gram-negative bacteria. Nucleic Acids Res. 2004;32:W394–W399. doi: 10.1093/nar/gkh351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bieker KL, Phillips GJ, Silhavy TJ. The sec and prl genes of Escherichia coli. J Bioenerg Biomembr. 1990;22:291–310. doi: 10.1007/BF00763169. [DOI] [PubMed] [Google Scholar]
- Bigelow HR, Petrey DS, Liu J, Przybylski D, Rost B. Predicting transmembrane beta-barrels in proteomes. Nucleic Acids Res. 2004;32:2566–2577. doi: 10.1093/nar/gkh580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bitto E, McKay DB. The periplasmic molecular chaperone protein SurA binds a peptide motif that is characteristic of integral outer membrane proteins. J Biol Chem. 2003;278:49316–49322. doi: 10.1074/jbc.M308853200. [DOI] [PubMed] [Google Scholar]
- Blatch GL, Lassle M. The tetratricopeptide repeat: a structural motif mediating protein–protein interactions. Bioessays. 1999;21:932–939. doi: 10.1002/(SICI)1521-1878(199911)21:11<932::AID-BIES5>3.0.CO;2-N. [DOI] [PubMed] [Google Scholar]
- Bolender N, Sickmann A, Wagner R, Meisinger C, Pfanner N. Multiple pathways for sorting mitochondrial precursor proteins. EMBO Rep. 2008;9:42–49. doi: 10.1038/sj.embor.7401126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bölter B, Soll J, Schulz A, Hinnah S, Wagner R. Origin of a chloroplast protein importer. Proc Natl Acad Sci USA. 1998;95:15831–15836. doi: 10.1073/pnas.95.26.15831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bos MP, Robert V, Tommassen J. Biogenesis of the gram-negative bacterial outer membrane. Annu Rev Microbiol. 2007;61:191–214. doi: 10.1146/annurev.micro.61.080706.093245. [DOI] [PubMed] [Google Scholar]
- Breyton C, Haase W, Rapoport TA, Kuhlbrandt W, Collinson I. Three-dimensional structure of the bacterial protein-translocation complex SecYEG. Nature. 2002;418:662–665. doi: 10.1038/nature00827. [DOI] [PubMed] [Google Scholar]
- Buchanan SK. Beta-barrel proteins from bacterial outer membranes: structure, function and refolding. Curr Opin Struct Biol. 1999;9:455–461. doi: 10.1016/S0959-440X(99)80064-5. [DOI] [PubMed] [Google Scholar]
- Bulieris PV, Behrens S, Holst O, Kleinschmidt JH. Folding and insertion of the outer membrane protein OmpA is assisted by the chaperone Skp and by lipopolysaccharide. J Biol Chem. 2003;278:9092–9099. doi: 10.1074/jbc.M211177200. [DOI] [PubMed] [Google Scholar]
- Burri L, Strahm Y, Hawkins CJ, Gentle IE, Puryer MA, Verhagen A, Callus B, Vaux D, Lithgow T. Mature DIABLO/Smac is produced by the IMP protease complex on the mitochondrial inner membrane. Mol Biol Cell. 2005;16:2926–2933. doi: 10.1091/mbc.E04-12-1086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burri L, Vascotto K, Gentle IE, Chan NC, Beilharz T, Stapleton DI, Ramage L, Lithgow T. Integral membrane proteins in the mitochondrial outer membrane of Saccharomycescerevisiae. FEBS J. 2006;273:1507–1515. doi: 10.1111/j.1742-4658.2006.05171.x. [DOI] [PubMed] [Google Scholar]
- Cavalier-Smith T. Rooting the tree of life by transition analyses. Biol Direct. 2006;1:19. doi: 10.1186/1745-6150-1-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan NC, Lithgow T. The peripheral membrane subunits of the SAM complex function codependently in mitochondrial outer membrane biogenesis. Mol Biol Cell. 2008;19:126–136. doi: 10.1091/mbc.E07-08-0796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan NC, Likic VA, Waller RF, Mulhern TD, Lithgow T. The C-terminal TPR domain of Tom70 defines a family of mitochondrial protein import receptors found only in animals and fungi. J Mol Biol. 2006;358:1010–1022. doi: 10.1016/j.jmb.2006.02.062. [DOI] [PubMed] [Google Scholar]
- Chen Y, Tai PC, Sui SF. The active ring-like structure of SecA revealed by electron crystallography: conformational change upon interaction with SecB. J Struct Biol. 2007;159:149–153. doi: 10.1016/j.jsb.2007.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clantin B, Delattre AS, Rucktooa P, Saint N, Meli AC, Locht C, Jacob-Dubuisson F, Villeret V. Structure of the membrane protein FhaC: a member of the Omp85-TpsB transporter superfamily. Science. 2007;317:957–961. doi: 10.1126/science.1143860. [DOI] [PubMed] [Google Scholar]
- Collins RF, Derrick JP. Wza: a new structural paradigm for outer membrane secretory proteins? Trends Microbiol. 2007;15:96–100. doi: 10.1016/j.tim.2007.01.002. [DOI] [PubMed] [Google Scholar]
- Cowan SW, Schirmer T, Rummel G, Steiert M, Ghosh R, Pauptit RA, Jansonius JN, Rosenbusch JP. Crystal structures explain functional properties of two E. coli porins. Nature. 1992;358:727–733. doi: 10.1038/358727a0. [DOI] [PubMed] [Google Scholar]
- Cullen PA, Haake DA, Adler B. Outer membrane proteins of pathogenic spirochetes. FEMS Microbiol Rev. 2004;28:291–318. doi: 10.1016/j.femsre.2003.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalbey RE, Lively MO, Bron S, van Dijl JM. The chemistry and enzymology of the type I signal peptidases. Protein Sci. 1997;6:1129–1138. doi: 10.1002/pro.5560060601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Andrea LD, Regan L. TPR proteins: the versatile helix. Trends Biochem Sci. 2003;28:655–662. doi: 10.1016/j.tibs.2003.10.007. [DOI] [PubMed] [Google Scholar]
- Dautin N, Bernstein HD. Protein secretion in gram-negative bacteria via the autotransporter pathway. Annu Rev Microbiol. 2007;61:89–112. doi: 10.1146/annurev.micro.61.080706.093233. [DOI] [PubMed] [Google Scholar]
- de Cock H, Overeem W, Tommassen J. Biogenesis of outer membrane protein PhoE of Escherichia coli. Evidence for multiple SecB-binding sites in the mature portion of the PhoE protein. J Mol Biol. 1992;224:369–379. doi: 10.1016/0022-2836(92)91001-6. [DOI] [PubMed] [Google Scholar]
- de Cock H, Struyve M, Kleerebezem M, van der Krift T, Tommassen J. Role of the carboxy-terminal phenylalanine in the biogenesis of outer membrane protein PhoE of Escherichia coli K-12. J Mol Biol. 1997;269:473–478. doi: 10.1006/jmbi.1997.1069. [DOI] [PubMed] [Google Scholar]
- de Kroon AI, Dolis D, Mayer A, Lill R, de Kruijff B. Phospholipid composition of highly purified mitochondrial outer membranes of rat liver and Neurospora crassa. Is cardiolipin present in the mitochondrial outer membrane? Biochim Biophys Acta. 1997;1325:108–116. doi: 10.1016/s0005-2736(96)00240-4. [DOI] [PubMed] [Google Scholar]
- de Kroon AI, Koorengevel MC, Goerdayal SS, Mulders PC, Janssen MJ, de Kruijff B. Isolation and characterization of highly purified mitochondrial outer membranes of the yeast Saccharomyces cerevisiae. Mol Membr Biol. 1999;16:205–211. doi: 10.1080/096876899294670. [DOI] [PubMed] [Google Scholar]
- Dekker C, de Kruijff B, Gros P. Crystal structure of SecB from Escherichia coli. J Struct Biol. 2003;144:313–319. doi: 10.1016/j.jsb.2003.09.012. [DOI] [PubMed] [Google Scholar]
- Doerrler WT, Raetz CR. Loss of outer membrane proteins without inhibition of lipid export in an Escherichia coli YaeT mutant. J Biol Chem. 2005;280:27679–27687. doi: 10.1074/jbc.M504796200. [DOI] [PubMed] [Google Scholar]
- Dolezal P, Likic V, Tachezy J, Lithgow T. Evolution of the molecular machines for protein import into mitochondria. Science. 2006;313:314–318. doi: 10.1126/science.1127895. [DOI] [PubMed] [Google Scholar]
- Driessen AJ. SecB, a molecular chaperone with two faces. Trends Microbiol. 2001;9:193–196. doi: 10.1016/s0966-842x(01)01980-1. [DOI] [PubMed] [Google Scholar]
- Driessen AJ, Nouwen N. Protein translocation across the bacterial cytoplasmic membrane. Annu Rev Biochem. 2007;77:643–667. doi: 10.1146/annurev.biochem.77.061606.160747. [DOI] [PubMed] [Google Scholar]
- Driessen AJ, Brundage L, Hendrick JP, Schiebel E, Wickner W. Preprotein translocase of Escherichia coli: solubilization, purification, and reconstitution of the integral membrane subunits SecY/E. Methods Cell Biol. 1991;34:147–165. doi: 10.1016/s0091-679x(08)61679-9. [DOI] [PubMed] [Google Scholar]
- Eddy SR. Profile hidden Markov models. Bioinformatics. 1998;14:755–763. doi: 10.1093/bioinformatics/14.9.755. [DOI] [PubMed] [Google Scholar]
- Elofsson A, von Heijne G. Membrane protein structure: prediction versus reality. Annu Rev Biochem. 2007;76:125–140. doi: 10.1146/annurev.biochem.76.052705.163539. [DOI] [PubMed] [Google Scholar]
- Emelyanov VV, Vyssokikh MY. On the nature of obligate intracellular symbiosis of rickettsiae –Rickettsia prowazekii cells import mitochondrial porin. Biochemistry (Moscow) 2006;71:730–735. doi: 10.1134/s0006297906070054. [DOI] [PubMed] [Google Scholar]
- Ernst F, Hoffschulte HK, Thome-Kromer B, Swidersky UE, Werner PK, Müller M. Precursor-specific requirements for SecA, SecB, and delta muH+ during protein export of Escherichia coli. J Biol Chem. 1994;269:12840–12845. [PubMed] [Google Scholar]
- Fekkes P, van der Does C, Driessen AJ. The molecular chaperone SecB is released from the carboxy-terminus of SecA during initiation of precursor protein translocation. EMBO J. 1997;16:6105–6113. doi: 10.1093/emboj/16.20.6105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuangthong M, Sallabhan R, Atichartpongkul S, Rangkadilok N, Sriprang R, Satayavivad J, Mongkolsuk S. The omlA gene is involved in multidrug resistance and its expression is inhibited by coumarins in Xanthomonas campestris pv. phaseoli. Arch Microbiol. 2008;189:211–218. doi: 10.1007/s00203-007-0310-1. [DOI] [PubMed] [Google Scholar]
- Fussenegger M, Facius D, Meier J, Meyer TF. A novel peptidoglycan-linked lipoprotein (ComL) that functions in natural transformation competence of Neisseria gonorrhoeae. Mol Microbiol. 1996;19:1095–1105. doi: 10.1046/j.1365-2958.1996.457984.x. [DOI] [PubMed] [Google Scholar]
- Gakh O, Cavadini P, Isaya G. Mitochondrial processing peptidases. Biochim Biophys Acta. 2002;1592:63–77. doi: 10.1016/s0167-4889(02)00265-3. [DOI] [PubMed] [Google Scholar]
- Gelis I, Bonvin AM, Keramisanou D, Koukaki M, Gouridis G, Karamanou S, Economou A, Kalodimos CG. Structural basis for signal-sequence recognition by the translocase motor SecA as determined by NMR. Cell. 2007;131:756–769. doi: 10.1016/j.cell.2007.09.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genevrois S, Steeghs L, Roholl P, Letesson JJ, van der Ley P. The Omp85 protein of Neisseria meningitidis is required for lipid export to the outer membrane. EMBO J. 2003;22:1780–1789. doi: 10.1093/emboj/cdg174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gentle I, Gabriel K, Beech P, Waller R, Lithgow T. The Omp85 family of proteins is essential for outer membrane biogenesis in mitochondria and bacteria. J Cell Biol. 2004;164:19–24. doi: 10.1083/jcb.200310092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gentle IE, Burri L, Lithgow T. Molecular architecture and function of the Omp85 family of proteins. Mol Microbiol. 2005;58:1216–1225. doi: 10.1111/j.1365-2958.2005.04906.x. [DOI] [PubMed] [Google Scholar]
- Gray MW, Burger G, Lang BF. Mitochondrial evolution. Science. 1999;283:1476–1481. doi: 10.1126/science.283.5407.1476. [DOI] [PubMed] [Google Scholar]
- Hardy PH, Jr, Levin J. Lack of endotoxin in Borrelia hispanica and Treponema pallidum. Proc Soc Exp Biol Med. 1983;174:47–52. doi: 10.3181/00379727-174-41702. [DOI] [PubMed] [Google Scholar]
- Henderson IR, Navarro-Garcia F, Desvaux M, Fernandez RC, Ala'Aldeen D. Type V protein secretion pathway: the autotransporter story. Microbiol Mol Biol Rev. 2004;68:692–744. doi: 10.1128/MMBR.68.4.692-744.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hennecke G, Nolte J, Volkmer-Engert R, Schneider-Mergener J, Behrens S. The periplasmic chaperone SurA exploits two features characteristic of integral outer membrane proteins for selective substrate recognition. J Biol Chem. 2005;280:23540–23548. doi: 10.1074/jbc.M413742200. [DOI] [PubMed] [Google Scholar]
- Holt SC. Anatomy and chemistry of spirochetes. Microbiol Rev. 1978;42:114–160. doi: 10.1128/mr.42.1.114-160.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoogenboom BW, Suda K, Engel A, Fotiadis D. The supramolecular assemblies of voltage-dependent anion channels in the native membrane. J Mol Biol. 2007;370:246–255. doi: 10.1016/j.jmb.2007.04.073. [DOI] [PubMed] [Google Scholar]
- Huang Y, Anderson R. Glucosyl diglyceride lipid structures in Deinococcus radiodurans. J Bacteriol. 1995;177:2567–2571. doi: 10.1128/jb.177.9.2567-2571.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ieva R, Skillman KM, Bernstein HD. Incorporation of a polypeptide segment into the beta-domain pore during the assembly of a bacterial autotransporter. Mol Microbiol. 2008;67:188–201. doi: 10.1111/j.1365-2958.2007.06048.x. [DOI] [PubMed] [Google Scholar]
- Jacob-Dubuisson F, Fernandez R, Coutte L. Protein secretion through autotransporter and two-partner pathways. Biochim Biophys Acta. 2004;1694:235–257. doi: 10.1016/j.bbamcr.2004.03.008. [DOI] [PubMed] [Google Scholar]
- Jain S, Goldberg MB. Requirement for YaeT in the outer membrane assembly of autotransporter proteins. J Bacteriol. 2007;189:5393–5398. doi: 10.1128/JB.00228-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jawad Z, Paoli M. Novel sequences propel familiar folds. Structure. 2002;10:447–454. doi: 10.1016/s0969-2126(02)00750-5. [DOI] [PubMed] [Google Scholar]
- Jong WS, ten Hagen-Jongman CM, den Blaauwen T, Slotboom DJ, Tame JR, Wickström D, de Gier JW, Otto BR, Luirink J. Limited tolerance towards folded elements during secretion of the autotransporter Hbp. Mol Microbiol. 2007;63:1524–1536. doi: 10.1111/j.1365-2958.2007.05605.x. [DOI] [PubMed] [Google Scholar]
- Kawasaki S, Moriguchi R, Sekiya K, Nakai T, Ono E, Kume K, Kawahara K. The cell envelope structure of the lipopolysaccharide-lacking gram-negative bacterium Sphingomonas paucimobilis. J Bacteriol. 1994;176:284–290. doi: 10.1128/jb.176.2.284-290.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S, Malinverni JC, Sliz P, Silhavy TJ, Harrison SC, Kahne D. Structure and function of an essential component of the outer membrane protein assembly machine. Science. 2007;317:961–964. doi: 10.1126/science.1143993. [DOI] [PubMed] [Google Scholar]
- Kleinschmidt JH. Membrane protein folding on the example of outer membrane protein A of Escherichia coli. Cell Mol Life Sci. 2003;60:1547–1558. doi: 10.1007/s00018-003-3170-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleinschmidt JH, Tamm LK. Secondary and tertiary structure formation of the beta-barrel membrane protein OmpA is synchronized and depends on membrane thickness. J Mol Biol. 2002;324:319–330. doi: 10.1016/s0022-2836(02)01071-9. [DOI] [PubMed] [Google Scholar]
- Kozjak V, Wiedemann N, Milenkovic D, Lohaus C, Meyer HE, Guiard B, Meisinger C, Pfanner N. An essential role of Sam50 in the protein sorting and assembly machinery of the mitochondrial outer membrane. J Biol Chem. 2003;278:48520–48523. doi: 10.1074/jbc.C300442200. [DOI] [PubMed] [Google Scholar]
- Krojer T, Sawa J, Schafer E, Saibil HR, Ehrmann M, Clausen T. Structural basis for the regulated protease and chaperone function of DegP. Nature. 2008;453:885–890. doi: 10.1038/nature07004. [DOI] [PubMed] [Google Scholar]
- Kutik S, Stojanovski D, Becker L, et al. Dissecting membrane insertion of mitochondrial beta-barrel proteins. Cell. 2008;132:1011–1024. doi: 10.1016/j.cell.2008.01.028. [DOI] [PubMed] [Google Scholar]
- Lang BF, Gray MW, Burger G. Mitochondrial genome evolution and the origin of eukaryotes. Annu Rev Genet. 1999;33:351–397. doi: 10.1146/annurev.genet.33.1.351. [DOI] [PubMed] [Google Scholar]
- Lazar SW, Kolter R. SurA assists the folding of Escherichia coli outer membrane proteins. J Bacteriol. 1996;178:1770–1773. doi: 10.1128/jb.178.6.1770-1773.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lecker SH, Driessen AJ, Wickner W. ProOmpA contains secondary and tertiary structure prior to translocation and is shielded from aggregation by association with SecB protein. EMBO J. 1990;9:2309–2314. doi: 10.1002/j.1460-2075.1990.tb07402.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macek B, Gnad F, Soufi B, Kumar C, Olsen JV, Mijakovic I, Mann M. Phosphoproteome analysis of E. coli reveals evolutionary conservation of bacterial Ser/Thr/Tyr phosphorylation. Mol Cell Proteomics. 2008;7:299–307. doi: 10.1074/mcp.M700311-MCP200. [DOI] [PubMed] [Google Scholar]
- Malinverni JC, Werner J, Kim S, Sklar JG, Kahne D, Misra R, Silhavy TJ. YfiO stabilizes the YaeT complex and is essential for outer membrane protein assembly in Escherichia coli. Mol Microbiol. 2006;61:151–164. doi: 10.1111/j.1365-2958.2006.05211.x. [DOI] [PubMed] [Google Scholar]
- Meng G, Surana NK, St Geme JW, III, Waksman G. Structure of the outer membrane translocator domain of the Haemophilus influenzae Hia trimeric autotransporter. EMBO J. 2006;25:2297–2304. doi: 10.1038/sj.emboj.7601132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misra R. First glimpse of the crystal structure of YaeT's POTRA domains. ACS Chem Biol. 2007;2:649–651. doi: 10.1021/cb700212p. [DOI] [PubMed] [Google Scholar]
- Mogensen JE, Otzen DE. Interactions between folding factors and bacterial outer membrane proteins. Mol Microbiol. 2005;57:326–346. doi: 10.1111/j.1365-2958.2005.04674.x. [DOI] [PubMed] [Google Scholar]
- Narita S, Matsuyama S, Tokuda H. Lipoprotein trafficking in Escherichia coli. Arch Microbiol. 2004;182:1–6. doi: 10.1007/s00203-004-0682-4. [DOI] [PubMed] [Google Scholar]
- Newman CL, Stathopoulos C. Autotransporter and two-partner secretion: delivery of large-size virulence factors by gram-negative bacterial pathogens. Crit Rev Microbiol. 2004;30:275–286. doi: 10.1080/10408410490499872. [DOI] [PubMed] [Google Scholar]
- Nikaido H. Molecular basis of bacterial outer membrane permeability revisited. Microbiol Mol Biol Rev. 2003;67:593–656. doi: 10.1128/MMBR.67.4.593-656.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ochsner UA, Vasil AI, Johnson Z, Vasil ML. Pseudomonas aeruginosa fur overlaps with a gene encoding a novel outer membrane lipoprotein, OmlA. J Bacteriol. 1999;181:1099–1109. doi: 10.1128/jb.181.4.1099-1109.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogata H, Audic S, Renesto-Audiffren P, et al. Mechanisms of evolution in Rickettsia conorii and R. prowazekii. Science. 2001;293:2093–2098. doi: 10.1126/science.1061471. [DOI] [PubMed] [Google Scholar]
- Onufryk C, Crouch ML, Fang FC, Gross CA. Characterization of six lipoproteins in the sigmaE regulon. J Bacteriol. 2005;187:4552–4561. doi: 10.1128/JB.187.13.4552-4561.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oomen CJ, van Ulsen P, van Gelder P, Feijen M, Tommassen J, Gros P. Structure of the translocator domain of a bacterial autotransporter. EMBO J. 2004;23:1257–1266. doi: 10.1038/sj.emboj.7600148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paetzel M, Dalbey RE, Strynadka NC. Crystal structure of a bacterial signal peptidase apoenzyme: implications for signal peptide binding and the Ser–Lys dyad mechanism. J Biol Chem. 2002;277:9512–9519. doi: 10.1074/jbc.M110983200. [DOI] [PubMed] [Google Scholar]
- Papanikou E, Karamanou S, Economou A. Bacterial protein secretion through the translocase nanomachine. Nat Rev Microbiol. 2007;5:839–851. doi: 10.1038/nrmicro1771. [DOI] [PubMed] [Google Scholar]
- Paschen SA, Waizenegger T, Stan T, Preuss M, Cyrklaff M, Hell K, Rapaport D, Neupert W. Evolutionary conservation of biogenesis of beta-barrel membrane proteins. Nature. 2003;426:862–866. doi: 10.1038/nature02208. [DOI] [PubMed] [Google Scholar]
- Paschen SA, Neupert W, Rapaport D. Biogenesis of beta-barrel membrane proteins of mitochondria. Trends Biochem Sci. 2005;30:575–582. doi: 10.1016/j.tibs.2005.08.009. [DOI] [PubMed] [Google Scholar]
- Pfanner N, Wiedemann N, Meisinger C, Lithgow T. Assembling the mitochondrial outer membrane. Nat Struct Mol Biol. 2004;11:1044–1048. doi: 10.1038/nsmb852. [DOI] [PubMed] [Google Scholar]
- Porcella SF, Schwan TG. Borrelia burgdorferi and Treponema pallidum: a comparison of functional genomics, environmental adaptations, and pathogenic mechanisms. J Clin Invest. 2001;107:651–656. doi: 10.1172/JCI12484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi HY, Hyndman JB, Bernstein HD. DnaK promotes the selective export of outer membrane protein precursors in SecA-deficient Escherichia coli. J Biol Chem. 2002;277:51077–51083. doi: 10.1074/jbc.M209238200. [DOI] [PubMed] [Google Scholar]
- Raetz CR, Whitfield C. Lipopolysaccharide endotoxins. Annu Rev Biochem. 2002;71:635–700. doi: 10.1146/annurev.biochem.71.110601.135414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Randall LL, Crane JM, Lilly AA, Liu G, Mao C, Patel CN, Hardy SJ. Asymmetric binding between SecA and SecB two symmetric proteins: implications for function in export. J Mol Biol. 2005;348:479–489. doi: 10.1016/j.jmb.2005.02.036. [DOI] [PubMed] [Google Scholar]
- Rapoport TA. Protein translocation across the eukaryotic endoplasmic reticulum and bacterial plasma membranes. Nature. 2007;450:663–669. doi: 10.1038/nature06384. [DOI] [PubMed] [Google Scholar]
- Robert V, Volokhina EB, Senf F, Bos MP, Van Gelder P, Tommassen J. Assembly factor omp85 recognizes its outer membrane protein substrates by a species-specific C-terminal motif. PLoS Biol. 2006;4:e377. doi: 10.1371/journal.pbio.0040377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz N, Falcone B, Kahne D, Silhavy TJ. Chemical conditionality: a genetic strategy to probe organelle assembly. Cell. 2005;121:307–317. doi: 10.1016/j.cell.2005.02.014. [DOI] [PubMed] [Google Scholar]
- Ruiz N, Kahne D, Silhavy TJ. Advances in understanding bacterial outer-membrane biogenesis. Nat Rev Microbiol. 2006;4:57–66. doi: 10.1038/nrmicro1322. [DOI] [PubMed] [Google Scholar]
- Sallstrom B, Andersson SG. Genome reduction in the Alphaproteobacteria. Curr Opin Microbiol. 2005;8:579–585. doi: 10.1016/j.mib.2005.08.002. [DOI] [PubMed] [Google Scholar]
- Scheffers DJ, Pinho MG. Bacterial cell wall synthesis: new insights from localization studies. Microbiol Mol Biol Rev. 2005;69:585–607. doi: 10.1128/MMBR.69.4.585-607.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schleiff E, Soll J. Membrane protein insertion: mixing eukaryotic and prokaryotic concepts. EMBO Rep. 2005;6:1023–1027. doi: 10.1038/sj.embor.7400563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulz GE. beta-barrel membrane proteins. Curr Opin Struct Biol. 2000;10:443–447. doi: 10.1016/s0959-440x(00)00120-2. [DOI] [PubMed] [Google Scholar]
- Schulz GE. The structure of bacterial outer membrane proteins. Biochim Biophys Acta. 2002;1565:308–317. doi: 10.1016/s0005-2736(02)00577-1. [DOI] [PubMed] [Google Scholar]
- Scott JR, Barnett TC. Surface proteins of gram-positive bacteria and how they get there. Annu Rev Microbiol. 2006;60:397–423. doi: 10.1146/annurev.micro.60.080805.142256. [DOI] [PubMed] [Google Scholar]
- Sklar JG, Wu T, Kahne D, Silhavy TJ. Defining the roles of the periplasmic chaperones SurA, Skp, and DegP in Escherichia coli. Genes Dev. 2007a;21:2473–2484. doi: 10.1101/gad.1581007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sklar JG, Wu T, Gronenberg LS, Malinverni JC, Kahne D, Silhavy TJ. Lipoprotein SmpA is a component of the YaeT complex that assembles outer membrane proteins in Escherichia coli. Proc Natl Acad Sci USA. 2007b;104:6400–6405. doi: 10.1073/pnas.0701579104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith MA, Clemons WM, Jr, DeMars CJ, Flower AM. Modeling the effects of prl mutations on the Escherichia coli SecY complex. J Bacteriol. 2005;187:6454–6465. doi: 10.1128/JB.187.18.6454-6465.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stenberg F, Chovanec P, Maslen SL, Robinson CV, Ilag LL, von Heijne G, Daley DO. Protein complexes of the Escherichia coli cell envelope. J Biol Chem. 2005;280:34409–34419. doi: 10.1074/jbc.M506479200. [DOI] [PubMed] [Google Scholar]
- Struyve M, Moons M, Tommassen J. Carboxy-terminal phenylalanine is essential for the correct assembly of a bacterial outer membrane protein. J Mol Biol. 1991;218:141–148. doi: 10.1016/0022-2836(91)90880-f. [DOI] [PubMed] [Google Scholar]
- Sugai R, Takemae K, Tokuda H, Nishiyama K. Topology inversion of SecG is essential for cytosolic SecA-dependent stimulation of protein translocation. J Biol Chem. 2007;282:29540–29548. doi: 10.1074/jbc.M704716200. [DOI] [PubMed] [Google Scholar]
- Tamm LK, Hong H, Liang B. Folding and assembly of beta-barrel membrane proteins. Biochim Biophys Acta. 2004;1666:250–263. doi: 10.1016/j.bbamem.2004.06.011. [DOI] [PubMed] [Google Scholar]
- Thompson BG, Murray RG. Isolation and characterization of the plasma membrane and the outer membrane of Deinococcus radiodurans strain Sark. Can J Microbiol. 1981;27:729–734. doi: 10.1139/m81-111. [DOI] [PubMed] [Google Scholar]
- Tokuda H, Matsuyama S. Sorting of lipoproteins to the outer membrane in E. coli. Biochim Biophys Acta. 2004;1694 IN1-9. [PubMed] [Google Scholar]
- Topping TB, Woodbury RL, Diamond DL, Hardy SJ, Randall LL. Direct demonstration that homotetrameric chaperone SecB undergoes a dynamic dimer-tetramer equilibrium. J Biol Chem. 2001;276:7437–7441. doi: 10.1074/jbc.M009584200. [DOI] [PubMed] [Google Scholar]
- Ureta AR, Endres RG, Wingreen NS, Silhavy TJ. Kinetic analysis of the assembly of the outer membrane protein LamB in Escherichia coli mutants each lacking a secretion or targeting factor in a different cellular compartment. J Bacteriol. 2007;189:446–454. doi: 10.1128/JB.01103-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van den Berg B, Clemons WM, Jr, Collinson I, Modis Y, Hartmann E, Harrison SC, Rapoport TA. X-ray structure of a protein-conducting channel. Nature. 2004;427:36–44. doi: 10.1038/nature02218. [DOI] [PubMed] [Google Scholar]
- Vanini MM, Spisni A, Sforca ML, Pertinhez TA, Benedetti CE. The solution structure of the outer membrane lipoprotein OmlA from Xanthomonasaxonopodis pv. citri reveals a protein fold implicated in protein–protein interaction. Proteins. 2008;71:2051–2064. doi: 10.1002/prot.21886. [DOI] [PubMed] [Google Scholar]
- Voulhoux R, Bos MP, Geurtsen J, Mols M, Tommassen J. Role of a highly conserved bacterial protein in outer membrane protein assembly. Science. 2003;299:262–265. doi: 10.1126/science.1078973. [DOI] [PubMed] [Google Scholar]
- Vuong P, Bennion D, Mantei J, Frost D, Misra R. Analysis of YfgL and YaeT interactions through bioinformatics, mutagenesis, and biochemistry. J Bacteriol. 2008;190:1507–1517. doi: 10.1128/JB.01477-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walton TA, Sousa MC. Crystal structure of Skp, a prefoldin-like chaperone that protects soluble and membrane proteins from aggregation. Mol Cell. 2004;15:367–374. doi: 10.1016/j.molcel.2004.07.023. [DOI] [PubMed] [Google Scholar]
- Wang YF, Dutzler R, Rizkallah PJ, Rosenbusch JP, Schirmer T. Channel specificity: structural basis for sugar discrimination and differential flux rates in maltoporin. J Mol Biol. 1997;272:56–63. doi: 10.1006/jmbi.1997.1224. [DOI] [PubMed] [Google Scholar]
- Webb CT, Gorman MA, Lazarou M, Ryan MT, Gulbis JM. Crystal structure of the mitochondrial chaperone TIM9.10 reveals a six-bladed alpha-propeller. Mol Cell. 2006;21:123–133. doi: 10.1016/j.molcel.2005.11.010. [DOI] [PubMed] [Google Scholar]
- Wiese A, Seydel U. Interaction of peptides and proteins with bacterial surface glycolipids: a comparison of glycosphingolipids and lipopolysaccharides. J Ind Microbiol Biotechnol. 1999;23:414–424. doi: 10.1038/sj.jim.2900709. [DOI] [PubMed] [Google Scholar]
- Wimley WC. Toward genomic identification of beta-barrel membrane proteins: composition and architecture of known structures. Protein Sci. 2002;11:301–312. doi: 10.1110/ps.29402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wimley WC. The versatile beta-barrel membrane protein. Curr Opin Struct Biol. 2003;13:404–411. doi: 10.1016/s0959-440x(03)00099-x. [DOI] [PubMed] [Google Scholar]
- Woese CR. Bacterial evolution. Microbiol Rev. 1987;51:221–271. doi: 10.1128/mr.51.2.221-271.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Work E, Griffiths H. Morphology and chemistry of cell walls of Micrococcus radiodurans. J Bacteriol. 1968;95:641–657. doi: 10.1128/jb.95.2.641-657.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu T, Malinverni J, Ruiz N, Kim S, Silhavy TJ, Kahne D. Identification of a multicomponent complex required for outer membrane biogenesis in Escherichia coli. Cell. 2005;121:235–245. doi: 10.1016/j.cell.2005.02.015. [DOI] [PubMed] [Google Scholar]
- Wu Y, Sha B. Crystal structure of yeast mitochondrial outer membrane translocon member Tom70p. Nat Struct Mol Biol. 2006;13:589–593. doi: 10.1038/nsmb1106. [DOI] [PubMed] [Google Scholar]
- Yang D, Oyaizu Y, Oyaizu H, Olsen GJ, Woese CR. Mitochondrial origins. Proc Natl Acad Sci USA. 1985;82:4443–4447. doi: 10.1073/pnas.82.13.4443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young JC, Hartl FU. A stress sensor for the bacterial periplasm. Cell. 2003;113:1–2. doi: 10.1016/s0092-8674(03)00192-2. [DOI] [PubMed] [Google Scholar]
- Young MJ, Bay DC, Hausner G, Court DA. The evolutionary history of mitochondrial porins. BMC Evol Biol. 2007;7:31. doi: 10.1186/1471-2148-7-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zahedi RP, Sickmann A, Boehm AM, Winkler C, Zufall N, Schonfisch B, Guiard B, Pfanner N, Meisinger C. Proteomic analysis of the yeast mitochondrial outer membrane reveals accumulation of a subclass of preproteins. Mol Biol Cell. 2006;17:1436–1450. doi: 10.1091/mbc.E05-08-0740. [DOI] [PMC free article] [PubMed] [Google Scholar]