Abstract
EDA, the gene mutated in anhidrotic ectodermal dysplasia, encodes ectodysplasin, a TNF superfamily member that activates NF-kB mediated transcription. To identify EDA target genes, we have earlier used expression profiling to infer genes differentially expressed at various developmental time points in Tabby (Eda-deficient) compared to wild-type mouse skin. To increase the resolution to find genes whose expression may be restricted to epidermal cells, we have now extended studies to primary keratinocyte cultures established from E19 wild-type and Tabby skin. Using microarrays bearing 44,000 gene probes, we found 385 preliminary candidate genes whose expression was significantly affected by Eda loss. By comparing expression profiles to those from Eda-A1 transgenic skin, we restricted the list to 38 “candidate EDA targets”, 14 of which were already known to be expressed in hair follicles or epidermis. We confirmed expression changes for 3 selected genes, Tbx1, Bmp7, and Jag1, both in keratinocytes and in whole skin, by Q-PCR and Western blotting analyses. Thus, by the analysis of keratinocytes, novel candidate pathways downstream of EDA were detected.
Keywords: Anhidrotic ectodermal dysplasia, Ectodysplasin, Edar, Tbx1, Bmp7, Jag1
1. Introduction
Ectodermal dysplasias (EDs) are a heterogeneous group of hereditary genetic disorders comprising nearly 200 clinically distinguishable forms, with a combined frequency of about 7 in 10,000 births (Itin and Fistarol, 2004). EDs are defined by the deficiency of at least two ectodermal derivatives among hair, sweat glands, teeth and nails (Priolo et al., 2000). Anhidrotic/hypohidrotic ectodermal dysplasia (EDA/HED) is the most frequent form of ED, affecting the development of sweat glands, hair follicles and teeth in human patients and in animals (reviewed in Cui and Schlessinger, 2006).
EDA is caused by mutations in any of several members of EDA signaling pathway genes. The pathway includes the ligand ectodysplasin, receptor EDAR, and receptor adaptor protein EDARADD (Kere et al., 1996; Headon and Overbeek, 1999; Headon et al., 2001). EDA signaling accesses the canonical NF-kB cascade through TRAF6, NEMO and IkBα, and thus represents a new TNF subfamily for skin appendage development (Cui and Schlessinger, 2006). Accordingly, mutations in EDA, EDAR and EDARADD cause deficiencies in skin appendages, with mutations in the downstream TRAF6, NEMO, IkBα, and NF-kB genes also causing additional immune malfunction (Döffinger et al., 2001; Naito et al., 2002; Courtois et al., 2003).
The regulatory hierarchy of the EDA signaling pathway has proven to be complex. Shh, Wnt/Dkk, Bmp and LTβ pathway genes were shown to be located downstream of EDA-NF-kB (Andl et al., 2002; Cui et al., 2006, 2007; Headon and Overbeek,1999; Mou et al., 2006; Närhi et al., 2008). Among various candidate target genes, some were down regulated in a wide range of skin appendages, some only in certain organs, and some only at delimitated times. This variety suggests that there are general and time- or organ specific targets of EDA for skin appendage development (Cui et al., 2006). However, none of the inferred target genes could carry out the entire range of EDA functions, and knowledge of the full spectrum of EDA targets and their cooperative interactions remains incomplete.
Genome-wide expression profiling of whole skin RNA from embryonic and adult mice has inferred a number of EDA target genes (Cui et al., 2002, 2006). In a complementary effort to discover target genes, we have now profiled gene expression pattern of cultured primary keratinocytes from wild-type and Tabby mice. This approach has revealed a number of candidate EDA target genes, including Tbx1, Bmp7 and Jag1, that were not previously detected.
2. Materials and methods
2.1. Primary keratinocyte isolation and in vitro culture
Timed mating was set up with C57BL/6J male and Tabby female mice (C57BL/6j-AW–J-Ta6J). Fresh skin was harvested from the backs of E19 embryos just before delivery, and the epidermis/upper-follicle segment was isolated from dermis by enzymatic digestion (CellnTec, Bern, Switzerland). At this stage, guard and awl hair follicles are growing and the highly prevalent zigzag hair follicles are being initiated. Thus, isolated keratinocytes are heterogeneous, including epidermal keratinocytes and epidermal cells from the various types of hair follicles.
Primary keratinocytes were cultured in CnT-07 medium (Chemicon International, MA, USA) at 35 °C in 5% CO2 atmosphere. Genotyping to confirm sex and Tabby mutation status was done by PCR and subsequent enzymatic digestion as previously described (Cui et al., 2006). Both wild-type and Tabby keratinocytes were morphologically heterogeneous and grew slowly until about passage 7, when growth accelerated with the expected spontaneous immortalization.
2.2. RNA isolation, gene expression profiling and Q-PCR
Total RNA was isolated from wild-type and Tabby primary keratinocytes at passage 4 using Trizol (Invitrogen, CA, USA). RNAs were then LiCl precipitated as previously described (Cui et al., 2005). Quality was checked by electrophoresis. RNAs were cyan-3-labeled and hybridized to the 44,000-feature 60-mer-oligo Agilent microarray (Carter et al., 2003). Duplicate hybridization data were obtained from each of two different keratinocyte cultures and analyzed by ANOVA, with the false discovery rate (FDR) set to ≤ 0.01, fold-differences ≥ 2, and log intensity ≥ 3.0 (Carter et al., 2003). To narrow down the list to candidate EDA targets with further support, the resultant 385 “preliminary candidate” genes were further compared to our previous expression profiles of adult stage Eda-A1 transgenic skin (Cui et al., 2006). We selected genes that were downregulated in Tabby keratinocytes but upregulated in Eda-A1 transgenic skin (or upregulated in Tabby keratinocytes and downregulated in Eda-A1 transgenic skin) as “candidate EDA targets”. We then categorized them by their possible functions indicated in GO term (Table 1).
Table 1.
“Candidate EDA target” genes from expression profiling of primary keratinocytes
| Altered genes | Fold-changes in PK a(Ta/WT) | Fold-changes in WTTG skin (TG/WT) | Sublocalization in skin b | References |
|---|---|---|---|---|
| Transcription factors | ||||
| Tbx1 | 0.1 ▼ | 2.0 ▲ | ORS | Zoupa et al. (2006) |
| Tsc22d3 | 2.2 ▲ | 0.3 ▼ | n.d. | |
| Ifi204 | 2.3 ▲ | 0.6 ▼ | n.d. | |
| Signal transduction | ||||
| Bmp7 | 0.1 ▼ | 1.5 ▲ | Mx, DP | Rendl et al. (2005) |
| Ror2 | 0.1 ▼ | 2.7 ▲ | n.d. | |
| Jag1 | 0.4 ▼ | 1.6 ▲ | Epi, ORS, Mx | Estrach et al. (2006) |
| Edn1 | 0.4 ▼ | 1.6 ▲ | Epi | Kim et al. (2007) |
| Inhba | 0.5 ▼ | 1.7 ▲ | Epi, DP | Cruise et al. (2004); Nakamura et al. (2003) |
| Cytokine/kinase/hormone | ||||
| Il23a | 0.2 ▼ | 2.5 ▲ | Epi | Piskin et al. (2006) |
| Areg | 0.3 ▼ | 2.0 ▲ | Epi | Yoshida et al. (2008) |
| Mark1 | 0.4 ▼ | 1.9 ▲ | n.d. | |
| Nek6 | 0.4 ▼ | 1.9 ▲ | n.d. | |
| Nppb | 0.4 ▼ | 1.7 ▲ | n.d. | |
| Enzyme/inhibitor | ||||
| Prss12 | 0.1 ▼ | 2.1 ▲ | DP | Rendl et al. (2005) |
| Pla2g7 | 0.2 ▼ | 1.6 ▲ | ORS | Rendl et al. (2005) |
| Stfa3 | 0.2 ▼ | 2.1 ▲ | ORS | Rendl et al. (2005) |
| Glul | 6.3 ▲ | 0.6 ▼ | n.d. | |
| Transporter | ||||
| Oabpl3 | 0.4 ▼ | 1.6 ▲ | n.d. | |
| Slc16a6 | 0.4 ▼ | 2.0 ▲ | n.d. | |
| Zinc finger protein | ||||
| Zfp518b | 0.4 ▼ | 1.6 ▲ | n.d. | |
| Zdhhc2 | 2.5 ▲ | 0.4 ▼ | n.d. | |
| Fhl1 | 9.6 ▲ | 0.6 ▼ | n.d. | |
| Ca++/protein binding | ||||
| S100a8 | 0.3 ▼ | 7.0 ▲ | Epi, Mdu | Schmidt et al. (2001) |
| Whrn | 0.3 ▼ | 2.0 ▲ | n.d. | |
| Pdzrn3 | 0.4 ▼ | 1.7 ▲ | n.d. | |
| Nrp2 | 0.4 ▼ | 2.1 ▲ | n.d. | |
| Cd44 | 0.4 ▼ | 1.7 ▲ | Epi, DP | Tuhkanen et al. (1999) |
| Tnnt2 | 0.5 ▼ | 2.1 ▲ | n.d | |
| Ahnak | 2.1 ▲ | 0.5 ▼ | Epi | Masunaga et al. (1995) |
| Tubulin/cell cycle/heat shock | ||||
| Tubb2a | 0.3 ▼ | 1.6 ▲ | n.d | |
| Mad1l1 | 0.4 ▼ | 1.6 ▲ | n.d | |
| Dnaja1 | 0.5 ▼ | 1.6 ▲ | n.d | |
| Unknown function | ||||
| Palmd | 0.3▼ | 1.5 ▲ | ORS | Rendl et al. (2005) |
| Ler5 | 0.4 ▼ | 1.5 ▲ | n.d. | |
| Prkrip1 | 0.5 ▼ | 1.6 ▲ | n.d. | |
| Herpud1 | 2.4 ▲ | 0.6 ▼ | n.d. | |
| Arrdc3 | 2.7 ▲ | 0.5 ▼ | n.d. | |
| 0610010D20Rik | 3.4 ▲ | 0.5 ▼ | n.d. | |
▼ and ▲ represent down- and upregulated genes in Ta keratinocytes or Eda-A1 transgenic (WTTG) skin. The false discovery rate for listed genes are <0.01, corresponding to P-values <0.0005.
ORS, outer root sheath of hair follicle; Mx, hair follicle matrix; DP, dermal papillae; Epi, epidermis; Mdu, medulla; n.d., newly-identified genes, sublocalization in skin not yet determined.
Selected candidate genes in resultant lists were confirmed by One-step Q-PCR with TaqMan “Assays on-Demand” probe/primers (Applied Biosystems, NJ, USA). The more readily available cells from passages 7 to 9 were used for this confirmation. To quantify the relative changes in gene expression, the −2ΔΔCT method (Livak and Schmittgen, 2001) was used and reactions were normalized to GAPDH expression levels. In addition, we isolated RNAs from skin samples of Tabby and wild-type littermates at embryonic stages E15.5, E16.5, E18.5 and postnatal day 1 (P1) for Real-Time PCR analysis.
2.3. Protein isolation and Western blot analysis
Proteins were isolated from primary keratinocytes by vigorous vortexing in ice-cold RIPA buffer [containing 150 mM NaCl, 1.0% NP-40, 0.5% sodium deoxycholate, 0.1% SDS, 50 mM Tris, pH 8.0.) (Sigma, MO, USA)] and centrifugation. The supernatant was designated as Ext 1. The pellet was then suspended in RIPA buffer +1% sodium dodecyl sulfate and sonicated to extract less soluble proteins (Ext 2). Protein concentrations were measured with the Bradford method, using a Bio-Rad protein assay system (Bio-Rad, CA, USA). Extracted proteins were then denatured by adding β-mercaptoethanol (5% in final volume) and boiling for 5 min. 40 µg of each extract was separated on a 12% Tris-glycine acrylamide gel (Invitrogen) and transferred to a nitrocellulose membrane (GE Healthcare, NJ, USA). The membrane was then blocked with 5% non-fat dry milk in 1 × phosphate buffered saline containing 0.1% Tween-20 and incubated with primary antibodies overnight at 4 °C. The antibodies included rabbit polyclonal anti-Tbx1 (Abcam, MA, USA), dilution 4 µg/ml; goat polyclonal anti-Pitx1 (N-15) (Santa Cruz Biotechnology, CA, USA), dilution 1:200; and rabbit polyclonal anti-Sox11 (H-290) (Santa Cruz Biotechnology), dilution 1:200. After washing in PBS containing 0.1% Tween-20, the membranes were incubated with HRP-conjugated donkey anti-goat or donkey anti-rabbit antibodies (Santa Cruz Biotechnology) for 2 h at room temperature. Immunoreactive bands were then visualized with an enhanced chemiluminescence (ECL) kit (Amersham Buckinghamshire, UK).
3. Results and discussion
EDA signaling regulates initiation and progression of skin appendages during early developmental stages and hair shaft formation at later stages. Shh, BMP, Wnt, and LTβ pathways have all been implicated in these processes downstream of EDA signaling (Cui and Schlessinger, 2006). Recent findings suggested that EDA signaling also regulates hair follicle cycling during postnatal life, through the apoptosis-related XIAP (X-linked inhibitor of apoptosis protein) (Fessing et al., 2006). Thus EDA signaling likely regulates a variety of genes in its action in different appendages at different stages.
A number of downstream targets of EDA signaling have been revealed by comparing gene expression profiles of whole skin samples from Eda-expressing (wild-type) and Eda-null (Tabby) animals (Cui et al., 2006). However, the expression of EDA pathway genes is restricted to epidermis and the epidermal part of skin appendages (Cui and Schlessinger, 2006); and because the epidermis comprises only about 1/10th of whole skin, we reasoned that RNA species from many EDA-responsive genes might be diluted in whole skin RNA preparations to a level too low to be seen easily. Furthermore, whole skin is difficult to manipulate for in vitro studies of EDA signaling. We therefore established primary keratinocytes from wild-type and Tabby skin as a possible cellular model to extend the studies with whole skin.
3.1. Expression profiling of wild-type and Tabby primary keratinocytes
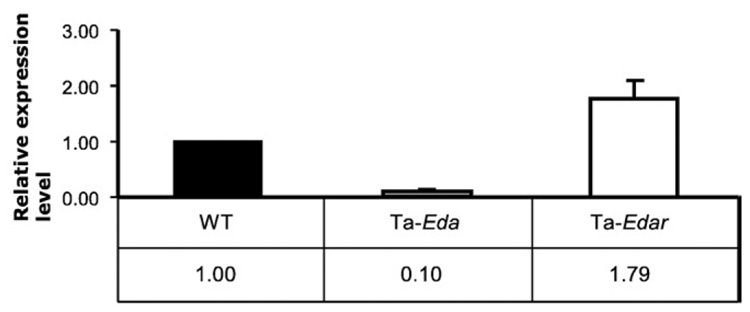
Before starting transcription profiling, we carried out Q-PCR for Eda and Edar to confirm that EDA pathway members are expressed in the primary keratinocytes. Both Eda and Edar were highly expressed in wild-type primary keratinocytes. As expected, Eda expression was significantly downregulated in Tabby keratinocytes; however, Edar expression level was comparable to or even slightly higher in Tabby than in wild-type (Fig. 1). Expression levels of further downstream genes, Edaradd, Nemo and Rela in Tabby keratinocytes were also comparable to wild-type in expression profiles (data not shown). These results suggested that the EDA pathway is active in wild-type keratinocytes and might be primed to function even in Tabby keratinocytes.
Fig. 1.
Expression level of Eda and Edar in primary keratinocytes from wild-type (set to 1.0) and Tabby mice. Eda expression was significantly downregulated, whereas Edar was slightly upregulated in Ta keratinocytes (Ta-Eda and Ta-Edar).
Microarray hybridization of RNAs from passage 4 cells yielded a list of 385 genes with significantly altered expression (see Materials and methods). The list included 208 genes downregulated and 177 genes upregulated in Tabby keratinocytes (Supplementary Table 1). We designated these genes as “preliminary candidate” genes.
Because of the differences of the cell system from intact skin and the possible drift during cell culture, additional criteria were applied to identify the most likely true targets of Eda action. The keratinocytes proved to be resistant to transfection procedures, so that we could not simply ask what expression differences were reversed by introduction of an Eda gene into Tabby cells. Also, although TNF-alpha activated NF-kB as expected, recombinant ectodysplasin from two commercial sources did not stimulate the NF-kB pathway in the cells, perhaps because of poor multimerization or post-translational modification of the recombinant protein.
We therefore further discriminated the candidate EDA targets based on in vivo results, comparing the keratinocyte profiles with previous expression profiles of adult stage transgenic skin in which Eda-A1 was expressed at a very high level (Cui et al., 2006). From this analysis, among the initial 385 genes, 38 were also upregulated at least 1.5-fold when the Eda-A1 transgene was over-expressed and downregulated at least 1.5-fold when the transgene was not expressed. The subsets are designated as “candidate EDA targets” (Table 1). The 38 selected genes, classified according to their known or probable functions, include transcription factors, signaling proteins and protease inhibitors (Table 1). Most had not been associated with the EDA pathway in earlier studies with whole skin.
3.2. Confirmation of expression changes in keratinocytes by Q-PCR and Western blot assays for selected genes
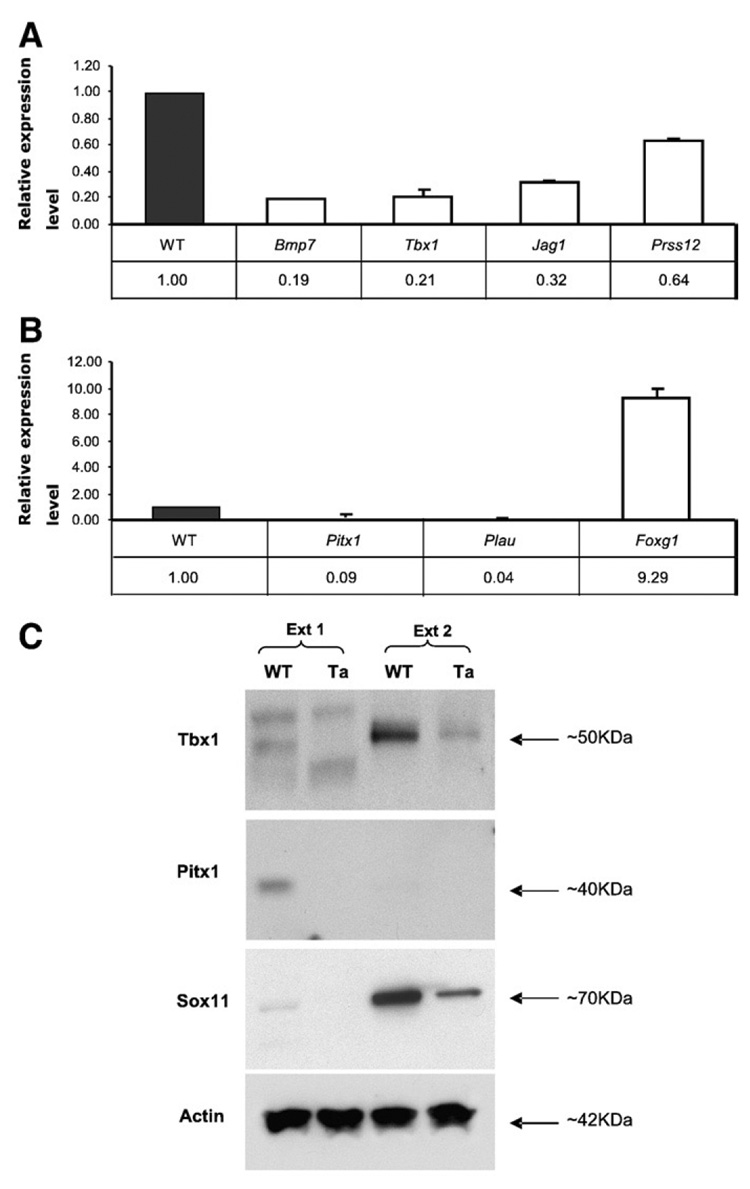
We selected 4 genes from the “candidate EDA target” group (Tbx1, Bmp7, Jag1 and Prss12), and 4 additional genes from the “preliminary candidate” group (Pitx1, Foxg1, Sox11 and Plau) for further tests by Q-PCR and/or Western blotting approaches with primary keratinocytes.
Q-PCR analyses confirmed microarray results for 7 of the genes, Tbx1, Bmp7, Jag1, Prss12, Pitx1, Foxg1 and Plau, representing both “candidate EDA target” and “preliminary candidate” groups (Figs. 2A, B). Sox11 was not efficiently amplified by Q-PCR, but Sox11 protein was downregulated in Tabby keratinocytes, consistent with the microarray results (Fig. 2C). Notably, although the differences between wild-type and Tabby were unequivocal, positive Q-PCR signals for most of genes were observed only after about 35 cycles rather than the 30 or fewer that are sufficient for highly expressed genes. The results thus support the notion that genes expressed at low level can be more easily scored in keratinocytes then in whole skin. As expected, Western blot analysis confirmed that Tbx1 and Pitx1 were also significantly downregulated in Tabby keratinocytes (Fig. 2C). Pitx1 protein was detectable, but only at very low levels even in wild-type keratinocytes (Fig. 2C). Thus, Q-PCR and Western blot assays corroborate the microarray results for both “preliminary candidate” and “candidate EDA target” groups in primary keratinocytes. However, the shorter candidate gene list was more reliably confirmed in Q-PCR assays on whole skin, as follows.
Fig. 2. Relative expression levels for selected genes in Ta primary keratinocytes.
(A) Bmp7, Tbx1, Jag1 and Prss12 from the “candidate EDA target” group showed significant downregulation in Ta keratinocytes, with WT set to 1.0. (B) Pitx1, Plau and Foxg1 from the “preliminary candidate” group also showed significant expression changes that were consistent with microarray results. (C) Western blotting assays showed that Tbx1 protein was significantly downregulated in Ta keratinocytes in the Ext 2 fraction (see Materials and methods). Pitx1 and Sox11 were also downregulated in Ta keratinocytes in Ext 1 and Ext 2 fractions, respectively. Actin was a loading control.
3.3. Q-PCR confirmed expression changes between wild-type and Tabby whole skin for selected genes from the “candidate EDA target” group
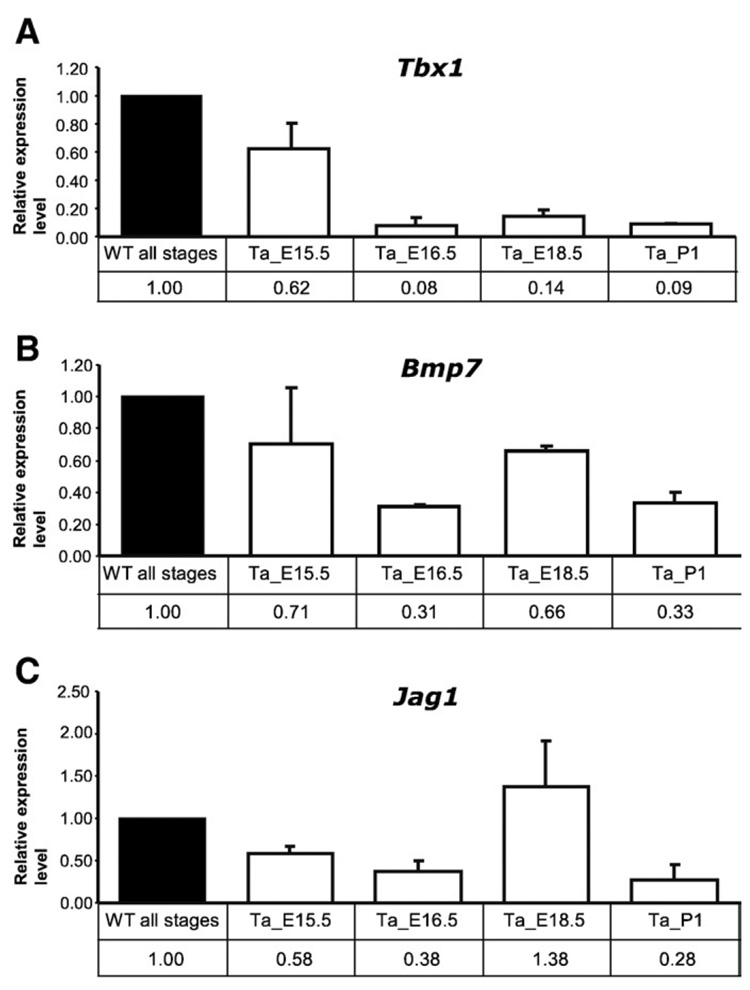
To assess expression levels of selected genes in vivo we carried out additional Q-PCR assays for 3 “candidate EDA target” genes, Tbx1, Bmp7 and Jag1, and 3 “preliminary candidate” genes Pitx1, Foxg1 and Sox11 using RNA samples from skin of littermate wild-type and Tabby embryos at E15.5, 16.5, 18.5 and postnatal day 1 (P1).
Tbx1 was slightly downregulated in E15.5 Tabby skin and more sharply at E16.5, 18.5 and p1 (Fig. 3A). Downregulation of Bmp7 in Tabby skin was less pronounced, but was significant at E16.5, 18.5 and p1 (Fig. 3B). Jag1 was slightly downregulated in Tabby skin at E15.5 and significantly at E16.5 and p1 (though it was similar to wild-type at E18.5) (Fig. 3C). These results suggested that all genes are affected over a considerable developmental period for hair follicles in Tabby mice.
Fig. 3. Relative expression levels of Tbx1, Bmp7 and Jag1 in E15.5, 16.5, 18.5 and postnatal day 1 skin, with WT set at 1.0.
(A) Tbx1 was slightly downregulated in Ta skin at E15.5, and significantly thereafter. (B) Bmp7 was significantly downregulated in Ta skin at E16.5 and thereafter. (C) Jag1 was significantly downregulated in Ta skin at E15.5, 16.5 and p1, but was comparable to wild-type level at E18.5.
In contrast, 3 genes from the “preliminary candidate” group showed confirmatory levels in keratinocytes, but were either too low to detect (Pitx1) or gave non-significant expression changes (Foxg1 and Sox11) in whole skin (data not shown). Further validation efforts will therefore be necessary to determine how many of the “preliminary candidate” genes from the list are truly involved in the response to EDA in vivo.
3.4. Implication of novel candidate target genes
The candidate target genes, especially those encoding transcription factors and signaling proteins, are most likely located downstream of the EDA-NF-kB cascade. NF-kB inactive mice showed phenotypes almost identical to Tabby mice (Schmidt-Ullrich et al., 2001, 2006; Cui et al., 2003), suggesting that NF-kB is the primary transcriptional player in EDA signaling pathway.
The newly associated transcription factors and signaling genes imply some further features of the mechanism of EDA action. Tbx1, a member of T-box transcription factor family, is involved in the development of many organs, and has been recently shown to be expressed in developing tooth germs and the outer root sheath of hair follicles, though with no known function (Zoupa et al., 2006; Rendl et al., 2005). Tbx1 was significantly downregulated in E16.5 and E18.5 Tabby skin in our previous whole skin expression profiling (Cui et al., 2006), and we here confirmed the expression changes in vivo and in vitro by Q-PCR and Western blot assays. Significant downregulation of Tbx1 in Tabby primary keratinocytes demonstrates that the in vivo expression changes are not a secondary effect of a different timing of hair follicle development in Tabby compared to wild-type mice. These results thus strongly suggest Tbx1 as a proximal target of EDA.
Bmp7 was previously shown to be involved in feather placode formation in chicken (Harris et al., 2004) and was found to be expressed both in epidermal matrix region and dermal papillae of mice (Rendl et al., 2005). It was recently shown that dermal Bmp7 was rapidly upregulated in skin organ cultures stimulated with ectodysplasin (Mou et al., 2006). Bmp7 did not show significant changes in our previous expression profiling of whole Tabby skin (Cui et al., 2006), but is now revealed as a likely target of EDA, providing an additional link between the EDA and BMP pathways.
We also found that Jag1, a Notch pathway ligand, the critical factor for late stage hair follicle development (Millar, 2002), is regulated by the EDA pathway, with an in vivo expression pattern that suggested stage-specific regulation. This is the first indication of a possible functional interaction between the EDA and Notch pathways.
Keratinocytes have several intrinsic limitations as a model system because they are heterogeneous; because their status compared to epidermal stem cell differentiation is not well defined; and because their gene expression profile may shift in culture compared to in vivo. However, they do express the Eda receptor, and compared to whole skin, they increase the sensitivity to recognize a number of additional Eda targets, particularly those expressed at a very low level in epidermal tissue. The cell system or cloned sublines may also provide a route to further analyses — for example, to discriminate primary vs. secondary targets of EDA action.
4. Conclusions
Defining the full spectrum of downstream target effectors is critical to understand EDA function in skin appendage development. Here we showed that primary keratinocytes from wild-type and mutant Tabby skin constitute an alternative source to explore EDA target genes. By genome-wide expression profiling, we inferred a list of 385 genes that are possible candidate EDA targets and a subset including Tbx1, Bmp7, Jag1 and another 35 genes that are likely EDA targets. Keratinocytes may thus provide an in vitro model to study details of events involved in EDA signaling.
Acknowledgments
We thank Drs. Kunisada M, Ko M, Piao Y and Nagaraja R for their helpful discussions and technical assistances. This work was supported by the IRP of the NIH, National Institute on Aging.
Abbreviations
- EDA
anhidrotic ectodermal dysplasia
- FDR
false discovery rate
- NF-kB
nuclear factor kappa B
- Q-PCR
quantitative real-time PCR
Appendix A. Supplementary data
Supplementary data associated with this article can be found, in the online version, at doi:10.1016/j.gene.2008.09.014.
References
- Andl T, Reddy ST, Gaddapara T, Millar SE. WNT signals are required for the initiation of hair follicle development. Dev. Cell. 2002;2:643–653. doi: 10.1016/s1534-5807(02)00167-3. [DOI] [PubMed] [Google Scholar]
- Carter MG, Hamatani T, Sharov AA, et al. In situ-synthesized novel microarray optimized for mouse stem cell and early developmental expression profiling. Genome Res. 2003;13:1011–1021. doi: 10.1101/gr.878903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Courtois G, et al. A hypermorphic IkappaBalpha mutation is associated with autosomal dominant anhidrotic ectodermal dysplasia and T cell immunodeficiency. J. Clin. Invest. 2003;112:1108–1115. doi: 10.1172/JCI18714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cruise BA, Xu P, Hall AK. Wounds increase activin in skin and a vasoactive neuropeptide in sensory ganglia. Dev. Biol. 2004;271:1–10. doi: 10.1016/j.ydbio.2004.04.003. [DOI] [PubMed] [Google Scholar]
- Cui CY, Schlessinger D. EDA signaling and skin appendage development. Cell Cycle. 2006;5(21):2477–2483. doi: 10.4161/cc.5.21.3403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui CY, Durmowicz M, Tanaka TS, et al. EDA targets revealed by skin gene expression profiles of wild-type, Tabby and Tabby EDA-A1 transgenic mice. Hum. Mol. Genet. 2002;11:1763–1773. doi: 10.1093/hmg/11.15.1763. [DOI] [PubMed] [Google Scholar]
- Cui CY, Durmowicz M, Ottolenghi C, et al. Inducible mEDA-A1 transgene mediates sebaceous gland hyperplasia and differential formation of two types of mouse hair follicles. Hum. Mol. Genet. 2003;12:2931–2940. doi: 10.1093/hmg/ddg325. [DOI] [PubMed] [Google Scholar]
- Cui CY, Smith JA, Schlessinger D, Chan CC. X-linked anhidrotic ectodermal dysplasia disruption yields a mouse model for ocular surface disease and resultant blindness. Am. J. Pathol. 2005;167:89–95. doi: 10.1016/S0002-9440(10)62956-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui CY, Hashimoto T, Grivennikov SI, Piao Y, Nedospasov SA, Schlessinger D. Ectodysplasin regulates the lymphotoxin-beta pathway for hair differentiation. Proc. Natl. Acad. Sci. U. S. A. 2006;103:9142–9147. doi: 10.1073/pnas.0509678103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui CY, Kunisada M, Esibizione D, et al. Lymphotoxin-beta regulates periderm differentiation during embryonic skin development. Hum. Mol.Genet. 2007;16:2583–2590. doi: 10.1093/hmg/ddm210. [DOI] [PubMed] [Google Scholar]
- Döffinger R, et al. X-linked anhidrotic ectodermal dysplasia with immunodeficiency is caused by impaired NF-kappaB signaling. Nat. Genet. 2001;27 doi: 10.1038/85837. 277-28. [DOI] [PubMed] [Google Scholar]
- Estrach S, Ambler CA, Lo Celso C, Hozumi K, Watt FM. Jagged 1 is a beta-catenin target gene required for ectopic hair follicle formation in adult epidermis. Development. 2006;133:4427–4438. doi: 10.1242/dev.02644. [DOI] [PubMed] [Google Scholar]
- Fessing MY, Sharova TY, Sharov AA, Atoyan R, Botchkarev VA. Involvement of the Edar signaling in the control of hair follicle involution (catagen) Am. J. Pathol. 2006;169:2075–2084. doi: 10.2353/ajpath.2006.060227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris MP, Linkhart BL, Fallon JF. Bmp7 mediates early signaling events during induction of chick epidermal organs. Dev. Dyn. 2004;231:22–32. doi: 10.1002/dvdy.20096. [DOI] [PubMed] [Google Scholar]
- Headon DJ, Overbeek PA. Involvement of a novel Tnf receptor homologue in hair follicle induction. Nat. Genet. 1999;22:370–374. doi: 10.1038/11943. [DOI] [PubMed] [Google Scholar]
- Headon DJ, Emmal SA, Ferguson BM, et al. Gene defect in ectodermal dysplasia implicates a death domain adapter in development. Nature. 2001;414:913–916. doi: 10.1038/414913a. [DOI] [PubMed] [Google Scholar]
- Itin PH, Fistarol SK. Ectodermal dysplasias. Am. J. Med. Genet. C. Semin. Med. Genet. 2004;131C:45–51. doi: 10.1002/ajmg.c.30033. [DOI] [PubMed] [Google Scholar]
- Kere J, Srivastava AK, Montonen O, et al. X-linked anhidrotic (hypohidrotic) ectodermal dysplasia is caused by mutation in a novel transmembrane protein. Nat. Genet. 1996;13:409–416. doi: 10.1038/ng0895-409. [DOI] [PubMed] [Google Scholar]
- Kim HJ, Choi CP, Uhm YK, et al. The association between endothelin-1 gene polymorphisms and susceptibility to vitiligo in a Korean population. Exp. Dermatol. 2007;16:561–566. doi: 10.1111/j.1600-0625.2007.00561.x. [DOI] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Masunaga T, Shimizu H, Ishiko A, Fujiwara T, Hashimoto T, Nishikawa T. Desmoyokin/AHNAK protein localizes to the non-desmosomal keratinocyte cell surface of human epidermis. J. Invest. Dermatol. 1995;104:941–945. doi: 10.1111/1523-1747.ep12606213. [DOI] [PubMed] [Google Scholar]
- Millar SE. WNTs: multiple genes, multiple functions. J. Invest. Dermatol. 2002;120:7–8. doi: 10.1046/j.1523-1747.2003.00001.x. [DOI] [PubMed] [Google Scholar]
- Mou C, Jackson B, Schneider P, Overbeek PA, Headon DJ. Generation of the primary hair follicle pattern. Proc. Natl. Acad. Sci. U. S. A. 2006;103:9075–9080. doi: 10.1073/pnas.0600825103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naito A, Yoshida H, Nishioka E, et al. TRAF6-deficient mice display hypohidrotic ectodermal dysplasia. Proc. Natl. Acad. Sci. U. S. A. 2002;99:8766–8771. doi: 10.1073/pnas.132636999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura M, Matzuk MM, Gerstmayer B, et al. Control of pelage hair follicle development and cycling by complex interactions between follistatin and activin. FASEB J. 2003;17:497–499. doi: 10.1096/fj.02-0247fje. [DOI] [PubMed] [Google Scholar]
- Närhi K, Järvinen E, Birchmeier W, Taketo MM, Mikkola ML, Thesleff I. Sustained epithelial beta-catenin activity induces precocious hair development but disrupts hair follicle down-growth and hair shaft formation. Development. 2008;135:1019–1028. doi: 10.1242/dev.016550. [DOI] [PubMed] [Google Scholar]
- Piskin G, Sylva-Steenland RM, Bos JD, Teunissen MB. In vitro and in situ expression of IL-23 by keratinocytes in healthy skin and psoriasis lesions: enhanced expression in psoriatic skin. J. Immunol. 2006;176:1908–1915. doi: 10.4049/jimmunol.176.3.1908. [DOI] [PubMed] [Google Scholar]
- Priolo M, Silengo M, Lerone M, Ravazzolo R. Ectodermal dysplasias: not only 'skin' deep. Clin. Genet. 2000;58:415–430. doi: 10.1034/j.1399-0004.2000.580601.x. [DOI] [PubMed] [Google Scholar]
- Rendl M, Lewis L, Fuchs E. Molecular dissection of mesenchymal-epithelial interactions in the hair follicle. PLoS Biol. 2005;3:e331. doi: 10.1371/journal.pbio.0030331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt M, Gillitzer R, Toksoy A, et al. Selective expression of calcium-binding proteins S100a8 and S100a9 at distinct sites of hair follicles. J. Invest. Dermatol. 2001;117:748–750. doi: 10.1046/j.0022-202x.2001.01485.x. [DOI] [PubMed] [Google Scholar]
- Schmidt-Ullrich R, Aebischer T, Hülsken J, Birchmeier W, Klemm U, Scheidereit C. Requirement of NF-kappaB/Rel for the development of hair follicles and other epidermal appendices. Development. 2001;128:3843–3853. doi: 10.1242/dev.128.19.3843. [DOI] [PubMed] [Google Scholar]
- Schmidt-Ullrich R, Tobin DJ, Lenhard D, Schneider P, Paus R, Scheidereit C. NF-kappaB transmits Eda A1/EdaR signalling to activate Shh and cyclin D1 expression, and controls post-initiation hair placode down growth. Development. 2006;133:1045–1057. doi: 10.1242/dev.02278. [DOI] [PubMed] [Google Scholar]
- Tuhkanen AL, Agren UM, Tammi MI, Tammi RH. CD44 expression marks the onset of keratinocyte stratification and mesenchymal maturation into fibrous dermis in fetal human skin. J. Histochem. Cytochem. 1999;47:1617–1624. doi: 10.1177/002215549904701213. [DOI] [PubMed] [Google Scholar]
- Yoshida A, Kanno H, Watabe D, Akasaka T, Sawai T. The role of heparin-binding EGF-like growth factor and amphiregulin in the epidermal proliferation of psoriasis in cooperation with TNFalpha. Arch. Dermatol. Res. 2008;300:37–45. doi: 10.1007/s00403-007-0809-y. [DOI] [PubMed] [Google Scholar]
- Zoupa M, Seppala M, Mitsiadis T, Cobourne MT. Tbx1 is expressed at multiple sites of epithelial-mesenchymal interaction during early development of the facial complex. Int. J. Dev. Biol. 2006;50:504–510. doi: 10.1387/ijdb.052116mz. [DOI] [PubMed] [Google Scholar]