Summary
Ageing is a complex, challenging phenomenon that will require multiple, interdisciplinary approaches to unravel its puzzles. To assist basic research on ageing, we developed the Human Ageing Genomic Resources (HAGR). This work provides an overview of the databases and tools in HAGR and describes how the gerontology research community can employ them. Several recent changes and improvements to HAGR are also presented. The two centrepieces in HAGR are GenAge and AnAge. GenAge is a gene database featuring genes associated with ageing and longevity in model organisms, a curated database of genes potentially associated with human ageing, and a list of genes tested for their association with human longevity. A myriad of biological data and information is included for hundreds of genes, making GenAge a reference for research that reflects our current understanding of the genetic basis of ageing. GenAge can also serve as a platform for the systems biology of ageing, and tools for the visualization of protein-protein interactions are also included. AnAge is a database of ageing in animals, featuring over 4,000 species, primarily assembled as a resource for comparative and evolutionary studies of ageing. Longevity records, developmental and reproductive traits, taxonomic information, basic metabolic characteristics, and key observations related to ageing are included in AnAge. Software is also available to aid researchers in the form of Perl modules to automate numerous tasks and as an SPSS script to analyse demographic mortality data. The Human Ageing Genomic Resources are available online at http://genomics.senescence.info.
Keywords: bioinformatics, comparative biology, functional genomics, genetics of longevity, systems biology
Introduction
Despite recent progress, human ageing is a largely controversial process. Many age-related changes have been described, yet there are multiple and conflicting theories regarding what mechanism(s) drive such changes (de Magalhaes, 2005). Moreover, we do not know why different species age at different paces and there is still no proven intervention capable of delaying or postponing the human ageing process (Olshansky et al., 2002). As such, it is clear that ageing is a complex, challenging phenomenon that will require extensive research using multiple, interdisciplinary approaches to unravel its puzzles.
To help researchers address the key questions in gerontology, we developed a collection of online resources entitled the Human Ageing Genomic Resources (HAGR). Previously, we described the databases and tools present in HAGR and how these resources were created and implemented (de Magalhaes et al., 2005a). Herein, our aim is to present a non-technical description of these resources to the gerontology community and provide an overview of how biogerontologists can employ them. Numerous recent updates and improvements are also described. We hope this work will help readers better realize what is on offer at HAGR and how its contents can be useful for their own research. The Human Ageing Genomic Resources are freely available online (no registration necessary) at http://genomics.senescence.info (Figure 1).
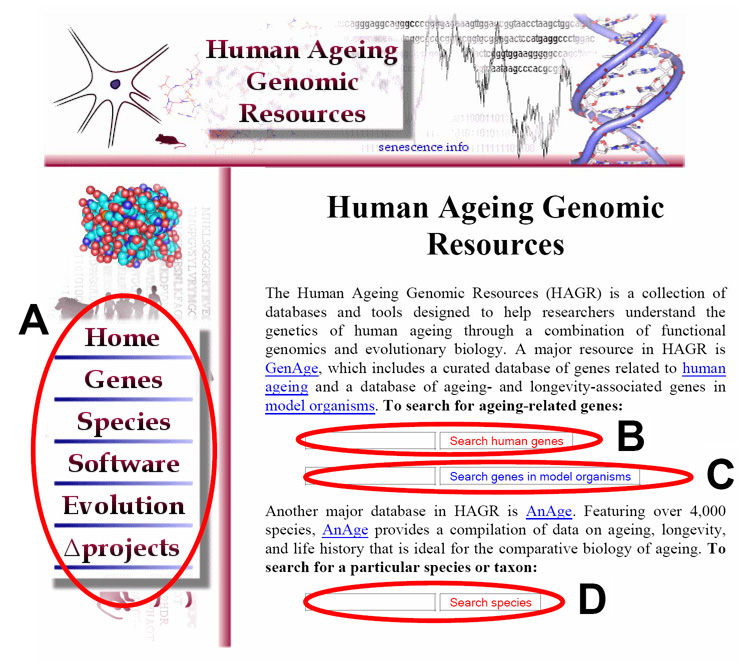
Figure 1.
From the main page of HAGR (http://genomics.senescence.info/) users have hyperlinks to the various databases and tools (A). Users may also search directly the HAGR databases: GenAge human genes dataset (B); GenAge model organisms’ gene dataset (C); and AnAge (D). HAGR pages are interconnected and feature hyperlinks to other relevant databases and tools as well as to the main menu.
GenAge: the ageing gene database
Philosophy and overview of resources
It is undisputed that genetic factors influence ageing. In a remarkable series of recent breakthroughs, a number of genes capable of altering the ageing process as a whole--or at least to a large degree--have been identified in animal models and even a few in humans (de Magalhaes, 2005; Finch & Ruvkun, 2001; Kenyon, 2005). Furthermore, multiple alleles have been examined for their association with human exceptional longevity (Vijg & Suh, 2005). This is a fascinating and important area of research, yet there are now so many genes being associated with ageing and longevity that keeping track of them all is becoming increasingly more difficult. Moreover, it is necessary now to study not only individual genes but their interactions with each other and with the environment, and how together genes give rise to a given phenotype: the so-called systems biology approach. To help researchers address these issues we created GenAge, a database of genes related to longevity and/or ageing.
GenAge consists of several searchable datasets. Considering the extraordinary discoveries in the genetics of ageing in model organisms, GenAge includes a dataset of genes associated with longevity and/or ageing in model organisms. We consider a given gene for inclusion in GenAge if genetic manipulations of the gene result in noticeable changes in the ageing phenotype and/or longevity. Most genes in GenAge are from the four typical model organisms: mice, worms, fruit flies, and yeast (Table 1). Strikingly, homologues of many genes--such as insulin receptors and sirtuins--have been shown to regulate ageing in model organisms separated by large evolutionary distances (Kenyon, 2005; Liu et al., 2005; Smith et al., 2008). Moreover, we have shown that genes associated with ageing and/or longevity in model organisms are evolutionary conserved in terms of having more homologues than predicted by chance (Budovsky et al., 2007; 2008) and exhibiting slower molecular evolution rates (de Magalhaes & Church, 2007). Therefore, it is now clear that at least some genes identified in model organisms may be relevant to human ageing.
Table 1.
Species in GenAge model organisms
| Organism (species) | Number of genes |
|---|---|
| Roundworm (Caenorhabditis elegans) | 555 |
| Baker’s yeast (Saccharomyces cerevisiae) | 87 |
| Fruit fly (Drosophila melanogaster) | 75 |
| Mouse (Mus musculus) | 68 |
| Filamentous fungus (Podospora anserine) | 2 |
| Golden hamster (Mesocricetus auratus) | 1 |
| Fission yeast (Schizosaccharomyces pombe) | 1 |
To allow researchers to focus specifically on human ageing, GenAge also features a dataset of genes that may regulate ageing in humans or that at least appear to be considerably associated with the human ageing phenotype. This dataset includes orthologues--derived from established databases, mainly InParanoid (O'Brien et al., 2005) but also HomoloGene (http://www.ncbi.nlm.nih.gov/HomoloGene)--of genes strongly associated with ageing in model organisms. Also included are genes in which mutations result in segmental progeroid syndromes, like the Werner’s syndrome gene, as well as genes critical in pathways previously related to ageing such as the insulin/insulin-like signalling pathway (de Magalhaes et al., 2005a). The main reason why each gene is included in the database is detailed in each entry (Table 2). Even though we are sure that GenAge will continue to change in the future as a result of new breakthroughs, this dataset represents the genes thus far most strongly associated with human ageing, which opens several opportunities for research (see below).
Table 2.
Criteria used to select entries for inclusion in the GenAge human dataset
| Main reason for selection | Number of genes |
|---|---|
| Evidence directly linking the gene product to ageing in humans | 3 |
| Evidence directly linking the gene product to ageing in a mammalian model organism | 27 |
| Evidence directly linking the gene product to ageing in a non-mammalian animal model | 25 |
| Evidence directly linking the gene product to ageing in a cellular model system | 14 |
| Evidence directly linking the gene product to the regulation or control of genes previously linked to ageing | 29 |
| Evidence linking the gene product to a pathway or mechanism linked to ageing | 64 |
| Indirect or inconclusive evidence linking the gene product to ageing or showing the gene product to be an effector of genes related to ageing | 99 |
Lastly, one of the latest additions to GenAge is a list of human genes analyzed for their possible association with human longevity (http://genomics.senescence.info/genes/longevity.html). All longevity association studies in humans we could find by the time of the latest update were added to this list. These include studies reporting negative results, which we see as essential since many genes display population-specific associations with longevity.
As of writing, build 14 of GenAge, updated on July 20, 2008, features 261 human genes and 789 genes in model organisms plus 258 longevity association experiments in humans. GenAge can be accessed at http://genomics.senescence.info/genes/.
Usage and applications
In the modern information age, even established experts in the field of ageing may find it difficult to keep up-to-date with all the data being generated. As such, the first use of GenAge is as a reference database, and GenAge is in its essence an organized information source on the genetics of ageing. In particular for the human dataset, a wealth of biological data is provided for each entry, including relevant information in the context of biogerontology (see example below) to an extent that is not available in larger, more generic databases like Entrez Gene. Therefore, to learn about the involvement of a given gene in ageing, a quick search in GenAge is the best place to start. GenAge features a sophisticated search engine and its user-friendly interface is easy to use by anyone with access to the Internet.
Searching GenAge can be done from its main page, which also includes hyperlinks to the human and models datasets. The established nomenclature of the human dataset is that of the Human Genome Nomenclature Committee (HGNC), but genes’ common names and frequently-used aliases--obtained from HGNC and organism-specific databases (described below)--can also be searched. For example, searching ‘ghbp’ (HAGR search pages are case-insensitive) in the human dataset will retrieve the growth hormone receptor (GHR) since GHR is frequently referred to as ‘GHBP’. In GHR’s entry (http://genomics.senescence.info/genes/entry.php?hgnc=GHR), apart from basic biological information, the potential link between GHR and ageing is described and key bibliographical references are cited: GHR gene products have been linked to ageing in mice--and this was the major reason for GHR’s inclusion in GenAge--but not in humans, even though patients with mutations in GHR have been documented. Searching for genes in the model organisms’ dataset is similar with users having the added option of being able to search for genes in a given species and, in fact, we provide quick hyperlinks to the four major model organisms.
Although the models dataset comprises all genes (to our knowledge) shown by the time of the latest update to statistically increase longevity or alter the ageing process in a noticeable way, in the human dataset we try to evaluate whether a given intervention is affecting the ageing process itself or not. For example, many mutations may increase longevity by decreasing the incidence of specific diseases, rather than by altering the basic process of ageing (de Magalhaes et al., 2005a; 2005b). Therefore, the human dataset is not merely an extension of the work conducted in model organisms and of its bibliography, but a manually selected list of the most pertinent human ageing candidate genes, each presented with a higher annotation level. We cite studies on whether the functions of ageing-associated genes in model organisms are conserved in their human orthologues. Likewise, we cite flaws in previous studies based on new published observations, though we have a neutral stance on conflicting findings from different research groups. Our policy is to cite all conflicting reports and let visitors make their own decisions on how to interpret them. By contrast, each entry in GenAge model organisms has only one reference: the first publication reporting an association of the gene with longevity or ageing. Moreover, one of the latest enhancements in the human dataset was the inclusion of Gene Ontology (GO) annotation. GO terms and annotation files were obtained from the Gene Ontology Consortium website (http://www.geneontology.org/) and provide an additional layer of description for the gene products in a cellular context (Ashburner et al., 2000).
Genes capable of delaying ageing or modulating multiple age-related changes represent important clues in advancing our knowledge of ageing. Therefore, a second use of GenAge is as a way to integrate different mechanisms of ageing. Numerous authors--including ourselves (Budovsky et al., 2007; 2008; de Magalhaes & Toussaint, 2004)--have attempted to reconstruct the different pathways linked with ageing and even study gene networks. In such works, GenAge is an adequate resource as it provides a framework for the functional genomics of ageing. For example, Xue et al. (2007) employed GenAge to construct a modular network of ageing and obtain insights into ageing, including the fact that genes connecting different modules are more likely to affect longevity and/or ageing, an hypothesis the authors validated experimentally in worms (Xue et al., 2007). In a sense, GenAge offers an overall view of what is presently known about the genetics of ageing in model organisms and in humans that can be used for numerous studies, including in contemporary functional genomics and systems biology methods.
By providing a list of candidate genes, the genes in GenAge can serve as basis for gene expression and genetic association longevity studies, including human studies, or even for clinical studies of interventions hypothesized to affect ageing. In fact, recent gene expression studies have used GenAge to focus on ageing-associated genes (Chen et al., 2008; Hardman & Ashcroft, 2008). Because researchers may have disparate opinions regarding the relevance of different model systems to understand human ageing, an important tool to investigate the human dataset as a whole is GenAge’s browser (http://genomics.senescence.info/genes/browser.php). The browser makes it possible to (among other things) retrieve only those entries that pass certain criteria related to the annotation in GenAge, such as selection process and gene function. For example, users can retrieve only genes associated with ageing in non-mammalian model organisms. Several criteria can be set, all of which must be passed for genes to be retrieved. Afterwards, users can select all or a sub-group of the genes retrieved, which can then be further analysed using simple statistical tools (http://genomics.senescence.info/genes/analyse.php). Cross-links between the different sections of GenAge make it easy to navigate between the different tools and datasets.
Another use of GenAge is for researchers to associate genes already under investigation with other, little-known genes, which can lead to new experimental designs. To do this, protein-protein interactions are one possible approach, and GenAge’s human dataset features 673 interactions, most of which manually curated obtained from the Human Protein Reference Database (HPRD) (Peri et al., 2003). In fact, one of our earliest applications of GenAge involved finding novel genes that may be linked to ageing by way of an analysis of protein-protein interactions. The principle being that proteins not previously thought to be related to ageing which interact with a large number of proteins directly linked to ageing might too be involved in ageing and are thus promising candidates for future studies (Budovsky et al., 2007; de Magalhaes & Toussaint, 2004). Similar works are made easy with GenAge. Protein-protein interactions with one or more genes as query can be visualized (Figure 2), or they can be downloaded for use with more advanced biological pathway analysis software.
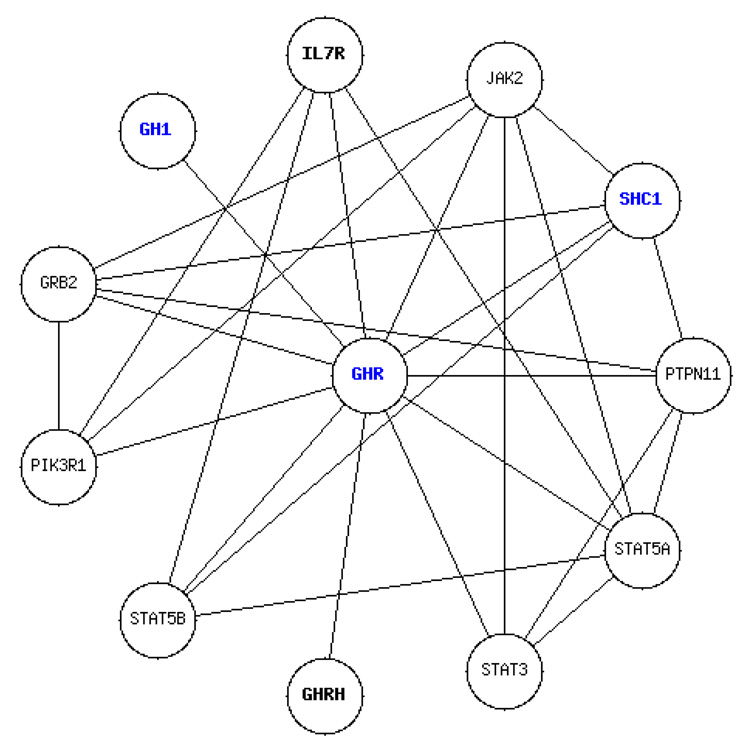
Figure 2.
Protein-protein interactions of GHR with one degree of freedom as generated by HAGR’s Interactions Graphical Display or IGD. Entries directly linked to mammalian ageing are highlighted in blue. Genes only associated with ageing indirectly or due to inconclusive evidence are displayed using a non-bold font.
GenAge can be helpful in more classical genetic studies of ageing and longevity. For example, if a given chromosomal region is identified as suspect in influencing longevity, it is now possible to look up which GenAge genes are present in that region (http://genomics.senescence.info/genes/map.php). This new feature of GenAge may assist researchers trying to map genes influencing ageing or longevity in human populations.
Although it was not our intention that GenAge became a bibliographic database, the manually-included literature in the human dataset (1,956 references in the latest build) may prove a valuable resource. In addition to providing a searchable bibliographic database where visitors can search titles, authors, and PubMed IDs (http://genomics.senescence.info/genes/biblio.php), and look up which references are most cited within GenAge, one novel feature in GenAge is bibliographic data-mining. The system employs the simplest method available, which is that of text co-occurrence (Stephens et al., 2001): the principle being that genes or proteins mentioned in the same academic work have higher chances of being related. Consequently, it is now possible to search through the bibliography of GenAge to find genes referred to in the same work to generate putative functional links (http://genomics.senescence.info/genes/mine_biblio.php). To use this feature, users must select--using the browser or search pages--or type the GenAge entries they wish to look up, and then set the threshold defining how many references two genes must have in common for them to be considered linked. This process can be useful, for instance, to discover novel associations between genes of interest--which may serve as inspiration to develop novel experiments--or to be kept up-to-date with the growing literature in gerontology.
Of course, GenAge has its limitations and while we aim to include the most relevant information (in particular in the context of genetics and ageing), not all data is available. For instance, searching ‘GHR’ in PubMed yields (at the time of writing) roughly 1,000 results. It would be unreasonable to include them all in GenAge. GenAge is an integrative database, however, that draws together data obtained from multiple other sources, including OMIM, SWISS-PROT, Entrez Gene, and HPRD (de Magalhaes et al., 2005a). Hyperlinks to these and other major databases are included to help researchers quickly locate additional information within other or broader biological scopes. Model organisms’ genes also have relevant hyperlinks specific to each organism: mouse genes link to the Mouse Genome Informatics website (http://www.informatics.jax.org/), fly genes link to FlyBase (http://flybase.bio.indiana.edu/), worm genes link to WormBase (http://www.wormbase.org/), and yeast genes link to the Saccharomyces Genome Database (http://www.yeastgenome.org/), in addition to hyperlinks to the AnAge database described below.
For the above reasons, the design and information in GenAge is prone to automated computational methods, and GenAge can be subject to a variety of data-mining algorithms. Since we included, if available, open reading frame, promoter, and protein sequences for each entry, a variety of bioinformatics approaches may be employed, such as studies of gene networks and transcriptional regulation. For example, we studied selection pressures on GenAge genes since humans and chimpanzees diverged to obtain novel insights, such as showing that ageing-related genes evolve under stronger functional constraints than expected by chance (de Magalhaes & Church, 2007). If researchers wish to apply sophisticated computational methods to data-mine GenAge, the whole database can be downloaded as tab-delimited file or as an XML file.
AnAge: the animal ageing and longevity database
Philosophy and overview of resources
As argued by many others (Austad, 2005; Finch, 1990; Miller, 2001), if we could understand why different species age at different rates, we would be able to gather key insights into the mechanisms of ageing. Comparative studies of ageing are thus one established way of conducting research on ageing, and multiple parameters can be compared between species (Finch, 1990; Frolkis & Muradian, 1991). With the comparative biology of ageing as its primary goal, AnAge is a database of longevity and ageing in animal species. Though commonly used model organisms like yeast are included, AnAge focuses primarily on chordates and a greater attention is given to species evolutionary closer to humans like primates and mammals (Table 3). Arguably, AnAge is the most extensive, complete, and accurate database of longevity records available in any format. In addition, observations on physiological or pathological changes with age in animals are (where available) featured.
Table 3.
Number of entries per taxa in AnAge
| Taxa (scientific name) | Number of entries |
|---|---|
| Mammals (Mammalia) | 1,331 |
| Birds (Aves) | 1,098 |
| Reptiles (Reptilia) | 539 |
| Amphibians (Amphibia) | 169 |
| Fishes (Actinopterygii, Chondrichthyes, etc.) | 962 |
| Non-chordates | 28 |
The main aim of AnAge is to provide data for comparative and evolutionary studies of ageing that allow researchers to study how and why the ageing phenotype and longevity vary across phylogeny and which factors influence such differences. Although demographic measurements of ageing are included in AnAge, these require detailed animal studies that are rarely available and thus represent only a small fraction of the data. Maximum lifespan as estimated from longevity records will continue to be the standard--even if not perfect--measure of ageing (de Magalhaes, 2006), and this is available for the large majority of AnAge entries. Recently, we incorporated the longevity information from FishBase (Froese & Pauly, 2008) and from Richard Weigl’s excellent compilation of mammalian longevity records in captivity (Weigl, 2005) as well as data from mammalian ageing expert Steven Austad, which includes personal correspondence with zoo personnel, field biologists, and veterinarians.
Included in AnAge is also extensive data for a number of factors that might bias comparative studies of ageing, such as body size, metabolic rates, and developmental schedules, so authors can analyze these factors in their studies. Otherwise, correlations with longevity may merely reflect a correlation between the parameter under study and, for example, body size which in turn tends to correlate with longevity (Charnov, 1993). Indeed, we recently conducted a number of analyses of factors related to longevity (de Magalhaes et al., 2007). Succinctly, we showed that metabolic rate, when corrected for the effects of body size and phylogeny, does not correlate with longevity or ageing rates. In contrast, developmental schedules such as age at maturity appear to be robust predictors of longevity and ageing rates. This work highlights the importance of taking potentially confounding traits in consideration in ageing studies that employ the comparative method. Consequently, a great effort has been made to include updated and extensive developmental and reproductive data in AnAge.
At the time of writing (build 10, updated on April 18, 2008), AnAge features 4,122 entries, including 1,331 mammals (Table 3). Our focus is on accuracy and quality, however, not quantity, and only species for which we have confidence in the data are featured. For instance, longevity records based on single or a few observations, particularly if not in captivity, are normally excluded, at least if much better data is available for similar species (e.g., for birds, bats, and cetaceans AnAge features extensive records from animals in the wild since very few records in captivity are available). Likewise, although we use data from websites, these have to come from authoritative sources like zoos, parks, and academic institutions. Unverified longevity records or those from unreliable sources may be cited as anecdotes in the observations section of each entry but do not become the established maximum lifespan for that species. AnAge is available at http://genomics.senescence.info/species/.
Usage and applications
Like GenAge, AnAge can be useful as an information source and web portal by allowing researchers to learn more about the ageing process of a particular species in a user-friendly environment. To use AnAge, its search engine--also accessible from AnAge’s main page--is the place to start (http://genomics.senescence.info/species/query.php). Species’ and common names can be searched (see example below). In fact, terms at any taxonomic level can be searched and a recent addition to AnAge has been a number of entries at higher taxonomic levels. The taxonomy of AnAge follows that of the Integrated Taxonomic Information System (http://www.itis.usda.gov).
AnAge provides an up-to-date synthesis of what is known about longevity and ageing of a given organism, which is particularly useful for those not familiar with it. Furthermore, species with unique ageing phenotypes or of special interest to biogerontologists, such as species with negligible senescence, are featured. Again, like GenAge, AnAge has its limits and external hyperlinks point users to further sources of information such as the Tree of Life (http://tolweb.org) and Animal Diversity Web (http://animaldiversity.ummz.umich.edu) websites. To obtain pictures of a given species the popular search engine Google (http://images.google.com) is used.
Like GenAge, AnAge features a browser (http://genomics.senescence.info/species/browser.php). AnAge’s browser is somewhat different, though, as it allows users to browse through the taxonomy of species in AnAge. Once a given set of entries has been selected it can be surveyed to gather simple descriptive statistics (http://genomics.senescence.info/species/survey.php). As in GenAge, it is easy to navigate between the different AnAge tools due to extensive cross-links. Several species of interest can be selected with a mouse click and the parameter of interest’s average ± standard deviation can be calculated. For instance, to search for turtles one would simply type ‘testudines’, the order to which turtles belong to. The list of 122 turtles in AnAge would then be displayed according to taxonomy or longevity, and one could then obtain the averages and standard deviations for longevity, adult body size, age at sexual maturity, or other trait present in AnAge. The phylogeny of the species selected can be inferred from their taxonomy using the Phylogenetic Tree Plotter (http://genomics.senescence.info/species/ptp.php), though its use is limited to fewer than 50 species and should not be employed as a replacement for established phylogenies--integrating published phylogenies in AnAge is, however, in our future plans.
Given the size and scope of AnAge, it is a major resource to study the evolution of ageing, longevity, and life-history. Indeed, several groups have taken advantage of AnAge to study the evolutionary events responsible for species differences in ageing and longevity. For example, a number of studies have employed AnAge to investigate how features of the mitochondrial DNA, such as mutation and substitution rates, relate to longevity and other life-history traits (Khaidakov et al., 2006; Lehmann et al., 2006; 2008; Moosmann & Behl, 2008; Nabholz et al., 2008; Welch et al., 2008). Others have employed AnAge to compare experimentally-determined parameters, such as cellular stress resistance (Harper et al., 2007), rates of free radical production in mitochondria (Lambert et al., 2007), and telomerase activity (Seluanov et al., 2007), with species longevity.
Given the size of AnAge, downloading the entire database as a tab-delimited or XML file may be necessary for some analyses and is also possible.
Software, tutorials, and additional tools
It is crucial that the information in HAGR is kept up-to-date and thus far AnAge and GenAge have been updated regularly. To update our databases we make use of computer scripts to automatically retrieve data from other online resources. As believers in open source and sharing, not only the databases but all these scripts, most of which written in Perl, are available for download as part of the Ageing Research Computational Tools (ARCT). Admittedly, a basic understanding of Perl is necessary for ARCT to be useful. This is by far the greatest weakness of the ARCT toolkit, which thus appears to be useful only to those in the community with a moderate-to-high level of computer knowledge. Still, it may be a valuable set of tools to those wishing to apply bioinformatics to study ageing. ARCT is available for download at http://genomics.senescence.info/software/.
Also available is a script for use with SPSS for demographic analysis. Succinctly, this script allows users with the SPSS statistical package (SPSS Inc., Chicago, IL) to determine whether the slope of the age-related mortality curve changes between two or more different cohorts, a method commonly employed to infer whether the rate of ageing was altered or not, as detailed previously (de Magalhaes et al., 2005b).
The Human Ageing Genomic Resources are part of the parent website senescence.info (http://www.senescence.info), which one of us (J. P. M.) started in 1997 as a mix of speculative essays, hypotheses, and reviews. Primarily, senescence.info is an educational resource on the science of ageing, including not only information on the biology of ageing and advice for students but discussions on relevant issues to gerontology, such as the sociological implications of ageing research and lifespan increase. Of interest, senescence.info features numerous tutorials regarding ageing research aimed at students and newcomers to the field.
Lastly, a non-exhaustive list of people and companies working on ageing and related disciplines is available at the WhosAge database (http://whoswho.senescence.info/). Presently, WhosAge features 171 researchers and 13 companies, including a brief description of the work of each company or individual, contact information, and hyperlinks to websites (if available) and further sources of information such as hyperlinks to PubMed and Google searches. WhosAge is searchable--it employs the engine of GenAge and AnAge--and can be used to find a specific individual or company, or search for researchers working at a given institution or on a particular topic.
Concluding remarks
We designed the databases and tools in HAGR primarily so they can be useful to researchers working on the science of ageing. The audience of HAGR ranges from beginners to experts in gerontology, from experimentalists to theorists, from computer illiterates to expert computational biologists. Generally, while GenAge and AnAge use simple and intuitive navigation tools, the ability to download the databases gives ample freedom to computational biologists to employ more powerful methods, as exemplified in the numerous works that have already taken advantage of these datasets.
Because our aim has always been to develop HAGR in collaboration with the gerontology community, nearly all pages have a hyperlink to our contact information or to an easy-to-use feedback form. Visitors are thus encouraged to send us feedback on errors they find, propose enhancements, and suggest features they would like to see in HAGR--and many have already done so. Understanding ageing is a collaborative effort. The Human Ageing Genomic Resources will hopefully reflect that spirit and continue to improve not only due to our efforts but thanks to the feedback from the research community working on the science of ageing. In the past 12 months, these online resources attracted roughly 15,000 ± 7,500 (SD) unique users per month. In the future, we hope to continually upgrade, update, and expand the resources in HAGR, as well as develop new tools that can benefit the gerontology community. Ideally, we would like HAGR to become a common platform for biogerontologists and thus invite everyone in the community to visit, use, and help us improve HAGR.
Acknowledgments
We are indebted to many people who have helped in this project, of which we highlight the assistance and opinions of Olivier Toussaint and Steven Austad. J. P. de Magalhães is supported by a NIH-NHGRI CEGS grant to G. Church. The work of V. Fraifeld’s lab is partially supported by the European Union FP7 Health Research Grant number HEALTH-F4-2008-202047 (to V.F.).
References
- Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Austad SN. Diverse aging rates in metazoans: targets for functional genomics. Mech. Ageing Dev. 2005;126:43–49. doi: 10.1016/j.mad.2004.09.022. [DOI] [PubMed] [Google Scholar]
- Budovsky A, Abramovich A, Cohen R, Chalifa-Caspi V, Fraifeld V. Longevity network: construction and implications. Mech. Ageing Dev. 2007;128:117–124. doi: 10.1016/j.mad.2006.11.018. [DOI] [PubMed] [Google Scholar]
- Budovsky A, Tacutu R, Yanai H, Abramovich A, Wolfson M, Fraifeld V. Common gene signature of cancer and longevity. Mech. Ageing Dev. 2008 doi: 10.1016/j.mad.2008.04.002. [DOI] [PubMed] [Google Scholar]
- Charnov EL. Life History Invariants: Some Explorations of Symmetry in Evolutionary Ecology. Oxford: Oxford University Press; 1993. [Google Scholar]
- Chen DP, Weber SC, Constantinou PS, Ferris TA, Lowe HJ, Butte AJ. Novel integration of hospital electronic medical records and gene expression measurements to identify genetic markers of maturation. Pac. Symp. Biocomput. 2008:243–254. [PMC free article] [PubMed] [Google Scholar]
- de Magalhaes JP, Toussaint O. GenAge: a genomic and proteomic network map of human ageing. FEBS Lett. 2004;571:243–247. doi: 10.1016/j.febslet.2004.07.006. [DOI] [PubMed] [Google Scholar]
- de Magalhaes JP. Open-minded scepticism: inferring the causal mechanisms of human ageing from genetic perturbations. Ageing Res. Rev. 2005;4:1–22. doi: 10.1016/j.arr.2004.05.003. [DOI] [PubMed] [Google Scholar]
- de Magalhaes JP, Costa J, Toussaint O. HAGR: the Human Ageing Genomic Resources. Nucleic Acids Res. 2005a;33:D537–D543. doi: 10.1093/nar/gki017. Database Issue. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Magalhaes JP, Cabralq JA, Magalhaes D. The influence of genes on the aging process of mice: a statistical assessment of the genetics of aging. Genetics. 2005b;169:265–274. doi: 10.1534/genetics.104.032292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Magalhaes JP. Species selection in comparative studies of aging and antiaging research. In: Conn PM, editor. Handbook of Models for Human Aging. Burlington, MA: Elsevier Academic Press; 2006. pp. 9–20. [Google Scholar]
- de Magalhaes JP, Church GM. Analyses of human-chimpanzee orthologous gene pairs to explore evolutionary hypotheses of aging. Mech. Ageing Dev. 2007;128:355–364. doi: 10.1016/j.mad.2007.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Magalhaes JP, Costa J, Church GM. An analysis of the relationship between metabolism, developmental schedules, and longevity using phylogenetic independent contrasts. J. Gerontol. A Biol. Sci. Med. Sci. 2007;62:149–160. doi: 10.1093/gerona/62.2.149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finch CE. Longevity, Senescence, and the Genome. Chicago and London: The University of Chicago Press; 1990. [Google Scholar]
- Finch CE, Ruvkun G. The genetics of aging. Annu. Rev. Genomics Hum. Genet. 2001;2:435–462. doi: 10.1146/annurev.genom.2.1.435. [DOI] [PubMed] [Google Scholar]
- Froese R, Pauly D. FishBase. World Wide Web electronic publication; 2008. www.fishbase.org version (04/2008) [Google Scholar]
- Frolkis VV, Muradian KK. Life Span Prolongation. Boca Raton: CRC Press; 1991. [Google Scholar]
- Hardman MJ, Ashcroft GS. Estrogen, not intrinsic aging, is the major regulator of delayed human wound healing in the elderly. Genome Biol. 2008;9:R80. doi: 10.1186/gb-2008-9-5-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harper JM, Salmon AB, Leiser SF, Galecki AT, Miller RA. Skin-derived fibroblasts from long-lived species are resistant to some, but not all, lethal stresses and to the mitochondrial inhibitor rotenone. Aging Cell. 2007;6:1–13. doi: 10.1111/j.1474-9726.2006.00255.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenyon C. The plasticity of aging: insights from long-lived mutants. Cell. 2005;120:449–460. doi: 10.1016/j.cell.2005.02.002. [DOI] [PubMed] [Google Scholar]
- Khaidakov M, Siegel ER, Shmookler Reis RJ. Direct repeats in mitochondrial DNA and mammalian lifespan. Mech. Ageing Dev. 2006;127:808–812. doi: 10.1016/j.mad.2006.07.008. [DOI] [PubMed] [Google Scholar]
- Lambert AJ, Boysen HM, Buckingham JA, Yang T, Podlutsky A, Austad SN, Kunz TH, Buffenstein R, Brand MD. Low rates of hydrogen peroxide production by isolated heart mitochondria associate with long maximum lifespan in vertebrate homeotherms. Aging Cell. 2007;6:607–618. doi: 10.1111/j.1474-9726.2007.00312.x. [DOI] [PubMed] [Google Scholar]
- Lehmann G, Budovsky A, Muradian KK, Fraifeld VE. Mitochondrial genome anatomy and species-specific lifespan. Rejuvenation Res. 2006;9:223–226. doi: 10.1089/rej.2006.9.223. [DOI] [PubMed] [Google Scholar]
- Lehmann G, Segal E, Muradian KK, Fraifeld VE. Do mitochondrial DNA and metabolic rate complement each other in determination of the mammalian maximum longevity? Rejuvenation Res. 2008;11:409–417. doi: 10.1089/rej.2008.0676. [DOI] [PubMed] [Google Scholar]
- Liu X, Jiang N, Hughes B, Bigras E, Shoubridge E, Hekimi S. Evolutionary conservation of the clk-1-dependent mechanism of longevity: loss of mclk1 increases cellular fitness and lifespan in mice. Genes Dev. 2005;19:2424–2434. doi: 10.1101/gad.1352905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller RA. A position paper on longevity genes. Sci. Aging Knowledge Environ. 2001;2001:vp6. doi: 10.1126/sageke.2001.9.vp6. [DOI] [PubMed] [Google Scholar]
- Moosmann B, Behl C. Mitochondrially encoded cysteine predicts animal lifespan. Aging Cell. 2008;7:32–46. doi: 10.1111/j.1474-9726.2007.00349.x. [DOI] [PubMed] [Google Scholar]
- Nabholz B, Glemin S, Galtier N. Strong variations of mitochondrial mutation rate across mammals--the longevity hypothesis. Mol. Biol. Evol. 2008;25:120–130. doi: 10.1093/molbev/msm248. [DOI] [PubMed] [Google Scholar]
- O'Brien KP, Remm M, Sonnhammer EL. Inparanoid: a comprehensive database of eukaryotic orthologs. Nucleic Acids Res. 2005;33:D476–D480. doi: 10.1093/nar/gki107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olshansky SJ, Hayflick L, Carnes BA. No truth to the fountain of youth. Sci. Am. 2002;286:92–95. doi: 10.1038/scientificamerican0602-92. [DOI] [PubMed] [Google Scholar]
- Peri S, Navarro JD, Amanchy R, Kristiansen TZ, Jonnalagadda CK, Surendranath V, Niranjan V, Muthusamy B, Gandhi TK, Gronborg M, et al. Development of human protein reference database as an initial platform for approaching systems biology in humans. Genome Res. 2003;13:2363–2371. doi: 10.1101/gr.1680803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seluanov A, Chen Z, Hine C, Sasahara TH, Ribeiro AA, Catania KC, Presgraves DC, Gorbunova V. Telomerase activity coevolves with body mass not lifespan. Aging Cell. 2007;6:45–52. doi: 10.1111/j.1474-9726.2006.00262.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith ED, Tsuchiya M, Fox LA, Dang N, Hu D, Kerr EO, Johnston ED, Tchao BN, Pak DN, Welton KL, et al. Quantitative evidence for conserved longevity pathways between divergent eukaryotic species. Genome Res. 2008;18:564–570. doi: 10.1101/gr.074724.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephens M, Palakal M, Mukhopadhyay S, Raje R, Mostafa J. Detecting gene relations from Medline abstracts. Pac. Symp. Biocomput. 2001:483–495. doi: 10.1142/9789814447362_0047. [DOI] [PubMed] [Google Scholar]
- Vijg J, Suh Y. Genetics of longevity and aging. Annu. Rev. Med. 2005;56:193–212. doi: 10.1146/annurev.med.56.082103.104617. [DOI] [PubMed] [Google Scholar]
- Weigl R. Longevity of Mammals in Captivity; from the Living Collections of the World. Stuttgart: Kleine Senckenberg-Reihe; 2005. [Google Scholar]
- Welch JJ, Bininda-Emonds OR, Bromham L. Correlates of substitution rate variation in mammalian protein-coding sequences. BMC Evol. Biol. 2008;8:53. doi: 10.1186/1471-2148-8-53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue H, Xian B, Dong D, Xia K, Zhu S, Zhang Z, Hou L, Zhang Q, Zhang Y, Han JD. A modular network model of aging. Mol. Syst. Biol. 2007;3:147. doi: 10.1038/msb4100189. [DOI] [PMC free article] [PubMed] [Google Scholar]