Abstract
Dietary selenium (Se) requirements in rats have been based largely upon glutathione peroxidase-1 (Gpx1) enzyme activity and Gpx1 mRNA levels can also be used to determine Se requirements. The identification of the complete selenoprotein proteome suggests that we might identify additional useful molecular biomarkers for assessment of Se status. To characterize Se regulation of the entire rat selenoproteome, weanling male rats were fed a Se-deficient diet (<0.01 μg Se/g) supplemented with graded levels of Se (0–0.8 μg/g diet) for 28 d, Se status was determined by tissue Se concentration and selenoenzyme activity, and selenoprotein mRNA abundance in liver, kidney, and muscle was determined by quantitative real-time-PCR. Tissue Se and selenoenzyme biomarkers indicated that minimal Se requirements were ≤0.1 μg Se/g diet for most biomarkers. Liver Gpx1 mRNA also decreased to <10% of Se-adequate levels, with a minimum Se requirement at 0.07 μg/g diet. Five selenoprotein mRNA in liver, 4 in kidney, and 2 in muscle decreased to <41% of Se-adequate levels, all with minimum Se requirements at ≤0.07 μg/g diet; the majority of selenoprotein mRNA in each tissue were not significantly regulated by Se status, and 1 selenoprotein, selenophosphate synthetase-2, was upregulated in Se-deficient kidney. Plateau breakpoints for all regulated selenoprotein mRNA were very similar, suggesting that 1 underlying mechanism is in play in Se regulation of selenoprotein mRNA. Lastly, we did not find any selenoprotein mRNA that could be used as biomarkers for super-nutritional/anticarcinogenic levels (up to 0.8 μg Se/g diet) of Se.
Introduction
Dietary selenium (Se) requirements for rats were first reported to be 0.04 μg/g diet based on prevention of liver necrosis (1) and 0.05 μg Se/g diet based on maintenance of growth in weanling rats (2). The discovery that Se is an essential cofactor for glutathione peroxidase (Gpx)5 and that Gpx activity drops dramatically in Se deficiency (3) provided a biochemical marker that was used to establish a dietary requirement of 0.1 μg Se/g diet in the rapidly growing rat (4,5). Discovery of additional selenoproteins, such as phospholipid hydroperoxide Gpx (Gpx4), selenoprotein P (Sepp1), iodothyronine deiodinase (Dio), thioredoxin reductase-1 (Txnrd1), selenoprotein W (Sepw1), and plasma Gpx (Gpx3), provided further biomarkers of Se status that also indicate that the dietary Se requirements are ≤0.1 μg Se/g diet (6–11). The current dietary Se requirements (12) now cite 0.15 μg Se/g diet as the requirement for growing rats; this level includes a safety factor and is based in part on prevention of cataract development in rats fed high-sucrose diets.
Gpx1 mRNA level also drops dramatically in Se deficiency in rats (13), increases sigmoidally with increasing dietary Se, and reaches well-defined plateaus (6,14–16), providing a molecular biology-based biomarker for Se status. Dietary Se requirements based on hepatic Gpx1 mRNA levels are ∼0.05 μg Se/g diet for both male and female rats, even though female rats have twice the level of liver Gpx1 mRNA as well as Gpx1 activity compared with male rats (14). In contrast, Gpx4 mRNA levels are not regulated by Se status in rats, eliminating the use of Gpx4 mRNA as a molecular biomarker for assessing Se requirements (6). Effects of Se status on mRNA levels for several other selenoproteins have also been studied, including Sepp1, Txnrd1, and Dio1 (6,9,14–17), but these mRNA do not fall as dramatically as does Gpx1 mRNA in liver and other tissues; requirements based on these molecular biomarkers are also near 0.05 μg Se/g diet and are generally less than the levels required for maximal enzyme activities (16).
Elegant use of the nucleotide (nt) sequences required for Se incorporation into all selenoproteins led Gladyshev et al. (18) to identify the complete selenoprotein proteome or selenoproteome in humans, rodents, and other species. This revealed that rats and mice have 24 selenoproteins, including 6 that were previously unidentified. These new selenoproteins and the availability of mouse commercial microarrays provided us with the tools to conduct an initial characterization of Se regulation of the complete selenoproteome in mice (19). We found in mice that most selenoprotein mRNA were not significantly regulated by Se status, but that 3 selenoproteins were highly downregulated in Se deficiency. This in turn suggested that an assessment of Se regulation of the complete selenoproteome in the well-characterized rat model had the potential to identify useful additional molecular biomarkers for Se status.
The probing of the complete rodent Se regulon has wider potential implications. High levels of dietary Se have been implicated in the prevention of cancer in animal models (20,21) and in humans (22). This anticarcinogenic activity is associated with dietary Se levels well above that needed for maximum selenoprotein activity for the well-characterized proteins (20,21), but this has not been studied for most of the other selenoproteins. Thus, our hypothesis was that characterization of Se regulation of the full selenoprotein proteome would identify new potential molecular biomarkers for Se status and might reveal novel regulation patterns that are distinct from the well-studied selenoproteins.
To characterize the complete selenoprotein regulon in rats, we conducted 2 experiments by feeding graded levels of dietary Se to young, rapidly growing rats. Our objectives were to expand the molecular biology biomarkers for Se status to include the full selenoproteome in the well-characterized rat model, to expand the selenoprotein regulon analysis to include multiple tissues, and to use super-nutritional levels of dietary Se in this characterization of the selenoprotein regulon.
Methods
Reagents
Molecular biology reagents were purchased from Promega, Invitrogen, or Sigma. All other chemicals were of molecular biology or reagent grade.
Animals and diets
Expt. 1.
Male weanling rats (n = 32; 21 d old) were obtained from Holtzman and housed individually in hanging wire-mesh cages. The basal diet was a Se-deficient torula-yeast diet (0.007 μg Se/g by analysis), supplemented with 100 mg/kg of all-rac-α-tocopherol acetate to ensure prevention of liver necrosis, and supplemented with 0.4% l-methionine to ensure adequate growth, as described previously (14,16). Rats were allocated randomly to dietary treatments and fed the basal diet supplemented with graded levels of Se (0, 0.02, 0.05, 0.075, 0.1, 0.15, 0.2, or 0.3 μg Se/g as Na2SeO3) for 28 d (4 rats per treatment).
Expt. 2.
Male weanling rats (n = 60; 21 d old) were obtained from Holtzman and treated as in Expt. 1. Rats were fed the basal Se-deficient diet (0.005 μg Se/g by analysis) supplemented with graded levels of Se (0, 0.016, 0.04, 0.06, 0.08, 0.12, 0.16, 0.24, 0.4, or 0.8 μg Se/g) as Na2SeO3 for 28 d (6 rats per treatment). Body weight was measured bi-weekly. Animals had free access to feed and water and the care and treatment protocols were approved by the Institutional Animal Care and Use Committee at the University of Missouri (Expt. 1) and University of Wisconsin (Expt. 2).
Tissue analysis
Rats were anesthetized with ether and blood was drawn by cardiac puncture using EDTA-containing syringes. Livers were perfused in situ with ice-cold 0.15 mol/L KCl and liver, kidney, and muscle were removed and quick-frozen in liquid nitrogen (16). Blood was centrifuged at 1500 × g, 15 min at 4°C (Eppendorf 5415R, F-45–24–11 rotor, Brinkmann) to separate plasma from red cells, which were reconstituted to original volume using saline phosphate buffer (76 mmol/L NaCl, 50 mmol/L sodium phosphate, pH 7.4). Liver and kidney (only Expt. 2) and diet Se concentrations (Expt. 1 and 2) were determined by neutron activation analysis (23).
Enzyme activity assays
Liver, kidney, and muscle were homogenized in 9 volumes of sucrose buffer (20 mmol/L Tris/HCl, pH 7.4, 0.25 mol/L sucrose, 1.1 mmol/L EDTA, and 0.1% peroxide-free Triton × 100) and centrifuged at 10,000 × g, 15 min at 4°C (model J2–21M, JA-21 rotor, Beckman Instruments) as described previously (6,16). Gpx1 activity in liver, kidney, muscle, and RBC (designated as Gpx1) and Gpx3 activity in plasma (designated as Gpx3) were measured by the coupled assay procedure (24) using 120 μmol/L H2O2. Gpx4 activity was measured by the coupled assay procedure (6) using 78 μmol/L phosphatidylcholine hydroperoxide, the specific substrate. For both assays, 1 enzyme unit is the amount of enzyme that will oxidize 1 μmol/min of GSH under these conditions. Txnrd was measured using the gold-inhibition assay with DTNB as a substrate (25). The protein concentration of each sample was determined by the method of Lowry et al. (26).
RNA isolation and analysis
Total RNA from liver and kidney (50–100 mg tissue; n = 3/diet group) was isolated using the guanidinium isothiocyanate method with TRIzol Reagent (catalog no. 15596–026, Invitrogen) following the manufacturer's protocol. The RNA pellet was dissolved in diethyl pyrocarbonate-treated water and quantitated using a ND-1000 UV-Vis spectrophotometer (NanoDrop Technologies).
Relative mRNA abundance was determined by quantitative RT-PCR. RNA (1 μg) was reverse transcribed to cDNA using the RETROscript kit (AM1710, Ambion) following the manufacturer's instructions. Gene-specific primers were designed to span a splice-junction and amplify an ∼150 base segment (Supplemental Table 1). The final 25-μL real-time reactions contained 10 ng reverse transcribed RNA, 0.2 mmol/L gene-specific forward and reverse primers, and 1× SybrGreen PCR Master mix (no. 4309155, Applied Biosystems). Reactions were followed in an ABI Prism 7000 (Applied Biosystems) with initial stages of 50°C for 2 min and 95°C for 10 min, followed by 50 cycles consisting of 95°C for 15 s and 60°C for 2 min. A dissociation curve was run for each plate to confirm the production of a single product. The amplification efficiency for each gene was determined using the DART-PCR program (27). The mRNA relative abundance was calculated according to Pfaffl (28), accounting for gene-specific efficiencies, normalized to the mean of β-actin and glyceraldehyde-3-phosphate dehydrogenase, and expressed as a percentage of Se-adequate levels (0.2 μg Se/g in Expt. 1; 0.24 μg Se/g or plateau level in Expt. 2).
Statistical analysis
Data are presented as means ± SEM. For Expt. 1 for all analyses, n = 3 per treatment. For Expt. 2, n = 5 or 6 per treatment for body weight and for all enzyme analyses except skeletal muscle Gpx4; n = 3/treatment for skeletal muscle Gpx4 activity, for Se concentrations, and for liver and kidney mRNA analyses; and n = 3 or 4 per treatment for skeletal muscle mRNA analysis. All data were analyzed by ANOVA using a fixed model testing the main effect of diet (SAS Institute). When the main effect of diet was significant, differences between means were assessed by Duncan's multiple range analysis (P < 0.05), with Kramer's modification for unequal class sizes where necessary (29). When variance equality was significant, as tested by Bartlett's test (α = 0.05), significant differences between means were assessed instead by the Scheffé F-test. For all tests, P < 0.05 was considered significant. The plateau breakpoint for each Se response curve, defined as the intersection of the line tangent to the point of steepest slope and the plateau, was calculated as described previously (14–16) using sigmoidal or hyperbolic regression analysis (Sigma Plot, Jandel Scientific) to estimate the minimum dietary Se necessary to obtain plateau responses.
Results
Animal growth.
In both experiments, initial body weights of the rats did not differ and dietary Se supplementation did not alter the growth rate in either Expt. 1 (not shown) or Expt. 2 (Supplemental Fig. 1), where the rats grew at an mean rate of 7.7 g/d. Thus, in these studies, neither Se deficiency nor Se supplementation affected growth.
Tissue Se analysis.
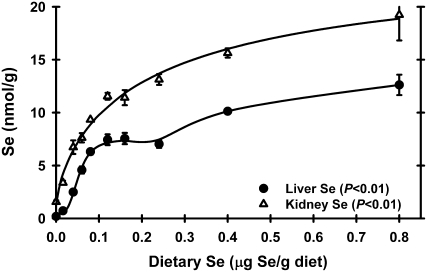
Tissue Se levels were not determined in Expt. 1. In Expt. 2, liver Se concentrations in rats fed the basal diet were 3% of levels in Se-adequate (0.24 μg Se/g diet) rats, showing that these rats were Se deficient (Fig. 1). Se supplementation resulted in a sigmoidal response in liver Se concentration, with a plateau breakpoint at 0.08 μg Se/g diet (Table 1). In this study using super-nutritional levels of 0.4 and 0.8 μg Se/g diet, however, liver Se levels increased above the plateau and were 70% higher in rats fed 0.8 μg Se/g than in rats fed 0.08–0.24 μg Se/g. Kidney Se levels had a similar response. Se-deficient kidney Se was 12% of levels in Se-adequate kidney, with a plateau breakpoint at 0.11 μg Se/g diet, and then kidney Se further increased in rats fed the 0.8 μg Se/g diet to levels 70% above Se-adequate levels.
FIGURE 1 .
Liver and kidney Se concentration in male weanling rats fed diets containing graded levels of Se for 28 d. Values are means ± SEM, n = 5 or 6. Detailed statistical analysis is provided in Supplemental Table 2.
TABLE 1.
Se requirement hierarchy in growing rats1
| Biomarker | Extent of regulation (P-value)2 | Minimum requirement3 |
|---|---|---|
| μg Se/g diet | ||
| Conventional biomarkers | ||
| Growth | Low (0.99 ) | <0.01 |
| Muscle Gpx4 activity | Low (0.83) | <0.01 |
| Plasma Gpx3 activity | Very high (<0.0001) | 0.06 |
| Liver Gpx4 activity | Moderate (<0.0001) | 0.06 |
| Liver Txnrd activity | Very high (<0.0001) | 0.06 |
| Kidney Gpx4 activity | Moderate (<0.0001) | 0.07 |
| Liver Se concentration | Very high (<0.0001) | 0.08 |
| RBC Gpx1 activity | High (<0.0001) | 0.08 |
| Liver Gpx1 activity | Very high (<0.0001) | 0.09 |
| Kidney Se concentration | High (<0.0001) | 0.11 |
| Kidney Gpx1 activity | Very high (<0.0001) | 0.12 |
| Muscle Gpx1 activity | Very high (<0.0001) | 0.13 |
| Molecular biomarkers | ||
| Liver Gpx4 mRNA | Low (0.10) | <0.01 |
| Kidney Gpx4 mRNA | Low (0.36) | <0.01 |
| Muscle Gpx4 mRNA | Low (0.99) | <0.01 |
| Kidney Sephs2 mRNA | Moderate up (0.008) | 0.02 |
| Muscle Gpx3 mRNA | Moderate (0.005) | 0.02 |
| Kidney Sepn1 mRNA | Moderate (0.04) | 0.02 |
| Liver Selk mRNA | High (<0.0001) | 0.03 |
| Liver Selt mRNA | Moderate (0.04) | 0.03 |
| Liver Gpx3 mRNA | High (0.005) | 0.04 |
| Liver Sepn1 mRNA | Moderate (0.007) | 0.04 |
| Liver Sepp1 mRNA | Moderate (0.0008) | 0.04 |
| Muscle Selh mRNA | Moderate (0.02) | 0.04 |
| Kidney Selh mRNA | High (<0.0001) | 0.05 |
| Muscle Sepw1 mRNA | High (0.04) | 0.05 |
| Muscle Gpx1 mRNA | High (0.14) | 0.05 |
| Liver Txnrd3 mRNA | Moderate (0.0004) | 0.05 |
| Kidney Selk mRNA | Moderate (0.0008) | 0.05 |
| Kidney Txnrd1 mRNA | Moderate (<0.0001) | 0.05 |
| Liver Selh mRNA | High (<0.0001) | 0.06 |
| Kidney Sepw1 mRNA | High (<0.0001) | 0.06 |
| Kidney Gpx1 mRNA | High (<0.0001) | 0.06 |
| Kidney Gpx3 mRNA | High (0.05) | 0.06 |
| Liver Sep15 mRNA | Moderate (0.01) | 0.06 |
| Liver Dio1 mRNA | Moderate (0.0002) | 0.06 |
| Liver Gpx1 mRNA | Very high (<0.0001) | 0.07 |
| Liver Sepw1 mRNA | High (<0.0001) | 0.07 |
Detailed statistical analyses are provided in Supplemental Tables 2 and 4 and values are shown for some biomarkers in Figures 1–3. Extent of Se regulation for all 24 rat selenoproteins for all tissues studied is provided in Supplemental Table 5.
Susceptibility to Se regulation of the indicated biomarker in Se-deficient rat tissue in Se-deficient compared with Se-adequate (0.24 μg Se/g diet) rats: very high = <10.9% of Se adequate; high = 11–40.9% of Se adequate; moderate = 41–70% of Se adequate; low = >70% of Se adequate; kidney Sephs2 mRNA is moderately upregulated in Se deficiency. Significance (P-value) of regulation indicated in parentheses.
Minimum dietary Se requirement for the growing rat as determined for each indicated biomarker in Expt. 2. Requirements are the minimum dietary Se necessary for the indicated parameter to reach plateau levels when Se-adequate weanling rats are fed these diets from weaning, as determined by breakpoint analysis as described in the text. Shown are all conventional biomarkers assessed in these studies, all molecular biomarkers that were significantly (P < 0.05) regulated by Se status, and all molecular biomarkers shown in Figure 3.
Enzyme activity analyses.
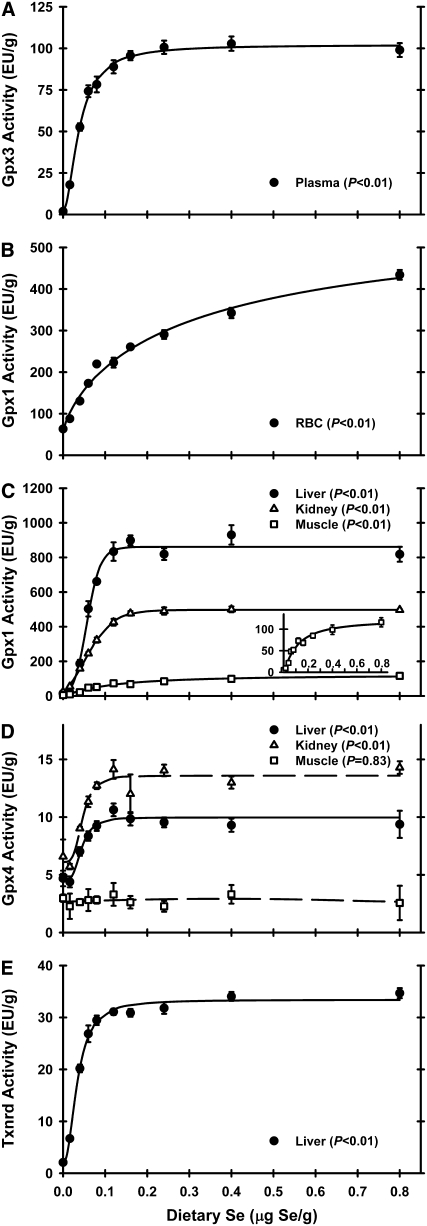
In Expt. 1, plasma Gpx3 activity in Se-deficient rats was 6% of Se-adequate levels (0.2 μg Se/g diet), with the plateau breakpoint at 0.075 μg Se/g diet. In Expt. 2, plasma Gpx3 activity in rats fed a Se-deficient diet was 2% of the activity in rats fed the Se-adequate (0.24 μg Se/g) diet (Fig. 2A). Graded dietary Se supplementation resulted in plasma Gpx3 activity increasing in a sigmoidal response curve with a plateau breakpoint at 0.06 μg Se/g diet (Table 1). Super-nutritional levels of Se supplementation (0.4 μg Se/g diet and higher) did not result in further increases in Gpx3 activity.
FIGURE 2 .
Plasma Gpx3 activity (A), RBC Gpx1 activity (B), tissue Gpx1 activity (C), tissue Gpx4 activity (D), and liver Txnrd (E) activities in male weanling rats fed diets containing graded levels of Se for 28 d. Activities are expressed as enzyme unit/g protein. Values are means ± SEM, n = 5 or 6, except muscle Gpx4, n = 3. Detailed analysis is provided in Supplemental Table 2.
In Expt. 1, RBC Gpx1 activity in Se-deficient rats was 28% of Se-adequate levels (0.2 μg Se/g diet), with the plateau breakpoint at 0.1 μg Se/g diet (data not shown). In Expt. 2, RBC Gpx1 activity in Se-deficient rats decreased to 24% of Se-adequate levels (Fig. 2B). With increasing dietary Se, RBC Gpx1 activity increased and reached a breakpoint at 0.08 μg Se/g diet. RBC Gpx1 activity continued to increase with increasing dietary Se above the 0.08 μg Se/g diet breakpoint, but the rate was less than one-half of the rate of increase before the ∼0.1 μg Se/g diet.
In Expt. 1, liver and kidney Gpx1 activities in Se-deficient rats were 2 and 7%, respectively, of Se-adequate levels (0.2 μg Se/g diet), with the plateau breakpoints at 0.08 μg Se/g diet (data not shown). In Expt. 2 in Se-deficient animals, liver, kidney, and muscle Gpx1 activities fell to 2, 5, and 6% of Se-adequate animals, respectively. Gpx1 activities in liver and kidney reached defined plateaus in this study with breakpoints at 0.09 and 0.12 μg Se/g diet, respectively (Fig. 2C). Plateau liver Gpx1 activity was the highest, with kidney and muscle Gpx1 plateau activities 53 and 8%, respectively, of liver plateau levels. Muscle Gpx1 activity response to increasing dietary Se supplementation was similar to RBC Gpx1 activity, with a muscle Gpx1 activity breakpoint at 0.13 μg Se/g diet.
In Expt. 1, liver Gpx4 activity in Se-deficient rats was 34% of Se-adequate levels (0.2 μg Se/g diet), with the plateau breakpoint at 0.075 μg Se/g diet (data not shown). In Expt. 2, tissue Gpx4 activities also were far less affected by Se deficiency (Fig. 2D). Relative to Se-adequate plateau levels, Se-deficient liver and kidney Gpx4 activities were 47 and 48% of Se-adequate animals, respectively. Muscle Gpx4 activity, however, was not significantly altered by dietary Se supplementation. Kidney had the highest Gpx4 activity, with liver and muscle Gpx4 plateau activities at 73 and 19% of plateau kidney levels; liver and kidney had Gpx4 activity breakpoints at 0.06 and 0.07 μg Se/g diet, respectively.
In Expt. 1, kidney Txnrd activity in Se-deficient rats was 22% of Se-adequate levels (0.2 μg Se/g diet), with the plateau breakpoint at 0.075 μg Se/g diet (data not shown). In Expt. 2 and similar to Gpx1, liver Txnrd activity decreased in Se-deficient rats to 7% of Se-adequate rats (Fig. 2E). Liver Txnrd activity increased sigmoidally with increasing dietary Se, with a breakpoint at 0.06 μg Se/g diet. As with all enzyme activities except RBC and muscle Gpx1, super-nutritional levels of Se supplementation (0.4 μg Se/g diet and higher) did not result in further increases in Txnrd activity.
Selenoprotein mRNA abundance.
In Expt. 1, total RNA from liver and kidney of rats fed graded levels of dietary Se from 0 to 0.3 μg Se/g were analyzed by quantitative real-time-PCR for selenoprotein mRNA levels for 9 and 7 selenoproteins, respectively, that we previously found to be highly or moderately downregulated in mice (19). In Se-deficient rat liver, mRNA levels for Gpx1, Sepw1, and selenoprotein H (Selh) were highly regulated by Se status, decreasing to 12, 19, and 31%, respectively, of Se-adequate levels (see Supplemental Table 3). Except as noted below, the plateau breakpoints and Se response curves (data not shown) in this study were very similar to the results described for Expt. 2.
In Expt. 2, all 24 selenoproteins in the complete rat selenoproteome were analyzed for potential dietary Se regulation in 3 tissues. Overall, the majority of selenoprotein genes were not regulated by dietary Se in any tissues. The selenoproteins were grouped by the extent of reduction in Se deficiency into 5 groups: very high regulation (<10.9% of Se adequate); high regulation (11–40.9% of Se adequate); moderate regulation (40.9–70% of Se adequate; P < 0.05); low regulation (present, > 70% of Se adequate but not significantly regulated; P > 0.05); and not expressed. In liver, 5 selenoprotein mRNA (Gpx1, Sepw1, Selh, Gpx3, and Selk) were very highly or highly regulated; 6 (Dio1, Sepn1, Sepp1, Selt, Sep15, and Txnrd3) were moderately regulated; 12 [Dio2, Dio3, Gpx2, Gpx4, Seli, Selm, Selo, methionine-R-sulfoxide reductase (Sepx1), Sels, selenophosphate synthetase-2 (Sepx2), Txnrd1, and Txnrd2] were expressed but not regulated (P > 0.05); and 1 (Selv) was not expressed (Table 1; Supplemental Tables 4 and 5). In kidney, 4 selenoproteins (Gpx1, Sepw1, Selh, and Gpx3) were highly regulated; 3 (Selk, Sepn1, and Txnrd1) were moderately downregulated; 1 (Sephs2) was moderately upregulated; 15 (Dio1, Dio2, Dio3, Gpx2, Gpx4, Seli, Selm, Selo, Sepp1, methionine-R-sulfoxide reductase, Sels, Selt, Sep15, Txnrd2, and Txnrd3) were expressed but not regulated; and 1 (Selv) was not expressed. In muscle, 1 selenoprotein was highly regulated (Sepw1), 1 (Gpx1) was highly but nonsignificantly regulated, 2 (Gpx3, Selh) were moderately regulated, and the remaining 20 were expressed but not regulated (Supplemental Table 5). For all Se-regulated genes, except for Sepn1 and Sephs2 in kidney and Gpx3 in muscle, the mRNA plateau breakpoints were between 0.03 and 0.07 μg Se/g diet (Table 1).
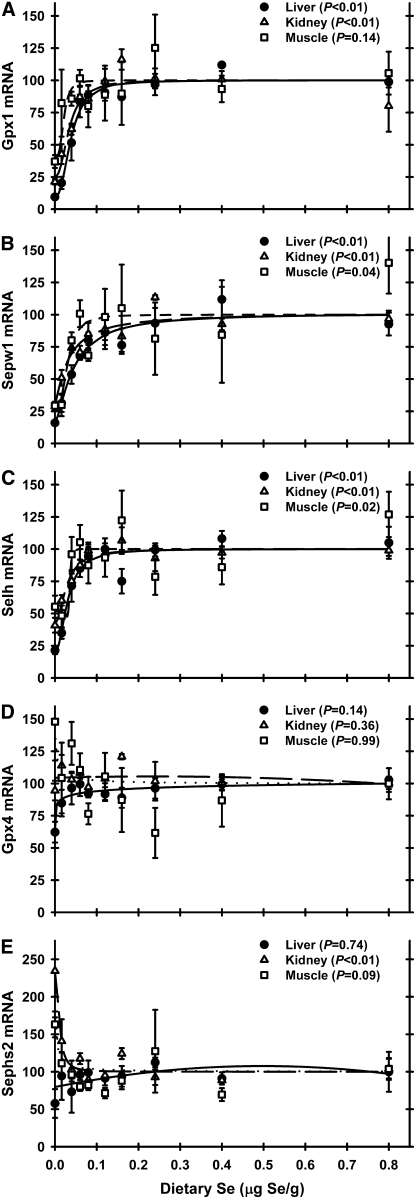
Gpx1 mRNA levels in Se deficiency were reduced to 10% in liver and 21% in kidney and tended to be reduced to 37% (P = 0.14) in muscle relative to Se-adequate levels (Fig. 3A). Plateau breakpoints in liver (0.07 μg Se/g diet) and kidney (0.06 μg Se/g diet) were very similar. The shape of the muscle Gpx1 mRNA curve was similar to those for liver and kidney, with a breakpoint of 0.05 μg Se/g diet. In Expt. 1, liver and kidney Gpx1 mRNA levels in Se-deficient rats were 12 and 34%, respectively, of Se-adequate rats (Supplemental Table 3), and breakpoints for liver and kidney Gpx1 mRNA were 0.07 and 0.06 μg Se/g diet and thus similar to Expt. 2.
FIGURE 3 .
Relative mRNA levels for Gpx1(A), Sepw1(B), Selh (C), Gpx4 (D), and Sephs2 (E) in male weanling rats fed diets containing graded levels of Se for 28 d, as determined by quantitative real-time-PCR on total RNA isolated from the indicated tissues. Values were determined in triplicate for each sample, normalized to the mean of glyceraldehyde-3-phosphate dehydrogenase and β-actin mRNA levels in each sample, expressed as a percentage of Se-adequate plateau levels, and plotted as mean ± SEM, n = 3. Detailed analysis is provided in Supplemental Table 4.
Gpx4 mRNA was not regulated by dietary Se in any tissue in Expt. 2 (Fig. 3D). In Expt. 1, Gpx4 mRNA was also not regulated in either liver or kidney (Supplemental Table 3).
Sepw1 was the only mRNA highly regulated in all 3 examined tissues (Fig. 3B). In Se-deficient rats, Sepw1 mRNA levels were 16, 29, and 29% of Se-adequate levels in liver, kidney, and muscle, respectively. Plateau breakpoints were 0.07, 0.06, and 0.05 μg Se/g diet for liver, kidney, and muscle, respectively. In Expt. 1, Sepw1 mRNA was also highly regulated in liver (19% of Se adequate) and moderately regulated in kidney (41% of Se adequate).
Selh mRNA was the 3rd most highly regulated selenoprotein identified in this rat study (Fig. 3C). In Se deficiency, liver and kidney Selh mRNA levels were reduced to 21 and 40.8%, respectively, of Se-adequate levels; in muscle, Selh was moderately regulated (54% of Se-adequate levels). Breakpoints for Selh mRNA were similar across tissues (0.06, 0.05, and 0.04 μg Se/g diet for liver, kidney, and muscle, respectively). In Expt. 1, Selh was also highly regulated (31% of Se adequate) in liver and moderately regulated (53% of Se adequate) in kidney (Supplemental Table 3).
Kidney Sephs2 was the only gene to exhibit a different pattern of Se regulation (Fig. 3E). In both Expt. 1 and 2, kidney Sephs2 was significantly upregulated in Se deficiency to 134 and 234% of Se-adequate rats, respectively (Supplemental Tables 3 and 4). The plateau breakpoints were 0.08 (not shown) and 0.02 μg Se/g diet for Expt. 1 and 2, respectively. In liver and muscle, in contrast, Sephs2 mRNA levels were not regulated significantly by dietary Se.
Gpx3 mRNA was highly regulated in liver and kidney with breakpoints of 0.04 and 0.06 μg Se/g diet, respectively, and moderately regulated in muscle. Selk mRNA was highly regulated in liver and kidney, with breakpoints of 0.03 and 0.05 μg Se/g diet, respectively, but not regulated in muscle (Table 1).
In addition to the genes discussed above, Se deficiency moderately downregulated mRNA levels in liver for Dio1, Sepn1, Sepp1, Selt, Sep15, and Txnrd3 (Table 1). In kidney, Sepn1 and Txnrd1 mRNA levels were moderately downregulated by Se deficiency (Table 1).
Discussion
In both studies reported here, male weanling rats were fed a Se-deficient diet for 28 d or supplemented with graded levels of Se up to 0.8 μg Se/g diet. Just as in a number of previous studies (6,14,15), regardless of level of Se supplementation, there was no significant effect of dietary Se on growth, indicating the Se requirement for growth was <0.01 μg Se/g diet. Liver and kidney Se concentrations at the end of the study, however, were 3 and 12%, respectively, of Se-adequate levels, clearly indicating that these rats were Se deficient. Similarly, liver and kidney Gpx1 activities were 2 and 5%, respectively, of Se-adequate levels. Thus, the Se-deficient young rats in the present study were biochemically Se deficient but otherwise physiologically indistinguishable from Se-supplemented rats. So the changes in molecular biomarkers in this study appear to be specific for altered Se status rather than secondary effects.
This characterization of the selenoprotein regulon found that Gpx1 mRNA was the mRNA species that was most dramatically regulated by Se status in rats. In addition, 2 selenoprotein mRNA, Sepw1 and Selh, were also highly regulated by Se status. Sepw1 in liver, kidney, and muscle was decreased to 16, 29, and 29% of Se-adequate levels, respectively. Previous studies (10,30,31) reported that Sepw1 was regulated by dietary Se in muscle, but these reports did not detect Sepw1 in liver or kidney. Selh mRNA in liver, kidney, and muscle was decreased to 21, 41, and 54%, respectively, of Se-adequate levels in Se deficiency. Regulation of Selh mRNA in rats by Se status has not been previously reported. Changes in Se distribution in Se-adequate mice due to knockout of Sepp1, however, were reported not to dramatically affect Selh mRNA levels in heart, lung, brain, or testes and were reported to not affect Sepw1 mRNA levels in heart or to raise Sepw1 mRNA levels in lung but were reported to dramatically decrease Sepw1 mRNA levels in brain and testes (32). This shows that genetic mouse models, ostensibly creating Se deficiency by impairing aspects of Se metabolism, may not result in the same effects as reducing total Se by dietary Se deficiency. It may be that specialized transport of Se by Sepp1 and specialized distribution due to Sepp1 receptors (33,34) alters Se flux and Se retention, causing some of these differences. In the present study, both Sepw1 and Selh mRNA reached well-defined plateaus with breakpoints at 0.05–0.07 μg Se/g diet. Thus, Gpx1, Sepw1, and Selh mRNA are biomarkers that dramatically decrease in vivo and thus should be considered when developing molecular biomarkers for Se status. These biomarkers can be detected in total RNA isolated from human blood but are not reduced in humans with plateau levels of plasma Gpx3 activity (35), similar to what was observed in this study with rats.
This study expanded the assessment of potential molecular biomarkers to kidney and muscle to better understand variations in expression and sensitivity to changes in Se status. Wide differences between tissues in Se concentration and sensitivity to Se depletion are well known, as observed in this study. In addition to Gpx1, Sepw1, and Selh, 2 more selenoprotein mRNA were highly regulated by Se status in at least 1 tissue; Selk and Gpx3 mRNA fell to <35% of Se-adequate levels in liver. In kidney, the major source of circulating plasma Gpx3, Se deficiency reduced Gpx3 mRNA levels to 40% of Se-adequate levels. In addition, 6 mRNA in liver, 3 mRNA in kidney, and 2 mRNA in muscle were moderately downregulated in this young, rapidly growing rat model. In other species, these differences may be more profound and, thus, these mRNA have the potential to be effective biomarkers and should be investigated.
An important aspect of this study is the inclusion of super-nutritional dietary Se concentrations. We did not find, however, any biomarkers that showed incisive breakpoints well above 0.1 μg Se/g diet and thus that could be used as biomarkers for super-nutritional or anticarcinogenic levels of Se. This further supports the idea that the anticarcinogenic activity of Se is mediated by effects not directly related to selenoprotein activity (20,21,36). The increased liver and kidney Se > 0.24 μg Se/g diet (Fig. 1), which are not accompanied by increases in selenoenzymes (Fig. 2) or by increases in selenoprotein mRNA levels, suggests that additional unknown forms of Se are accumulating in these tissues in rats fed super-nutritional levels of Se.
Breakpoint analysis was used to calculate minimum dietary Se requirements. These requirements based on liver Gpx1, Gpx4, and Txnrd activity were 0.09, 0.07 and 0.06 μg Se/g diet, respectively, and thus are close to the values of 0.1, 0.05, and 0.07 μg Se/g diet reported previously (6,9,15). The inclusion of higher levels of Se supplementation in the present study, however, allowed more careful calculation of the breakpoints in kidney and muscle and resulted in minimum dietary Se requirements of 0.12 and 0.06 μg Se/g diet based on kidney Gpx1 and Gpx4 activity, respectively, and a minimum dietary Se requirement of 0.13 μg Se/g diet based on muscle Gpx1 activity. A striking feature of these Se response curves is that dietary Se over just a small range (0.03–0.07 μg Se/g diet) raised virtually all selenoprotein mRNA levels to Se-adequate levels with the dramatic impact on selenoprotein mRNA levels occurring at <0.05 μg Se/g diet. This strongly suggests that there is 1 underlying mechanism at play in Se regulation of selenoprotein mRNA levels.
One important point should not be overlooked: the majority of selenoprotein mRNA in each tissue were not significantly regulated by Se status. The lack of Se regulation of Gpx4 mRNA has been long reported (6), but this study now clearly shows that Se regulation of mRNA level is not universal for all selenoprotein mRNA. The mechanism involved in Se regulation of selenoprotein mRNA appears to be related to nonsense-mediated decay (37,38) and is thought to require a nonsense codon, or selenocysteine codon in a Se-deficient organism, sufficiently up-stream (the >55-nt rule) of a splice junction (39–41). The present study and a previous mouse microarray study (19), however, clearly show that this rule alone is not sufficient to explain Se regulation of selenoprotein mRNA degradation, because highly regulated Sepw1, Selk, and Gpx3 mRNA have UGA positioned 15, 5, and 22 nt, respectively, upstream of a splice junction, whereas unregulated Gpx4 and Dio1 mRNA are positioned 105 and 103 nt, respectively, upstream of a splice junction. The present study thus provides a number of new examples showing that the >55 nt rule does not explain susceptibility to degradation.
This study did find 1 selenoprotein with a unique pattern of regulation. Sephs2 was significantly upregulated in Se-deficient rat kidney in both Expt. 1 and 2. Although not significant, skeletal muscle Sephs2 mRNA in Expt. 2 had a similar pattern (Se deficient was 185% of Se adequate). Sephs2 protein has been detected in rat kidney (42) and recently Xu et al. (43) reported that Sephs2 was required for selenoprotein biosynthesis and that Sephs1 cannot substitute for Sephs2 activity. Potentially, upregulation of Sephs2 mRNA in Se deficiency may facilitate enhanced recycling of limiting Se. The mechanism is unclear and may be due to enhanced transcription rather than reduced mRNA stability in Se deficiency, similar to increases in expression of glutathione-S-transferase in Se deficiency (44) or to increases in Dio1 mRNA in thyroid in Se deficiency due to TSH feedback (8).
The present study affirms that the minimum Se requirement for growing rats is 0.1 μg Se/g diet based on liver Se, liver Gpx1 activity, and RBC Gpx1 activity, and affirms slightly lower requirements based on plasma Gpx3 activity, liver Txnrd activity, and liver and kidney Gpx4 activity (Table 1). Using the response curve breakpoints of the highly regulated mRNA in liver and kidney to estimate minimum dietary requirements, the resulting minimum dietary requirements are grouped between 0.04 and 0.07 μg Se/g diet (Table 1). Apparent requirements in muscle are slightly lower, ranging between 0.03 and 0.05 μg Se/g diet. Thus, these requirements based on molecular biology biomarkers are, generally, slightly lower than the requirements based on biochemical markers and slightly higher than the earlier reported requirements of 0.04 and 0.05 μg Se/g diet based on prevention of disease or maintenance of growth (1,2). Unlike the early studies, the Se requirement today for growth is <0.01 μg Se/g diet using pups from Se-adequate dams with diets supplemented with vitamin E and sulfur amino acids. Higher breakpoints of up to 0.13 μg Se/g diet were observed in this young, rapidly growing rat model based on kidney and muscle Gpx1 activity; the basis for these higher breakpoints is unclear but may be related to a role of Gpx1 as a Se store or buffer (45) or slower metabolism in these tissues, such that these storage pools are not fully saturated in the young developing rodent. In summary, the current NRC Se requirement (12) of 0.15 μg Se/g diet includes a safety factor, because it is well above the minimum dietary Se requirement of 0.07 μg Se/g diet based on molecular biomarkers, above the minimum dietary Se requirement of 0.1 μg Se/g diet based on biochemical biomarkers, and much higher than minimum requirements based on growth and prevention of disease (1,2,4).
In conclusion, these studies covering the entire rat selenoproteome have expanded the rat Se regulon to include several additional, highly regulated selenoprotein mRNA, including Sepw1, Selh, Selk, and Gpx3, as well as Gpx1. These studies, however, did not detect any selenoprotein mRNA that decreased to a greater extent than did Gpx1 or that required a higher dietary Se concentration to reach maximum levels than did Gpx1. Also identified was 1 selenoprotein, Sephs2, that is upregulated in Se-deficient kidney and perhaps muscle. Finally, the plateau breakpoints for all of the selenoprotein mRNA were very similar, suggesting that 1 underlying mechanism is at play in Se regulation of selenoprotein mRNA. Thus, this analysis of the full selenoproteome did not identify any selenoprotein molecular biomarkers that might help to assess higher Se status associated with cancer prevention or super-nutritional levels of Se supplementation. Studies on the regulation of nonselenoprotein mRNA by super-nutritional levels of dietary Se may, however, have the potential to identify good molecular biomarkers associated with high Se status.
Supplementary Material
Supported in part by the NIH grant DK74184 and training grant T32-DK07665 (supporting K. M. B.) and by the University of Wisconsin Agricultural Experiment Station grant WIS04909.
Author disclosures: K. M. Barnes, J. K. Evenson, A. M. Raines, and R. A. Sunde, no conflicts of interest.
Supplemental Figure 1 and Supplemental Tables 1–5 are available with the online posting of this paper at jn.nutrition.org.
Abbreviations used: Dio, iodothyronine deiodinase; Gpx1, glutathione peroxidase-1; Gpx3, plasma glutathione peroxidase; Gpx4, phospholipid hydroperoxide glutathione peroxidase; nt, nucleotide; Sephs2, selenophosphate synthetase-2; Selh, selenoprotein H; Sepw1, selenoprotein W; Sepp1, selenoprotein P; Sepx1, methionine-R-sulfoxide reductase; Txnrd, thioredoxin reductase.
References
- 1.Schwarz K, Foltz CM. Selenium as an integral part of factor 3 against dietary necrotic liver degeneration. J Am Chem Soc. 1957;79:3292–3. [PubMed] [Google Scholar]
- 2.Hurt HD, Cary EE, Visek WJ. Growth, reproduction, and tissue concentrations of selenium in the selenium-depleted rat. J Nutr. 1971;101:761–6. [DOI] [PubMed] [Google Scholar]
- 3.Rotruck JT, Pope AL, Ganther HE, Swanson AB, Hafeman DG, Hoekstra WG. Selenium: biochemical role as a component of glutathione peroxidase. Science. 1973;179:588–90. [DOI] [PubMed] [Google Scholar]
- 4.Hafeman DG, Sunde RA, Hoekstra WG. Effect of dietary selenium on erythrocyte and liver glutathione peroxidase in the rat. J Nutr. 1974;104:580–7. [DOI] [PubMed] [Google Scholar]
- 5.NRC. Nutrient requirements of laboratory animals. 3rd ed. Washington, DC: National Academy Press; 1978.
- 6.Lei XG, Evenson JK, Thompson KM, Sunde RA. Glutathione peroxidase and phospholipid hydroperoxide glutathione peroxidase are differentially regulated in rats by dietary selenium. J Nutr. 1995;125:1438–46. [DOI] [PubMed] [Google Scholar]
- 7.Yang JG, Hill KE, Burk RF. Dietary selenium intake controls rat plasma selenoprotein P concentration. J Nutr. 1989;119:1010–2. [DOI] [PubMed] [Google Scholar]
- 8.Bermano G, Nicol F, Dyer JA, Sunde RA, Beckett GJ, Arthur JR, Hesketh JE. Tissue-specific regulation of selenoenzyme gene expression during selenium deficiency in rats. Biochem J. 1995;311:425–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hadley KB, Sunde RA. Selenium regulation of thioredoxin reductase activity and mRNA levels in rat liver. J Nutr Biochem. 2001;12:693–702. [DOI] [PubMed] [Google Scholar]
- 10.Yeh JY, Vendeland SC, Gu Q, Butler JA, Ou BR, Whanger PD. Dietary selenium increases selenoprotein W levels in rat tissues. J Nutr. 1997;127:2165–72. [DOI] [PubMed] [Google Scholar]
- 11.Cohen HJ, Chovaneic ME, Mistretta D, Baker SS. Selenium repletion and glutathione peroxidase- differential effects on plasma and red blood cell enzyme activity. Am J Clin Nutr. 1985;41:735–47. [DOI] [PubMed] [Google Scholar]
- 12.NRC. Nutrient requirements of laboratory animals. 4th ed. Washington, DC: National Academy Press; 1995.
- 13.Saedi MS, Smith CG, Frampton J, Chambers I, Harrison PR, Sunde RA. Effect of selenium status on mRNA levels for glutathione peroxidase in rat liver. Biochem Biophys Res Commun. 1988;153:855–61. [DOI] [PubMed] [Google Scholar]
- 14.Weiss SL, Evenson JK, Thompson KM, Sunde RA. The selenium requirement for glutathione peroxidase mRNA level is half of the selenium requirement for glutathione peroxidase activity in female rats. J Nutr. 1996;126:2260–7. [DOI] [PubMed] [Google Scholar]
- 15.Weiss SL, Evenson JK, Thompson KM, Sunde RA. Dietary selenium regulation of glutathione peroxidase mRNA and other selenium-dependent parameters in male rats. J Nutr Biochem. 1997;8:85–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sunde RA, Evenson JK, Thompson KM, Sachdev SW. Dietary selenium requirements based on glutathione peroxidase-1 activity and mRNA levels and other selenium parameters are not increased by pregnancy and lactation in rats. J Nutr. 2005;135:2144–50. [DOI] [PubMed] [Google Scholar]
- 17.Weiss Sachdev S, Sunde RA. Selenium regulation of transcript abundance and relative translational efficiency of glutathione peroxidase 1 and 4 in rat liver. Biochem J. 2001;357:851–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kryukov GV, Castellano S, Novoselov SV, Lobanov AV, Zehtab O, Guigo R, Gladyshev VN. Characterization of mammalian selenoproteins. Science. 2003;300:1439–43. [DOI] [PubMed] [Google Scholar]
- 19.Sunde RA, Raines AM, Barnes KM, Evenson JK. Selenium status highly-regulates selenoprotein mRNA levels for only a subset of the selenoproteins in the selenoproteome. Biosci Repts. In press 2009. [DOI] [PMC free article] [PubMed]
- 20.Ip C. Selenium inhibition of chemical carcinogenesis. Fed Proc. 1985;44:2573–8. [PubMed] [Google Scholar]
- 21.Novoselov SV, Calvisi DF, Labunskyy VM, Factor VM, Carlson BA, Fomenko DE, Moustafa ME, Hatfield DL, Gladyshev VN. Selenoprotein deficiency and high levels of selenium compounds can effectively inhibit hepatocarcinogenesis in transgenic mice. Oncogene. 2005;24:8003–11. [DOI] [PubMed] [Google Scholar]
- 22.Clark LC, Combs GF, Turnbull BW, Slate EH, Chalker DK, Chow J, Davis LS, Glover RA, Graham GF, et al. Effects of selenium supplementation for cancer prevention in patients with carcinoma of the skin. JAMA. 1996;276:1957–63. [PubMed] [Google Scholar]
- 23.McKown DM, Morris JS. Rapid measurement of selenium in biological samples using instrumental neutron activation analysis. J Radioanal Nucl Chem. 1978;43:411–20. [Google Scholar]
- 24.Lawrence RA, Sunde RA, Schwartz GL, Hoekstra WG. Glutathione peroxidase activity in rat lens and other tissues in relation to dietary selenium intake. Exp Eye Res. 1974;18:563–9. [DOI] [PubMed] [Google Scholar]
- 25.Hill KE, McCollum GW, Burk RF. Determination of thioredoxin reductase activity in rat liver supernatant. Anal Biochem. 1997;253:123–5. [DOI] [PubMed] [Google Scholar]
- 26.Lowry OH, Rosebrough NJ, Farr AL, Randall RJ. Protein measurement with the Folin phenol reagent. J Biol Chem. 1951;193:265–75. [PubMed] [Google Scholar]
- 27.Peirson SN, Butler JN, Foster RG. Experimental validation of novel and conventional approaches to quantitative real-time PCR data analysis. Nucleic Acids Res. 2003;31:e73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pfaffl MW. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001;29:e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Steel RGD, Torrie JH. Principles and procedures of statistics. New York: McGraw-Hill Book; 1960.
- 30.Vendeland SC, Beilstein MA, Yeh JY, Ream W, Whanger PD. Rat skeletal muscle selenoprotein W: cDNA clone and mRNA modulation by dietary selenium. Proc Natl Acad Sci USA. 1995;92:8749–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hooven LA, Butler J, Ream LW, Whanger PD. Microarray analysis of selenium-depleted and selenium-supplemented mice. Biol Trace Elem Res. 2006;109:173–9. [DOI] [PubMed] [Google Scholar]
- 32.Hoffmann PR, Hoge SC, Li PA, Hoffmann FW, Hashimoto AC, Berry MJ. The selenoproteome exhibits widely varying, tissue-specific dependence on selenoprotein P for selenium supply. Nucleic Acids Res. 2007;35:3963–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Olson GE, Winfrey VP, Nagdas SK, Hill KE, Burk RF. Apolipoprotein E receptor-2 (ApoER2) mediates selenium uptake from selenoprotein P by the mouse testis. J Biol Chem. 2007;282:12290–7. [DOI] [PubMed] [Google Scholar]
- 34.Olson GE, Winfrey VP, Hill KE, Burk RF. Megalin mediates selenoprotein P uptake by kidney proximal tubule epithelial cells. J Biol Chem. 2008;283:6854–60. [DOI] [PubMed] [Google Scholar]
- 35.Sunde RA, Paterson E, Evenson JK, Barnes KM, Lovegrove JA, Gordon MH. Longitudinal selenium status in healthy British adults: assessment using biochemical and molecular biomarkers. Br J Nutr. 2008;99 Suppl 3:S37–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Combs GF Jr. Current evidence and research needs to support a health claim for selenium and cancer prevention. J Nutr. 2005;135:343–7. [DOI] [PubMed] [Google Scholar]
- 37.Weiss SL, Sunde RA. Cis-acting elements are required for selenium regulation of glutathione peroxidase-1 mRNA levels. RNA. 1998;4:816–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Moriarty PM, Reddy CC, Maquat LE. Selenium deficiency reduces the abundance of mRNA for Se-dependent glutathione peroxidase 1 by a UGA-dependent mechanism likely to be nonsense codon-mediated decay of cytoplasmic mRNA. Mol Cell Biol. 1998;18:2932–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Low SC, Berry MJ. Knowing when not to stop: selenocysteine incorporation in eukaryotes. Trends Biochem Sci. 1996;21:203–8. [PubMed] [Google Scholar]
- 40.Nagy E, Maquat LE. A rule for termination-codon position within intron-containing genes: when nonsense affects RNA abundance. Trends Biochem Sci. 1998;23:198–9. [DOI] [PubMed] [Google Scholar]
- 41.Maquat LE. Nonsense-mediated mRNA decay in mammals. J Cell Sci. 2005;118:1773–6. [DOI] [PubMed] [Google Scholar]
- 42.Kim IY, Stadtman TC. Selenophosphate synthetase: detection in extracts of rat tissues by immunoblot assay and partial purification of the enzyme from the archaean Methanococcus vannielii. Proc Natl Acad Sci USA. 1995;92:7710–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Xu XM, Carlson BA, Irons R, Mix H, Zhong N, Gladyshev VN, Hatfield DL. Selenophosphate synthetase 2 is essential for selenoprotein biosynthesis. Biochem J. 2007;404:115–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Reiter R, Wendel A. Selenium and drug metabolism. II: independence of glutathione peroxidase and reversibility of hepatic enzyme modulations in deficient mice. Biochem Pharmacol. 1984;33:1923–8. [DOI] [PubMed] [Google Scholar]
- 45.Sunde RA. Molecular biology of selenoproteins. Annu Rev Nutr. 1990;10:451–74. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.