Abstract
DNA double-strand breaks (DSBs) are repaired by two principal mechanisms: non-homologous end-joining (NHEJ) and homologous recombination (HR)1. HR is the most accurate DSB repair mechanism but is generally restricted to the S and G2 phases of the cell cycle, when DNA has been replicated and a sister chromatid is available as a repair template2-5. By contrast, NHEJ operates throughout the cell cycle but assumes most importance in G1 (refs 4, 6). The choice between repair pathways is governed by cyclin-dependent protein kinases (CDKs)2,3,5,7, with a major site of control being at the level of DSB resection, an event that is necessary for HR but not NHEJ, and which takes place most effectively in S and G2 (refs 2, 5). Here we establish that cell-cycle control of DSB resection in Saccharomyces cerevisiae results from the phosphorylation by CDK of an evolutionarily conserved motif in the Sae2 protein. We show that mutating Ser 267 of Sae2 to a non-phosphorylatable residue causes phenotypes comparable to those of a sae2Δ null mutant, including hypersensitivity to camptothecin, defective sporulation, reduced hairpin-induced recombination, severely impaired DNA-end processing and faulty assembly and disassembly of HR factors. Furthermore, a Sae2 mutation that mimics constitutive Ser 267 phosphorylation complements these phenotypes and overcomes the necessity of CDK activity for DSB resection. The Sae2 mutations also cause cell-cycle-stage specific hypersensitivity to DNA damage and affect the balance between HR and NHEJ. These findings therefore provide a mechanistic basis for cell-cycle control of DSB repair and highlight the importance of regulating DSB resection.
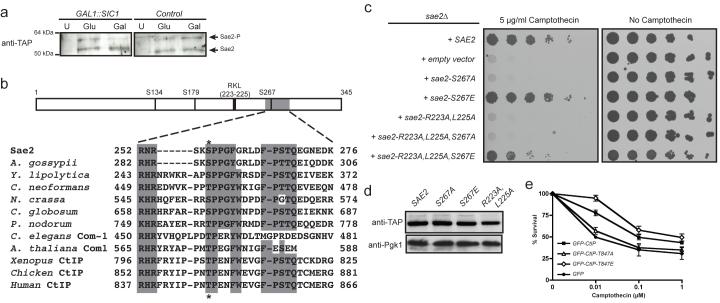
To initiate HR, one strand of the broken DNA duplex is resected in the 5′→3′ direction, generating single-stranded DNA (ssDNA) that can anneal with a homologous DNA duplex8. In S. cerevisiae, effective resection and HR require sustained Cdc28/Clb (Cdk1/cyclin B) kinase activity2,3,5, although the CDK targets mediating this control are still unknown. One potential target is Sae2, a protein first identified as being required for meiotic recombination. Sae2 controls the initiation of DNA-end resection in meiotic and mitotic cells9-12 and was recently shown to be a DNA endonuclease13. Previous work has shown that Sae2 is targeted by the Mec1 and Tel1 kinases in response to DNA damage, generating forms of Sae2 with decreased mobility in SDS-polyacrylamide gels14. Such alterations in Sae2 gel-mobility also occurred in unperturbed cycling cells, specifically in S and G2, indicating that Sae2 might be a Cdc28 target (data not shown, Fig. 2b and Supplementary Fig. 2a). In accord with this idea, the amount of slower-migrating Sae2 was diminished when Cdc28 was inactivated in G2-synchronized cultures by galactose-driven expression of the Cdc28/Clb repressor, Sic1 (ref. 15) (Fig. 1a).
Figure 2. Sae2 is phosphorylated by Cdc28 on Ser 267.
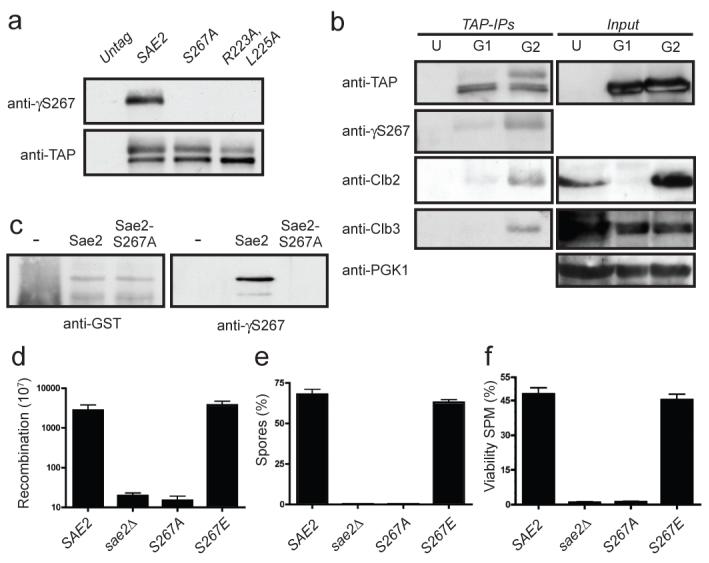
a, TAP-tagged Sae2 derivatives were immunoprecipitated and detected as indicated. b, TAP-tagged Sae2 was purified from G1 or G2 arrested cultures. U, G2 arrested untagged control cells. Immunoprecipitated samples and inputs (5%) were immunoblotted as indicated. c, Glutathione S-transferase (GST)-fused Sae2 and Sae2-S267A were purified, incubated with recombinant Cdk2/Cyclin A and ATP, resolved by 10% SDS–PAGE and immunoblotted as indicated. d, Recombination frequencies of strains in a hairpin-containing recombination system22. e, Spores after 24 h in sporulation medium. f, Spore viability 24 h after the addition of sporulation medium (SPM)10. Error bars in d-f represent s.d. (n = 2).
Figure 1. Ser 267 mutation impairs Sae2 function.
a, Left: TAP-tagged Sae2 was purified from cells expressing galactose-inducible SIC1 (Gal) or not expressing SIC1 (Glc). U, control untagged strain. Right: as above, but the strain lacked galactose-inducible SIC1. b, Sae2 diagram and homology to orthologues (see Methods for full alignment). S. cerevisiae, Saccharomyces cerevisiae, A. gossypii, Ashbya gossypii; Y. lipolytica, Yarrowia lipolytica; C. neoformans, Cryptococcus neoformans; N. crassa, Neurospora crassa; C. globosum, Chaetomium globosum; P. nodorum, Phaeosphaeria nodorum; C. elegans, Caenorhabditis elegans; A. thaliana, Arabidopsis thaliana; Xenopus, Xenopus laevis; chicken, Gallus gallus; human, Homo sapiens. c, Fivefold serial dilutions of sae2Δ cultures containing the indicated SAE2 genes plated on medium lacking or containing camptothecin (5 μg ml−1). d, Extracts of cells harbouring TAP-tagged Sae2 variants were western immunoblotted as indicated. e, Survival of U2OS cells expressing GFP–CtIP fusions to 1 h treatments with the indicated doses of camptothecin. Error bars indicate s.d. (n = 2).
Sae2 contains three potential CDK phosphorylation sites (Fig. 1b and Supplementary Fig. 1); two of these—Ser 267 and Ser 134—received the highest scores for predicted phosphorylation sites in the protein (Supplementary Table 1). Ser 267 maps to the Sae2 region most highly conserved with its non-yeast orthologues (Fig. 1b), which include human CtIP, Caenorhabditis elegans Com1 and Arabidopsis thaliana Com1 (refs 16-18). To address the possible function(s) of Ser 267 and other potential target sites for CDK in Sae2, we generated yeast strains in which each site was individually mutated to a non-phosphorylatable alanine residue. The sae2-S267A mutant showed strong hypersensitivity towards the topoisomerase I inhibitor camptothecin (Fig. 1c and Supplementary Fig. 2b; Supplementary Fig. 2c shows that this mutant is nearly as sensitive as the sae2Δ strain). By contrast, sae2-S134A, sae2-S179A and sae2-S134A,S179A cells did not show detectable hypersensitivity to camptothecin, and combining sae2-S267A with these other mutations showed no synergistic effect (Supplementary Fig. 2b). When we mutated Ser 267 to glutamic acid to mimic constitutive phosphorylation, the resulting strain displayed no detectable hypersensitivity to camptothecin (Fig. 1c and Supplementary Fig. 2c). Together with the fact that mutation of Ser 267 did not alter Sae2 protein expression (Fig. 1d), these data suggested that Ser 267 phosphorylation is required for Sae2 function. Furthermore, as the major cytotoxic lesions for camptothecin are DSBs arising when replication forks encounter trapped topoisomerase I–DNA complexes19, these results suggested that phosphorylation of Sae2 on Ser 267 is important for responses to DSBs generated during S phase. Indeed, the sae2-null and sae2-S267A strains, but not the sae2-S267E strain, showed hypersensitivity to methyl methanesulphonate (MMS), which also yields DSBs in S phase (Supplementary Fig. 2d). Consistent with analogous residues controlling the activity of Sae2-related proteins in other species, human U2OS cells downregulated for endogenous CtIP and expressing short interfering RNA (siRNA)-resistant GFP–CtIP-T847A were as sensitive to camptothecin as control cells expressing GFP (green fluorescent protein) alone, whereas cells expressing a phospho-mimicking CtIP derivative (GFP–CtIP-T847E) showed higher resistance to camptothecin (Fig. 1e; for expression levels and downregulation see Supplementary Fig. 2e).
Efficient phosphorylation of CDK substrates in vivo often requires the binding of cyclin to an Arg-X-Leu (RXL) motif in the target20. Such a motif is present upstream of Ser 267 in Sae2 (Fig. 1b and Supplementary Fig. 1), and mutating this motif (Sae2-R223A,L225A) caused hypersensitivity to camptothecin as severe as that of sae2-S267A or sae2Δ cells (Fig. 1c) even though the mutated proteins were expressed at normal levels (Fig. 1d). Furthermore, the camptothecin hypersensitivity caused by the Sae2-R223A,L225A mutation was largely suppressed when the protein also contained the phospho-mimicking Sae2 S267E mutation (Fig. 1c). Collectively, these findings strongly suggested that Sae2 function requires its modification by CDK–cyclin complexes.
To examine the phosphorylation of Sae2 on Ser 267 directly, we raised a phosphospecific antibody against this site (γS267). Western immunoblotting revealed that this antibody specifically detected immunoprecipitated wild-type Sae2 but not the Sae2-S267A or Sae2-R223A,L225A proteins (Fig. 2a). Furthermore, the antibody detected the slower-migrating form of Sae2 that was present at elevated levels in G2-synchronized cultures (Fig. 2b), indicating that phosphorylation of Sae2 on Ser 267 is subject to cell-cycle control. Notably, Sae2 immunoprecipitation recovered cyclins Clb3 and Clb2 from extracts prepared from G2-synchronized cells but not from G1 cells (Fig. 2b). Also consistent with Sae2 being a direct target of CDK, incubation of purified glutathione S-transferase-fused Sae2 protein—but not the S267A mutant—with recombinant CDK-cyclin complexes and ATP produced Ser 267 phosphorylation (Fig. 2c).
Further analyses suggested that all aspects of Sae2 function require phosphorylation at Ser 267. Thus, like sae2Δ cells, sae2-S267A cells were severely compromised in hairpin-induced recombination21, whereas sae2-S267E cells behaved similarly to the wild-type (Fig. 2d). Furthermore, whereas homozygous diploid sae2-S267E cells produced viable spores at levels similar to those produced by the wild-type strain, like sae2Δ cells10,12, the sae2-S267A homozygous mutant strain showed a severe sporulation defect and almost no spore viability (Figs 2e and 2f). A defect in spore viability due to mutations in Ser 267 or Pro 268 has also been reported recently18.
Sae2 regulates resection of chromosomal DSBs formed by the HO endonuclease9. To determine whether Sae2 Ser 267 controls this function, we generated an irreparable HO-induced DSB in the MAT locus and analysed resulting samples with a neutral dot-blot approach that detected only resected DNA22. For this we used three probes: one directly adjacent to the HO cleavage site, one 5 kilobases (kb) downstream and one at the LEU2 locus, about 100 kb distal from the HO site, as a negative control. Significantly, sae2Δ cells and cells bearing the sae2-S267A mutation were impaired in resection close to the HO site, and this impairment was even more pronounced when assayed 5 kb away (Fig. 3a). By contrast, the sae2-S267E mutant carried out resection almost as efficiently as the wild-type strain (Fig. 3a and Supplementary Fig. 3a).
Figure 3. DNA-end resection is controlled by Sae2.
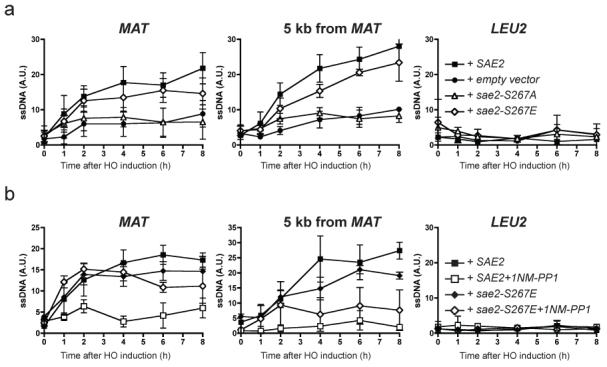
a, Resection-mediated ssDNA formation at an HO DSB in wild-type SAE2 (filled squares), sae2-S267A (open triangle), sae2-S267E (open diamonds) or empty vector (solid circles) at indicated times after HO induction at the MAT locus (left), 5 kb downstream of MAT (centre) or LEU2 locus (right). b, Wild-type SAE2 (squares) or sae2-S267E (diamonds) strains containing Cdc28-as1 were grown as in a but in the presence of dimethylsulphoxide (filled symbols) or 1NM-PP1 (open symbols). Results are shown as means ± s.d. (n = 5).
To test whether Cdc28/Cdk1-mediated Sae2 phosphorylation promotes DSB resection, we assessed resection in a strain expressing a Cdc28 derivative (cdc28-as1) that can be specifically inhibited by the ATP analogue 1NM-PP1 (ref. 23). Whereas inhibition of Cdk1 markedly curtailed end resection in a strain expressing wild-type Sae2, it had little effect in the sae2-S267E mutant when resection was measured close to the HO site (Fig. 3b and Supplementary Fig. 3b). Nevertheless, the inhibitor still had some effect on the sae2-S267E strain when resection was assessed at the 5 kb distal site (Fig. 3b and Supplementary Fig. 3b). We therefore conclude that phosphorylation of Sae2 on Ser 267 by Cdc28/Cdk1 is required for effective DSB resection but that additional Cdk1 target sites are required for resection to take place optimally. One candidate for such an additional CDK target is Rad9, which is phosphorylated by Cdc28 and was recently shown to affect DSB resection24.
S. cerevisiae Mre11 is recruited quickly to DSB sites, then replaced by the HR protein Rad52 as ssDNA is formed in S and G2 cells24. We found that, like SAE2 deletion25, the sae2-S267A mutation caused Mre11 foci to persist longer than in wild-type cells (Fig. 4a) and delayed Rad52 focus formation in S/G2 after X-ray treatment (Fig. 4b). In contrast, the sae2-S267E mutant displayed Mre11 focus disassembly kinetics similar to the wild-type strain (Fig. 4a), and in fact reproducibly formed Rad52 foci faster in S and G2 than the wild-type strain (Fig. 4b). As expected, essentially no Rad52 foci were detected in wild-type G1 cells, because DSBs are not efficiently resected. By contrast—and unlike the sae2-S267A mutant—the sae2-S267E mutant formed Rad52 foci in G1 at later time points (Fig. 4c), confirming that DNA ends are processed to some degree in this mutant even in the absence of active CDK (Fig. 3b).
Figure 4. Sae2 mutations affect Mre11 and Rad52 dynamics, and DSB repair.
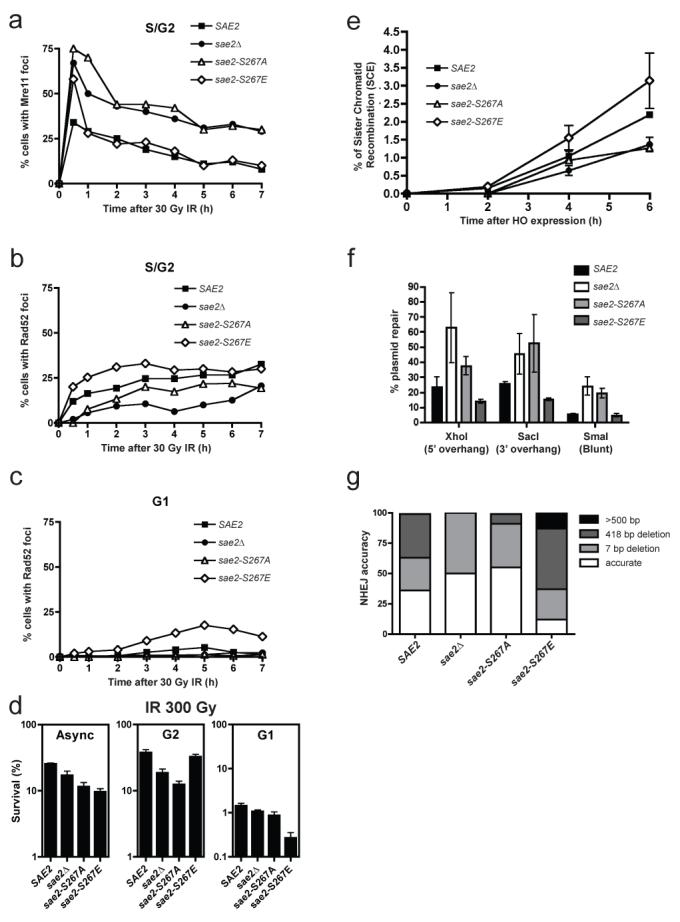
a, b, Percentages of S/G2 cells containing Mre11 (a) or Rad52 (b) foci. c, Percentage of G1 cells containing a Rad52 focus. d, Survival of sae2Δ mutants containing wild-type SAE2, sae2-S267A, sae2-S267E or empty vector grown asynchronously (Async) or arrested in G1 or G2, after irradiation with 30 Gy. Error bars represent s.d. (n = 2). e, Sister-chromatid recombination measured as described previously27. Standard deviations of two independent experiments are shown (see Supplementary Fig. 4 for details and representative blot). f, Plasmid cleaved by Xho I, Sac I or Sma I was transformed into strains and NHEJ efficiency was measured. Means and s.d. of three independent experiments are shown. g, Classes of plasmid rejoining products from 50 independent clones of each strain transformed with the Xho I-cut plasmid.
To address the impact of aberrant DSB processing on DSB repair, we irradiated asynchronous, G2-arrested and G1-arrested cell cultures, then kept cells in the same state (asynchronous, G2 or G1) for 6 h to allow DNA repair. Cells were subsequently plated and colony formation was used to determine survival. When we analysed asynchronous cultures, the sae2Δ strain showed moderate hypersensitivity to radiation (as shown previously26) and similar, although slightly more pronounced, hypersensitivities were displayed by the sae2-S267A and sae2-S267E strains (Fig. 4d). Moreover, the two Ser 267 mutations had markedly different effects in G1 and G2 (Fig. 4d). Thus, whereas the sae2-S267A strain was more sensitive to radiation than the control strain in G2, little or no G2 hypersensitivity was shown by the sae2-S267E strain. In contrast, sae2-S267E cells showed marked hypersensitivity to radiation in G1, whereas the sae2-null and sae2-S267A cells did not.
The above data suggested that the sae2-S267A and sae2-null strains, but not the sae2-S267E strain, are defective in HR because of impaired DSB resection in G2, and also suggested that the hypersensitivity of the sae2-S267E mutant to radiation in G1 reflects aberrant DSB resection, thus impairing NHEJ and/or triggering futile attempts to carry out HR in the absence of a sister chromatid. To test these ideas, we determined HR and NHEJ efficiencies in various sae2 mutant backgrounds. When we used an assay in which HR intermediates were monitored by Southern blot analysis27, the sae2-null and sae2-S267A mutants showed delayed HR, whereas the sae2-S267E mutant showed slightly accelerated recombination (Fig. 4e and Supplementary Fig. 4). Furthermore, by measuring NHEJ with an in vivo plasmid-recircularization assay28, we found that cells lacking Sae2 or bearing the S267A mutation had enhanced (P < 0.05 compared with wild-type) NHEJ efficiencies, regardless of whether the DSB contained a 5′ overhang, a 3′ overhang or a blunt end (Fig. 4f). In addition, and in agreement with an increased propensity for DSB resection that would impair NHEJ, sae2-S267E mutant cells showed a decrease in NHEJ efficiency when overhang substrates were used (P < 0.05; Fig. 4f). By retrieving repaired plasmids from independent clones and sequencing them, we found that in wild-type cells most repair took place accurately or by micro-homology-mediated end-joining involving pairs of 4-bp repeats separated by 7 or 418 bp to create small or moderate deletions (Fig. 4g). In agreement with less efficient resection taking place in the sae2-null and sae2-S267A mutant strains, such cells favoured accurate repair or repair involving small (7-bp) deletions (P < 10−8; Fig. 4g; similar data were reported for sae2Δ cells29). In contrast, sae2-S267E mutant cells showed little accurate NHEJ and, instead, most repair products contained larger deletions of up to 2 kb (P < 10−4; Fig. 4g).
Thus, Cdc28/Cdk1-mediated Sae2 phosphorylation modulates the balance between NHEJ and HR during the cell cycle. These results lend strong support to models in which the commitment to DSB resection is highly regulated to ensure that the cell engages the most appropriate DNA repair pathway, thereby optimizing genome stability. As Sae2 has endonuclease activity13, we favour a model in which Sae2, possibly in cooperation with the Mre11-Rad50-Xrs2 (MRX) complex, facilitates resection in S/G2 by mediating an endonucleolytic cleavage close to the DNA break, thus generating a clean end that can serve as an efficient substrate for nucleases such as MRX and Exo1. Sae2 activity might be particularly important to initiate resection at DSBs that contain covalently bound proteins that would otherwise resist exonuclease action; indeed, this would explain why deletion of SAE2 causes defective removal of Spo11-DNA adducts during meiosis and marked hypersensitivity to camptothecin. Sae2 might also initiate resection at radiation-induced DSBs that are resistant to exonucleases because they bear protein–DNA crosslinks or complex damage to bases at their termini. By contrast, at sites of clean DSBs, SAE2 deletion would only slow down resection and ensuing HR, thus explaining why sae2 mutants are not as sensitive to radiation as other HR mutants22. Finally, we note that the motif encompassing Ser 267 of Sae2 is highly conserved in Sae2 counterparts in higher eukaryotes, and that mutation of the analogous Thr 847 site in human CtIP to Ala (but not to Glu) yields hypersensitivity to camptothecin. This suggests that analogous CDK-control mechanisms for DSB resection operate in many other organisms. One exception to this, however, is likely to be provided by Schizosaccharomyces pombe, whose Sae2/CtIP homologue, Ctp1, lacks a CDK site analogous to Ser 267 of Sae2. In this case, it seems that, rather than controlling Ctp1 phosphorylation, the CDK machinery instead regulates the protein expression of Ctp1 (ref. 30). Nevertheless, although some species-specific variations undoubtedly exist, we speculate that Sae2/CtIP/Com1/Ctp1 proteins will turn out to have ubiquitous functions in facilitating DSB resection in S and G2 and modulating the choice of DSB repair pathway in eukaryotic cells.
METHODS SUMMARY
A sae2Δ strain in W303 background9,14 was transformed with plasmids harbouring the indicated SAE2 mutant and used in all experiments except those listed below. For Figs 1a and 2b, a Sae2-TAP strain (Open Biosystems) was used. In Fig. 2e, f, a strain harbouring the indicated Sae2 mutant at its chromosomal locus in the SK1 background was used. For Fig. 3, we deleted SAE2 in a strain harbouring the cdc28as1 allele in the JKM179 background5. The W5573-15D strain was used in Fig. 4a-c (ref. 25). A sae2-deleted OIS-15 strain was used in Fig. 4e (ref. 27). Yeasts were grown with standard procedures. When indicated, cells were arrested in G1 with α-factor and in G2 with nocodazole. DNA resection assays22, focus formation25, recombination between sister chromatids27 and NHEJ assays30 were as described previously. CtIP downregulation was as previously reported17. Western blotting was by standard methods.
Supplementary Material
Acknowledgements
We thank M. P. Longhese, R. Rothstein, K. Lobachev, M. Lichten and M. Foiani for providing strains, and R. Driscoll, S. Gravel, K. Dry and K. Miller for helpful discussions and comments on the manuscript. P.H. is the recipient of a Long-Term EMBO Fellowship. A.A.S. is supported by a Swiss National Foundation Grant. The S.P.J. laboratory is supported by grants from Cancer Research UK and the European Community (Integrated Project DNA repair, grant LSHG-CT-2005-512113). The A.A. laboratory is supported by grants from the Spanish Ministry of Science and Education (BFU2006-05260 and CDS2007-0015) and Junta de Andalucia (CVI624).
Appendix
METHODS
TAP-tagged Sae2 immunoprecipitation
TAP complexes were purified by a variation of previously described methods31. Cultures (250 ml) of TAP-tagged Sae2 variants were collected by centrifugation at 4 °C and resuspended in 1 volume of 50 mM Tris-HCl pH 7.5, 100 mM NaCl, 1.5 mM MgCl2, 0.15% Nonidet P40 in the presence of protease inhibitor (Roche) and phosphatase inhibitors. Extracts were prepared with a One-Shot cell disruptor (Constant Systems) and centrifuged for 1 h at 3,000 r.p.m. (1400 g) and 4 °C. Next, samples were incubated for 2 h at 4 °C with IgG-Sepharose (Amersham) pre-equilibrated in the same buffer. The matrix was then packed in a column and washed with 50 ml of the same buffer at 4 °C. Next, the resin was resuspended in 100 μl of 10 mM Tris-HCl pH 8.0, 150 mM NaCl, 0.1% Nonidet P40, 0.5 mM EDTA, 1 mM dithiothreitol, transferred to a microcentrifuge tube, incubated for 2 h at 16 °C and then overnight at 4 °C with 10 U of TEV protease (Qiagen) to release Sae2 complexes from the beads. Through this procedure, Sae2 retained half of the TAP tag that could then be detected with anti-TAP antibody (Open Biosystems). Samples were centrifuged for 1 min at maximum speed (2900 g) at 4 °C; the supernatant was transferred to a new tube and 100 μl of sample loading buffer was added followed by immunoblot analysis by SDS–PAGE with the following antibodies: anti-TAP, anti- γS267 (custom made; Eurogentec), anti-PGK1 (Molecular Probes), Cdc28, Clb2 and Clb3 (Santa Cruz).
Human cell survival assays
Human U2OS cells expressing siRNA-resistant wild-type or mutant GFP–CtIP fusions were downregulated for endogenous CtIP with a previously published siRNA17, and 72 h afterwards were exposed to doses of camptothecin for 1 h. Survivals represent the number of colonies formed after 12 days normalized with an unirradiated control.
Sporulation efficiency
Homozygous diploids were grown overnight in YPAD medium, washed twice with warm sporulation medium, left in sporulation medium for 24 h at 30 °C, then fixed with 50% ethanol and stained with 4,6-diamidino-2-phenylindole (DAPI). The percentage of sporulated cells was determined by microscopy10.
DNA-end resection assay
Cultures of sae2Δ cdc28-as1 GAL1::HO strain transformed with wild-type SAE2, sae2-S267A, sae2-S267E or empty vector were grown to mid-exponential phase in raffinose. Samples were taken at indicated times after inducing HO by the addition of galactose. DNA was isolated, of which 1 mg was blotted in neutral and denaturing conditions with a dot-blot manifold as described previously23, then hybridized with radioactively labelled probes against the MAT locus, 5 kb downstream of the MAT locus or LEU2 locus. Signals were quantified with a FLA-5000 instrument (Fuji) and values obtained in neutral conditions were normalized to those obtained under denaturing conditions (see Supplementary Fig. 3a). When indicated, 2 h before the addition of galactose, the culture was split in two and dimethylsulphoxide or Cdc28-as1 inhibitor 1NM-PP1 (5 μM final concentration; Calbiochem) was added.
Rad52 and Mre11 foci analyses
Mre11-YFP Rad52-RFP sae2Δ strain transformed with SAE2, sae2-S267A, sae2-S267E or empty vector was irradiated (30 Gy) with a Faxitron (Faxitron X-ray Corporation). Samples were taken, fixed by the addition of 0.1 volume of formaldehyde, washed three times with PBS, sonicated for 10 s and mixed 1:1 with DAPI-containing mounting medium (Vector Laboratories Inc.). Microscopy was with a DeltaVision microscope (Applied Precision). A minimum of 50 G1 (unbudded) and 50 S/G2 (budded) cells were counted at each time point and for each sample.
Survival of irradiation
sae2Δ mutants transformed with wild-type SAE2, sae2-S267A, sae2-S267E or an empty vector were grown asynchronously or were arrested with α-factor (G1) or nocodazole (G2); they were then irradiated with 300 Gy (Faxitron), kept for 6 h in the same cell-cycle stage and then plated. Colonies arising were normalized with respect to non-irradiated samples and plotted.
NHEJ assays
A pRS416 vector restricted with Xho I, Sac I or Sma I was transformed into cells harbouring various sae2 mutations. The number of colonies formed after 3 days was normalized with the number of colonies obtained in a parallel transformation with a circular pRS416 plasmid. Plasmids from 50 independent clones of each strain transformed with a Xho I-restricted plasmid as described previously were isolated and sequenced.
References
- 1.Shrivastav M, De Haro LP, Nickoloff JA. Regulation of DNA double-strand break repair pathway choice. Cell Res. 2008;18:134–147. doi: 10.1038/cr.2007.111. [DOI] [PubMed] [Google Scholar]
- 2.Aylon Y, Liefshitz B, Kupiec M. The CDK regulates repair of double-strand breaks by homologous recombination during the cell cycle. EMBO J. 2004;23:4868–4875. doi: 10.1038/sj.emboj.7600469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Caspari T, Murray JM, Carr AM. Cdc2-cyclin B kinase activity links Crb2 and Rqh1-topoisomerase III. Genes Dev. 2002;16:1195–1208. doi: 10.1101/gad.221402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hinz JM, Yamada NA, Salazar EP, Tebbs RS, Thompson LH. Influence of double-strand-break repair pathways on radiosensitivity throughout the cell cycle in CHO cells. DNA Repair (Amst.) 2005;4:782–792. doi: 10.1016/j.dnarep.2005.03.005. [DOI] [PubMed] [Google Scholar]
- 5.Ira G, et al. DNA end resection, homologous recombination and DNA damage checkpoint activation require CDK1. Nature. 2004;431:1011–1017. doi: 10.1038/nature02964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Karathanasis E, Wilson TE. Enhancement of Saccharomyces cerevisiae end-joining efficiency by cell growth stage but not by impairment of recombination. Genetics. 2002;161:1015–1027. doi: 10.1093/genetics/161.3.1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Esashi F, et al. CDK-dependent phosphorylation of BRCA2 as a regulatory mechanism for recombinational repair. Nature. 2005;434:598–604. doi: 10.1038/nature03404. [DOI] [PubMed] [Google Scholar]
- 8.Aylon Y, Kupiec M. DSB repair: the yeast paradigm. DNA Repair (Amst.) 2004;3:797–815. doi: 10.1016/j.dnarep.2004.04.013. [DOI] [PubMed] [Google Scholar]
- 9.Clerici M, Mantiero D, Lucchini G, Longhese MP. The Saccharomyces cerevisiae Sae2 protein promotes resection and bridging of double strand break ends. J. Biol. Chem. 2005;280:38631–38638. doi: 10.1074/jbc.M508339200. [DOI] [PubMed] [Google Scholar]
- 10.McKee AH, Kleckner N. A general method for identifying recessive diploid-specific mutations in Saccharomyces cerevisiae, its application to the isolation of mutants blocked at intermediate stages of meiotic prophase and characterization of a new gene SAE2. Genetics. 1997;146:797–816. doi: 10.1093/genetics/146.3.797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Neale MJ, Pan J, Keeney S. Endonucleolytic processing of covalent protein-linked DNA double-strand breaks. Nature. 2005;436:1053–1057. doi: 10.1038/nature03872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Prinz S, Amon A, Klein F. Isolation of COM1, a new gene required to complete meiotic double-strand break-induced recombination in Saccharomyces cerevisiae. Genetics. 1997;146:781–795. doi: 10.1093/genetics/146.3.781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lengsfeld BM, Rattray AJ, Bhaskara V, Ghirlando R, Paull TT. Sae2 is an endonuclease that processes hairpin DNA cooperatively with the Mre11/Rad50/Xrs2 complex. Mol. Cell. 2007;28:638–651. doi: 10.1016/j.molcel.2007.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Baroni E, Viscardi V, Cartagena-Lirola H, Lucchini G, Longhese MP. The functions of budding yeast Sae2 in the DNA damage response require Mec1- and Tel1-dependent phosphorylation. Mol. Cell. Biol. 2004;24:4151–4165. doi: 10.1128/MCB.24.10.4151-4165.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mendenhall MD, Hodge AE. Regulation of Cdc28 cyclin-dependent protein kinase activity during the cell cycle of the yeast Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 1998;62:1191–1243. doi: 10.1128/mmbr.62.4.1191-1243.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Penkner A, et al. A conserved function for a Caenorhabditis elegans Com1/Sae2/CtIP protein homolog in meiotic recombination. EMBO J. 2007;26:5071–5082. doi: 10.1038/sj.emboj.7601916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sartori AA, et al. Human CtIP promotes DNA end resection. Nature. 2007;450:509–514. doi: 10.1038/nature06337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Uanschou C, et al. A novel plant gene essential for meiosis is related to the human CtIP and the yeast COM1/SAE2 gene. EMBO J. 2007;26:5061–5070. doi: 10.1038/sj.emboj.7601913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pommier Y. Topoisomerase I inhibitors: camptothecins and beyond. Nature Rev. Cancer. 2006;6:789–802. doi: 10.1038/nrc1977. [DOI] [PubMed] [Google Scholar]
- 20.Chen J, Saha P, Kornbluth S, Dynlacht BD, Dutta A. Cyclin-binding motifs are essential for the function of p21CIP1. Mol. Cell. Biol. 1996;16:4673–4682. doi: 10.1128/mcb.16.9.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lobachev KS, Gordenin DA, Resnick MA. The Mre11 complex is required for repair of hairpin-capped double-strand breaks and prevention of chromosome rearrangements. Cell. 2002;108:183–193. doi: 10.1016/s0092-8674(02)00614-1. [DOI] [PubMed] [Google Scholar]
- 22.Sugawara N, Haber JE. Repair of DNA double strand breaks: in vivo biochemistry. Methods Enzymol. 2006;408:416–429. doi: 10.1016/S0076-6879(06)08026-8. [DOI] [PubMed] [Google Scholar]
- 23.Bishop AC, et al. A chemical switch for inhibitor-sensitive alleles of any protein kinase. Nature. 2000;407:395–401. doi: 10.1038/35030148. [DOI] [PubMed] [Google Scholar]
- 24.Lazzaro F, et al. Histone methyltransferase Dot1 and Rad9 inhibit single-stranded DNA accumulation at DSBs and uncapped telomeres. EMBO J. 2008;27:1502–1512. doi: 10.1038/emboj.2008.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lisby M, Barlow JH, Burgess RC, Rothstein R. Choreography of the DNA damage response: spatiotemporal relationships among checkpoint and repair proteins. Cell. 2004;118:699–713. doi: 10.1016/j.cell.2004.08.015. [DOI] [PubMed] [Google Scholar]
- 26.Rattray AJ, McGill CB, Shafer BK, Strathern JN. Fidelity of mitotic double-strand-break repair in Saccharomyces cerevisiae: a role for SAE2/COM1. Genetics. 2001;158:109–122. doi: 10.1093/genetics/158.1.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cortes-Ledesma F, Aguilera A. Double-strand breaks arising by replication through a nick are repaired by cohesin-dependent sister-chromatid exchange. EMBO Rep. 2006;7:919–926. doi: 10.1038/sj.embor.7400774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Boulton SJ, Jackson SP. Saccharomyces cerevisiae Ku70 potentiates illegitimate DNA double-strand break repair and serves as a barrier to error-prone DNA repair pathways. EMBO J. 1996;15:5093–5103. [PMC free article] [PubMed] [Google Scholar]
- 29.Lee K, Lee SE. Saccharomyces cerevisiae Sae2- and Tel1-dependent single-strand DNA formation at DNA break promotes microhomology-mediated end joining. Genetics. 2007;176:2003–2014. doi: 10.1534/genetics.107.076539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Limbo O, et al. Ctp1 is a cell-cycle-regulated protein that functions with Mre11 complex to control double-strand break repair by homologous recombination. Mol. Cell. 2007;28:134–146. doi: 10.1016/j.molcel.2007.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Puig O, et al. The tandem affinity purification (TAP) method: a general procedure of protein complex purification. Methods. 2001;24:218–229. doi: 10.1006/meth.2001.1183. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.