Abstract
Mouse ES cells can differentiate into all three germ layers of the embryo but are generally excluded from the trophoblast lineage. Here we show that ES cells deficient in DNA methylation can differentiate efficiently into trophoblast derivatives. In a genome-wide screen we identify the transcription factor Elf5 as methylated and repressed in ES cells, and hypomethylated and expressed in TS and methylation-deficient ES cells. Elf5 creates a positive feedback loop with TS cell determinants Cdx2 and Eomes that is restricted to the trophoblast lineage by epigenetic regulation of Elf5. Importantly, the late-acting function of Elf5 allows initial plasticity and regulation in the early blastocyst. Thus, Elf5 acts downstream of initial lineage determination as a gatekeeper to reinforce commitment to the trophoblast lineage, or to abort this pathway in epiblast cells. This epigenetic restriction of cell lineage fate provides a molecular mechanism for Waddington’s concept of canalization of developmental pathways.
Formation of the first two cell populations of the blastocyst, the trophectoderm (TE) and the inner cell mass (ICM), represents the earliest differentiation event in mammalian development. TE cells are restricted in their differentiation potential towards the trophoblast lineage that is essential for formation of the placenta. By contrast, cells of the inner cell mass (ICM) lose their capacity to differentiate into trophoblast derivatives, but remain pluripotent and give rise to all cell types of the embryo proper as well as to extraembryonic endoderm and mesoderm1-4. Embryonic stem (ES) cells are derived from the ICM of blastocyst-stage embryos5. Like ICM cells, they are extremely efficient in colonising all germ layers of the embryo6, but are generally excluded from the trophoblast lineage7. Conversely, trophoblast stem (TS) cells, derived from the TE, differentiate into all trophoblast subtypes of the placenta, but are excluded from embryonic tissues8,9. Thus, the stem cell populations derived from the blastocyst stably retain the cell lineage restrictions that are imposed by this stage.
The initial cell lineage specification is established by key transcription factors such as Oct4, Nanog, Sox2, as well as Cdx2 and Eomes, that are capable of defining embryonic and trophoblast lineage identity, respectively10. Once fixed, lineage determination must be accompanied by epigenetic modifications that ensure the stable inheritance of cell fate11. It is particularly important to determine how lineage-specific transcription factor networks interact with different epigenetic marking systems in order to achieve this stability. One of the earliest global epigenetic marks that distinguishes the ICM and TE lineages is DNA methylation: ICM cells become de novo methylated in early blastocysts while TE cells remain hypomethylated12, and this global difference in methylation between embryonic and extraembryonic tissues is maintained throughout development13,14. Indicating a functional role of this differential methylation pattern, expression of few markers of differentiated trophoblast has been observed in embryoid bodies derived from Dnmt-deficient (and therefore hypomethylated) ES cells15. Here, we have examined in detail the role of DNA methylation in the stability of embryonic lineage determination. We show that ES cells and cells of the embryo proper that are deficient in methylation can differentiate into trophoblast derivatives, progressing in an orderly fashion from stem cells towards giant cells. DNA methylation-mediated silencing of a trophoblast-specific transcription factor, Elf5, normally restricts this differentiation pathway. The identification of this epigenetically regulated gatekeeper provides an explanation for the early, definitive nature of embryonic and trophoblast cell lineage separation.
Results
Trophoblast from Dnmt1-/- embryonic cells
To evaluate whether DNA methylation is essential for the stability of embryonic cell lineage determination, we examined whether trophoblast differentiation occurs from ES cells deficient in DNA methylation due to deletion of the maintenance DNA methyltransferase, Dnmt1. Formation of trophoblast giant cells, a morphologically distinct placental cell type, was observed only sporadically when Dnmt1-/- ES cells were grown in standard ES cell medium. In sharp contrast, ∼ 25% of all Dnmt1-/- ES cells differentiated into trophoblast giant cells upon culture in TS cell conditions as assessed by morphological criteria and by marker gene expression (Fig. 1a, b). In a time course experiment where Dnmt1-/- ES cells were subjected to TS cell conditions for 2-12 days, trophoblast stem cell markers Eomes and Cdx2 were the first to be detected, followed by ectoplacental cone and spongiotrophoblast markers Ascl2 and Tpbpa, and subsequently giant cell-expressed genes Pl1 (Prl3d1) and Pl2 (Prl3b1; Fig. 1b). This sequence of expression was comparable to that observed when TS cells undergo differentiation8. No significant trophoblast differentiation or marker expression was observed with wildtype ES cells grown under TS cell conditions. Trophoblast differentiation was not restricted to methylation-deficient ES cells, but also occurred in Dnmt1-/- embryos in vivo. Quantitative RT-PCR analysis showed a striking up-regulation of markers for intermediate diploid trophoblast (Ascl2, Tpbpa) and trophoblast giant cells (Pl1, Pl2) in E9.5 Dnmt1-deficient embryos (Fig. 1c), thus reflecting a mid-to-late stage in the time course of trophoblast transdifferentiation from embryonic cells. The spatial distribution and morphology of these ectopically differentiating trophoblast cells was determined by in situ hybridization where cells positive for Tpbpa, Plf (Prl2c2), and Pl1 were detected in Dnmt1-/- but not wildtype embryos, with giant cell marker-expressing cells exhibiting an enlarged cell size compared to surrounding embryonic cells (Fig. 1d, e). These data clearly demonstrate the ectopic differentiation of trophoblast derivatives in Dnmt1-depleted embryos and ES cells, showing that appropriate DNA methylation levels are crucial for maintenance of the embryonic lineage.
Fig. 1.
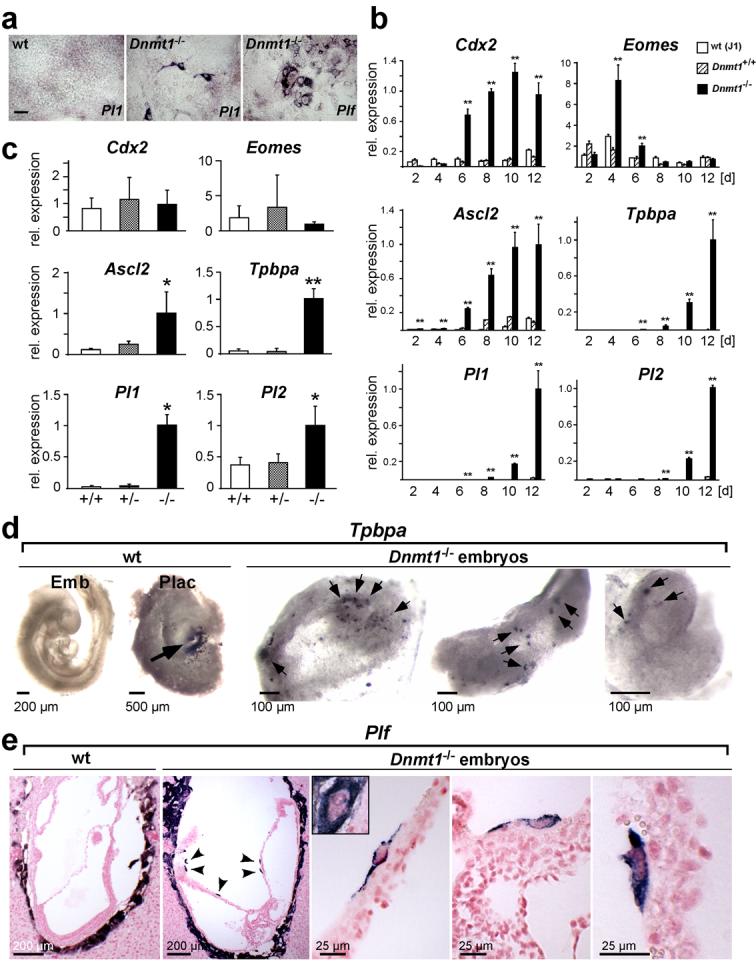
Dnmt1-deficiency enables trophoblast differentiation from cells committed to the embryonic cell lineage.
(a) Trophoblast giant cells differentiate from Dnmt1-/- ES cells, but not wildtype (wt) ES cells, in vitro when cultured in TS cell conditions. In situ hybridization with trophoblast giant cell-specific markers Pl1 (after 12d) and Plf (after 8d). (b) Time course of trophoblast marker activation in ES cells upon culture in TS cell medium for 2-12 days. Significant expression levels are only observed in Dnmt1-deficient ES cells (data are mean ± s.d., **P<0.005; n=3). Trophoblast stem cell markers (Cdx2, Eomes) are the first to be up-regulated, followed by intermediate diploid trophoblast markers Ascl2 and Tpbpa, and trophoblast giant cell markers Pl1 and Pl2. (c) Quantitative RT-PCR analysis of trophoblast markers on E9.5 embryos from Dnmt1+/- intercrosses. Trophoblast-restricted genes Ascl2, Tpbpa, Pl1 and Pl2 are detected at significant levels only in Dnmt1-/- embryos (data are mean ± s.d., *P<0.05, **P<0.005; n=7). (d) Whole mount in situ hybridization with the trophoblast marker Tpbpa on E9.5 Dnmt1-/- embryos and littermate control (wt) embryos and placentas. Positive staining (dark purple-black) is seen in the ectoplacental cone/spongiotrophoblast area (arrow) of the placenta (Plac). No staining is observed in the wt embryo (Emb). In Dnmt1-/- embryos, numerous Tpbpa-positive cells are present (arrows). (e) In situ hybridization with the giant cell marker Plf on E8.5 wildtype (wt) and Dnmt1-deficient conceptuses. Arrowheads point towards Plf-positive cells (blue-purple) that are observed within embryonic structures in Dnmt1-/- conceptuses only. Higher magnification views show the morphology of these cells; they are larger than surrounding cells but do not reach the size of parietal trophoblast giant cells (shown in inset). Scale bar in a represents 100 μm, scale bars in d and e are as indicated.
The major difference between ES and TS cell culture conditions is the absence of LIF and presence of FGF4 in TS cell medium. Therefore, we investigated whether the FGF signalling receptor in trophoblast stem cells, Fgfr2c8,16,17, was expressed concomitantly with trophoblast differentiation. Like Cdx2, Fgfr2c was activated in Dnmt1-/-, but not wildtype, ES cells upon growth in TS cell medium. Importantly, wildtype ES cells could be induced to express trophoblast genes at levels similar to those observed in Dnmt1-/- ES cells by addition of the methylation inhibitor 5-azacytidine (Fig. 2a, b). As in Dnmt1-/- ES cells, activation of trophoblast markers coincided with the appearance of giant cells (Fig. 2c). Interestingly, azacytidine-treatment of Dnmt1-/- ES cells did not lead to a further increase in trophoblast stem cell-specific Cdx2 and Fgfr2c expression but instead caused an up-regulation of markers of differentiated trophoblast (Fig. 2b), suggesting critical methylation threshold levels for trophoblast stem cell maintenance.
Fig. 2.
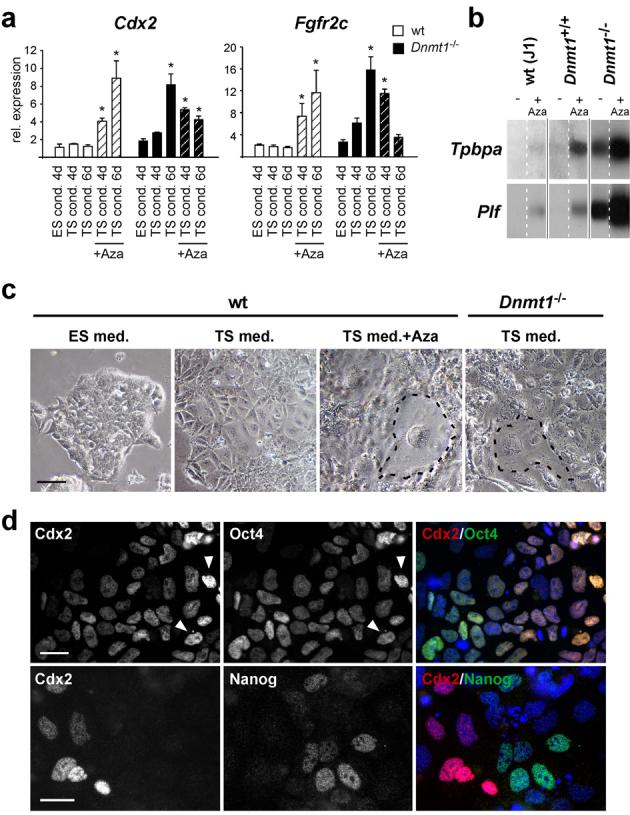
Trophoblast differentiation is induced in wildtype ES cells upon inhibition of DNA methylation and is independent of Oct4 down-regulation.
(a) Expression levels of Cdx2 and Fgfr2c in wildtype and Dnmt1-/- ES cells cultured in ES-, TS- and TS cell medium with 5-azacytidine over the indicated time periods. Culture in TS cell medium alone is sufficient to induce expression of Cdx2 and Fgfr2c from Dnmt1-/- ES cells, but expression levels of these trophoblast stem cell markers cannot be further enhanced by 5-azacytidine treatment. In wildtype ES cells, Cdx2 and Fgfr2c expression can be induced by inhibition of DNA methylation with 5-azacytidine. Data are mean ± s.d., *P<0.05; n=3. (b) Northern blot hybridization with Tpbpa and Plf on wildtype (J1 and Dnmt1+/+) and Dnmt1-/- ES cells after 12d culture in TS cell medium in the presence (+ Aza) or absence (-) of 5-azacytidine. 5-azacytidine treatment induces differentiation of trophoblast derivatives in wildtype ES cells and further enhances the amount of differentiated trophoblast subtypes from Dnmt1-/- ES cells. The separated lanes are parts of the same original blot; full scans of blots are provided in Supplemental Information, Fig. S5. (c) Wildtype and Dnmt1-/- ES cells grown for 4-6 days under the indicated conditions. Wildtype ES cells cultured in TS cell conditions undergo differentiation into embryonic lineage derivatives, but rarely form trophoblast giant cells. Trophoblast-like morphology and the appearance of giant cells (encircled) are evident in wildtype ES cells treated with 5-azacytidine and in Dnmt1-/- ES cells cultured in TS cell conditions. (d) Double immunofluorescence labelling of Dnmt1-/- ES cells grown for 6 days in TS cell conditions for Oct4 (Pou5f1), Nanog and Cdx2. Trophoblast differentiation is not triggered by down-regulation of Oct4. Arrows point to cells expressing high amounts of both Cdx2 and Oct4. A more reciprocal expression pattern is observed for Nanog and Cdx2. Scale bar in c represents 100 μm, scale bars in d represent 25 μm.
We next asked if trophoblast differentiation was triggered by down-regulation of Oct4, since loss of Oct4 expression in ES cells leads to transdifferentiation into trophoblast18. Interestingly, even after six days of culture in TS cell medium most cells co-expressed Cdx2 and Oct4 (Fig. 2d). This pattern suggests that demethylation of ES cells has expanded their developmental plasticity to include both extraembryonic and embryonic fate, rather than switching fate to an exclusive extraembryonic one. It may also reflect the pattern in compacting morulae where Oct4 levels remain high in both Cdx2+ and Cdx2- cells19. A more reciprocal pattern was observed with Cdx2 and Nanog (Fig. 2d).
To correlate the temporal progression and spatial distribution of transdifferentiating trophoblast derivatives in situ, Dnmt1-/- ES cells were co-stained for Cdx2 and Plf 2-6 days after culture in TS cell medium. Cdx2-positive cells were somewhat enlarged compared to surrounding ES cells and typically emerged at the margins of ES cell colonies (Fig. 3a, i). At the earliest time points of giant cell differentiation, all Plf-positive giant cells were located directly adjacent or in very close spatial proximity to Cdx2-positive cell groups (Fig. 3a, ii). In addition, some cells were observed in a transition state of differentiation, staining weakly positive for both Cdx2 and Plf (Fig. 3a, iii). The non-random appearance of giant cells indicates a clonal origin of trophoblast derivatives in Dnmt1-/- ES cells and suggests that they recapitulate a differentiation pathway comparable to that of the trophoblast lineage proper (Fig. 3a).
Fig. 3.
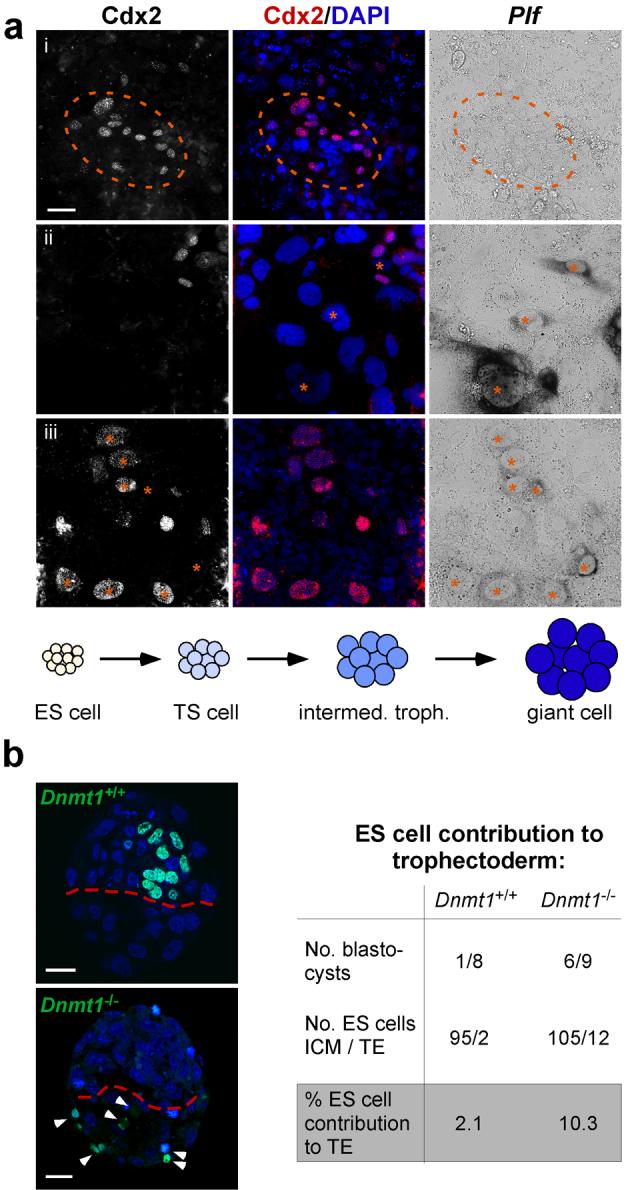
Dnmt1-/- ES cells transdifferentiate into functional trophoblast derivatives.
(a) Double labelling of Dnmt1-/- ES cells grown in TS cell conditions for 6 days and stained by in situ hybridization for the giant cell marker Plf and by immunofluorescence for the TS cell marker Cdx2. Cdx2-positive cells are observed in small groups consisting on average of 5-15 cells. At this early time point of transdifferentiation, giant cells (Plf) are either absent (i) or emerging in direct vicinity to Cdx2-positive TS cell clusters (ii). On rare occasions, cells can be captured at an intermediate stage of concomitant low Cdx2 and Plf expression (iii; marked with asterisks), demonstrating that giant cells differentiate from pre-existing trophoblast stem cells. Thus, trophoblast differentiation from Dnmt1-/- ES cells proceeds via a trophoblast stem cell stage from which other trophoblast subtypes differentiate sequentially, recapitulating the differentiation steps within the trophoblast lineage proper (graph). (b) Blastocysts derived from aggregation of wildtype embryos with H2B-GFP labelled Dnmt1+/+ or Dnmt1-/- ES cells that were pre-conditioned in TS cell medium. Dnmt1-/- ES cells differentiate into functional trophoblast and exhibit a 5-fold increased efficacy to contribute to trophectoderm. Arrowheads point to individual GFP-positive Dnmt1-/- cells within the mural trophoectoderm. Scale bar in a represents 20 μm, scale bars in b represent 25 μm.
Dnmt1-/- ES cells contribute to the TE
Since hypomethylation caused loss of embryonic lineage restriction in embryos and ES cells, we examined whether Dnmt1-/- ES cells could directly contribute to the TE in aggregation chimeras. For this experiment, histone H2B-GFP labelled Dnmt1+/+ and Dnmt1-/- ES cells were ‘pre-conditioned’ for transdifferentiation by culture in TS cell medium for 1-3 days prior to aggregation with 8-cell embryos. Comparable to original studies7, control ES cells contributed extremely rarely to the TE. Dnmt1-/- ES cells, however, were observed in the TE of two-thirds of all blastocysts and exhibited a 5-fold increase in contribution to the TE compared to their wildtype counterparts (Fig. 3b).
Elf5 is differentially methylated
Our finding that DNA methylation prevents trophoblast differentiation in ES cells and embryos suggested that cell lineage stability is achieved by methylating trophoblast determinants in the embryonic lineage. Hence, we analyzed the key determinants of the trophoblast lineage for promoter methylation in ES cells. Although Cdx2, Eomes and Fgfr2 were induced in differentiating Dnmt1-/- ES cells, we found that these genes were unmethylated in both ES and TS cells (Supplementary Information, Fig. S1). We thus carried out a genome-wide screen for promoter methylation by meDIP array hybridization20, comparing ES with TS cells. We screened for promoters that exhibited higher methylation levels in ES cells compared to TS cells. A very small number of candidate promoters of this type were identified (see Methods); intriguingly, however, after validation by Sequenom® mass array technology only one gene with a robust difference in methylation was confirmed, Elf5 (Fig. 4a; Supplementary Information, Fig. S2). Elf5 encodes a transcription factor of the Ets family that plays a pivotal role in the trophoblast lineage21. Upon ablation of Elf5, the TE lineage can be specified and blastocysts implant into the uterus, but the proliferative capacity of trophoblast stem cells is not maintained and conceptuses die shortly after implantation due to a lack of extraembryonic ectoderm21.
Fig. 4.
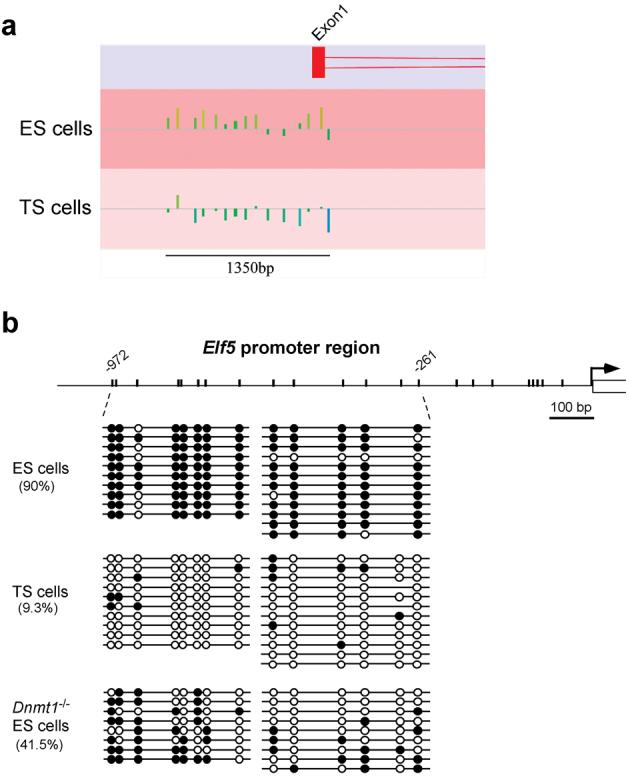
Global promoter methylation screen identifies Elf5 as the key gene that is methylated in ES cells and unmethylated in TS cells.
(a) meDIP chip screen reveals strikingly differential methylation at the Elf5 promoter. ChipMonk profile of the Elf5 upstream region in ES and TS cells. The line represents the median signal intensity of the array; vertical bars above indicate relative hypermethylation and vertical bars below hypomethylation at individual oligonucleotide probes. (b) Bisulphite analysis of the 1 kb promoter region of Elf5 in ES, TS, and Dnmt1-/- ES cells. The promoter is almost fully DNA methylated (filled circles) in ES cells, but largely hypomethylated (open circles) in TS cells. Dnmt1-/- ES cells exhibit intermediate DNA methylation levels. Note that the CpG site at position -355 bp is polymorphic.
Detailed characterization of the Elf5 promoter methylation profile by bisulphite sequencing analysis revealed that the vast majority of CpG residues in the 1 kb upstream region were methylated in ES cells but unmethylated in TS cells (Fig. 4b). Importantly, Dnmt1-deficient ES cells exhibited hypomethylation of the Elf5 promoter compared to wildtype ES cells, especially closer to the transcriptional start site. This methylation pattern was stable as it did not change over a 10-day time course of culture in TS cell conditions (Supplementary Information, Fig. S2). The mosaic methylation pattern of Elf5 in Dnmt1-/- ES cells provides a likely explanation why not all, but typically a total of 40-60% of all Dnmt1-/- cells were found to undergo transdifferentiation into trophoblast derivatives (i.e. stem, intermediate, spongio- and giant trophoblast cells) during this time period.
Elf5 marks diploid trophoblast
Consistent with the trophoblast-specific expression of Elf5 in early mouse embryos21, Elf5 was detected in undifferentiated TS cells but was absent from ES cells (Fig. 5a, b). Upon differentiation of TS cells, Elf5 expression was strongly down-regulated (Fig. 5b). Elf5 staining mostly overlapped with the TS cell marker Cdx2, but was also observed in some additional diploid trophoblast cells that were Cdx2-negative (Fig. 5c). Analysis of the temporal regulation in vivo showed that Elf5 expression was initiated later than Cdx2. In contrast to Cdx2, only marginal amounts of Elf5 transcripts and no protein were detected in blastocysts (data not shown). However, Elf5 was readily detected in blastocyst outgrowths where a central core of Cdx2+ cells was overlaid by Cdx2+ and Elf5+ double-positive and then Cdx2-negative, Elf5-positive diploid, proliferative trophoblast (Fig. 5d). Similarly, Elf5 marked the extraembryonic ectoderm of conceptuses immediately after implantation from E5.5 onwards (Fig. 5e). These data show that Cdx2 precedes Elf5 expression in the emerging TE, but that Elf5 expression is sustained longer in diploid proliferating trophoblast of the extraembryonic ectoderm (Fig. 5f).
Fig.5.
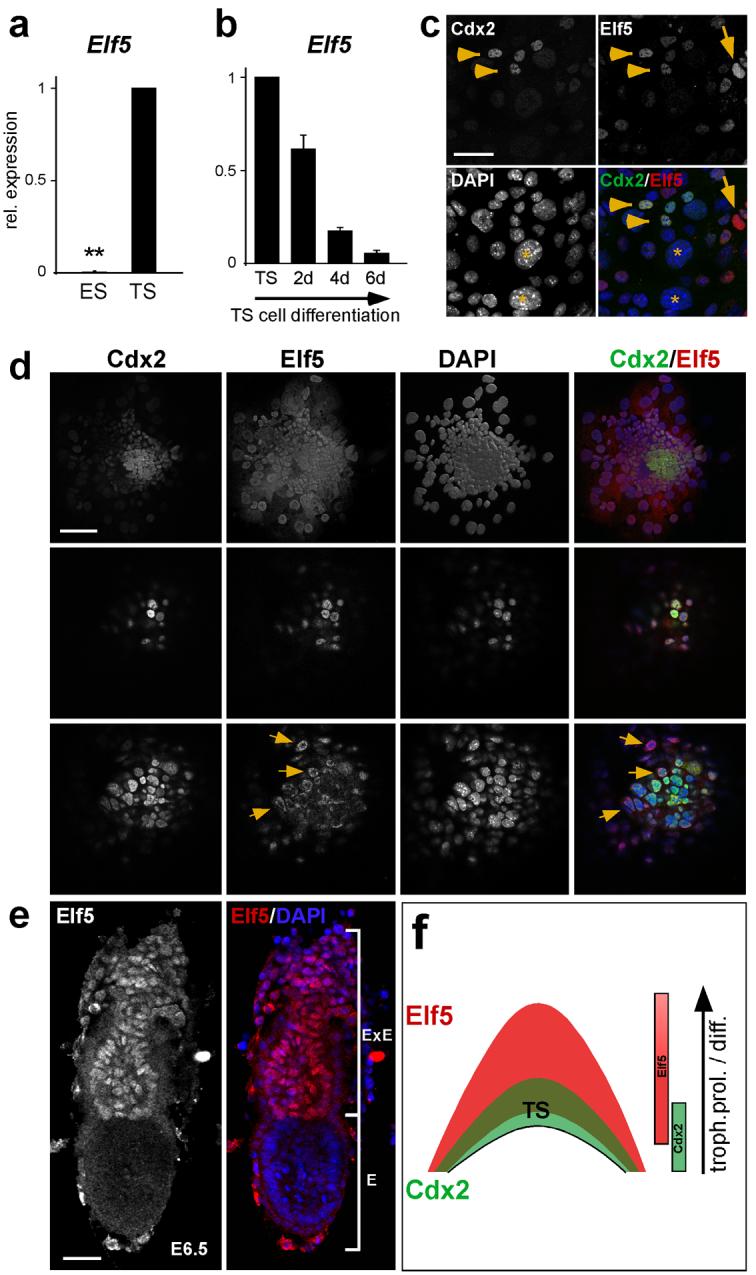
Elf5 is a trophoblast stem cell transcription factor specific to proliferative trophoblast.
(a) Elf5 is not expressed in ES cells but exhibits high expression levels in TS cells (Data are mean ± s.d., **P=1.12E-12; n=3). (b) Elf5 expression is specific to the stem cell state of TS cells and is down-regulated upon differentiation of TS cells. (c) Double immunofluorescence staining for Elf5 and Cdx2 on cultured TS cells shows co-localization of both transcription factors in most stem cells (arrowheads). Some diploid trophoblast cells only express Elf5 (arrow), while giant cell nuclei are mostly negative for both factors (asterisks). (d) Characterization of Elf5 expression in blastocyst outgrowths. Selected confocal section planes are shown. Compared to Cdx2, Elf5 marks a larger population of trophoblast cells (top row). A few central cells only express Cdx2, followed by a core of double positive trophoblast stem cells (middle row). Elf5 staining extends beyond this core into surrounding trophoblast cells (bottom row). Nuclear localization adopts a cortical distribution where diploid trophoblast starts to differentiate into giant cells (arrows). Giant cells are devoid of nuclear Elf5. (e) Whole mount staining of an E6.5 embryo for Elf5. Elf5 expression is strictly trophoblast lineage-specific and is confined to the extraembryonic ectoderm (where trophoblast stem cells are located) and to the ectoplacental cone. E=embryonic portion, ExE=extraembryonic portion. (f) Schematic diagram representing the temporal and spatial distribution of Cdx2 and Elf5. A small Cdx2+ population is overlaid by a Cdx2+ Elf+ trophoblast stem cell core. Elf5 extends from there into the surrounding diploid trophoblast population that undergoes proliferation (prol.) and differentiation (diff.) at the margins of the ectoplacental cone. Scale bar in c represents 50 μm, scale bars in d and e represent 100 μm.
Elf5 activates Cdx2 and Eomes
To analyze whether hypomethylation of the Elf5 promoter was correlated with its expression in cells of the embryonic lineage, we first assessed Dnmt1-/- embryos and ES cells for ectopic activation of Elf5 and found that they expressed high levels of this transcription factor. In fact, Elf5 was the earliest and most significantly induced trophoblast marker in the time course of ES-to-trophoblast transdifferentiation and was expressed in derivatives of every single cell (Fig. 6a-c; Supplementary Information, Fig. S4). Elf5 was also strongly activated upon treatment of wildtype ES cells with 5-azacytidine (Supplementary Information, Fig. S4). To further test the direct relationship between hypomethylation and expression of Elf5 and the activation of trophoblast markers, we examined other hypomethylated ES cell models null for Np95, Dnmt3a/b or Cxxc122-24. In all three cell lines, the Elf5 promoter was extremely hypomethylated resulting in high expression of Elf5 and up-regulation of other trophoblast markers (Supplementary Information, Fig. S3). Thus, Elf5 expression is associated with a loss of embryonic cell lineage determination irrespective of how promoter demethylation is achieved. Consistent with the observations of Cdx2 and Oct4 co-expression shown above, activation of Elf5 at earliest stages in the transdifferentiation cascade was independent of Oct4 repression (Fig. 6d).
Fig. 6.
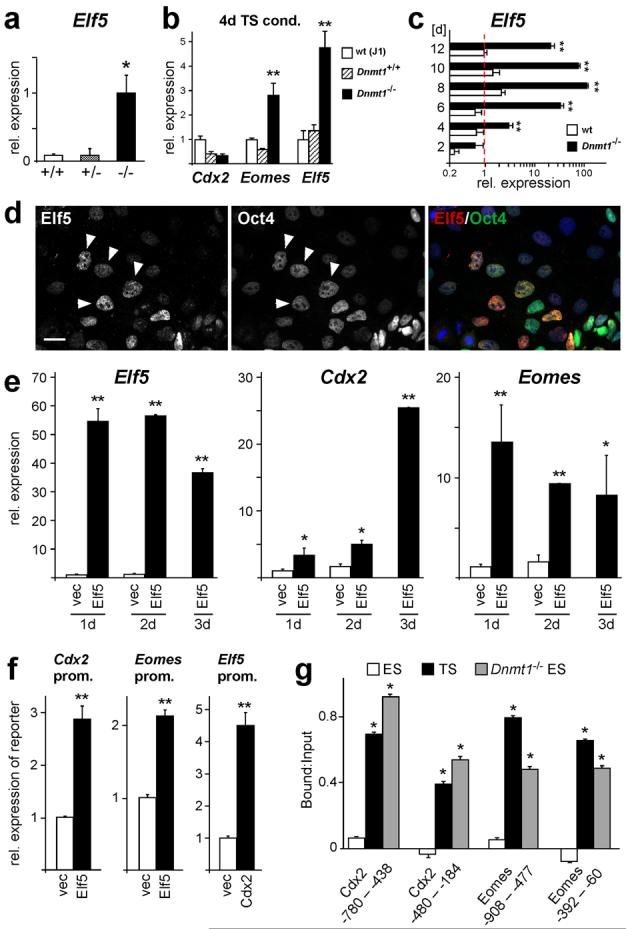
Epigenetically controlled Elf5 expression acts in a positive feedback loop to reinforce trophoblast identity.
(a) Elf5 expression is strongly induced in Dnmt1-/- embryos, but not in wildtype and heterozygous littermate embryos (data are mean ± s.d., *P=0.026; n≥6). (b) Elf5 is the earliest, most strongly activated trophoblast marker in Dnmt1-/- ES cells cultured for 4 days in TS cell conditions (data are mean ± s.d., **P=0.00185 [Eomes] and P=0.00051 [Elf5]; n=3). (c) Elf5 expression is further enhanced in Dnmt1-deficient ES cells during the following days of culture in TS cell conditions (data are mean ± s.d., **P<0.005; n=3). (d) Double labelling of Dnmt1-/- ES cells grown for 4 days in TS cell conditions for Elf5 and Oct4. Oct4 repression is not required for activation of Elf5. (e) Elf5 expression in wildtype ES cells causes activation of TS cell markers Cdx2 and Eomes. Wildtype ES cells were transfected with an Elf5-GFP expression construct, FACS sorted and assessed by qRT-PCR. The construct confers high levels of Elf5 expression to transfected cells (Elf5) compared to control transfected cells (vec) that do not express Elf5. Elf5 induces high expression of Eomes. Cdx2 expression is significantly elevated after 1 and 2 days (>3-fold), and reaches highest activation levels after 3 days of Elf5 transfection (data are mean ± s.d., *P<0.05, **P<0.005; n=3). (f) Elf5, Cdx2, or empty vector control, were co-transfected with 2 kb Cdx2, Eomes or Elf5 promoter-reporter constructs. Expression of the reporter gene was analyzed by qRT-PCR 24 hours after transfection. Elf5 can induce both Cdx2 and Eomes promoters. Cdx2 can activate the Elf5 promoter (data are mean ± s.d., **P<0.005; n=3). (g) Chromatin immunoprecipitation assays with an anti-Elf5 antibody. Elf5 binds directly to the Cdx2 and Eomes promoters in TS cells and 3-day transdifferentiated Dnmt1-/- ES cells, but not in wildtype ES cells. Values are represented as Bound:Input and normalized against mock control (data are mean ± s.d., *P<0.05; n=3). Scale bar in d represents 25 μm.
If expression of hypomethylated Elf5 was responsible for trophoblast differentiation from embryonic cells, Elf5 might directly activate the trophoblast stem cell determinants Cdx2 and Eomes. Indeed both the Cdx2 and Eomes promoters contain the core binding motif for the transcription factor Elf525. Consistent with this hypothesis, we found that forced expression of Elf5 in wildtype ES cells resulted in a strong induction of Eomes as early as 24h after transfection. Likewise, Cdx2 expression was significantly (>3-fold) activated after 24h but peaked somewhat later after 2-3 days of transfection with Elf5 (Fig. 6e). We confirmed that Elf5 was able to activate the Cdx2 and Eomes promoters in co-transfection experiments of Elf5 with reporter constructs, and that conversely, Cdx2 (and likely Eomes) could also activate the Elf5 promoter (Fig. 6f, Supplementary Information, Fig. S3). Chromatin immunoprecipitation (ChIP) assays demonstrated that Elf5 directly interacted with the Cdx2 and Eomes promoters and that this interaction occurred exclusively in TS cells and in Dnmt1-/- ES cells, but not in wildtype ES cells (Fig. 6g). Hence, ectopic expression of Elf5 directly induces trophoblast stem cell determinants and thereby initiates the trophoblast differentiation cascade in cells of the embryonic lineage. Importantly, this differentiation pathway depends on Elf5 since trophoblast marker activation is significantly decreased in hypomethylated ES cells in which Elf5 expression is diminished (Fig. 7a).
Fig. 7.
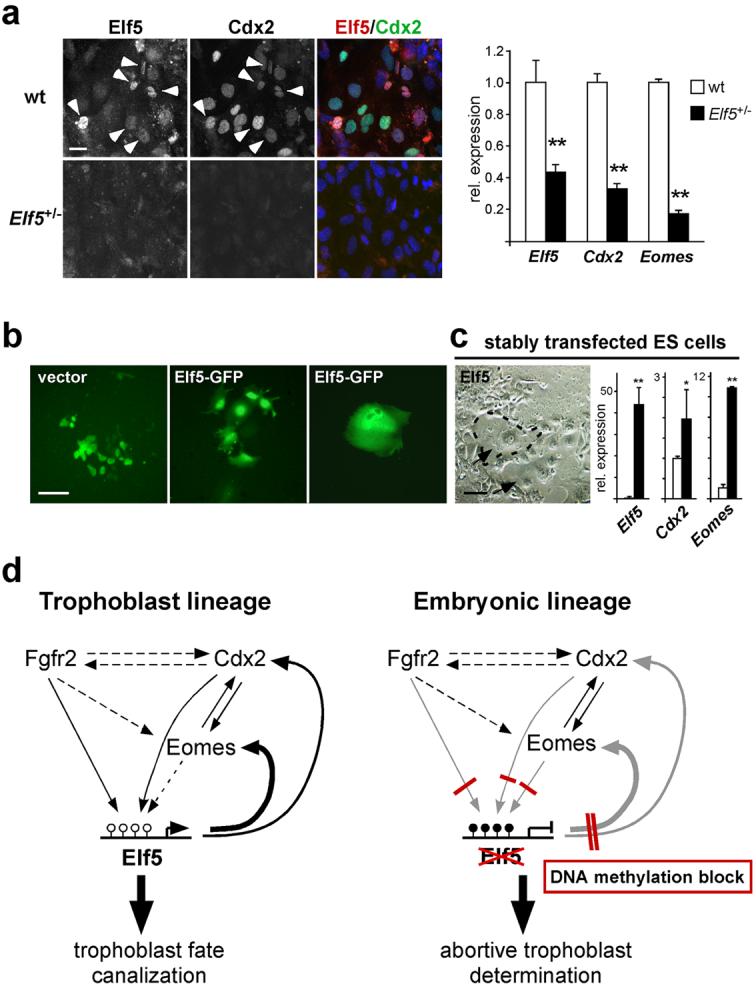
Gatekeeper function of Elf5 in cell lineage specification.
(a) Initiation of the trophoblast differentiation cascade depends on Elf5. ES cells heterozygous for a gene-trap insertion at the Elf5 locus grown for 5 days in TS medium with 5-azacytidine and assessed for Elf5, Cdx2 and Eomes expression. The gene-trap insertion reduces Elf5 mRNA levels to 44% and further reduces protein levels. Elf5 and Cdx2 staining is abundant in wild-type ES cells (arrowheads); almost no Elf5 immunostaining is detected in Elf5+/- cells. Low Elf5 expression is associated with a dramatic decrease in Cdx2 protein and Cdx2 and Eomes mRNA levels. Images were taken at identical exposure settings; qRT-PCR data are mean ± s.d., **P<0.005; n=3. (b) Wildtype ES cells transfected with Elf5-GFP or empty vector were FACS sorted and re-plated in TS cell medium for 4 days. Control cells proliferate and form colonies; Elf5-transfected cells cease to proliferate and differentiate into trophoblast giant cells. (c) Wildtype ES cells stably transfected with a Cre-inducible Elf5 expression construct were transfected with Cre-GFP, FACS-sorted, and re-plated in TS cell medium for 5 days. Elf5 expression leads to trophoblast differentiation and appearance of trophoblast giant cell clusters (arrows, examples encircled). White and black bars represent cells before and after Cre-mediated Elf5 induction, respectively (data are mean ± s.d., *P<0.05, **P<0.005; n=3). (d) Model of Elf5 function in lineage canalization. Developmental onset of expression and genetic data place Elf5 downstream of Cdx2, Eomes and Fgfr2. The developmental position of Elf5 coincides with the definitive fixation of cell lineage fate at the late blastocyst stage. Elf5 is regulated by DNA methylation, but the upstream players Cdx2, Eomes and Fgfr2 are not. Elf5 creates an essential feedback loop to reinforce Cdx2 and Eomes expression in trophoblast stem cells. Epigenetic silencing of Elf5 by DNA methylation interrupts this trophoblast-specific reinforcement loop in the embryonic lineage, and thereby safeguards embryonic cells from differentiating into trophoblast derivatives. Scale bar in a represents 25 μm, scale bar in b and c represents 100 μm.
These data establish that Elf5 is essential to reinforce the expression of critical trophoblast stem cell factors, which provides an explanation for the observed loss of Cdx2 expression in Elf5-mutant conceptuses21. As indicated by the immunostaining patterns of blastocyst outgrowths and early conceptuses, however, this transcriptional activator function of Elf5 is limited to a relatively narrow spatio-temporal window since Elf5 expression is sustained in diploid trophoblast cells that have ceased to express Cdx2 (Fig. 5). Consequently, stable expression of Elf5 in wildtype ES cells, as well as continued culture of Dnmt1-/- ES cells in TS cell conditions, resulted in the widespread differentiation of cells with trophoblast (giant) cell morphology, but did not allow the establishment of self-renewing TS cell lines (Fig. 7b, c; Supplementary Information, Fig. S4). Thus, Elf5 creates a cell lineage-restricting niche at the critical interface between promotion and loss of the self-renewing capacity of trophoblast stem cells.
Discussion
In this study we have made three significant observations: First, ES cells deficient in DNA methylation can differentiate efficiently into the trophoblast lineage. We demonstrate that the trophoblast differentiation programme occurs in vivo as well as in vitro and involves expression of TS cell transcription factors Cdx2, Eomes, and Elf5 and progression in an orderly fashion from stem cells and intermediate trophoblast to giant cells. Second, the transcription factor Elf5 binds to and positively regulates the Cdx2 and Eomes promoters, establishing a positive feedback loop which is critical for the maintenance of the trophoblast stem cell population. Third, the Elf5 gene is hypomethylated in the trophoblast lineage, so that the feedback loop is sustained and leads to expansion of the stem cell compartment and subsequent differentiation. By contrast, Elf5 is stably repressed by DNA methylation in the embryonic lineage; as a result any induction of trophoblast cell fate mediated by stochastic expression of Cdx2 or Eomes cannot be sustained in this compartment.
Several studies have demonstrated that epigenetic regulation through Polycomb group proteins is essential to maintain the pluripotent state within the embryonic lineage in ES cells26-28. Here, we show that DNA methylation functions as a lineage barrier between the embryonic and trophoblast lineage compartments (Fig. 7d). Consequently, hypomethylated ES cells can adopt a trophoblast cell fate. It is important to note, however, that methylation-deficient ES cells have not simply switched from an ES to a trophoblast fate, but have broadened their potency to include both an embryonic and extraembryonic fate. Thus, they adopt a state comparable to an earlier developmental stage prior to lineage commitment, and this interpretation is consistent with the observed overlapping staining patterns of lineage markers. While methylation-deficient ES cells are able to express trophoblast determinants Cdx2 and Eomes (as well as Fgfr2), the promoters of none of these genes are methylated in wildtype ES cells. Therefore, we performed a genome-wide screen for promoter methylation differences between ES and TS cells. This analysis revealed that the global methylation asymmetry observed between the embryonic and trophoblast lineage is not reflected by the methylation profile of gene promoters on a large scale. The major differences in methylation between the two lineages are therefore likely to be located in non-genic regions such as, for example, centromeric heterochromatin. These observations are similar to the X chromosome where global hypermethylation but promoter-specific hypomethylation is observed on the active X29. However, by specifically screening for gene promoters with higher methylation levels in ES cells, we identified Elf5 as the gatekeeper gene that is methylated in ES cells but unmethylated in TS cells. Elf5 is activated through Fgf-Fgfr signalling30 and in turn activates Eomes and Cdx2. When the Elf5 promoter is methylated, low levels of expression of Eomes and/or Cdx2 (Fig. 1a) remain inconsequential because the transcriptional loop is interrupted at the level of Elf5. The robust positive feedback loop created by Elf5 in the trophoblast stem cell niche allows this compartment to expand in a temporally defined window. Consequently, this cell population is lost and Cdx2 expression cannot be sustained in Elf5-mutants21. This compartment can also be established by overexpression of Cdx2 or Eomes in ES cells31 which we show is independent of Elf5 demethylation at early stages, but over prolonged periods leads to Elf5 demethylation and expression thereby maintaining the feedback loop and allowing progression of the trophoblast differentiation pathway (Supplementary Information, Fig. S4). Normally, the feedback loop established by Elf5 is temporally restricted to a few cell divisions, thereby positioning Elf5 at the interface between reinforcing trophoblast fate and onset of differentiation. This may be due to the gradual loss of other regulatory factors required for Cdx2 (or Eomes) expression. Consequently, Elf5 can induce trophoblast differentiation from ES cells but cannot, unlike Cdx2, convert them into continuously self-renewing TS cell lines. We cannot exclude that there may be additional gatekeeper genes such as Elf5 which are methylated in ES cells and the epiblast, but our screen was exhaustive and validation studies show that robust methylation differences are detected reliably32.
In the absence of silencing by DNA methylation or in a force-expression system, Elf5 has trophoblast-determining functions due its capacity to activate Cdx2 and Eomes. Within the normal developmental context, however, Elf5 acts downstream of the trophoblast determinants Cdx2 and Eomes as well as the recently identified Tead4 which has been shown to induce Cdx2, making it the gene currently at the top of the extraembryonic fate cascade33-35. A gatekeeper function for such a relatively late-acting gene may at first appear counterintuitive. However, the temporal position of Elf5 in the trophoblast determination cascade correlates perfectly with the developmental fixation of lineage restriction and specifically allows the plasticity and regulation that is characteristic of early mammalian development3,36,37. Thus, individual blastomeres are still able to cross lineages and expression of lineage ‘markers’ remains mosaic until the mid-blastocyst stage19. Only slightly later, Elf5 expression in the extraembryonic ectoderm reinforces extraembryonic cell fate, while the inability to activate Elf5 in the epiblast due to promoter methylation restricts its descendants to the embryonic cell fate. The late-acting function of Elf5 is in line with its position downstream of lineage-determining transcription factors at the interface of stem cell self-renewal and onset of differentiation.
To what extent blastomeres of early mouse cleavage-stage embryos are pre-determined towards their future fate has been a matter of some debate38. It is now well accepted that position, cell polarization and relative threshold levels of a few interacting transcription factors bias a blastomere’s fate towards the embryonic or extraembryonic cell lineage39. Irreversible fixation of cell fate only occurs at the late blastocyst stage, however, and our findings provide, for the first time, a molecular mechanism for this transition from initial plasticity to lineage-restricted potency in development. This regulative view of development was most famously expressed by C. H. Waddington, who likened the path of a cell lineage towards terminal differentiation to a ball travelling downwards along branching valleys; once it has entered its final valley it cannot easily cross the mountain into the neighbouring one or return to the beginning. This creates canalization of developmental pathways so that they become stable and potentially irreversible40. Our observations give rise to a molecular model of regulation and canalization in early mammalian development. Manipulation of gatekeeper genes such as Elf5 or their epigenetic regulation may help with strategies in regenerative medicine that aim to generate appropriate cell types by transdifferentiation or by reprogramming of somatic cells.
Methods
Animals and tissue preparation
Mice heterozugous for the Dnmt1c allele41 were maintained on a C57BL/6 background. Homozygous null Dnmt1c/c conceptuses were obtained by heterozygous matings, counting the day of the vaginal plug as E0.5. For histology, E8.5 conceptuses were fixed with 4% paraformaldehyde and processed for routine paraffin histology. For RNA preparation, embryos free of any trophoblast tissues were carefully dissected and snap frozen.
Tissue culture
ES cell lines used were: wildtype J1 and R1, ES cells derived from 129Sv x (M. cast. x 129 Sv) blastocysts either wildtype (Dnmt1+/+) or homozygous for the mutant s allele (Dnmt1-/-) at the DNA methyltransferase 1 locus41, Np95-/-, Dnmt3a/b double mutant and Cxxc1-/-. For normal growth and expansion, ES cells were grown on gelatinized dishes on embryonic feeder cell layers in standard ES cell medium. For transdifferentiation experiments, 1 × 105 cells were plated in gelatin-coated 6-well dishes and grown in standard TS cell conditions (20% fetal bovine serum, 1 mM Na-pyruvate, 50 U/ml penicillin, 50 μg/ml streptomycin, 50 μM 2-mercaproethanol, 25 ng/ml bFGF (Sigma), and 1 μg/ml heparin in RPMI1640 (Invitrogen), with 70% of the medium pre-conditioned on embryonic feeder cells8). TS cell differentiation medium consisted of the unconditioned medium without bFGF and heparin. Culture was for the indicated time periods, with media changes every two days. The TS-GFP line8 was used as control. For Elf5 overexpression and generation of stable cell lines, the full sequence-verified open reading frame was cloned into the hrGFP1a-IRES-EGFP (Stratagene), pcDNA3.1 (Stratagene) and pCALL42 vectors. Promoter constructs consisted of the 2 kb upstream regions of Cdx2, Eomes and Elf5 in the pTimer-1 (Clontech) vector. Transfections were carried out with Lipofectamine 2000 (Invitrogen) reagent according to the manufacturer’s instructions. FACS sorting of GFP-positive cells was carried out on a FACSAria cell sorter.
Aggregation chimeras and blastocyst outgrowths
For aggregation experiments, Dnmt1+/+ and Dnmt1-/- ES cells stably transfected with an H2B-GFP expression vector were cultured for 1-3 days in TS cell medium and then aggregated with 8-cell C57BL/6 embryos. Aggregated embryos were cultured in KSOM or DMEM containing 10% FBS for 2-3 days. Blastocysts were embedded in fibrin clots and analyzed on a Zeiss 510 Meta confocal microscope. The duration of preconditioning of ES cells in TS medium did not affect the rate of TE contribution. Blastocyst outgrowths were obtained by culturing C57BL/6 blastocysts in DMEM with 10% FBS for 3-5 days.
Staining of histological sections and cultured ES cells
In situ hybridizations on paraffin sections were carried out according to a standard protocol43 using digoxigenin-labelled riboprobes. For detection of ectopic giant cells, serial sections 42-70 μm apart were hybridized with the pan-trophoblast giant cell marker Plf. Counterstaining was performed with nuclear fast red. For in situ hybridization on cultured cells, ES cells plated on gelatinized coverslips were fixed for 20 min with 4% paraformaldehyde and processed using a standard protocol44. Immunostainings were performed after blocking in 0.1% serum, 0.5% BSA for 15 min using the following antibodies and dilutions: anti-Cdx2 (BioGenex) at 1:300-1:400; anti-Eomes (Abcam) 1:200; anti-Elf5 (Santa Cruz Biotechnology) 1:250; anti-Oct3/4 (Santa Cruz Biotechnology) 1:400; anti-Nanog (Abcam) 1:250. Secondary antibodies were Alexa fluorophores from Molecular Probes. Photographs were taken on an Olympus BX41 epifluorescence microscope and a Zeiss 510 Meta confocal microscope.
Quantitative gene expression analysis
Total RNAs were isolated from cultured cells or embryos using Trizol reagent and digested with 20U DNase for 30 min at room temperature. cDNA synthesis was performed with Powerscript reverse transcriptase (BD Biosciences) according to the manufacturer’s protocol. 1:10 diluted cDNAs were used for quantitative PCR on an ABI PRISM 7700 Sequence Detector and Stratagene Mx3005P lightcycler using SYBR Green I mastermix (Applied Biosystems) and intron-spanning primer pairs. Ct values were normalized against 3 housekeeping genes. All samples were analyzed at least in triplicate.
Northern and Southern blotting
For Northern blots, 20 μg total RNA was electrophoresed and blotted onto Nytran Supercharge Nylon membrane (Schleicher & Schuell). For Southern blots, 20 μg DNA was digested with NsiI or BglII at 37°C, precipitated and digested with MspI or HpaII. Digests were electrophoresed on 0.8-2% agarose gels and blotted in 0.4M NaOH onto Nytran Supercharge Nylon membrane. Random-primed DNA labelling was carried out with 25-40 μCi 32P-dCTP, followed by hybridization at 65°C according to standard protocols45.
Methyl-DIP chip
Genomic DNA from ES and TS cells was sheared to an average size of 300-1000 bp and immunoprecipitated with an anti-5-methylcytosine specific antibody (Eurogentec). The quality of the immunoprecipitation was determined by analysing enrichment of standard genes with known methylation patterns. 4 μg of immunoprecipitated DNA and 3 μg of sonicated input DNA were hybridized to NimblGen oligonucleotide promoter tiling arrays (2005-03-31_MM5) containing 35497 mouse promoter fragments. The hybridizations were carried out in triplicate with one dye swap using three independent biological samples. The microarray raw data were analyzed using the in-house developed software ChipMonk. Microarray data comparison was performed using an empirically determined algorithm32 where candidate regions containing 5 or more oligonucleotide probes were filtered to give a CpG proportion of 2-9% and to ensure that the differences were statistically significant (p<0.01). We confirmed that this algorithm was a robust predictor of methylation state with 88% accuracy (22/25 calls correct).
Sequenom analysis
Two regions were identified as candidates for higher methylation in ES versus TS cells: Elf5 and Sca10. The methylation pattern of these loci was analyzed by Sequenom® mass array technology (Sequenom, San Diego, CA). 1 μg of genomic DNA from ES and TS cells was bisulphite treated using the Zymo EZ DNA methylation kit (Zymo Research). Promoter regions were selected based on the position of the oligonucleotides on the NimbleGen promoter array and primer pairs were designed using the MethPrimer program (http://www.urogene.org/methprimer/index1.html). Amplification of the bisulphite converted DNA and preparation of the PCR product for quantitative analysis of the promoter methylation detected by the Mass Array system is according to the protocol provided by the manufacturer. The number of differentially methylated CpG dinucleotides for these promoters was: 9/10 for Elf5 (methylated CpG’s in ES cells: 45.5%; methylated CpG’s in TS cells: 5.36%) and 2/3 for Sca10 (ES 24.33%; TS 7.67%). We then examined correlation of methylation status with gene expression levels. Sca10 exhibited equal expression levels in ES and TS cells; thus, the very limited number of differentially methylated CpG’s was non-functional and did not affect transcriptional activity. Only Elf5 was expressed at significantly higher levels in TS cells as predicted from the methylation pattern. Thus, Elf5 was the only differentially methylated gene with functional relevance of the modification in the ES versus TS cell screen.
Bisulphite genomic sequencing
1 μg of DNA was processed for bisulphite sequencing analysis using the EpiTect Bisulphite kit (Qiagen) according to the manufacturer’s protocol. Nested PCR reactions were performed for all genes analyzed. Primer sequences are available upon request. PCR products were eluted, sub-cloned and sequenced.
Chromatin immunoprecipitation
Cells from 4 92mm dishes were formaldehyde-crosslinked according to a standard protocol46. 150 μg chromatin was pre-cleared and incubated with 2 μg goat anti-Elf5 antibody (Santa Cruz Biotechnology) overnight at 4°C, and then precipitated with Protein G sepharose. Incubation with goat IgG or with protein G sepharose only was used as mock control. Bound, unbound and input fractions were analyzed by standard and qPCR for Cdx2 and Eomes promoter regions, and normalized against mock control. ChIP was performed in triplicate from independent samples.
Supplementary Material
Acknowledgements
We would like to thank Drs. Fatima Santos and Annabelle Lewis for expert help with confocal microscopy and qPCR design, respectively. We also thank Dr. En Li for Dnmt1- mice and ES cells, and Drs. Haruhiko Koseki, Nick Gilbert and David Skalnik for ES cell lines. This work was supported by an MRC Career Development Award to M.H., by the Croucher Foundation Fellowship to R.K.N., and by BBSRC, MRC, EU NoE The Epigenome, CellCentric, and DIUS.
References
- 1.Gardner RL. Clonal analysis of early mammalian development. Philos Trans R Soc Lond B Biol Sci. 1985;312:163–78. doi: 10.1098/rstb.1985.0186. [DOI] [PubMed] [Google Scholar]
- 2.Dyce J, George M, Goodall H, Fleming TP. Do trophectoderm and inner cell mass cells in the mouse blastocyst maintain discrete lineages? Development. 1987;100:685–98. doi: 10.1242/dev.100.4.685. [DOI] [PubMed] [Google Scholar]
- 3.Fleming TP. A quantitative analysis of cell allocation to trophectoderm and inner cell mass in the mouse blastocyst. Dev Biol. 1987;119:520–31. doi: 10.1016/0012-1606(87)90055-8. [DOI] [PubMed] [Google Scholar]
- 4.Johnson MH, McConnell JM. Lineage allocation and cell polarity during mouse embryogenesis. Semin Cell Dev Biol. 2004;15:583–97. doi: 10.1016/j.semcdb.2004.04.002. [DOI] [PubMed] [Google Scholar]
- 5.Bradley A, Robertson E. Embryo-derived stem cells: a tool for elucidating the developmental genetics of the mouse. Curr Top Dev Biol. 1986;20:357–71. doi: 10.1016/s0070-2153(08)60675-4. [DOI] [PubMed] [Google Scholar]
- 6.Bradley A, Evans M, Kaufman MH, Robertson E. Formation of germ-line chimaeras from embryo-derived teratocarcinoma cell lines. Nature. 1984;309:255–6. doi: 10.1038/309255a0. [DOI] [PubMed] [Google Scholar]
- 7.Beddington RS, Robertson EJ. An assessment of the developmental potential of embryonic stem cells in the midgestation mouse embryo. Development. 1989;105:733–7. doi: 10.1242/dev.105.4.733. [DOI] [PubMed] [Google Scholar]
- 8.Tanaka S, Kunath T, Hadjantonakis AK, Nagy A, Rossant J. Promotion of trophoblast stem cell proliferation by FGF4. Science. 1998;282:2072–5. doi: 10.1126/science.282.5396.2072. [DOI] [PubMed] [Google Scholar]
- 9.Uy GD, Downs KM, Gardner RL. Inhibition of trophoblast stem cell potential in chorionic ectoderm coincides with occlusion of the ectoplacental cavity in the mouse. Development. 2002;129:3913–24. doi: 10.1242/dev.129.16.3913. [DOI] [PubMed] [Google Scholar]
- 10.Rossant J. Stem cells and lineage development in the mammalian blastocyst. Reprod Fertil Dev. 2007;19:111–8. doi: 10.1071/rd06125. [DOI] [PubMed] [Google Scholar]
- 11.Reik W. Stability and flexibility of epigenetic gene regulation in mammalian development. Nature. 2007;447:425–32. doi: 10.1038/nature05918. [DOI] [PubMed] [Google Scholar]
- 12.Santos F, Hendrich B, Reik W, Dean W. Dynamic reprogramming of DNA methylation in the early mouse embryo. Dev Biol. 2002;241:172–82. doi: 10.1006/dbio.2001.0501. [DOI] [PubMed] [Google Scholar]
- 13.Chapman V, Forrester L, Sanford J, Hastie N, Rossant J. Cell lineage-specific undermethylation of mouse repetitive DNA. Nature. 1984;307:284–6. doi: 10.1038/307284a0. [DOI] [PubMed] [Google Scholar]
- 14.Rossant J, Sanford JP, Chapman VM, Andrews GK. Undermethylation of structural gene sequences in extraembryonic lineages of the mouse. Dev Biol. 1986;117:567–73. doi: 10.1016/0012-1606(86)90325-8. [DOI] [PubMed] [Google Scholar]
- 15.Jackson M, et al. Severe global DNA hypomethylation blocks differentiation and induces histone hyperacetylation in embryonic stem cells. Mol Cell Biol. 2004;24:8862–71. doi: 10.1128/MCB.24.20.8862-8871.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ciruna BG, Rossant J. Expression of the T-box gene Eomesodermin during early mouse development. Mech Dev. 1999;81:199–203. doi: 10.1016/s0925-4773(98)00243-3. [DOI] [PubMed] [Google Scholar]
- 17.Haffner-Krausz R, Gorivodsky M, Chen Y, Lonai P. Expression of Fgfr2 in the early mouse embryo indicates its involvement in preimplantation development. Mech Dev. 1999;85:167–72. doi: 10.1016/s0925-4773(99)00082-9. [DOI] [PubMed] [Google Scholar]
- 18.Niwa H, Miyazaki J, Smith AG. Quantitative expression of Oct-3/4 defines differentiation, dedifferentiation or self-renewal of ES cells. Nat Genet. 2000;24:372–6. doi: 10.1038/74199. [see comments] [DOI] [PubMed] [Google Scholar]
- 19.Dietrich JE, Hiiragi T. Stochastic patterning in the mouse pre-implantation embryo. Development. 2007;134:4219–31. doi: 10.1242/dev.003798. [DOI] [PubMed] [Google Scholar]
- 20.Weber M, et al. Distribution, silencing potential and evolutionary impact of promoter DNA methylation in the human genome. Nat Genet. 2007;39:457–66. doi: 10.1038/ng1990. [DOI] [PubMed] [Google Scholar]
- 21.Donnison M, et al. Loss of the extraembryonic ectoderm in Elf5 mutants leads to defects in embryonic patterning. Development. 2005;132:2299–308. doi: 10.1242/dev.01819. [DOI] [PubMed] [Google Scholar]
- 22.Sharif J, et al. The SRA protein Np95 mediates epigenetic inheritance by recruiting Dnmt1 to methylated DNA. Nature. 2007;450:908–12. doi: 10.1038/nature06397. [DOI] [PubMed] [Google Scholar]
- 23.Chen T, Ueda Y, Dodge JE, Wang Z, Li E. Establishment and maintenance of genomic methylation patterns in mouse embryonic stem cells by Dnmt3a and Dnmt3b. Mol Cell Biol. 2003;23:5594–605. doi: 10.1128/MCB.23.16.5594-5605.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Carlone DL, et al. Reduced genomic cytosine methylation and defective cellular differentiation in embryonic stem cells lacking CpG binding protein. Mol Cell Biol. 2005;25:4881–91. doi: 10.1128/MCB.25.12.4881-4891.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Choi YS, Sinha S. Determination of the consensus DNA-binding sequence and a transcriptional activation domain for ESE-2. Biochem J. 2006;398:497–507. doi: 10.1042/BJ20060375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Azuara V, et al. Chromatin signatures of pluripotent cell lines. Nat Cell Biol. 2006;8:532–8. doi: 10.1038/ncb1403. [DOI] [PubMed] [Google Scholar]
- 27.Boyer LA, et al. Polycomb complexes repress developmental regulators in murine embryonic stem cells. Nature. 2006;441:349–53. doi: 10.1038/nature04733. [DOI] [PubMed] [Google Scholar]
- 28.Bernstein BE, et al. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 2006;125:315–26. doi: 10.1016/j.cell.2006.02.041. [DOI] [PubMed] [Google Scholar]
- 29.Weber M, et al. Chromosome-wide and promoter-specific analyses identify sites of differential DNA methylation in normal and transformed human cells. Nat Genet. 2005;37:853–62. doi: 10.1038/ng1598. [DOI] [PubMed] [Google Scholar]
- 30.Metzger DE, Xu Y, Shannon JM. Elf5 is an epithelium-specific, fibroblast growth factor-sensitive transcription factor in the embryonic lung. Dev Dyn. 2007;236:1175–92. doi: 10.1002/dvdy.21133. [DOI] [PubMed] [Google Scholar]
- 31.Niwa H, et al. Interaction between Oct3/4 and Cdx2 determines trophectoderm differentiation. Cell. 2005;123:917–29. doi: 10.1016/j.cell.2005.08.040. [DOI] [PubMed] [Google Scholar]
- 32.Farthing CR, et al. Global mapping of DNA methylation in mouse promoters reveals epigenetic reprogramming of pluripotency genes. PLoS Genet. 2008;4:e1000116. doi: 10.1371/journal.pgen.1000116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yagi R, et al. Transcription factor TEAD4 specifies the trophectoderm lineage at the beginning of mammalian development. Development. 2007;134:3827–36. doi: 10.1242/dev.010223. [DOI] [PubMed] [Google Scholar]
- 34.Strumpf D, et al. Cdx2 is required for correct cell fate specification and differentiation of trophectoderm in the mouse blastocyst. Development. 2005;132:2093–102. doi: 10.1242/dev.01801. [DOI] [PubMed] [Google Scholar]
- 35.Russ AP, et al. Eomesodermin is required for mouse trophoblast development and mesoderm formation. Nature. 2000;404:95–9. doi: 10.1038/35003601. [DOI] [PubMed] [Google Scholar]
- 36.Rossant J, Lis WT. Potential of isolated mouse inner cell masses to form trophectoderm derivatives in vivo. Dev Biol. 1979;70:255–61. doi: 10.1016/0012-1606(79)90022-8. [DOI] [PubMed] [Google Scholar]
- 37.Rossant J, Vijh KM. Ability of outside cells from preimplantation mouse embryos to form inner cell mass derivatives. Dev Biol. 1980;76:475–82. doi: 10.1016/0012-1606(80)90395-4. [DOI] [PubMed] [Google Scholar]
- 38.Zernicka-Goetz M. The first cell-fate decisions in the mouse embryo: destiny is a matter of both chance and choice. Curr Opin Genet Dev. 2006;16:406–12. doi: 10.1016/j.gde.2006.06.011. [DOI] [PubMed] [Google Scholar]
- 39.Yamanaka Y, Ralston A, Stephenson RO, Rossant J. Cell and molecular regulation of the mouse blastocyst. Dev Dyn. 2006;235:2301–14. doi: 10.1002/dvdy.20844. [DOI] [PubMed] [Google Scholar]
- 40.Waddington CH. Organisers and Genes. Cambridge: Cambridge University Press; 1940. [Google Scholar]
- 41.Lei H, et al. De novo DNA cytosine methyltransferase activities in mouse embryonic stem cells. Development. 1996;122:3195–205. doi: 10.1242/dev.122.10.3195. [DOI] [PubMed] [Google Scholar]
- 42.Lobe CG, et al. Z/AP, a double reporter for cre-mediated recombination. Dev. Biol. 1999;208:281–92. doi: 10.1006/dbio.1999.9209. [DOI] [PubMed] [Google Scholar]
- 43.Hemberger M, Nozaki T, Masutani M, Cross JC. Differential expression of angiogenic and vasodilatory factors by invasive trophoblast giant cells depending on depth of invasion. Dev Dyn. 2003;227:185–91. doi: 10.1002/dvdy.10291. [DOI] [PubMed] [Google Scholar]
- 44.Hemberger M, Hughes M, Cross JC. Trophoblast stem cells differentiate in vitro into invasive trophoblast giant cells. Dev Biol. 2004;271:362–71. doi: 10.1016/j.ydbio.2004.03.040. [DOI] [PubMed] [Google Scholar]
- 45.Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual. 3rd Edition Cold Spring Harbor, N.Y.: Cold Spring Harbor Press; 2001. [Google Scholar]
- 46.Lewis A, et al. Imprinting on distal chromosome 7 in the placenta involves repressive histone methylation independent of DNA methylation. Nat Genet. 2004;36:1291–5. doi: 10.1038/ng1468. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


