Abstract
Methylation of lysine residues on histone H3 tails regulates transcription. A recent addition to the list of known methylated histone binding modules is the plant homeodomain (PHD) finger, which is usually found in nuclear proteins with chromatin-related functions. Autoimmune regulator (AIRE) protein contains two PHD fingers and mutations in AIRE gene cause the monogenic disease autoimmune polyendocrinopathy-candidiasis-ectodermal dystrophy (APECED). AIRE is expressed in thymic medullary epithelial cells where it promotes the expression of tissue-specific antigens. However the mechanism by which AIRE controls gene expression is currently unknown and the function of its domains, in particular of its PHD fingers is still elusive and controversial. In this review we discuss recent works on AIRE PHD finger(s) providing a new link between the status of histone modifications and the regulation of tissue-specific antigen expression in thymus.
Keywords: AIRE, PHD finger, histone methylation, structure, tissue-specific antigen expression
The core histones of nucleosomes are subject to several forms of modifications at their N-terminal ends. These include acetylation of lysines, methylation of lysines and arginines, ubiquitination of lysines and other modifications.1 The N-terminal amino acids of histones can be modified to variable extent, for example the lysines can be methylated as mono-, di- or trimethylated forms. Usually these modifications are reversible and can be dynamically changed by specific enzymes. Because of the many modifications, it has been proposed that the combination of histone modifications forms a histone code that determines many aspects of chromatin accessibility and transcriptional regulation. For example, methylation of histone H3 lysine 4 (H3K4) or lysine 36 (H3K36) is usually associated with transcriptional activation whereas methylation of histone H3 lysine 9 (H3K9) or lysine 27 (H3K27) generally causes gene silencing. How the histone code is translated to specific chromatin-associated activity is currently under active investigation.2 In principle, histone modifications may change the chromatin accessibility by neutralizing the charge of histones (acetylation) or they may serve as binding sites for chromatin associated proteins. These proteins will be recruited to the chromatin via their specific domains.
Several recent studies have implicated plant homeodomain (PHD) finger as one of the chromatin binding domains. The PHD finger is a domain of ∼60 amino acids found in over 400 eukaryotic proteins, many of which act as nucleosome interaction determinants playing a fundamental role in histone recognition and epigenetic mechanisms.3 The physiological relevance of PHD modules is highlighted by the occurrence of pathological PHD finger mutants in genes such as ATRX, NSD1, RAG2, ING, triggering different human diseases including neurological and developmental diseases, cancer and immunological disorders.4
Mutations in Autoimmune Regulator (AIRE) cause the monogenic autoimmune disease autoimmune polyendocrinopathy candidiasis ectodermal dystrophy (APECED).5,6 AIRE has two PHD fingers and several other domains characteristic to transcriptional regulators such as N-terminal CARD domain, SAND domain and nuclear hormone binding LXXLL motifs. Currently, approximately 60 mutations in AIRE gene have been characterized. The clinical spectrum of APECED is variable and includes many autoimmune endocrine disorders such as autoimmune adrenocortical and gonadal failure, type 1 diabetes, autoimmune thyroid disease, autoimmune hepatitis and others. As a hallmark of autoimmunity, APECED patients and AIRE-deficient mice develop lymphocytic infiltrations in multiple tissues and autoantibodies to self-antigens.7 These antigens are often intracellular enzymes belonging to distinct protein families expressed in tissue-specific manner.
The highest expression of AIRE gene is seen in thymic medullary epithelial cells, which are important for selection of autoreactive T cells.8 Medullary thymic epithelial cells are able to express many self-antigens that are otherwise only expressed in specific tissues.9 This self-antigen expression, albeit at low level, is a basis for a so-called negative selection of T cells that recognize self-antigens. To avoid autoimmunity, the T cells that react to self-antigens are deleted by induction of apoptosis. Several studies have demonstrated that AIRE acts as a transcriptional regulator and is a crucial factor in the regulation of tissue-specific gene expression in thymic medullary epithelium.10,11
Two AIRE PHD fingers are targets of several point mutations or truncations in several APECED patients, supporting the crucial role of this domain both in healthy and in pathological conditions.5 AIRE PHD fingers strongly contribute to AIRE nuclear function as they are implicated in transcriptional activation and nuclear dot formations.12,13 Phylogenetic analysis suggests different functions for the two PHD fingers,14 however their respective role is still controversial and needs further investigations. AIRE PHD fingers seem also to play a role in correct cellular targeting as pathological PHD mutants lead to AIRE altered subcellular distribution.13
To gain more insight into the structure-function relationship of AIRE PHD finger(s) and their role in the pathogenesis of APECED, the solution structure of the first PHD finger of AIRE (AIRE-PHD1) was recently solved and the structural impact of pathological mutations was also characterized.15 AIRE-PHD1 presents the typical ββα-topology stabilized by conserved hydrophobic interactions centered around a well conserved tryptophan. The domain coordinates two zinc ions via the C4HC3 signature in a cross-brace coordination scheme similar to that of RING fingers domains. Based on sequence and structural similarity to RING fingers, PHD fingers have been proposed to function as E3 ligase in the ubiquitin-mediated proteosomal degradation pathway,16 although this hypothesis is hotly debated in the literature.15 Uchida et al.16 demonstrated the ability of AIRE-PHD1 to ubiquitylate substrates in a rabbit reticulocyte lysate system, however subsequent biochemical and biophysical experiments could not find any evidence for AIRE-PHD1 E3 ligase activity in autoubiquitilation assay, nor detect any direct interaction between AIRE-PHD1 and its putative cognate E2.15 AIRE-PHD fingers were also proposed to bind sequence specifically DNA in electrophoretic mobility shift assays,17,18 but NMR based titrations using PHD fingers and oligonucleotides containing putative PHD binding sequences (ATTGGTTA) did not confirm direct binding (Musco G, unpublished results).
The different functions that have been ascribed to the PHD finger module clearly show that this fold constitutes a robust structural scaffold which allows diversified activities. Because of the functional plasticity of the PHD fold, until very recently the real function of AIRE PHD finger(s) appeared extremely elusive and controversial. New perspectives in the field have been opened by recent studies which have clearly shown that two distinct subclasses of PHD fingers can differentially recognize either methylated (H3K4me3, H3K9me3)19-23 or unmethylated (H3K4me0) lysine residues on histone H3,24,25 raising the possibility that chromatin might be a common nuclear ligand of PHD fingers. Indeed, biochemical and biophysical data demonstrated that AIRE binds directly to histone H3 through its first PHD finger, thus activating gene expression. AIRE-PHD1 is a specialized histone tail reader module which is able to sense the methylation status of histone H3: it binds most tightly to H3K4me0 (Kd ∼ 5 μM) and with almost one order of magnitude less tightly to H3K4me1, with the binding affinities decreasing by two and three orders of magnitude for the di- and tri-methylated forms of H3K4, respectively. Since there is a 1,000-fold difference in binding affinity between un- and trimethylated forms of H3, it appears that the methylation state of H3 may act as an on/off switch for AIRE-PHD1 binding. Notably the opposite mechanism has been observed for an other subclass of PHD finger containing proteins including ING2,26 BPTF,19 RAG2,27,41 Taf3,28 Pygopus,29 whose PHD fingers specifically bind to trimethylated K4, strongly discriminating against the unmethylated histone H3 forms.
NMR experiments, mutational analysis and molecular dynamics calculations provided a detailed three-dimensional model of AIRE-PHD1 in complex with a unmethylated peptide comprising the first 10 residues of histone H3 N-terminal tail (H3K4me0). Similarly to what observed for other PHD fingers in complex with methylated or unmethylated histone tails, H3K4me0 snugly fits in the PHD binding pocket forming an additional β-strand onto the existing antiparallel β-sheet of the domain. Formation of the complex does not induce significant conformational changes in the protein, which constitutes a stable preformed binding platform (Fig. 1A). On the other hand complex formation imposes via an induced fit mechanism a β-strand conformation on the histone tail, which is otherwise unstructured in the absence of the PHD finger. This entropy penalty is largely compensated by the favorable enthalpic contribution (-9.2 Kcal/mol)30 generated by the interaction between the positively charged H3 tail and an extensive negatively charged binding surface (∼700 A2). Charge complementarity and negative enthalpic contributions appear to be a major driving force in both unmethylated or methylated H3 recognition mechanism,19-26 as demonstrated by the involvement of large negatively charged surface in other PHD-H3 complexes.
Figure 1.
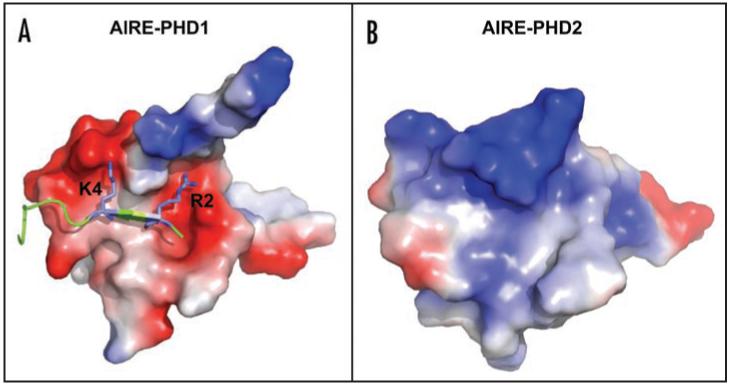
Electrostatic surface plots of AIRE1-PHD1 in complex with H3K4me0 (A) and of AIRE1-PHD2 (B). Blue and red colors indicate positive and negative electrostatic surface potentials, respectively. H3K4me0 peptide is represented in blue, the side-chains of K4 and R2 are explicitly shown in Blue. The model of AIRE-PHD1 in complex with H3K4me0 has been generated as described in Org et al.30 The model of AIRE-PHD2 has been generated as described in Bottomley et al.15
Remarkably, a homology model of the second PHD finger of AIRE (AIRE-PHD2) displays a positive electrostatic surface (Fig. 1B) suggesting repulsive interaction with the positively charged histone tails. Indeed, GST pull-down assays and NMR based titrations confirmed that AIRE-PHD2 is not suited for H3 tail interactions30 and belongs to a different subclass of PHD fingers with yet unidentified function. The two tandem PHD domains in AIRE might constitute a double effector module, which could simultaneously recognize H3K4me0 and possibly other histones, histone modifications or chromatin components thus interpreting the “multivalency” of chromatin patterns.2,4
At variance to what observed for the BPTF and ING subclass AIRE-PHD1 does not present the typical conserved aromatic side chains used to coordinate the tri- or di-methyl ammonium ion of H3K4me3 via Π-cation interactions.19-23,26 The key elements of the methylated lysine-binding aromatic cage are substituted by negatively charged residues in AIRE-PHD1 which can favourably interact with unmethylated H3K4me0, providing an alternative to the recognition of H3 via aromatic caging (Fig. 2). Also BHC80 PHD domain and Cys-rich domain of DNMT3L specifically interact with unmodified H3K4me0 through an electrostatic bridge between the unmodified epsilon aminogroup of H4K4me0 and an acidic residue (Asp489 and Asp90 in BHC80 and DNMT3L, respectively).24,25 Binding experiments and computational studies have clearly shown that methylation of the K4 sterically excludes this interaction with the consequent breakage of the additional β-strand.30 Notably this acidic hallmark is conserved in other PHD finger-containing transcription factors including Sp110, Sp140, NSD1 which might constitute together with AIRE-PHD1, BHC80 and DNMTL3 a subset of unmethylated histone H3 readers. Interestingly the PHD fingers of SP110 and NSD1 are hotspots for mutations causing the familial hepatic veno-occlusive disease with immunodeficiency (VODI)31 and the overgrowth Weaver syndrome32 respectively, thus strengthening the hypothesis that misinterpretation of the epigenetic marks strongly correlates with pathological conditions.4
Figure 2.
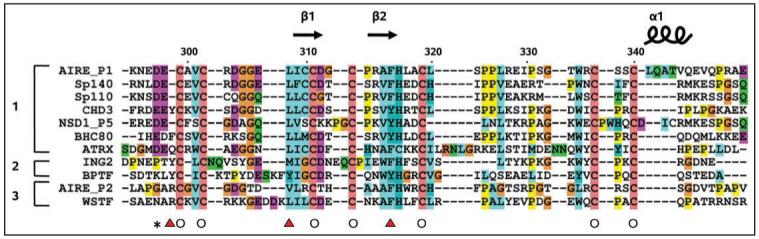
Sequence alignment of AIRE-PHD1 with other PHD fingers. A structure-guided multiple sequence alignment of PHD fingers, showing grouping for the proposed sub-families typified by (1) AIRE-PHD1, (2) ING/BPTF and (3) AIRE-PHD2. Secondary structure elements are shown (top) according to AIRE-PHD1 structure (PDB 1XWH). Empty circles identify residues chelating zinc ions 1 (empty circle). Red triangles highlight residues making the aromatic/hydrophobic cage that accommodates the trimethylated side chain of H3K4 in the ING2 and BPTF complex structures (PDB 2G6Q and 2FUU, respectively). The black star highlights AIRE residue D297 which is involved in binding K4 side-chain of H3K4me0. AIRE-P1: first PHD finger of Autoimmune Regulator; AIRE-P2: second PHD finger of Autoimmune Regulator; SP110: nuclear antigen Sp-110; SP140: nuclear antigen Sp-140; BHC80: BRAF35-HDAC complex protein; ATRX: ATP-dependent helicase; NSD1_P5: Fifth PHD finger of Nuclear receptor-binding SET domain-containing protein 1; CHD3: Chromodomain-helicase-DNA-binding protein 3; WSTF: Williams syndrome transcription factor.
How the interaction of AIRE-PHD1 with H3K4me0 would explain AIRE-mediated expression of tissue-specific antigens in thymus? Microarray experiments with AIRE-deficient medullary thymic epithelial cells have showed the decreased expression of several hundreds of tissue-specific genes.10,11 This result indicates that AIRE activity is more complex than to function as a simple DNA motif-binding transcription factor. As one explanation, AIRE might have a unique capacity to activate genes that have the H3K4me0 mark in their promoters. These genes are usually silent or expressed only at low levels. Indeed, when we analyzed the transcriptional activity of AIRE in tissue culture cells, we observed that AIRE preferentially activates genes which are expressed at low level and lack H3K4me3 modifications in their promoter regions.30 AIRE functions in thymus medullary epithelial cells that are needed to establish central tolerance by expressing a diverse set of tissue-specific genes.8 It can be speculated that the majority of tissue-specific genes in medullary thymic epithelial cells lacks H3K4me3 mark and is expressed at low levels. Thus the low expression and lack of H3K4me3 mark might be characteristic features for the AIRE dependent self-antigen gene activation outside of tissues where they are normally expressed, such as thymic epithelial cells. In this way, the expression of tissue-specific genes is not activated before the start of AIRE expression during the differentiation of medullary thymic epithelial cells (Fig. 3).
Figure 3.
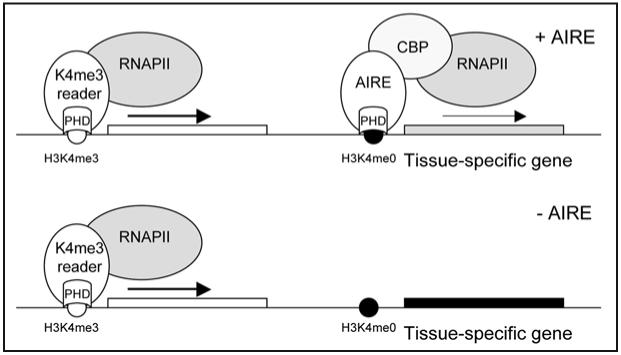
Schematic view of proposed AIRE function in regulation of tissue-specific gene expression in thymic medullary epithelial cells. The majority of active genes are activated by proteins that have PHD domain that reads H3K4me3 modification (left). AIRE is able to activate genes, which have H3K4me0 modifications and are normally silent or are expressed at low levels (right). Tissue-specific genes have high expression level and trimethylation of H3K4 mark in the corresponding tissues. Outside of their corresponding tissues, these genes have low methylation level of H3K4 and are silenced. In thymic medullary epithelial cells (+AIRE), AIRE is sufficient to increase the expression of multiple tissue-specific genes through the interaction with CBP and/or elongational factors. In AIRE deficient thymus (-AIRE), such as in APECED patients, tissue-specific genes are not expressed and are not presented as self-antigens to developing T cells.
AIRE is a potent transcriptional activator and functions on chromatin as a multimer of a large protein complex.13 This suggests that other proteins of the complex participate in activation of tissue-specific genes with low methylation mark of H3K4. AIRE-mediated transactivation is strongly enhanced by AIRE-interacting CREB-binding protein (CBP).12,33 CBP functions as a common transcriptional coactivator for a variety of transcription factors, including nuclear receptors, Jun, Fos, nuclear factor κB and STAT protein family members.34 Therefore, it is conceivable that CBP forms the link between AIRE binding to H3K4me0 and the basal transcriptional machinery. For example, CBP also has an intrinsic histone acetyltransferase activity and is a coactivator that modulates the transcription by changing chromatin accessibility to transcriptional complexes. In light of AIRE interaction with CBP as a histone acetyltransferase, it remains to be studied whether the AIRE binding and activation of promoters with H3K4me0 modifications has an impact on the epigenetic marks on target gene promoters, for example by changing the levels of histone H3 and H4 acetylation or H3K4 methylation.
Alternatively, AIRE may act in elongational processes as it has been recently shown to interact with positive transcription elongation factor b (P-TEFb) complex proteins CycT1 and Cdk9.35 The P-TEFb is a cyclin-dependent kinase that is required to control the transition of RNA polymerase II (RNAPII) from the initiation into productive elongation phase. After transcription initiation, RNAPII activity is inhibited by negative transcription elongation factors and the elongation complex stops near the promoter. The activity of RNAPII is restored by P-TEFb that overcomes the influences of the negative factors and RNAPII enters productive elongation.36 P-TEFb is recruited to gene promoters by a number of cellular transcription factors such as NFκB, CIITA and Brd4. Similarly to these transcriptional factors, AIRE recruits P-TEFb to the target gene promoters resulting in enhanced transcriptional elongation.35 It should be noted that P-TEFb functions are not limited to elongation of transcription as the complex has been shown to integrate mRNA synthesis with histone modifications.37 For example, recruitment of P-TEFb requires deubiquitination of histone H2B and phosphorylation of H3S10,37 and the recruitment of P-TEFb complex by HIV-1 Tat protein strongly induce RNAPII phosphorylation at serine 2, histone acetylation, and methylation of H3K4 on HIV-1 gene promoters.38 Whether the recruitment of P-TEFb to AIRE is associated with AIRE binding to H3K4me0 remains to be studied but considering the large number and diversity of AIRE-regulated genes, the enhancement of elongation and following induction of post-transcriptional events can be seen as one mechanism of AIRE action.
Since the first reports on BPTF and ING2 as founding members of methyllysine PHD finger subclass several structures describing the details of PHD finger interaction with histone H3K4me3 have been published.27-29,39 Conversely until now very few structural and biochemical data have been reported on H3K4me0 recognition.24,25 The detailed structural characterization of AIRE-PHD1 in complex with H3K4me0, complemented by other structural and functional investigations on PHD fingers belonging to this subclass will shed significant light in dynamic chromatin regulation and in transcriptional regulation.
Acknowledgements
P.P. and G.M. would like to thank their group members for fruitful discussions and comments. P.P. was supported by The Wellcome Trust, EU FP6 project Thymaide, European Regional Development Fund and Estonian Science Foundation (#6663). G.M. was supported by Fondazione Telethon, Fondazione Cariplo and Compagnia S. Paolo.
Footnotes
While this manuscript was under revision a paper was published reporting that AIRE-PHD1 is a histone binding module.40
Previously published online as an Epigenetics E-publication: http://www.landesbioscience.com/journals/epigenetics/article/7182
References
- 1.Kouzarides T. Chromatin modifications and their function. Cell. 2007;128:693–705. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- 2.Ruthenburg AJ, Li H, Patel DJ, Allis CD. Multivalent engagement of chromatin modifications by linked binding modules. Nat Rev Mol Cell Biol. 2007;8:983–94. doi: 10.1038/nrm2298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mellor J. It takes a PHD to read the histone code. Cell. 2006;126:22–4. doi: 10.1016/j.cell.2006.06.028. [DOI] [PubMed] [Google Scholar]
- 4.Baker LA, Allis CD, Wang GG. PHD fingers in human diseases: Disorders arising from misinterpreting epigenetic marks. Mutat Res. 2008 doi: 10.1016/j.mrfmmm.2008.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Peterson P, Peltonen L. Autoimmune polyendocrinopathy syndrome type 1 (APS1) and AIRE gene: New views on molecular basis of autoimmunity. J Autoimmun. 2005;25:49–55. doi: 10.1016/j.jaut.2005.09.022. [DOI] [PubMed] [Google Scholar]
- 6.Perheentupa J. Autoimmune polyendocrinopathy-candidiasis-ectodermal dystrophy. J Clin Endocrinol Metab. 2006;91:2843–50. doi: 10.1210/jc.2005-2611. [DOI] [PubMed] [Google Scholar]
- 7.Mathis D, Benoist C. A decade of AIRE. Nat Rev Immunol. 2007;7:645–50. doi: 10.1038/nri2136. [DOI] [PubMed] [Google Scholar]
- 8.Heino M, Peterson P, Kudoh J, Nagamine K, Lagerstedt A, Ovod V, Ranki A, Rantala I, Nieminen M, Tuukkanen J, Scott HS, Antonarakis SE, Shimizu N, Krohn K. Autoimmune regulator is expressed in the cells regulating immune tolerance in thymus medulla. Biochem Biophys Res Commun. 1999;257:821–5. doi: 10.1006/bbrc.1999.0308. [DOI] [PubMed] [Google Scholar]
- 9.Kyewski B, Klein L. A central role for central tolerance. Annu Rev Immunol. 2006;24:571–606. doi: 10.1146/annurev.immunol.23.021704.115601. [DOI] [PubMed] [Google Scholar]
- 10.Anderson MS, Venanzi ES, Klein L, Chen Z, Berzins SP, Turley SJ, von Boehmer H, Bronson R, Dierich A, Benoist C, Mathis D. Projection of an immunological self shadow within the thymus by the aire protein. Science. 2002;298:1395–401. doi: 10.1126/science.1075958. [DOI] [PubMed] [Google Scholar]
- 11.Derbinski J, Gabler J, Brors B, Tierling S, Jonnakuty S, Hergenhahn M, Peltonen L, Walter J, Kyewski B. Promiscuous gene expression in thymic epithelial cells is regulated at multiple levels. J Exp Med. 2005;202:33–45. doi: 10.1084/jem.20050471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pitkanen J, Doucas V, Sternsdorf T, Nakajima T, Aratani S, Jensen K, Will H, Vahamurto P, Ollila J, Vihinen M, Scott HS, Antonarakis SE, Kudoh J, Shimizu N, Krohn K, Peterson P. The autoimmune regulator protein has transcriptional transactivating properties and interacts with the common coactivator CREB-binding protein. J Biol Chem. 2000;275:16802–9. doi: 10.1074/jbc.M908944199. [DOI] [PubMed] [Google Scholar]
- 13.Halonen M, Kangas H, Ruppell T, Ilmarinen T, Ollila J, Kolmer M, Vihinen M, Palvimo J, Saarela J, Ulmanen I, Eskelin P. APECED-causing mutations in AIRE reveal the functional domains of the protein. Hum Mutat. 2004;23:245–57. doi: 10.1002/humu.20003. [DOI] [PubMed] [Google Scholar]
- 14.Saltis M, Criscitiello MF, Ohta Y, Keefe M, Trede NS, Goitsuka R, Flajnik MF. Evolutionarily conserved and divergent regions of the autoimmune regulator (aire) gene: A comparative analysis. Immunogenetics. 2008;60:105–14. doi: 10.1007/s00251-007-0268-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bottomley MJ, Stier G, Pennacchini D, Legube G, Simon B, Akhtar A, Sattler M, Musco G. NMR structure of the first PHD finger of autoimmune regulator protein (AIRE1). insights into autoimmune polyendocrinopathy-candidiasis-ectodermal dystrophy (APECED) disease. J Biol Chem. 2005;280:11505–12. doi: 10.1074/jbc.M413959200. [DOI] [PubMed] [Google Scholar]
- 16.Uchida D, Hatakeyama S, Matsushima A, Han H, Ishido S, Hotta H, Kudoh J, Shimizu N, Doucas V, Nakayama KI, Kuroda N, Matsumoto M. AIRE functions as an E3 ubiquitin ligase. J Exp Med. 2004;199:167–72. doi: 10.1084/jem.20031291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kumar PG, Laloraya M, Wang CY, Ruan QG, Davoodi-Semiromi A, Kao KJ, She JX. The autoimmune regulator (AIRE) is a DNA-binding protein. J Biol Chem. 2001;276:41357–64. doi: 10.1074/jbc.M104898200. [DOI] [PubMed] [Google Scholar]
- 18.Ruan QG, Tung K, Eisenman D, Setiady Y, Eckenrode S, Yi B, Purohit S, Zheng WP, Zhang Y, Peltonen L, She JX. The autoimmune regulator directly controls the expression of genes critical for thymic epithelial function. J Immunol. 2007;178:7173–80. doi: 10.4049/jimmunol.178.11.7173. [DOI] [PubMed] [Google Scholar]
- 19.Li H, Ilin S, Wang W, Duncan EM, Wysocka J, Allis CD, Patel DJ. Molecular basis for site-specific read-out of histone H3K4me3 by the BPTF PHD finger of NURF. Nature. 2006;442:91–5. doi: 10.1038/nature04802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shi X, Hong T, Walter KL, Ewalt M, Michishita E, Hung T, Carney D, Pena P, Lan F, Kaadige MR, Lacoste N, Cayrou C, Davrazou F, Saha A, Cairns BR, Ayer DE, Kutateladze TG, Shi Y, Cote J, Chua KF, Gozani O. ING2 PHD domain links histone H3 lysine 4 methylation to active gene repression. Nature. 2006;442:96–9. doi: 10.1038/nature04835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wysocka J, Swigut T, Xiao H, Milne TA, Kwon SY, Landry J, Kauer M, Tackett AJ, Chait BT, Badenhorst P, Wu C, Allis CD. A PHD finger of NURF couples histone H3 lysine 4 trim-ethylation with chromatin remodelling. Nature. 2006;442:86–90. doi: 10.1038/nature04815. [DOI] [PubMed] [Google Scholar]
- 22.Li H, Ilin S, Wang W, Duncan EM, Wysocka J, Allis CD, Patel DJ. Molecular basis for site-specific read-out of histone H3K4me3 by the BPTF PHD finger of NURF. Nature. 2006;442:91–5. doi: 10.1038/nature04802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Iwase S, Lan F, Bayliss P, de la Torre-Ubieta L, Huarte M, Qi HH, Whetstine JR, Bonni A, Roberts TM, Shi Y. The X-linked mental retardation gene SMCX/JARID1C defines a family of histone H3 lysine 4 demethylases. Cell. 2007;128:1077–88. doi: 10.1016/j.cell.2007.02.017. [DOI] [PubMed] [Google Scholar]
- 24.Lan F, Collins RE, De Cegli R, Alpatov R, Horton JR, Shi X, Gozani O, Cheng X, Shi Y. Recognition of unmethylated histone H3 lysine 4 links BHC80 to LSD1-mediated gene repression. Nature. 2007;448:718–22. doi: 10.1038/nature06034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ooi SK, Qiu C, Bernstein E, Li K, Jia D, Yang Z, Erdjument-Bromage H, Tempst P, Lin SP, Allis CD, Cheng X, Bestor TH. DNMT3L connects unmethylated lysine 4 of histone H3 to de novo methylation of DNA. Nature. 2007;448:714–7. doi: 10.1038/nature05987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pena PV, Davrazou F, Shi X, Walter KL, Verkhusha VV, Gozani O, Zhao R, Kutateladze TG. Molecular mechanism of histone H3K4me3 recognition by plant homeodomain of ING2. Nature. 2006;442:100–3. doi: 10.1038/nature04814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Matthews AG, Kuo AJ, Ramon-Maiques S, Han S, Champagne KS, Ivanov D, Gallardo M, Carney D, Cheung P, Ciccone DN, Walter KL, Utz PJ, Shi Y, Kutateladze TG, Yang W, Gozani O, Oettinger MA. RAG2 PHD finger couples histone H3 lysine 4 trimethylation with V(D)J recombination. Nature. 2007;450:1106–10. doi: 10.1038/nature06431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.van Ingen H, van Schaik FM, Wienk H, Ballering J, Rehmann H, Dechesne AC, Kruijzer JA, Liskamp RM, Timmers HT, Boelens R. Structural insight into the recognition of the H3K4me3 mark by the TFIID subunit TAF3. Structure. 2008;16:1245–56. doi: 10.1016/j.str.2008.04.015. [DOI] [PubMed] [Google Scholar]
- 29.Fiedler M, Sanchez-Barrena MJ, Nekrasov M, Mieszczanek J, Rybin V, Muller J, Evans P, Bienz M. Decoding of methylated histone H3 tail by the pygo-BCL9 wnt signaling complex. Mol Cell. 2008;30:507–18. doi: 10.1016/j.molcel.2008.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Org T, Chignola F, Hetenyi C, Gaetani M, Rebane A, Liiv I, Maran U, Mollica L, Bottomley MJ, Musco G, Peterson P. The autoimmune regulator PHD finger binds to non-methylated histone H3K4 to activate gene expression. EMBO Rep. 2008;9:370–6. doi: 10.1038/embor.2008.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Roscioli T, Cliffe ST, Bloch DB, Bell CG, Mullan G, Taylor PJ, Sarris M, Wang J, Donald JA, Kirk EP, Ziegler JB, Salzer U, McDonald GB, Wong M, Lindeman R, Buckley MF. Mutations in the gene encoding the PML nuclear body protein Sp110 are associated with immunodeficiency and hepatic veno-occlusive disease. Nat Genet. 2006;38:620–2. doi: 10.1038/ng1780. [DOI] [PubMed] [Google Scholar]
- 32.Kurotaki N, Imaizumi K, Harada N, Masuno M, Kondoh T, Nagai T, Ohashi H, Naritomi K, Tsukahara M, Makita Y, Sugimoto T, Sonoda T, Hasegawa T, Chinen Y, Tomita Ha HA, Kinoshita A, Mizuguchi T, Yoshiura Ki K, Ohta T, Kishino T, Fukushima Y, Niikawa N, Matsumoto N. Haploinsufficiency of NSD1 causes sotos syndrome. Nat Genet. 2002;30:365–6. doi: 10.1038/ng863. [DOI] [PubMed] [Google Scholar]
- 33.Pitkanen J, Rebane A, Rowell J, Murumagi A, Strobel P, Moll K, Saare M, Heikkila J, Doucas V, Marx A, Peterson P. Cooperative activation of transcription by autoimmune regulator AIRE and CBP. Biochem Biophys Res Commun. 2005;333:944–53. doi: 10.1016/j.bbrc.2005.05.187. [DOI] [PubMed] [Google Scholar]
- 34.Kalkhoven E. CBP and p300: HATs for different occasions. Biochem Pharmacol. 2004;68:1145–55. doi: 10.1016/j.bcp.2004.03.045. [DOI] [PubMed] [Google Scholar]
- 35.Oven I, Brdickova N, Kohoutek J, Vaupotic T, Narat M, Peterlin BM. AIRE recruits P-TEFb for transcriptional elongation of target genes in medullary thymic epithelial cells. Mol Cell Biol. 2007;27:8815–23. doi: 10.1128/MCB.01085-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Peterlin BM, Price DH. Controlling the elongation phase of transcription with P-TEFb. Mol Cell. 2006;23:297–305. doi: 10.1016/j.molcel.2006.06.014. [DOI] [PubMed] [Google Scholar]
- 37.Bres V, Yoh SM, Jones KA. The multi-tasking P-TEFb complex. Curr Opin Cell Biol. 2008;20:334–40. doi: 10.1016/j.ceb.2008.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhou M, Deng L, Lacoste V, Park HU, Pumfery A, Kashanchi F, Brady JN, Kumar A. Coordination of transcription factor phosphorylation and histone methylation by the P-TEFb kinase during human immunodeficiency virus type 1 transcription. J Virol. 2004;78:13522–33. doi: 10.1128/JVI.78.24.13522-13533.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ruthenburg AJ, Allis CD, Wysocka J. Methylation of lysine 4 on histone H3: Intricacy of writing and reading a single epigenetic mark. Mol Cell. 2007;25:15–30. doi: 10.1016/j.molcel.2006.12.014. [DOI] [PubMed] [Google Scholar]
- 40.Kuo AJ, Park SY, Cheung P, Abramson J, Bua D, Carney D, Shoelson SE, Gozani O, Kingston RE, Benoist C, Mathis D. Aire employs a histone-binding module to mediate immunological tolerance, linking chromatin regulation with organ-specific autoimmunity. Proc Natl Acad Sci USA. 2008;105:15878–83. doi: 10.1073/pnas.0808470105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Matthews AGW, Kuo A, Ramo’n-Maiques S, Han S, Champagne KS, Ivanov D, et al. RAG2 PHD finger couples histone H3 lysine 4 trimethylation with V(D)J recombination. Nature. 2007;450:1106–1111. doi: 10.1038/nature06431. [DOI] [PMC free article] [PubMed] [Google Scholar]


