Abstract
The Spindle Assembly Checkpoint (SAC) is required to block sister chromatid separation until all chromosomes are properly attached to the mitotic apparatus. The SAC prevents cells entering anaphase by inhibiting the ubiquitination of cyclin B1 and securin by the Anaphase Promoting Complex/Cyclosome (APC/C) ubiquitin ligase. The target of the SAC is the essential APC/C activator, Cdc20. It is unclear how the SAC inactivates Cdc20 but current models mostly involve Cdc20 forming a stable complex with the Mad2 checkpoint protein. Here we show that most Cdc20 is not in a complex with Mad2; instead Mad2 is required for Cdc20 to form a complex with another checkpoint protein, BubR1. We further show that during the SAC the APC/C ubiquitinates Cdc20 to target it for degradation. Thus, ubiquitination of human Cdc20 is not required to release it from the checkpoint complex, but to degrade it to maintain mitotic arrest.
Introduction
The proper control of mitosis depends on ubiquitin-mediated proteolysis of key regulators at the correct time 1. Crucially, the anaphase inhibitor, securin, and Cyclin B1, which maintains cells in mitosis, must not be degraded until all the chromosomes are properly attached to the spindle. To achieve this a conserved mechanism called the ‘Spindle Checkpoint’ (SAC) is activated by improperly attached kinetochores, and prevents the APC/C ubiquitin ligase recognising Cyclin B1 and securin (reviewed in 2). A number of conserved proteins have been identified as components of the SAC, including Mad1, Mad2, Mad3/BubR1, Bub1, Bub3, Mps1 and the Aurora B kinase. The primary target of the SAC is the Cdc20/fizzy protein 3, 4 that is an essential APC/C activator (reviewed in 5). However, it is unclear how the SAC regulates Cdc20. Current models of the checkpoint propose that the Mad2 protein has a crucial role either to sequester Cdc20, or act in conjunction with the BubR1 and Bub3 proteins to form an inhibitor called the ‘Mitotic Checkpoint Complex’ (MCC) reviewed in 2. The prominence given to the Mad2-Cdc20 complex is understandable because the crystal structure of Mad2 bound to a Cdc20 mimicking-peptide predicts that Mad2 changes conformation 6-8 to bind Cdc20 tightly via a ‘safety belt’ mechanism 6, 8, although another inhibitory complex comprised of BubR1 and Bub3 has also been identified 9, 10.
Recently, it has been suggested that inhibitory complexes have to be actively dissociated by the ubiquitination of Cdc20 mediated by the APC/C to turn off the SAC 11, 12. By contrast, in budding yeast Cdc20 is ubiquitinated during the spindle checkpoint to target it for destruction 13, 14, and this degradation is important because overexpressing Cdc20 overcomes the SAC 13.
Here, we have studied the mechanism by which the human SAC regulates Cdc20. In contrast to the prevailing models, we find that Cdc20 does not accumulate in a stoichiometric complex with Mad2, but primarily in a complex with BubR1 and Bub3. Moreover, although Cdc20 is ubiquitinated by the APC/C this ubiquitination is needed to target Cdc20 for destruction to maintain the checkpoint and not to release Cdc20 from a checkpoint complex: indeed, a non-ubiquitinatable form of Cdc20 overcomes the checkpoint. We suggest that the SAC is maintained through BubR1-Bub3 presenting Cdc20 to the APC/C as a substrate, and that the primary role of Mad2 is to generate the BubR1-Bub3-Cdc20 complex.
Results
Cdc20 is degraded by the Spindle Assembly Checkpoint
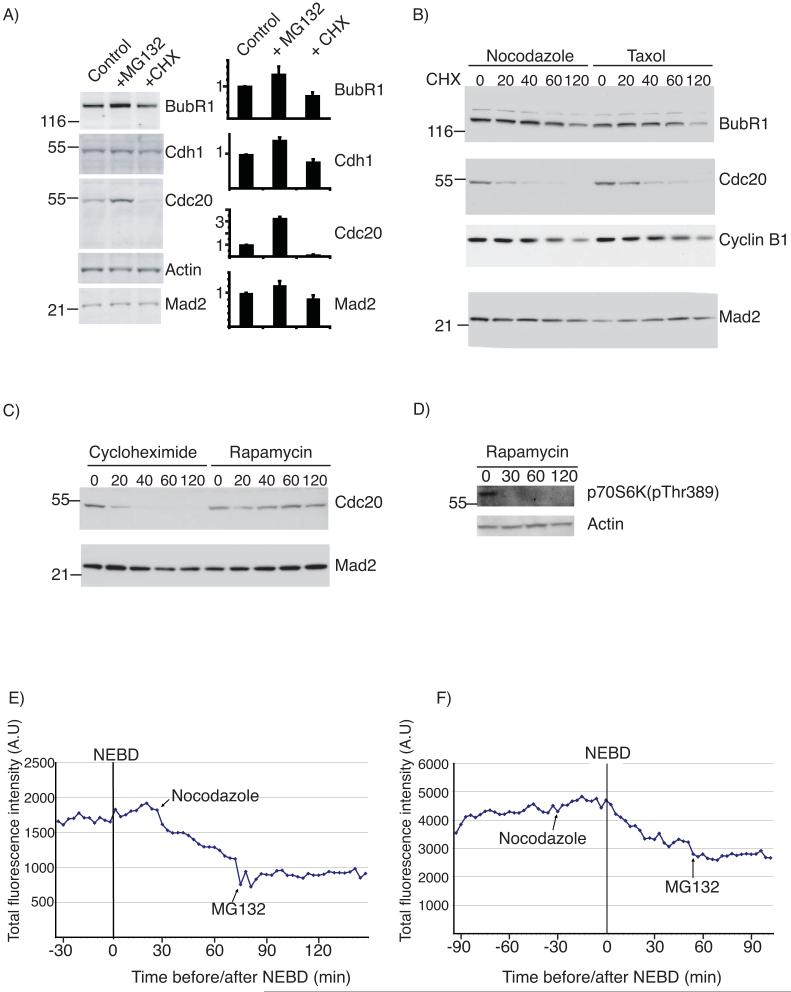
We set out to determine how the SAC inactivated Cdc20 in human cells and noticed that although Cdc20 levels apparently remained constant during a SAC arrest, they increased almost three fold on adding the proteasome inhibitor MG132 (Figs 1A and S1A), indicating that Cdc20 might be continually synthesised and degraded during the SAC. To test this we inhibited protein synthesis by adding cycloheximide and found the level of Cdc20 dropped substantially (Figs 1A & B, and S1B). A time-course showed that the half-life of Cdc20 in nocodazole- or taxol-arrested cells was ∼30 min (Fig 1B). Cyclin B1 (Fig 1B) and securin (not shown) were also degraded in SAC-arrested cells with a half-life of ∼ 1 hr, as previously shown 15, 16. Cdc20 levels did not change when we added rapamycin, which only inhibits cap (eIF4E)-dependent translation (Fig 1C), meaning that during mitosis Cdc20 may be synthesised from an internal ribosome entry site (IRES). Blotting for phospho-S6 kinase showed that rapamycin had inhibited the mTOR pathway and was thus likely to have inhibited eIF4E-dependent translation (Fig 1D). We confirmed the checkpoint-dependent degradation of Cdc20 in a live-cell assay 15 using a YFP-Cdc20 fusion protein. YFP-Cdc20 retained the functional properties of Cdc20 because it rescued cells depleted of Cdc20 by siRNA, localised to kinetochores in mitosis 17, 18 and was degraded as cells exited mitosis. YFP-Cdc20 fluorescence levels immediately began to decline in prometaphase or metaphase cells when we activated the SAC with nocodazole (Fig. 1E). When we added nocodazole or taxol to interphase cells, YFP-Cdc20 only began to be degraded at nuclear envelope breakdown (NEBD) (Fig 1F), and was stabilised by MG132 (Fig 1E and F). Identical results were obtained when we assayed Cdc20 degradation in RPE cells (Supplemental Fig S1B).
Figure 1.
Cdc20 is degraded in checkpoint-arrested cells
A) HeLa cells were pre-synchronised in S phase and subsequently arrested in mitosis with nocodazole for 6 hrs. Cells were left untreated (control) or treated with either MG132 or cycloheximide (CHX) for 2 hrs and the level of BubR1, Cdh1, Cdc20 and Mad2 was determined by quantitative immunoblotting. Protein levels were normalised against actin and the bar diagrams indicate the levels of the indicated proteins with the untreated sample set to 1. B) The levels of BubR1, Cdc20, Cyclin B1 and Mad2 at 0, 20, 40, 60 and 120 min after addition of cycloheximide to either nocodazole- or taxol-arrested cells (synchronised as in A) were analyzed by ECL western blot (whole blot shown in Fig S7). C) The levels of Cdc20 and Mad2 at 0, 20, 40, 60 and 120 min after addition of cycloheximide or rapamycin to nocodazole arrested cells (whole blot shown in Fig S7). D) The level of p70S6K phoshorylated on Thr389 at 0, 30, 60 and 120 min following addition of rapamycin to asynchronous HeLa cells. E and F) Degradation of YFP-Cdc20 in single cells. G2 phase HeLa cells were microinjected with a plasmid encoding YFP-Cdc20 and the fluorescence level assayed through mitosis. Note that the YFP-Cdc20 was encoded by cap-dependent mRNA, whose translation was repressed in mitosis, therefore, we could assay degradation without cycloheximide. Either nocodazole or MG132 was added at the times indicated. Nocodazole was added in prometaphase in E) and in G2 phase in F). Cells are representative of 10 cells in 3 independent experiments.
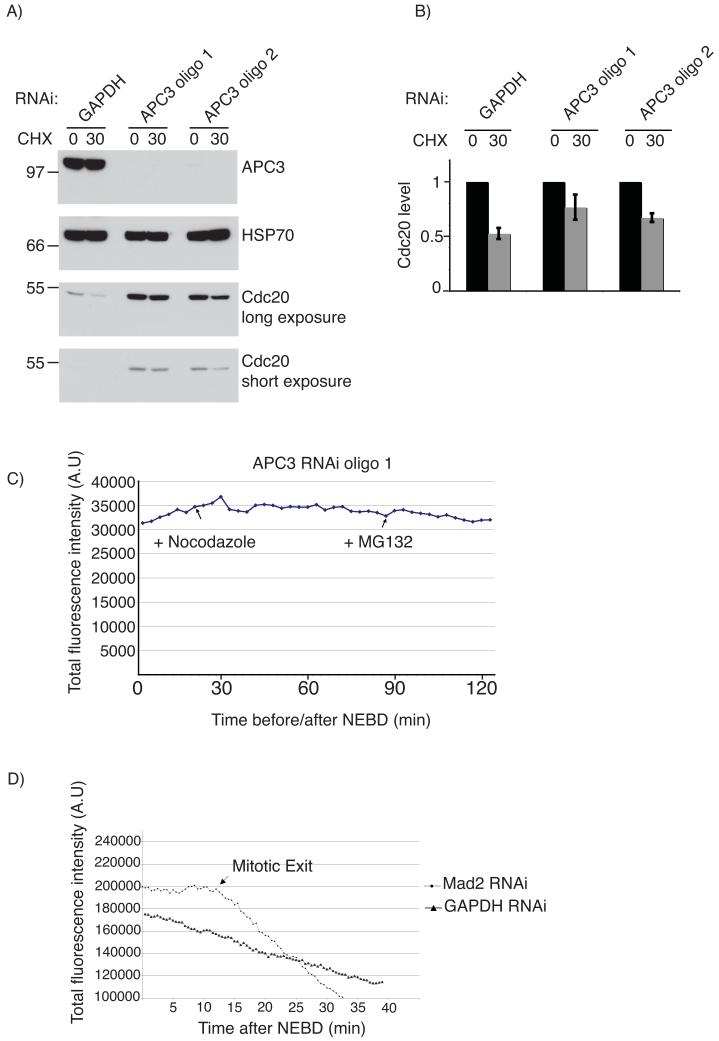
Our observation that YFP-Cdc20 began to be degraded only after NEBD indicated that in response to the SAC Cdc20 might be ubiquitinated by the APC/C, since this is the time when the APC/C recognises its first mitotic substrates 19, 20 21 We tested this by depleting APC3 by siRNA (Fig 2A & B), which inactivated the APC/C as evidenced by stabilising cyclin A, cyclin B1 and securin (D. Izawa and JP, manuscript in preparation) and blocked cells in mitosis. In APC/C-depleted cells endogenous Cdc20 and YFP-Cdc20 were stable in the presence of an active SAC (Fig 2A and C). We confirmed that the SAC itself was required for Cdc20 degradation by depleting Mad2 by siRNA, which stabilised Cdc20 until cells exited mitosis (Fig 2D).
Figure 2.
Cdc20 degradation is dependent on APC/C activity and a functional SAC.
A) Cells were treated with siRNA oligos against either GADPH (as in Fig. 1) or APC3 and after 72 hrs the stability of Cdc20 was determined in nocodazole-arrested cells by adding cycloheximide for 30 min and analysis by quantitative immunoblotting (whole blots shown in Fig S7). B) The level of Cdc20 was normalized to Hsp70 and the mean and standard deviation of Cdc20 levels in 3 experiments are shown with the untreated sample set to 1. C) The level of YFP-Cdc20 fluorescence in a mitotic cell treated with APC3 siRNA oligos (which blocked cells in mitosis for several hours in >95% of cells). The cell shown is representative of 6 cells in 2 experiments. D) The level of YFP-Cdc20 fluorescence in mitotic cells treated with siRNA targeting Mad2 or GAPDH in the presence of nocodazole. The data shown are representative of 10 and 6 cells in 2 experiments for cells treated with siRNA against, respectively, GAPDH and Mad2,
An inactive Cdc20 cannot be degraded in response to the SAC
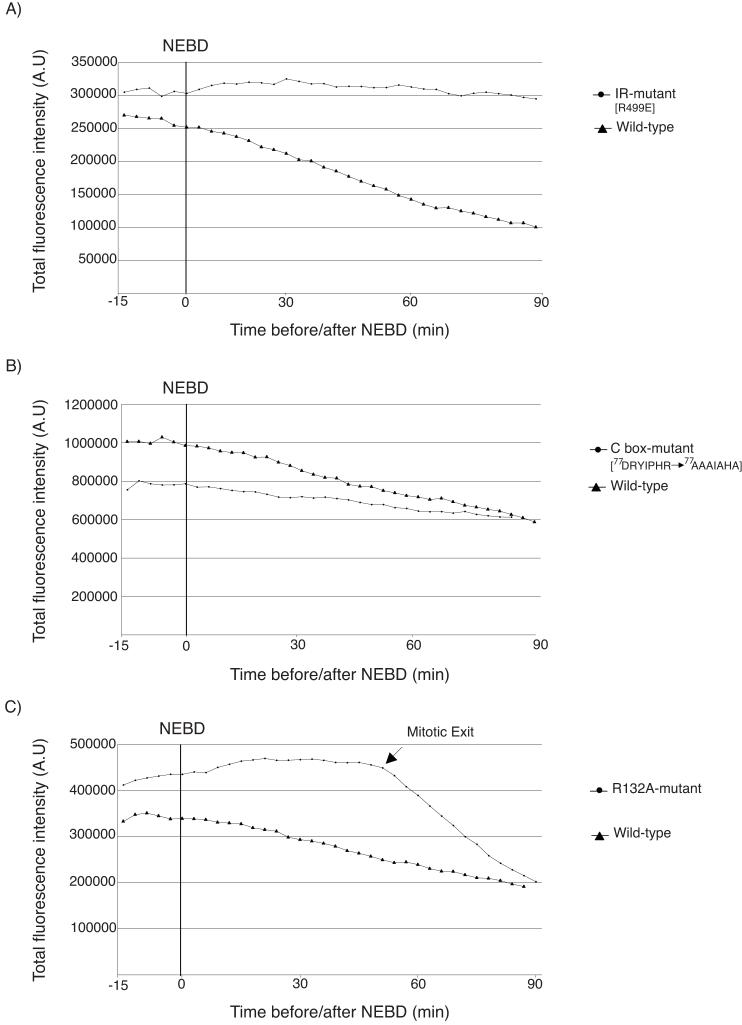
Our observation that Cdc20 was targeted for degradation by the APC/C in response to the SAC raised the interesting question of whether Cdc20 activated its own destruction, or whether it was recognised by another APC/CCdc20 complex. To analyse this we ectopically expressed various Venus-tagged Cdc20 mutants and measured their degradation in nocodazole-treated cells (Fig 3). To bind the APC/C, Cdc20 requires Isoleucine-Arginine at its C-terminus and the C-box motif 22, 23 and mutating either of these motifs prevented (IR mutant) or considerably slowed down (C-box mutant) its degradation in SAC-arrested cells (Fig 3A & B). Similarly, Cdc20 with a mutation in the Mad2-binding site (R132A) abrogated the SAC and was not degraded in the presence of nocodazole (Fig 3C).
Figure 3.
Cdc20 requires both APC/C and Mad2 binding motifs to be degraded in the SAC
HeLa cells were microinjected with plasmids encoding YFP- wild-type or mutant Cdc20 as indicated and the fluorescence level assayed through mitosis in the presence of nocodazole. Data are representative of 3 independent experiments for each mutant. A) IR mutant (R499E) 9 cells B) C-box mutant (D77RYIPHR83 to A77AAIAHA83) 13 cells C) Mad2-binding site R132A, 12 cells.
BubR1 and not Mad2 is the predominant partner of Cdc20 in checkpoint-arrested cells
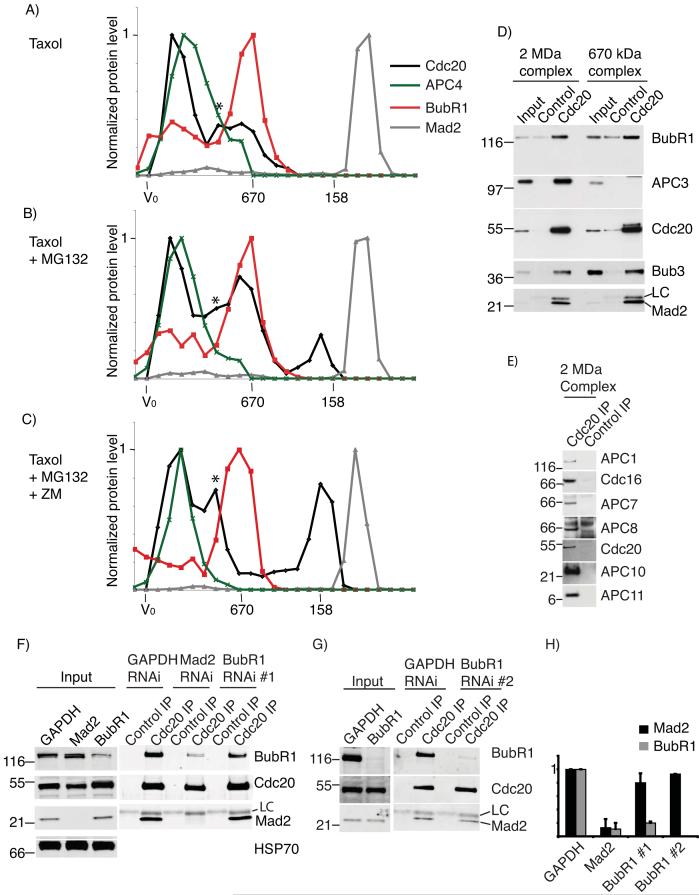
Our interpretation of these results was that the SAC caused Cdc20 to activate its own ubiquitination by the APC/C. To test this we analysed the interactions between Cdc20, the APC/C, and checkpoint proteins by size-exclusion chromatography and quantitative immunoblotting. (We used nitrogen cavitation to lyse the cells, which solubilised >95% of Cdc20, the APC/C and the checkpoint proteins (Fig S4 and S5), and measured the stoichiometry of protein complexes by calibrating our antibodies using recombinant proteins (Fig S1 and S2) We used fluorescently-labelled secondary antibodies and measured the signal on a LiCOR Odyssey scanner, which is linear over a 104 range and thus far more quantitative than ECL, although errors could be introduced in the protein transfer, recognition by the primary antibody and binding between the primary and secondary antibodies.).
In either nocodazole or taxol-arrested cells the bulk of Cdc20 migrated in two large complexes, one of ∼2 MDa and one of ∼670 kDa (Figs 4A and S3 and S4). Quantitative immunoblotting (Fig 4A and S3A & B) and immunoprecipitations from peak fractions (Fig 4D and FigS2) showed that the 2 MDa complex contained the APC/C, BubR1 and Bub3, whereas the 670 kDa complex contained BubR1 and Bub3, but no APC/C. Both complexes had very little Mad2, the bulk of which migrated at a size likely corresponding to free Mad2 (Figs 4 and S3 and S4). From the amounts of proteins in immunoprecipitates of Cdc20 from the 2 MDa and 670 kDa complexes, we estimate that only ∼5% of Cdc20 was bound to Mad2 in cells with an active checkpoint, whereas the molar ratio of BubR1 to Cdc20 was between 0.5 and 1 (Fig S2). Thus, the 670 kDa complex we detected probably corresponded to the Bub3-BubR1-Cdc20 checkpoint complex identified by Tang et al. (2001) 9 and Fang (2002) 10 rather than the MCC complex where Mad2, BubR1, Bub3 and Cdc20 were proposed to be present with 1:1:1:1 stoichiometry 24.
Figure 4.
Analysis of checkpoint complexes by gel-filtration chromatography
Taxol-arrested cells (A) or taxol-arrested cells treated with MG132 for 2 hrs (B), or taxol-arrested cells treated with MG132 and ZM447439 for 2 hrs (C) were fractionated on a Superose 6 column. Fractions were probed for BubR1 (red), APC4 (green), Cdc20 (black) and Mad2 (grey), and analyzed by quantitative immunoblotting. Values were normalized to the peak fraction value to obtain the profiles shown. (The novel peak of Cdc20 indicated by an asterisk that appears between the APC/C and BubR1 peaks may be Cdc20 bound to the CCT chaperone, since it is not always seen, for example, see Supplemental Fig 4 and 5.) Results are representative of 4 or more independent experiments. (Whole blot for taxol-arrested cells shown in Fig S7. Note that this is probed sequentially for Cdc20, then BubR1 and then Mad2.) D & E) Cdc20 was immunoprecipitated from each of the two peak fractions from nocodazole-arrested cells (fraction 22 and fraction 28 see Fig S4) and analyzed for the presence of BubR1, APC3, Bub3 and Mad2, and for APC/C subunits in the 2 MDa complex (E). F-H) Cells were treated with siRNA oligos against GAPDH, Mad2 or BubR1 for 72 hrs. During this time cells were released from a double thymidine block and treated 9 hrs later with nocodazole and MG132 for 2hrs. Cdc20 was immunoprecipitated from mitotic cells harvested by shake-off. The amount of BubR1 and Mad2 associating with Cdc20 was determined by quantitative immunoblotting and normalized to the amount of Cdc20. The levels of BubR1 and Mad2 in GAPDH treated cells were set to 1. Data are the mean+/-SD of 3 independent experiments in F using BubR1 RNAi oligo #1, and the average of 2 independent experiments in G using RNAi oligo #2. The whole blot is shown in Fig S7. In this case the blot was cut into 3 and the different panels probed for BubR1 or Cdc20 or Mad2.
When we blocked Cdc20 degradation with MG132, Cdc20 accumulated in both the 2 MDa and 670 kDa complexes (Fig 4B) and immunoprecipitating the APC/C verified the increase in binding to Cdc20 (Fig S4). In addition a third complex appeared running at approximately 200 kDa, which was likely to be free Cdc20 since it did not co-migrate with any of the known Cdc20 partners: the Mad and Bub proteins, the APC/C, and the CCT chaperone complex that migrates at ∼800 kDa 25. Moreover, when we released the checkpoint by adding the Aurora B inhibitor ZM447439 to taxol- or nocodazole-treated cells 26, but retained cells in mitosis by adding MG132, Cdc20 levels in the 670 kDa complex dropped and more migrated as ‘free’ Cdc20 (Fig. 4C). Identical results were obtained when we analysed cells in which we released the checkpoint by washing out the spindle poison in the presence of MG132 (Fig S4). The appearance of some Cdc20 not bound to checkpoint proteins upon the addition of MG132 indicated that preventing Cdc20 degradation caused Cdc20 to exceed the level of the BubR1-Bub3 complex, in agreement with there being similar amounts of Cdc20 and BubR1 in checkpoint-arrested cells (1:1:1:3, Cdc20:BubR1:APC/C:Mad2, Fig S2).
After releasing the checkpoint the amount of BubR1 migrating with the APC/C decreased but the amount of Cdc20 bound to the APC/C remained the same (Fig. 4C), likely corresponding to active APC/CCdc20.
These results were consistent with Cdc20 binding first to BubR1 to form the 670 kDa complex when the checkpoint was active, and subsequently being presented to the APC/C as a substrate for ubiquitination, thereby forming the 2 MDa complex. By inhibiting degradation we likely blocked this pathway, causing both the 2 MDa and the intermediate 670 kDa complex to accumulate. In support of this, the amount of Cdc20 and BubR1 bound to the APC/C increased when the proteasome was inhibited, whereas it decreased on adding cycloheximide (Fig. S4).
Since we did not find significant amounts of Mad2 bound to Cdc20 we asked whether, as in yeast, Mad2 was required for Cdc20 to bind to BubR1 27, but BubR1 was not required for binding between Mad2 and Cdc20. Consistent with this, siRNA against Mad2 caused a substantial drop in the amount of Cdc20 bound to BubR1 in SAC-arrested cells maintained in mitosis with MG132, whereas BubR1 siRNA did not significantly alter the amount of Mad2 bound to Cdc20 (Fig 4F-H).
A non-ubiquitinatable Cdc20 overrides the spindle assembly checkpoint
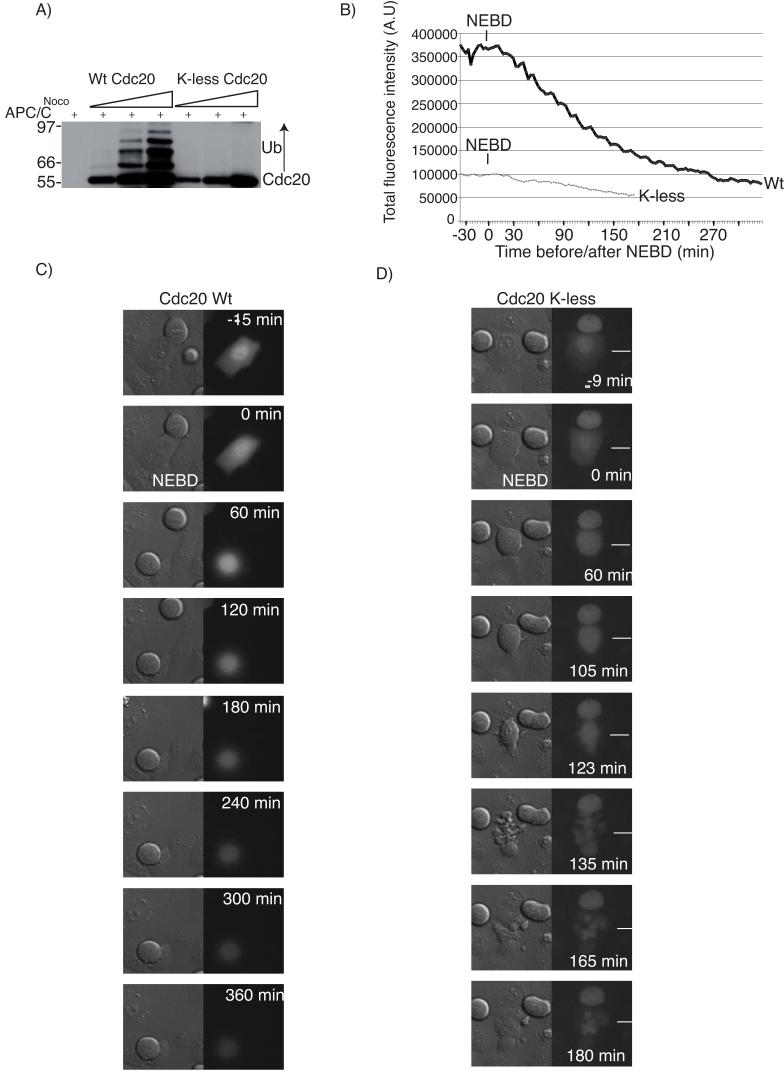
Our data indicated that the ubiquitin-mediated degradation of human Cdc20 catalysed by the APC/C is an important component to maintain the SAC. In contrast, ubiquitination has recently been proposed not to target Cdc20 for degradation but to inactivate the SAC by releasing Cdc20 from a checkpoint complex 11, 12. To determine whether ubiquitination of Cdc20 is required to maintain or inactivate the SAC, we made a mutant of Cdc20 that could not be ubiquitinated by changing all the lysines to arginines. This ‘K-less’ mutant of Cdc20 could not be ubiquitinated by the APC/C in vitro (Fig 5A) but was functional because it could activate the APC/C (Fig S5A) and rescue the mitotic arrest induced by siRNA targeting Cdc20 (21 of 21 cells in 3 independent experiments), with a similar timing to wild-type Cdc20 (16 of 16 cells in 3 independent experiments, Fig S5). (The siRNA depleted Cdc20 levels by greater than 95% and caused 90% of cells to arrest in mitosis 28.) Furthermore, the K-less mutant was able appropriately to bind and dissociate from checkpoint complexes (see below). The K-less mutant was more stable than wild-type Cdc20 (Fig 5), and consistent with Cdc20 degradation being an integral part of the SAC, it was able to drive checkpoint-arrested HeLa (Fig 5, Table and movie 1) and RPE (Fig S6, Table and movie 2) cells out of mitosis, whereas wild-type Cdc20 expressed at similar levels could not. Furthermore, cells expressing the K-less mutant at lower levels than endogenous Cdc20 could initially establish the checkpoint but were unable to maintain it, whereas cells overexpressing wild-type Cdc20 ∼10 fold remained arrested. Depleting Mad2 by siRNA abolished this delay with either wild-type or K-less Cdc20 (data not shown). These results pointed towards an important role for ubiquitination and degradation of Cdc20 in maintaining, but not initially imposing, the SAC.
Figure 5.
A Cdc20 mutant without lysines overrides the spindle checkpoint
A) Wild-type or the K-less mutant of Cdc20 were expressed and purified from baculovirus-infected SF9 cells, and increasing amounts incubated with APC/C purified from nocodazole-arrested cells. Reactions were analyzed by immunoblotting for Cdc20.. B) G2 phase HeLa cells treated with siRNA to deplete endogenous Cdc20 were injected with plasmids encoding siRNA resistant YFP-tagged wild-type Cdc20 or the K-less mutant and treated with nocodazole. Cells were monitored by time-lapse DIC and fluorescence microscopy at 3 min intervals and the fluorescence levels measured and quantified. The slow decrease in YFP-Cdc20 K-less is due to photo-bleaching. C and D) DIC and fluorescence images of the cells analysed in (B). The mitotic cell (arrowed) expressing the YFP-Cdc20 K-less protein (D) exits the nocodazole block after ∼120 min whereas the cell expressing wild-type YFP-Cdc20 remains arrested for more than 5.5 hrs and degraded the YFP-Cdc20 protein (C). Cells are representative of 17 (wild-type) and 16 (K-less mutant) cells in 2 independent experiments.
TABLE.
Timing of Nuclear Envelope Breakdown to anaphase and ability to overcome the SAC for HeLa and RPE cells expressing wild-type or the K-less mutant of Cdc20
| Time NEBD-Anaphase | Exit from mitosis in nocodazole | Exit from mitosis in taxol | Exit from mitosis in DMA | |
|---|---|---|---|---|
| HeLa + wt Cdc20 | 90 - 180 (n=16) | 0% (n = 17) | 0% (n = 8) | |
| HeLa + K-less | 24 - 90 (n=21) | 100% (n = 16) | 100% (n=14) | |
| RPE + wt Cdc20 | 0% (n = 10) | |||
| RPE + K-less | 100% (n = 15) |
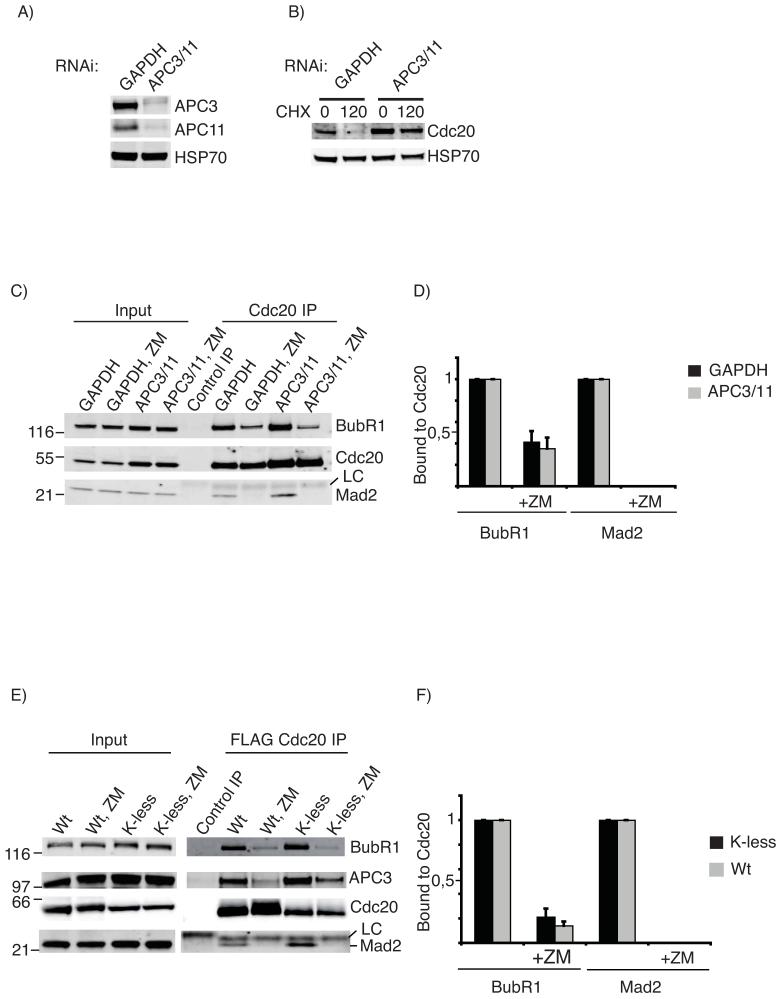
To test directly whether APC/C-dependent ubiquitination of Cdc20 was required to release the checkpoint proteins we depleted the APC3 and APC11 subunits from cells by siRNA (Fig 6A). As previously mentioned, this stabilised Cdc20 in SAC-arrested cells, consistent with the APC/C targeting Cdc20 for destruction (Figs 2 and 6B). When we immunoprecipitated Cdc20 from control or APC/C-depleted cells with an active SAC, Cdc20 was bound to BubR1 and some Mad2, and inactivating the checkpoint with ZM447439 showed that there was no difference in the ability of cells with or without APC/C activity to release Cdc20 from Mad2 and BubR1 (Fig 6C and D). Congruent with this, we found no difference in the ability of wild-type and the lysine-less Cdc20 mutant to be released from checkpoint complexes. We generated cell lines stably expressing FLAG-tagged wild-type or K-less Cdc20 under a tetracyclin inducible promoter and immunoprecipitated FLAG-Cdc20 from cells arrested with nocodazole plus MG132 using an anti-FLAG antibody. Although the K-less mutant bound more Mad2, BubR1 and APC3 (see discussion) than did wild-type Cdc20, there was no difference in the ability of the wild-type and K-less mutant to dissociate from Mad2 and BubR1 upon addition of ZM447439 (Fig 6E and F). Thus, we conclude that Cdc20 is ubiquitinated as part of the SAC to target it for destruction and not to release it from a checkpoint complex.
Figure 6.
Dissociation of checkpoint proteins from Cdc20 does not require ubiquitination of Cdc20.
A) Cells were treated with siRNA against GAPDH or siRNAs against APC3 and APC11 and immunoblotted for APC3, APC11, and Hsp70 as a loading control. B) Cells were treated as in A), arrested in mitosis with taxol and the half-life of Cdc20 assayed by adding cycloheximide (CHX). C) Cells treated as in A) were incubated with Taxol for 6 hrs, the samples divided in two and MG132 or MG132 plus ZM447439 added. After 2 hrs Cdc20 was immunopurified, and the amount of BubR1 and Mad2 bound to Cdc20 determined by quantitative immunobloting. (LC= antibody light chain). The whole blot is shown in Fig S7. Note the blot was cut into 3 and panels probed for BubR1 or Cdc20 or Mad2. D) Quantification of the experiment shown in C: the level of bound BubR1 and Mad2 is normalized to the amount of Cdc20, and the level of BubR1 and Mad2 set to 1 in the samples without ZM447439. The mean +/- SD of 3 independent experiments is shown.
E) HeLa cell lines stably expressing FLAG-tagged wt or K-less Cdc20 were released from S phase, MG132 added 9 hrs later for 2 hrs and mitotic cells harvested by shake-off. Samples were split in two and 100 nM Taxol plus MG132 added to both. After 1 hr FLAG-Cdc20 was immunopurified from one and 4 μM ZM447439 added to the other 1.5 hrs before immunoprecipitating FLAG-Cdc20. The amount of BubR1, APC3 and Mad2 bound to Cdc20 was determined by quantitative immunobloting. F) Quantification of the experiment shown in E) with the level of BubR1 and Mad2 normalized to the amount of Cdc20 and the amount of BubR1 and Mad2 bound to Cdc20 set to 1 in experiments without ZM447439. Note that the K-less mutant of Cdc20 co-immunopurifies from taxol-treated cells with more BubR1 and Mad2 than does wt Cdc20. The mean +/- SD of 3 independent experiments is shown. The amount of APC3 associated with K-less and wt Cdc20 dropped by 75% when the checkpoint was inactivated.
Discussion
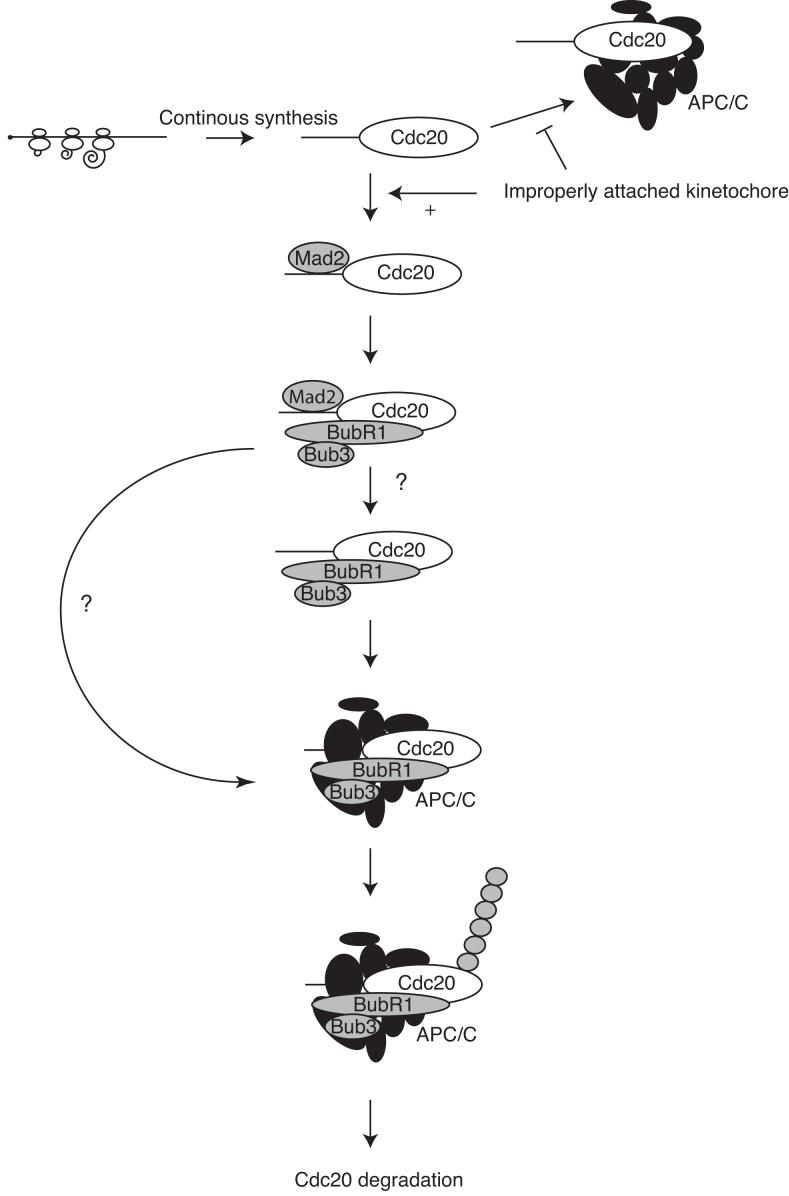
Here we have shown that the SAC maintains mitotic arrest by degrading Cdc20 as an APC/C substrate. To do this the SAC requires Mad2 but, in contrast to most current models of the checkpoint, we find that in human cells the majority of Mad2 does not form a stable complex with Cdc20. Instead, Cdc20 mostly accumulates in a complex with BubR1 and Bub3, and this complex binds to the APC/C. In the absence of Mad2, however, Cdc20 does not bind to BubR1. Thus, our working model (Fig 7) is that Mad2 acts catalytically to promote Cdc20 binding to BubR1, which in turn presents Cdc20 to the APC/C as a substrate, targeting it for degradation. Once the checkpoint is inactivated, Cdc20 no longer binds BubR1 and is free to activate the APC/C. This agrees with data from budding yeast that show that Cdc20 is degraded during the spindle checkpoint 13, 14, that the BubR1 homologue Mad3 has the greatest effect on Cdc20 stability, and that Mad2 is required for Cdc20 to bind to Mad3 27. Furthermore, overexpressing Cdc20 in budding yeast to 3 fold more than the endogenous level allows cells initially to arrest but not to remain in mitosis in the presence of spindle poisons 13. Thus, maintaining the SAC through degrading Cdc20 may be conserved through evolution.
Figure 7.
Spindle Checkpoint Model
Cdc20 is continually synthesized in mitosis and acts as an activator of the APC/C. However, in the presence of improperly attached kinetochores, Mad2 binds Cdc20 and this allows the loading of Cdc20 onto BubR1. Once BubR1 is bound to Cdc20, Mad2 is not required to sustain the binding and leaves the complex before or after the complex is presented to the APC/C. The APC/C then promotes ubiquitination and degradation of Cdc20 to sustain the checkpoint.
Why Cdc20 has to be degraded to maintain the checkpoint is not clear. The simplest explanation is that it prevents Cdc20 exceeding the level of the BubR1-Bub3 complex. This is feasible because Cdc20 and BubR1 are present at similar levels in the cell, and free Cdc20 appears when we add MG132 to checkpoint-arrested cells. It may not be as simple as this, however, because overexpressing wild-type Cdc20 to ∼10 fold endogenous levels does not override the checkpoint, and co-expressing BubR1 did not prevent the K-less mutant driving cells out of mitosis (although BubR1 might require Bub3 and/or another checkpoint component to act). Since even small amounts of K-less Cdc20 that are unlikely to exceed the amount of Bub3-BubR1 do overcome the checkpoint, this might indicate that ubiquitination directly inactivates Cdc20. (Although we cannot exclude the possibility that mutating all the lysines also altered other regulatory post-translational modifications such as phosphorylation, a mutant version of Cdc20 in which all the Cdk consensus phosphorylation sites were altered to alanine did not override the SAC, not shown). An alternative explanation might relate to our observation that metaphase substrates of the APC/C can only be degraded if they are able to localise to specific places in the cell (F.Cooke, A. Hagting and JP, in preparation). Therefore, overexpressing Cdc20 throughout the cell may not saturate the ability of the degradation machinery if it is only required to keep the levels of Cdc20 low in a specific location, whereas the K-less mutant could locally exceed the level of available checkpoint complexes. We have previously shown that the APC/C itself is recruited to improperly attached kinetochores by the checkpoint proteins 29, and the data we present here could indicate that it is at unattached kinetochores that BubR1 presents Cdc20 to the APC/C as a substrate.
Our model has the advantage that it makes the checkpoint a dynamic system. Previous work by ourselves and others has shown that the SAC and APC/C activity are very tightly coupled: cyclin B1 destruction begins almost immediately after the checkpoint is turned off, and destruction stops almost immediately if the checkpoint is re-imposed (see ref 15). Since Cdc20 is constantly synthesised during the SAC, this means that activating or inactivating Mad2 will very rapidly alter APC/C activity by determining whether Cdc20 binds to BubR1 and becomes inactive, or remains free to activate the APC/C.
We propose that Mad2 catalyses the binding between Cdc20 and BubR1. (A similar conclusion was reached by Davenport et al. but they used overexpressed protein in asynchronous cells 30). However, Mad2 does not have a recognisable catalytic domain and the crystal structures of Mad2 do not give immediate clues to what this catalytic activity might be. The counter evidence, however, that Mad2 alone is a Cdc20 inhibitor, or forms a stoichiometric part of the Cdc20-inhibitory complex, is not strong. Mad2 is a very poor inhibitor in vitro, and in fission yeast, Mad3/BubR1 is essential for Mad2 to block cells in mitosis 31. There are also caveats to the method that defined the stoichiometry of the Mitotic Checkpoint Complex 24 as 1:1:1:1 Mad2:BubR1:Bub3:Cdc20, where cells were only labelled with 35S-methionine for 6 hours, which is insufficient to label long-lived proteins to equilibrium.
Since the APC/C requires an activator to recognise its substrates, it is an interesting question whether the same molecule of Cdc20 acts as both activator and substrate during the SAC, or whether Cdc20 bound to BubR1 requires another APC/C activator to be degraded. We cannot yet distinguish between these possibilities but favour the idea that Cdc20 acts as both activator and substrate on the following evidence. Firstly, siRNA treatment shows the other APC/C activator, Cdh1, is not required for the SAC; indeed the mouse knock-out shows that Cdh1 is not required for embryonic or somatic cell division 32. Secondly, those motifs required for Cdc20 degradation during the checkpoint map to two classes (Fig 3): the first are motifs in the Mad2-binding region (R132), and the second are the C-box and the C-terminal ‘IR” motifs that are required for Cdc20 to bind and activate the APC/C.
We propose that ubiquitination and degradation are used to inactivate human Cdc20 and maintain the SAC, and not to turn off the checkpoint as recently proposed 11, 12. Although we note that the K-less Cdc20 binds more Mad2, this is probably because we increased its affinity for Mad2 by changing a conserved lysine in the Mad2 binding site of Cdc20, since an arginine was preferred in this position in a phage-display screen for Mad2-binding peptides 8. We find no evidence that Cdc20 ubiquitination is important to inactivate the checkpoint because the K-less form of Cdc20 that cannot be ubiquitinated is able to substitute for wild-type Cdc20 to promote mitotic exit with apparently normal timing, and is not impaired in its ability to be released from either Mad2 or BubR1 when the checkpoint is turned off.
Ubiquitination and degradation cannot be the only means by which Cdc20 is inactivated since cells expressing the K-less mutant initially delay in mitosis in the presence of microtubule poisons, and they progress at a similar rate to cells with wild-type Cdc20 through unchallenged mitosis, whereas cells without a SAC are greatly accelerated through mitosis 33. Thus, Cdc20 may initially be inactivated when it is bound by Mad2 and incorporated into the BubR1 complex, but to maintain the arrest Cdc20 must be ubiquitinated and degraded. The inactivation may be related to the way in which BubR1 binds and presents Cdc20 to the APC/C such that Cdc20 targets itself for destruction: it is unlikely that in this state Cdc20 could activate the APC/C against another substrate.
Lastly, we find that Cdc20 is continuously synthesised in mitosis to replace the protein that is degraded. Cdc20 synthesis is insensitive to rapamycin, indicating that it might be translated from an IRES. There is considerable variation in the ability of tumour cell lines to maintain a checkpoint arrest 34 and it will be interesting to see whether this variability arises from differences in Cdc20 synthesis or degradation.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Daisuke Izawa for pioneering APC3 and APC11 siRNA depletion, Paola Marco for help with the BubR1 constructs, Stephen Taylor, Gowei Fang and Andrea Musacchio for reagents, Athanassios Giannis for Dimethylanastron, to Mercedes Pardo and Lu Yu of the Mass Spectroscopy group at the Sanger Centre for mass spectroscopy, and Andrea Musacchio and members of our laboratory for helpful discussions. MY is particularly grateful to Takahiro Matsusaka for help with microscopy and microinjection. JN was supported by fellowships from the Carlsberg Foundation and the Danish Cancer Society, MY was supported by a Yousef Jameel scholarship. This work was supported by a Program Grant (C29/A3211) from Cancer Research UK to JP.
Appendix
MATERIALS AND METHODS
Antibodies
The following antibodies were used at the indicated dilution. Cdc20 (ab26483, Abcam) 1:200, Cdc20 (sc5296, Santa Cruz, for immunoprecipitations only), Mad2 (Clone AS55-A12 a kind gift from Dr Andrea Musacchio, IFOM, Milan)1:500 Mad1 (Clone BB3-J a kind gift from Dr. Andrea Musacchio)1:250, p31 (Clone E29.19.14 a kind gift from Dr Andrea Musacchio) 1:200 BubR1 (SBR1.1 a kind gift from Dr Stephen Taylor, Manchester University) 1:1000, Bub3 (611730, BD Transduction Laboratories) 1:500, Cyclin B1 (mAb GNS-1, BD Pharmingen) 1:2000, FLAG (F3165, Sigma) 1:1000, APC3 (610455, BD Transduction Laboratories) 1:500. APC4 (monoclonal antibody raised against a C-terminal peptide) 1:500, APC11 (monoclonal antibody raised against a C-terminal peptide) 1:500. APC7 (Abcam 4171) 1:500, APC8 (Biolegend) 1:500, Cdc16 (Santa Cruz Biotechnology) 1:500, phospho-APC1 (a kind gift from Dr. Jan-Michael Peters), APC10 (raised against full length protein) 1:2000
Secondary antibodies used for LiCor: Alexa Flour 680 rabbit anti-goat (A21088), Alexa Fluor 680 goat anti-mouse (A21057), Alexa Fluor 680 goat anti-rabbit (A21076) all used at 1:5000.
Gel filtration column chromatography
Cells were harvested by mitotic shake off and washed twice with ice-cold PBS. Cells were resuspended in buffer A (140 mM NaCl, 30 mM Hepes pH 7.8, 6 mM MgCl2, 5 % Glycerol, 1 mM DTT, 1 μg/μl Leupeptin, 1 μg/μl chymostatin, 0,2 μM microcystin, 3 mM ATP) at a 1:1 ratio of buffer to cells and opened by nitrogen cavitation (1000 PSI, 30 min, Parr Instrument Company, USA). Lysed cells were centrifuged at 20000g, 10 min and 259000g, 10 min before loading on Superose 6 PC 3.2/30 (GE Healthcare, USA). The column was run at a flow of 25 μl/min in buffer B (140 mM NaCl, 30 mM Hepes 7.8, 5 % Glycerol, 1 mM DTT) and 50 μl fractions collected.
Quantitative immunoblotting
Primary antibodies were incubated with fluorescently labelled secondary antibodies and the fluorescence measured using a LI-COR Odyssey CCD scanner according to the manufacturer’s instructions (LI-COR Biosciences, NE, USA). We have tested this scanner and it is linear over a range of at least 103, and we calibrated our antibodies over a range from 5 to 150 ng, see Fig S2B.
RNAi
The following ON-TARGETplus (Dharmacon, CO, USA) oligos were used APC3-1 (GGAAAUAGCCGAGAGGUAAUU) and APC3-2 (CAAAAGAGCCUUAGUUUAAUU), APC11 (UCUGCAGGAUGGCAUUUAAUU) Mad2 (GGAAGAGUCGGGACCACAGUU), BubR1-1 (CAGAAACGGGCAUUUGAAUUU), BubR1-2 (GAUGGUGAAUUGUGGAAUA) Cdc20 (CGGAAGACCUGCCGUUACAUU) and GAPDH (D-001830-01). Cells were transfected with 100 nM of oligo using oligofectamin (Invitrogen, USA). Cells were harvested 72 hrs after transfection for IPs or analyzed after 40 hrs by microscopy. In experiments were both APC3 and APC11 were knocked down 75 nM of APC3-1 and 50 nM APC11 was used and cells transfected 72 hrs and 48 hrs before harvest of cells.
Cell Culture
HeLa cells were maintained in Advanced D-MEM with 10% FBS. RPE cells were maintained in D-MEM (SIGMA) with 10% FBS. For synchronization at the beginning of S phase HeLa cells were treated with 2.5 mM Thymidine or 2.5 mM Thymidine followed by 2.5 μg/ml aphidicolin as previously described 35. To block HeLa cells in prometaphase, nocodazole or taxol was added to a final concentration of 0.08 ng/μl. RPE cells were blocked in prometaphase using the Eg5 inhibitor Dimethylanastron at 10 μM (kind gift of Dr Athanassios Giannis, University of Leipzig, Germany). Cells used for immunoprecipitation or gel filtration analysis were treated for 12 hrs with nocodazole or taxol at 0.08 ng/μl after a single or a double thymidine block. Stable cell lines expressing FLAG - Cdc20 wt and FLAG-Cdc20 K-less were made using the Flp-In system (Invitrogen). The HeLa FRT cell line was a kind gift from Stephen Taylor (Manchester). Protein synthesis was inhibited by adding cycloheximide to a final concentration of 20 μg/ml, or rapamycin to 20 ng/ml. To block the proteasome MG132 was added to 10 μM.
In vivo dissociation of checkpoint proteins
APC3 and APC11 were knocked down and cells were syncronized by a double Thymidine protocol and released into Taxol for 6 hrs. MG132 was added to cells and to 1 dish ZM447439 was added for 2 hrs. Cells were processed for Cdc20 immunoprecipitation as described.
Stable cell lines expressing FLAG-Cdc20 wt or FLAG-Cdc20 K-less were synchronized by a double Thymidine approach and 9 hrs after release from last Thymidine treatment MG132 was added for 2 hrs and mitotic cells were harvested by shake off. Cells were replated into Taxol and after 1 hr first FLAG IP was performed. To remaining half of cells ZM447439 was added for 1,5 hrs and second FLAG IP was performed.
Microscopy
Cells were incubated on the microscope using the delta T system (Bioptechs, PA, USA) and imaged by time-lapse fluorescence and DIC microscopy on a Leica DMIRBE or DMIR2 microscope equipped with a 40× 1.2 NA oil immersion lens. Cerulean/CFP and Venus/YFP were visualised using a JP5 filter set (Chroma, VE, USA) with excitation and emission filters in filter wheels (Lambda 10-3, Sutter Instrument Co, CA, USA) and a Cascade 512B or QuantEM CCD camera (Photometrics, AZ, USA).
Multiple cell positions were captured using a Corvus (Marzhauser, Germany) or H117 (Prior, UK) stage. Shutters (Smart shutter, Sutter Instrument Co, CA, USA), filter wheels, stages, microscopes and cameras were all controlled by SlideBook software (Intelligent Imaging Innovations, CO, USA). Images were captured at 3 min intervals and analysed using Slidebook. Images were exported to ImageJ to assemble into movies.
Immunoprecipitation
Complexes were immunoprecipitated with antibodies covalently coupled to Dynabeads (Invitrogen) using buffer B for incubation and washing. Cells for immunoprecipitation were lysed with 0.1% NP40 in buffer B for 30 min on ice and clarified by a 20000 × g spin for 10 min.
Constructs
pET30a-Mad2: Mad2 was cloned into BamHI and HindIII of pET30a. pET30a-TEV-Securin: TEV-Securin was digested with BamHI and XhoI and inserted into BamHI and XhoI digested pET30a. pET30a-UbcH10: UbcH10 digested with BamHI and HindIII and cloned into pET30a digested with BamHI and HindIII. pET30a-APC10: APC10 digested with BamHI and EcoRI and cloned into pET30a digested with BamHI and EcoRI. pFAST-BAC BubR1: BubR1 digested with BamHI and SmaI was ligated into pFAST BAC B digested with BamHI and Stu I. pFAST-BAC Cdc20 wt and K-less: Cdc20 digested with BglII and BamHI and cloned into BamHI digested pFAST-BAC. pPICZ His-Cdc20: Cdc20 cloned into EcoRI and Not I sites of pPICZ A. YFP-Cdc20: Cdc20 cloned into EcoRI and BamHI of pYFP-C1. pcDNA5/FRT/TO FLAG-Cdc20 was cloned into BamHI site of modified pcDNA5/FRT/TO vector. Details of all clones are available on request.
Protein Expression
Mad2, APC10, Securin and UbcH10 were expressed in BL21 (DE3) RIL at 37 °C and purified by Nickel affinity chromatography (Qiagen). APC10 was purified under denaturing conditions. Following Nickel affinity chromatography the His-Tag was removed from Securin by TEV cleavage followed by removal of Tag by Nickel affinity chromatography followed by chromatography on Superdex 75. Mad2 and UbcH10 were further purified by chromatography on Superdex 75. Cdc20 was expressed from pPICZ His-Cdc20 in Pichia pastoris and purified by Nickel affinity chromatography followed by chromatography on Superdex 200. BubR1, wt Cdc20 and K-less Cdc20 were expressed in Sf9 cells according to the manufacturer’s instructions (Invitrogen) and purified by Nickel affinity chromatography.
In vitro ubiquitination
The APC/C complex was purified with the APC4 antibody and eluted with the peptide antigen. Reactions were performed in 15 μl for 30 mins at 37°C in QA buffer (100 mM NaCl, 30 mM Hepes-KOH 7,8, 2 mM ATP, 2 mM MgCl2, 0,1 μl/μg BSA, 1mM DTT) and contained 0.02 μg/μl securin, 1,3 μg/μl ubiquitin, 0,05 μg/μl UbcH10, 0.015 μg/μl E1, 0,3 ng/μl Cdc20 and 1 μl APC/C.
RNAi resistant Cdc20 constructs
To rescue the RNAi-mediated depletion of Cdc20 we constructed wild-type Cdc20 and Cdc20 with all lysines changed to arginines by assembling chemically synthesised oligonucleotides as previously described 36. DNA sequences were designed to be maximally different from the wild-type using Gene Designer software 37.
References
- 1.Pines J. Mitosis: a matter of getting rid of the right protein at the right time. Trends Cell Biol. 2006;16:55–63. doi: 10.1016/j.tcb.2005.11.006. [DOI] [PubMed] [Google Scholar]
- 2.Musacchio A, Salmon ED. The spindle-assembly checkpoint in space and time. Nat Rev Mol Cell Biol. 2007;8:379–393. doi: 10.1038/nrm2163. [DOI] [PubMed] [Google Scholar]
- 3.Hwang LH, et al. Budding yeast Cdc20: a target of the spindle checkpoint. Science (New York, N.Y. 1998;279:1041–1044. doi: 10.1126/science.279.5353.1041. [DOI] [PubMed] [Google Scholar]
- 4.Kim SH, Lin DP, Matsumoto S, Kitazono A, Matsumoto T. Fission yeast Slp1: an effector of the Mad2-dependent spindle checkpoint. Science (New York, N.Y. 1998;279:1045–1047. doi: 10.1126/science.279.5353.1045. [DOI] [PubMed] [Google Scholar]
- 5.Peters JM. The anaphase promoting complex/cyclosome: a machine designed to destroy. Nat Rev Mol Cell Biol. 2006;7:644–656. doi: 10.1038/nrm1988. [DOI] [PubMed] [Google Scholar]
- 6.Sironi L, et al. Crystal structure of the tetrameric Mad1-Mad2 core complex: implications of a ‘safety belt’ binding mechanism for the spindle checkpoint. The EMBO journal. 2002;21:2496–2506. doi: 10.1093/emboj/21.10.2496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mapelli M, Massimiliano L, Santaguida S, Musacchio A. The mad2 conformational dimer: structure and implications for the spindle assembly checkpoint. Cell. 2007;131:730–743. doi: 10.1016/j.cell.2007.08.049. [DOI] [PubMed] [Google Scholar]
- 8.Luo X, Tang Z, Rizo J, Yu H. The Mad2 spindle checkpoint protein undergoes similar major conformational changes upon binding to either Mad1 or Cdc20. Molecular cell. 2002;9:59–71. doi: 10.1016/s1097-2765(01)00435-x. [DOI] [PubMed] [Google Scholar]
- 9.Tang Z, Bharadwaj R, Li B, Yu H. Mad2-Independent inhibition of APCCdc20 by the mitotic checkpoint protein BubR1. Developmental cell. 2001;1:227–237. doi: 10.1016/s1534-5807(01)00019-3. [DOI] [PubMed] [Google Scholar]
- 10.Fang G. Checkpoint protein BubR1 acts synergistically with Mad2 to inhibit anaphase-promoting complex. Molecular biology of the cell. 2002;13:755–766. doi: 10.1091/mbc.01-09-0437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Reddy SK, Rape M, Margansky WA, Kirschner MW. Ubiquitination by the anaphase-promoting complex drives spindle checkpoint inactivation. Nature. 2007;446:921–925. doi: 10.1038/nature05734. [DOI] [PubMed] [Google Scholar]
- 12.Stegmeier F, et al. Anaphase initiation is regulated by antagonistic ubiquitination and deubiquitination activities. Nature. 2007;446:876–881. doi: 10.1038/nature05694. [DOI] [PubMed] [Google Scholar]
- 13.Pan J, Chen RH. Spindle checkpoint regulates Cdc20p stability in Saccharomyces cerevisiae. Genes & development. 2004;18:1439–1451. doi: 10.1101/gad.1184204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.King EM, van der Sar SJ, Hardwick KG. Mad3 KEN Boxes Mediate both Cdc20 and Mad3 Turnover, and Are Critical for the Spindle Checkpoint. PLoS ONE. 2007;2:e342. doi: 10.1371/journal.pone.0000342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Clute P, Pines J. Temporal and spatial control of cyclin B1 destruction in metaphase. Nature cell biology. 1999;1:82–87. doi: 10.1038/10049. [DOI] [PubMed] [Google Scholar]
- 16.Brito DA, Rieder CL. Mitotic checkpoint slippage in humans occurs via cyclin B destruction in the presence of an active checkpoint. Curr Biol. 2006;16:1194–1200. doi: 10.1016/j.cub.2006.04.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kallio MJ, Beardmore VA, Weinstein J, Gorbsky GJ. Rapid microtubule-independent dynamics of Cdc20 at kinetochores and centrosomes in mammalian cells. The Journal of cell biology. 2002;158:841–847. doi: 10.1083/jcb.200201135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Howell BJ, et al. Spindle checkpoint protein dynamics at kinetochores in living cells. Curr Biol. 2004;14:953–964. doi: 10.1016/j.cub.2004.05.053. [DOI] [PubMed] [Google Scholar]
- 19.den Elzen N, Pines J. Cyclin A is destroyed in prometaphase and can delay chromosome alignment and anaphase. The Journal of cell biology. 2001;153:121–136. doi: 10.1083/jcb.153.1.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Geley S, et al. Anaphase-promoting complex/cyclosome-dependent proteolysis of human cyclin A starts at the beginning of mitosis and is not subject to the spindle assembly checkpoint. The Journal of cell biology. 2001;153:137–148. doi: 10.1083/jcb.153.1.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hayes MJ, et al. Early mitotic degradation of Nek2A depends on Cdc20-independent interaction with the APC/C. Nature cell biology. 2006;8:607–614. doi: 10.1038/ncb1410. [DOI] [PubMed] [Google Scholar]
- 22.Schwab M, Neutzner M, Mocker D, Seufert W. Yeast Hct1 recognizes the mitotic cyclin Clb2 and other substrates of the ubiquitin ligase APC. The EMBO journal. 2001;20:5165–5175. doi: 10.1093/emboj/20.18.5165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vodermaier HC, Gieffers C, Maurer-Stroh S, Eisenhaber F, Peters JM. TPR subunits of the anaphase-promoting complex mediate binding to the activator protein CDH1. Curr Biol. 2003;13:1459–1468. doi: 10.1016/s0960-9822(03)00581-5. [DOI] [PubMed] [Google Scholar]
- 24.Sudakin V, Chan GK, Yen TJ. Checkpoint inhibition of the APC/C in HeLa cells is mediated by a complex of BUBR1, BUB3, CDC20, and MAD2. The Journal of cell biology. 2001;154:925–936. doi: 10.1083/jcb.200102093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Camasses A, Bogdanova A, Shevchenko A, Zachariae W. The CCT chaperonin promotes activation of the anaphase-promoting complex through the generation of functional Cdc20. Molecular cell. 2003;12:87–100. doi: 10.1016/s1097-2765(03)00244-2. [DOI] [PubMed] [Google Scholar]
- 26.Ditchfield C, et al. Aurora B couples chromosome alignment with anaphase by targeting BubR1, Mad2, and Cenp-E to kinetochores. The Journal of cell biology. 2003;161:267–280. doi: 10.1083/jcb.200208091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hardwick KG, Johnston RC, Smith DL, Murray AW. MAD3 encodes a novel component of the spindle checkpoint which interacts with Bub3p, Cdc20p, and Mad2p. The Journal of cell biology. 2000;148:871–882. doi: 10.1083/jcb.148.5.871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wolthuis R, et al. Cdc20 and Cks direct the spindle checkpoint-independent destruction of cyclin a. Molecular cell. 2008;30:290–302. doi: 10.1016/j.molcel.2008.02.027. [DOI] [PubMed] [Google Scholar]
- 29.Acquaviva C, Herzog F, Kraft C, Pines J. The anaphase promoting complex/cyclosome is recruited to centromeres by the spindle assembly checkpoint. Nature cell biology. 2004;6:892–898. doi: 10.1038/ncb1167. [DOI] [PubMed] [Google Scholar]
- 30.Davenport J, Harris LD, Goorha R. Spindle checkpoint function requires Mad2-dependent Cdc20 binding to the Mad3 homology domain of BubR1. Exp Cell Res. 2006;312:1831–1842. doi: 10.1016/j.yexcr.2006.02.018. [DOI] [PubMed] [Google Scholar]
- 31.Millband DN, Hardwick KG. Fission yeast Mad3p is required for Mad2p to inhibit the anaphase-promoting complex and localizes to kinetochores in a Bub1p-, Bub3p-, and Mph1p-dependent manner. Mol Cell Biol. 2002;22:2728–2742. doi: 10.1128/MCB.22.8.2728-2742.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Garci-Higuera I, et al. Genomic stability and tumour suppression by the APC/C cofactor Cdh1. Nature cell biology. 2008;10:802–811. doi: 10.1038/ncb1742. [DOI] [PubMed] [Google Scholar]
- 33.Meraldi P, Draviam VM, Sorger PK. Timing and checkpoints in the regulation of mitotic progression. Developmental cell. 2004;7:45–60. doi: 10.1016/j.devcel.2004.06.006. [DOI] [PubMed] [Google Scholar]
- 34.Brito DA, Yang Z, Rieder CL. Microtubules do not promote mitotic slippage when the spindle assembly checkpoint cannot be satisfied. The Journal of cell biology. 2008 doi: 10.1083/jcb.200805072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Di Fiore B, Pines J. Emi1 is needed to couple DNA replication with mitosis but does not regulate activation of the mitotic APC/C. J. Cell Biol. 2007;177:425–437. doi: 10.1083/jcb.200611166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kodumal SJ, et al. Total synthesis of long DNA sequences: synthesis of a contiguous 32-kb polyketide synthase gene cluster. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:15573–15578. doi: 10.1073/pnas.0406911101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Villalobos A, Ness JE, Gustafsson C, Minshull J, Govindarajan S. Gene Designer: a synthetic biology tool for constructing artificial DNA segments. BMC Bioinformatics. 2006;7:285. doi: 10.1186/1471-2105-7-285. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.