Abstract
The target of rapamycin (TOR) proteins regulate various cellular processes including autophagy1, which may play a protective role in certain neurodegenerative and infectious diseases2. Here we show that a primary small-molecule screen in yeast yields novel small-molecules modulators of mammalian autophagy. We first identified novel small-molecule enhancers (SMER) and inhibitors (SMIR) of the cytostatic effects of rapamycin in Saccharomyces cerevisiae. Three SMERs induced autophagy independently of rapamycin in mammalian cells, enhancing the clearance of autophagy substrates like mutant huntingtin and A53T α-synuclein, associated with Huntington’s disease (HD) and familial Parkinson’s disease, respectively3-5. These SMERs, which appear to act either independently, or downstream, of TOR, attenuated mutant huntingtin-fragment toxicity in HD cell and Drosophila models, suggesting therapeutic potential. We also screened structural analogs of these SMERs and identified additional candidate drugs enhancing autophagy. Thus, we have demonstrated proof-of-principle for a novel approach for discovery of small-molecule modulators of mammalian autophagy.
The autophagy/lysosome and ubiquitin/proteasome pathways are the two major routes for protein clearance in eukaryotic cells. Proteasomes predominantly degrade short-lived nuclear and cytosolic proteins, which need to be unfolded to pass through the narrow pore of the proteasome barrel, precluding clearance of large membrane proteins and protein complexes (including oligomers and aggregates). Mammalian lysosomes, on the other hand, can degrade substrates like protein complexes and organelles. The bulk degradation of cytoplasmic proteins or organelles is largely mediated by macroautophagy, generally referred to as autophagy1. It involves the formation of double-membrane structures called autophagosomes/autophagic vacuoles (AVs), which fuse with lysosomes to form autolysosomes (also called autophagolysosomes) where their contents are then degraded by acidic lysosomal hydrolases. Autophagosomes are generated by elongation of small membrane structures, whose precise origins have yet to be elucidated1. Autophagy can be induced under physiological stress conditions such as starvation. Several protein kinases regulate autophagy, the best characterised being the mammalian target of rapamycin (mTOR), which negatively regulates the pathway in organisms from yeast to man1. However, the targets of mTOR-dependent and - independent signalling in the autophagy apparatus are not well understood in mammalian systems. Recently, we described an mTOR-independent pathway where autophagy is induced by agents that lower inositol (1) or inositol-1,4,5-triphosphate (IP3) (2) levels6.
Autophagy is an important process in a variety of human diseases caused by toxic, aggregate-prone, intracytosolic proteins, which become inaccessible to the proteasome when they oligomerise2-5. These include Huntington’s disease (HD), an autosomal-dominant neurodegenerative disorder caused by a CAG trinucleotide repeat expansion (>35 repeats) that encodes an abnormally long polyglutamine (polyQ) tract in the N-terminus of the huntingtin protein7. Mutant huntingtin toxicity is thought to be exposed after it is cleaved to form N-terminal fragments comprising the first 100-150 residues with the expanded polyQ tract, which are also the toxic species found in aggregates. Thus, HD pathogenesis is frequently modelled with exon 1 fragments containing expanded polyQ repeats that cause aggregate formation and toxicity in cell models and in vivo7.
In addition to mutant huntingtin, autophagy also regulates the clearance of other aggregate-prone disease-causing proteins, like those causing spinocerebellar ataxia type 3, forms of tau (which cause fronto-temporal dementias) and the A53T and A30P α-synuclein mutants [which cause familial Parkinson’s disease (PD)]3-6, 8, 9. Autophagy induction reduces mutant huntingtin levels and protects against its toxicity in cell, Drosophila and mouse models3, 4. Similar effects are seen with other polyQ-containing proteins and tau in cells and flies9. Certain bacterial and viral infections may also be treatable by autophagy upregulation, since the pathogens can be engulfed by autophagosomes and transferred to lysosomes for degradation. These include Mycobacterium tuberculosis (that causes tuberculosis), Group A Streptococcus, and viruses like herpes simplex virus type I2, 10-12.
Currently, the only small molecule known to upregulate autophagy in mammalian brains is rapamycin (3), which, when complexed to its intracellular binding protein FKBP12, is a specific TOR inhibitor2, 4. However, TOR proteins control several cellular processes besides autophagy in organisms from yeast to man, including repression of ribosome biogenesis and protein translation, and transcriptional induction of compensatory metabolic pathways13. These probably contribute to the complications seen with long-term rapamycin use, like immunosuppression, which is not compatible with therapy for infectious diseases. Accordingly, we are aiming to identify safer ways of inducing autophagy, either by identifying mTOR substrates regulating autophagy, compounds that enhance the activity of rapamycin, or compounds acting via mTOR-independent pathways.
Here we have identified novel enhancers of mammalian autophagy by starting with a small-molecule screen in yeast14. We reasoned that a small-molecule screen would uncover enhancers and suppressors of the physiological state induced by rapamycin in yeast, and that the activities of at least some of these modifiers as single agents would be conserved in mammalian systems. From 50,729 compounds screened, we observed suppressors of the cytostatic effects of rapamycin in yeast - small-molecule inhibitors of rapamycin (SMIRs) - in a temporal window that spans 48-72 h; we discerned both fast- and slow-acting SMIRs, but hereafter do not distinguish between them. Later, at 96 h, we observed enhancers of the cytostatic effects of rapamycin - small-molecule enhancers of rapamycin (SMERs) - in wells containing yeast that failed to exit rapamycin-induced growth arrest.
We retested 72 total primary assay positives, from which we identified a structurally non-redundant set of 21 SMIRs (4-24) and 12 SMERs (25-36) (Supplementary Fig. 1a online). Further characterisation of these SMIRs and SMERs in yeast is described in Supplementary Methods and Supplementary Fig. 1b-f online.
Although these SMIRs and SMERs were discovered as modulators of the effects of rapamycin on yeast growth, we tested if they were also modulators of mammalian autophagy. We screened all the yeast primary assay positives in mammalian cells (including hits that were excluded from the above analyses in yeast due to their poor potency) in the absence of rapamycin for their potential to induce clearance of the autophagy substrate A53T α-synuclein. We used a stable doxycycline-inducible PC12 cell line expressing A53T mutant α-synuclein, where the transgene expression is first induced by adding doxycycline and then switched off by removing doxycycline from the medium5. If the transgene expression level is measured at 24 h after switching off expression after an initial induction period of 48 h, one can assess if specific agents alter the clearance of the transgene product, as the amount of transgene product decays when synthesis is stopped5. Interestingly, we observed that 13 SMIRs (14-18, 19a, 19b, 22, 23, 29a, 29b, 30, 31) slowed the clearance of this autophagy substrate, while 4 SMERs (10, 16, 18 and 28) enhanced its clearance (Supplementary Fig. 2a-d online). We categorised SMIRs as possible inhibitors of autophagy if they increased A53T α-synuclein levels by 40-50 % or more. Likewise, we categorised SMERs as possible enhancers of autophagy if they reduced A53T α-synuclein levels to 50 % or lower than the control (Supplementary Fig. 2a-d online).
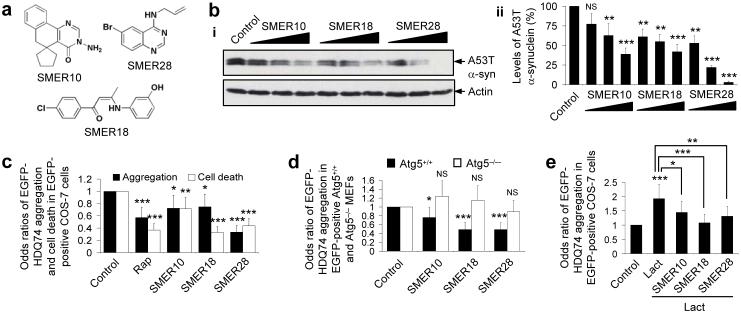
We then concentrated on the autophagy-inducing SMERs, which may have the greatest immediate utility as drug leads for neurodegenerative diseases (Fig. 1a). We confirmed that SMERs 10 (27), 18 (30) and 28 (36) significantly enhanced A53T α-synuclein clearance in PC12 cells independent of rapamycin treatment at the doses used in the yeast screen, and also at lower concentrations (Fig. 1b). In all subsequent experiments with these compounds alone, we have used the concentrations used in the yeast screen.
Figure 1. SMERs 10, 18 and 28 enhance the clearance of mutant aggregate-prone proteins by autophagy in mammalian cell models of Huntington’s and Parkinson’s disease, independent of rapamycin.
(a) Chemical structures of SMERs 10, 18 and 28.
(b) A stable inducible PC12 cell line expressing A53T α-synuclein was induced with 1 μg ml-1 doxycycline for 48 h, and expression of the transgene was switched off for 24 h. Cells were treated with DMSO (control), or with 0.94 μM, 4.7 μM, 47 μM of SMER10, 0.86 μM, 4.3 μM, 43 μM of SMER18 or 0.9 μM, 4.7 μM, 47 μM of SMER28 added in the switch-off period as 1:20000, 1:4000 or 1:400 dilutions of 5 mg ml-1 stock solution (in DMSO), respectively. A53T α-synuclein (α-syn) levels were analysed by immunoblotting with antibody against HA (i) and densitometry analysis relative to actin (ii). Error bars denote S.E.M. p=0.0917, p=0.009, p=0.0001 (for increasing concentrations of SMER10); p=0.0068, p=0.0023, p=0.0002 (for increasing concentrations of SMER18); p=.0016, p<0.0001, p<0.0001 (for increasing concentrations of SMER28).
(c) COS-7 cells transfected with EGFP-HDQ74 construct for 4 h were treated with DMSO (control), 0.2 μM rapamycin (rap), 47 μM SMER10, 43 μM SMER18 or 47 μM SMER28 for 48 h. The effects of treatment on the percentage of EGFP-positive cells with EGFP-HDQ74 aggregates or apoptotic morphology (cell death) were expressed as odds ratios and the control was taken as 1. Error bars: 95 % confidence interval. p<0.0001 (rap and SMER28), p=0.013 (SMER10), p=0.019 (SMER18) (aggregation); p<0.0001 (rap, SMER18 and SMER28), p=0.004 (SMER10) (cell death). Odds ratios were used to determine pooled estimates for the changes in aggregate formation or cell death, resulting from perturbations assessed in multiple independent experiments (see Supplementary Methods online).
(d) Wild-type (ATG5+/+) and knock-out (ATG5−/−) Atg5 mouse embryonic fibroblasts (MEFs) were transfected with EGFP-HDQ74 for 4 h and treated with DMSO (control), 47 μM SMER10, 43 μM SMER18 or 47 μM SMER28 for 24 h. The effects of treatment on the percentage of EGFP-positive cells with EGFP-HDQ74 aggregates were expressed as odds ratios and the control (DMSO-treated) values were fixed at 1 for both cell lines. Error bars: 95 % confidence interval. p=0.039 (SMER10), p<0.0001 (SMER18 and SMER28) (in ATG5+/+ cells); p=0.092 (SMER10), p=0.271 (SMER18), p=0.358 (SMER28) (in ATG5−/− cells). Note that EGFP-HDQ74 aggregation was higher in ATG5−/− cells compared to ATG5+/+ cells (Supplementary Fig. 3a online).
(e) The percentage of EGFP-positive COS-7 cells with EGFP-HDQ74 aggregates as in Fig. 1c, treated with DMSO (control), 10 μM lactacystin (proteasome inhibitor), or both 10 μM lactacystin along with either 47 μM SMER10, 43 μM SMER18 or 47 μM SMER28 for 48 h, were expressed as odds ratios. Error bars: 95 % confidence interval. p<0.0001 (Control vs Lact), p=0.014 (SMER10 vs Lact), p<0.0001 (SMER18 vs Lact), p=0.001 (SMER28 vs Lact).
***, p<0.001; **, p<0.01; *, p<0.05; NS, Non-significant.
We next studied the effect of these SMERs on another autophagy substrate, mutant huntingtin3, 4. SMERs 10, 18 and 28 reduced aggregation and cell death caused by EGFP-tagged huntingtin exon 1 with 74 polyQ repeats (EGFP-HDQ74) in COS-7 cells (Fig. 1c). We excluded SMER 16 (subsequently redesignated SMIR 33 because additional retesting revealed it to be a suppressor of the cytostatic effects of rapamycin) from our subsequent experiments as it was toxic in COS-7 and other cell lines at the concentration that enhanced the clearance of A53T α-synuclein in PC12 cells (data not shown). No overt toxicity was observed with SMERs 10, 18 and 28 (see Supplementary Methods online).
We confirmed that this reduction of EGFP-HDQ74 aggregation is through autophagy, using autophagy-competent mouse embryonic fibroblasts (MEFs) (ATG5+/+) or matched MEFs lacking the essential autophagy gene Atg5 (ATG5−/−)15. EGFP-HDQ74 aggregation was significantly increased in untreated ATG5−/− (autophagy-deficient) cells compared to untreated ATG5+/+ cells, as mutant huntingtin is an autophagy substrate (Supplementary Fig. 3a online). SMERs 10, 18 and 28 significantly reduced EGFP-HDQ74 aggregation in ATG5+/+ cells, but not in ATG5−/− cells (Fig. 1d). Thus, these SMERs can only reduce mutant huntingtin aggregation in autophagy-competent cells. Indeed, these SMERs decreased EGFP-HDQ74 aggregation in cells constitutively treated with the proteasome inhibitor lactacystin16 (37) (which targets the major alternative route for mutant huntingtin clearance) (Fig. 1e).
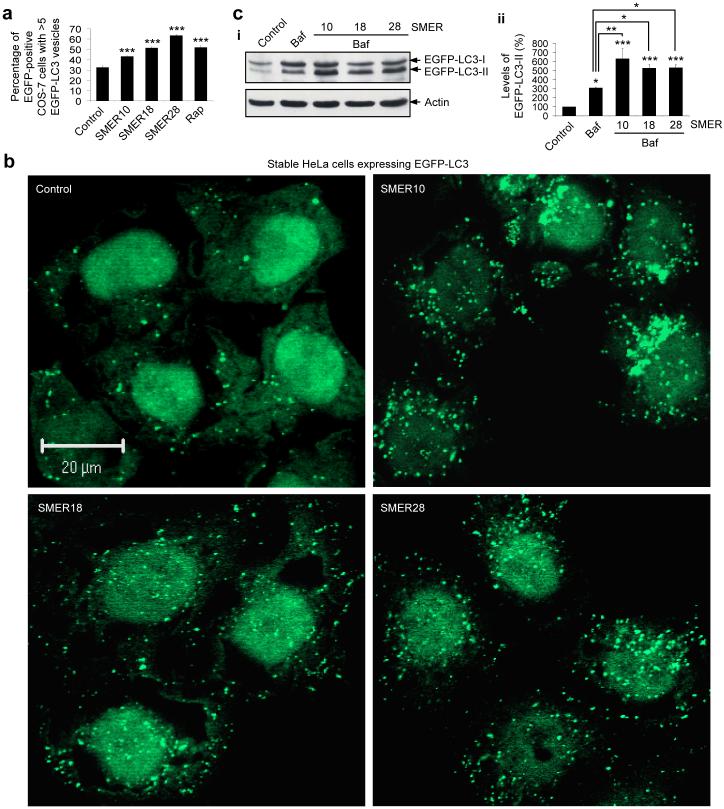
We next assessed the effect of these SMERs on autophagy by transfecting COS-7 cells with the microtubule-associated protein 1 light chain 3 (LC3) fused to EGFP (EGFP-LC3). LC3 (and EGFP-LC3) localizes only to autophagic membranes but not on other membrane structures and serves as a specific marker for autophagosomes17. As EGFP-LC3 overexpression does not affect autophagic activity, the number of EGFP-LC3 vesicles has frequently been used to assess autophagosome number, and therefore to make inferences about autophagic activity18. SMERs 10, 18 and 28 significantly increased the proportion of cells with EGFP-LC3 vesicles, compared to control (DMSO-treated) cells (Fig. 2a).
Figure 2. SMERs 10, 18 and 28 induce autophagy in mammalian cells.
(a) COS-7 cells transfected with EGFP-LC3 construct for 4 h were treated with DMSO (control), 0.2 μM rapamycin (rap) (positive control), 47 μM SMER10, 43 μM SMER18 or 47 μM SMER28 for 16 h, and analysed by fluorescence microscopy. The effects of treatment on the percentage of EGFP-positive cells with >5 EGFP-LC3 vesicles are shown. Error bars denote S.E.M. p<0.0001 (all SMERs).
(b) HeLa cells stably expressing EGFP-LC3 were treated with DMSO (control), 47 μM SMER10, 43 μM SMER18 or 47 μM SMER28 for 24 h. Confocal sections show cells containing EGFP-positive autophagic vesicles. Nuclei are stained with DAPI. Bar, 20 μM.
(c) HeLa cells stably expressing EGFP-LC3 were treated for 4 h with DMSO (control) or 200 nM bafilomycin A1 (baf), or with 200 nM bafilomycin A1 and 47 μM SMER10, 43 μM SMER18 or 47 μM SMER28. Cells were left untreated or pre-treated with SMERs for 24 h before adding bafilomycin A1. Levels of EGFP-LC3-II were determined by immunoblotting with antibody against EGFP (i) and densitometry analysis relative to actin (ii). Error bars denote S.E.M. p=0.0259 (baf), p<0.0001 (SMER10), p=0.0003 (SMER18 and SMER28) vs control; p=0.0025 (SMER10), p=0.0218 (SMER18), p=0.0195 (SMER28) vs bafilomycin A1.
***, p<0.001; **, p<0.01; *, p<0.05.
We then tested for autophagy induction by the SMERs in an EGFP-LC3 expressing stable HeLa cell line19. Treatment of these cells with SMERs 10, 18 and 28 led to overt EGFP-LC3 vesicle formation compared to the control cells (Fig. 2b). (Note that some cell types, e.g. HeLa cells, have more autophagosomes per cell than other cell types, e.g. COS-7 cells).
Endogenous LC3 is processed post-translationally into LC3-I, which is cytosolic. LC3-I is in turn converted to LC3-II, which associates with autophagosome membranes17. LC3-II can accumulate due to increased upstream autophagosome formation, but also if there is impaired downstream autophagosome-lysosome fusion. In order to distinguish between those two possibilities, we assayed LC3-II in the presence of bafilomycin A1 (38), which blocks downstream autophagosome-lysosome fusion20. Bafilomycin A1 increased EGFP-LC3-II levels in stable HeLa cells, as expected (Fig. 2c). The dose of bafilomycin A1 used is saturating for LC3-II levels in this assay and no further increases in LC3-II are observed when we treat cells with bafilomycin A1 and agents that block autophagosome-lysosome fusion via independent mechanisms [like the dynein inhibitor, erythro-9-[3-(2-hydroxynonyl)] adenine (EHNA)21] (39) (data not shown). However, autophagy inducers increase LC3-II levels even when cells are constitutively treated with this dose of bafilomycin A18. SMERs 10, 18 and 28 significantly increased EGFP-LC3-II levels in the presence of bafilomycin A1, compared to bafilomycin A1 alone, strongly arguing that the increased numbers of autophagosomes induced by these SMERs are due to enhanced autophagosome formation (Fig. 2c).
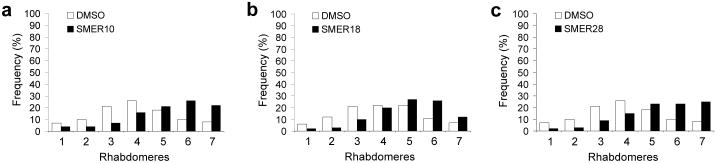
We tested the potential of the three autophagy-enhancing SMERs to function in vivo using a Drosophila model of HD expressing the first 171 residues of mutant huntingtin with 120 polyQ repeats in photoreceptors, using the pseudopupil technique (see Methods). The compound eyes in flies consist of several hundred ommatidia, each containing eight photoreceptor neurons with light-gathering parts called rhabdomeres, seven of which can be visualised using the pseudopupil technique. This method assesses the number of visible rhabdomeres by light microscopy and has been widely used to quantify the toxicity of proteins with long polyQs in the fly eye4, 22, 23. The number of visible rhabdomeres in each ommatidium decreases over time in the mutant Drosophila expressing mutant huntingtin with 120 polyQ repeats in photoreceptors, compared to the wild-type flies or transgenic flies expressing otherwise identical huntingtin with 23 polyQ (wild-type) repeats (where there is no degeneration). SMERs 10, 18 and 28 protected against neurodegeneration in Drosophila expressing mutant huntingtin, compared to flies treated with the vehicle (DMSO) (Fig. 3a-c). Thus, these SMERs protect against polyglutamine toxicity in vivo in neurons. These SMERs are not toxic for Drosophila development or for Drosophila photoreceptors at the relevant concentrations we have used to protect against polyQ toxicity (see Supplementary Methods online).
Figure 3. SMERs 10, 18 and 28 protect against neurodegeneration in Drosophila model of Huntington’s disease.
(a-c) Flies treated with 100 μM SMER10 (a), 200 μM SMER18 (b) or 100 μM SMER28 (c) show a shift in the distribution of the number of rhabdomeres compared to flies treated with DMSO (control) alone (2 days after eclosion). Rhabdomere counts from all 3 independent experiments are included. n=600 ommatidia (SMER10), n=1500 ommatidia (SMER18) and n=600 ommatidia (SMER28). Mann-Whitney test values p<0.0001 (all SMERs). Student’s t-test (1 tailed) p=0.005 (SMER10), p=0.004 (SMER18), p=0.03 (SMER28) compared distributions of means of independent experiments. These SMER concentrations cause no overt toxicity to flies (see Supplementary Methods online). Distributions of DMSO-treated flies may vary when SMERs were treated in different experiments at different times. For instance, an individual experiment may have lasted slightly longer or shorter and photoreceptor degeneration is progressive and time-dependent.
We next tested whether these autophagy-inducing SMERs acted primarily via the pathway that is negatively regulated by mTOR. mTOR kinase activity can be inferred by the levels of phosphorylation of its substrates, ribosomal S6 protein kinase (S6K1, also known as p70S6K) and eukaryotic initiation factor 4E-binding protein 1 (4E-BP1) at Thr389 and Thr37/46, respectively24. While rapamycin dramatically reduced the amounts of total p70S6K and 4E-BP1 that were phosphorylated, SMERs 10, 18 and 28 had no such effects (Supplementary Fig. 3b,c online). However, the proportion of total S6K1 that was phosphorylated in the presence of SMER 28 may be slightly less than the control-treated cells (although the effect is diminished relative to that seen with rapamycin).
The SMERs did not affect the levels of various autophagy regulators, such as Beclin-1 (Atg6), Atg5, Atg7 and Atg12, and did not enhance conjugation of Atg12 to Atg5, which is a critical step in autophagosome assembly preceding LC3 conjugation (Supplementary Fig. 3d-h online). Our data suggest that these SMERs induce mammalian autophagy in an mTOR-independent fashion. However, it is also conceivable that the SMERs impinge upon a hitherto unknown component of the mTOR autophagy pathway downstream of mTOR. Whichever the case may be, the increased sensitivity afforded by using cellular growth as a screening readout may be one reason why not all SMERs or SMIRs that emerged from the primary screen modulate autophagy, which is but one of many cellular pathways downstream of TOR. Unfortunately, it is difficult to test these hypotheses directly, as the mTOR substrate(s) and the relevant downstream effectors that mediate mammalian autophagy have not been identified.
These SMERs did not cause accumulation of the UbG76V-EGFP degron, a specific proteasome substrate25, in contrast to the proteasome inhibitor lactacystin (Supplementary Fig. 3i online). Thus, these SMERs do not induce autophagy by causing major impairments in the ubiquitin/proteasome pathway26.
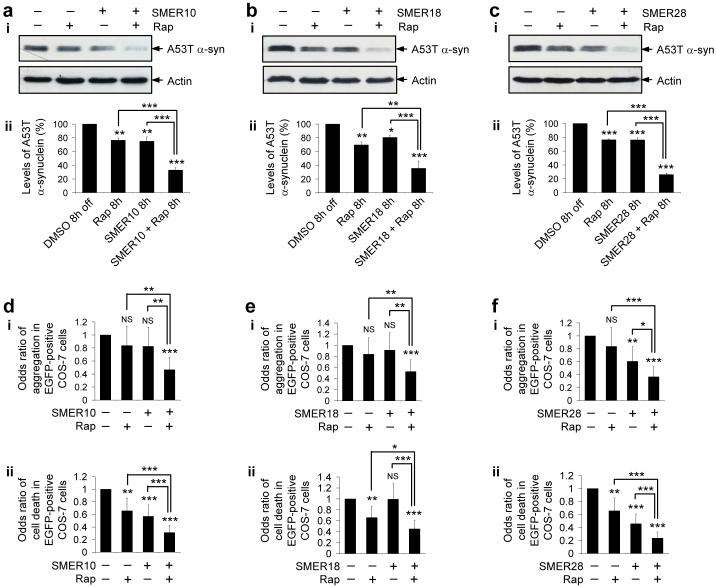
We next studied if these SMERs, which enhance rapamycin’s cytostatic effects in yeast, could also enhance the clearance of mutant proteins by rapamycin in mammalian cells. We assessed the clearance of A53T α-synuclein in stable PC12 cells at an early time-point of 8 h (instead of 24 h as shown earlier), where we do not see dramatic reductions of the levels of this autophagy substrate when the cells are treated with either of the compounds alone. SMERs 10, 18, 28 or rapamycin had small but significant effects even at this early time-point (Fig. 4a-c). Combined treatment of SMERs 10, 18 or 28 with rapamycin dramatically enhanced the effects of rapamycin (or the SMER) alone on A53T α-synuclein clearance (Fig. 4a-c). Here, we have used saturating doses of rapamycin6 to demonstrate the combined effects.
Figure 4. Rapamycin and SMERs have additive protective effects on the clearance and toxicity of mutant aggregate-prone proteins.
(a-c) Clearance of A53T α-synuclein (α-syn) in stable PC12 cells as in Fig. 1b, treated with DMSO (control), or with 0.2 μM rapamycin alone, SMER alone [140 μM SMER10 (a), 43 μM SMER18 (b) or 47 μM SMER28 (c)] or both for the 8 h switch-off period, was analysed by immunoblotting with antibody against HA (i) and densitometry analysis relative to actin (ii). The concentration of rapamycin is saturating for its effect on the clearance of A53T α-synuclein. Error bars denote S.E.M. p=0.0025 (rap), p=0.0018 (SMER10), p<0.0001 (SMER10+rap), p<0.0001 (rap or SMER10 vs SMER10+rap) (a); p=0.0069 (rap), p=0.0498 (SMER18), p<0.0001 (SMER18+rap), p=0.0038 (rap vs SMER18+rap), p=0.0007 (SMER18 vs SMER18+rap) (b); p<0.0001 (rap, SMER28, rap vs SMER28+rap, SMER28 vs SMER28+rap) (c).
(d-f) The percentage of EGFP-positive cells with EGFP-HDQ74 aggregates (i) and cell death (ii) in COS-7 cells as in Fig. 1c, treated with DMSO (control), or with 0.2 μM rapamycin alone, SMER alone [140 μM SMER10 (d), 43 μM SMER18 (e) or 47 μM SMER28 (f)] or both for 24 h, were expressed as odds ratios. Error bars: 95 % confidence interval. (i), For aggregation: p=0.248 (rap), p=0.217 (SMER10), p<0.0001 (SMER10+rap), p<0.001 (rap or SMER10 vs SMER10+rap) (d); p=0.248 (rap), p=0.543 (SMER18), p<0.0001 (SMER18+rap), p=0.008 (rap vs SMER18+rap), p=0.002 (SMER18 vs SMER18+rap) (e); p=0.248 (rap), p=0.002 (SMER28), p<0.0001 (SMER28+rap), p<0.0001 (rap vs SMER28+rap), p=0.012 (SMER28 vs SMER28+rap) (f). (ii), For cell death: p=0.002 (rap), p<0.0001 (SMER10, SMER10+rap, rap or SMER10 vs SMER10+rap) (d); p=0.002 (rap), p=0.948 (SMER18), p<0.0001 (SMER18+rap), p=0.015 (rap vs SMER18+rap), p<0.0001 (SMER18 vs SMER18+rap) (e); p=0.002 (rap), p<0.0001 (SMER28, SMER28+rap, rap or SMER28 vs SMER28+rap) (f). Note that we have treated cells for a shorter time in this experiment (24 h), compared to Fig. 1c (48 h) - this probably accounts for the failure of the protective trends of rapamycin and some of the SMERs to reach significance for aggregation, on their own in this experiment.
***, p<0.001; **, p<0.01; *, p<0.05; NS, Non-significant.
We further confirmed this enhanced protective effect due to dual treatment in mutant huntingtin-expressing COS-7 cells. Treatment with SMERs 10, 18 or 28 and rapamycin had an additive effect in reducing EGFP-HDQ74 aggregates and toxicity, compared to the single treatments of rapamycin or the SMERs (Fig. 4d-f).
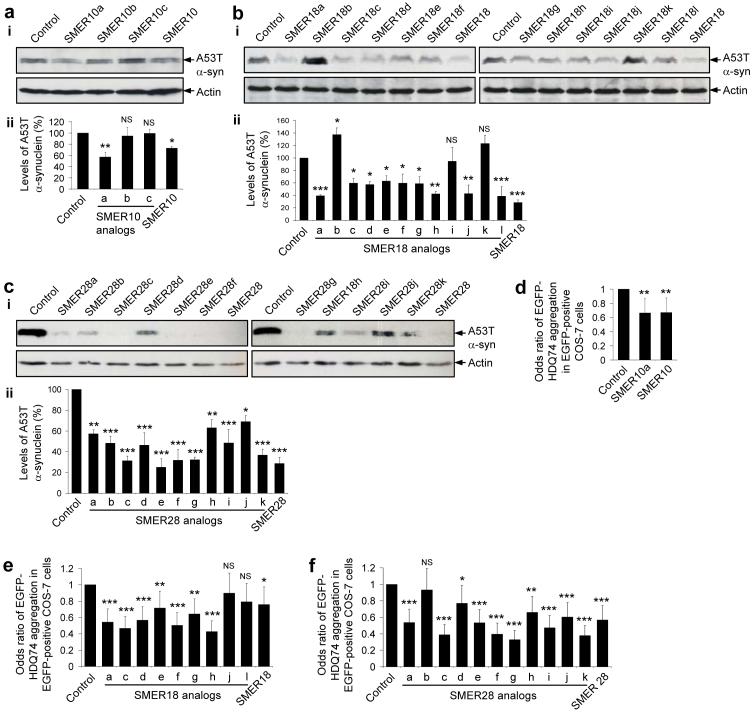
We next performed limited structure/activity relationship (SAR) analyses on SMERs 10, 18 and 28. We tested three commercially available structural analogs of SMER10 (40-42), twelve commercially available structural analogs of SMER18 (43-54), and we synthesized eleven structural analogs of SMER28 (55-64). Note that SMER28e is the same as the parent compound. We first screened for clearance of A53T α-synuclein in stable PC12 cells treated with these structural analogs alone. Structural analogs that retained the ability to induce clearance of A53T α-synuclein were then further tested for their ability to reduce mutant huntingtin aggregation in COS-7 cells. We found that one SMER10 analog (SMER10a), seven SMER18 analogs (SMER18a, c-h), and ten SMER28 analogs (SMER28a, c-k) both significantly enhanced the clearance of the A53T α-synuclein, and reduced huntingtin aggregation, though in most cases not significantly better than the parent compounds (Fig. 5a-f). Thus, we have identified further candidates having potential for therapeutic development.
Figure 5. Screen of chemical analogs of autophagy-inducing SMERs for their protective effects on the clearance and aggregation of mutant proteins.
(a-c) Clearance of A53T α-synuclein (α-syn) in stable PC12 cells as in Fig. 1b, treated for 24 h with either DMSO (control), or with 47 μM SMER10 and its analogs (SMER10a-c) (a), 43 μM SMER18 and its analogs (SMER18a-l) (b), or 47 μM SMER28 and its analogs (SMER28a-k) (c), was analysed by immunoblotting with anti-HA antibody (i) and densitometry analysis relative to actin (ii). All the analogs were used in the cell culture media at 1:400 dilution of 5 mg ml-1 stock solution (in DMSO). Error bars denote S.E.M. p=0.0058 (SMER10a), p=0.6736 (SMER10b), p=0.9507 (SMER10c), p=0.0481 (SMER10) (a); p=0.0006 (SMER18a), p=0.0249 (SMER18b), p=0.0167 (SMER18c), p=0.0117 (SMER18d), p=0.0269 (SMER18e), p=0.0165 (SMER18f), p=0.0148 (SMER18g), p=0.0011 (SMER18h), p=0.7369 (SMER18i), p=0.0012 (SMER18j), p=0.1531 (SMER18k), p=0.0006 (SMER18l), p=0.0001 (SMER18) (b); p=0.0014 (SMER28a), p=0.0002 (SMER28b), p=0.0001 (SMER28d), p=0.0048 (SMER28h), p=0.0002 (SMER28i), p=0.0162 (SMER28j), p<0.0001 (SMER28c, e-g, k, SMER28) (c).
(d-f) The percentage of EGFP-positive cells with EGFP-HDQ74 aggregates in COS-7 cells as in Fig. 1c, treated for 48 h with either DMSO (control), or with 47 μM SMER10 and its analog (SMER10a) (d), 43 μM SMER18 and its analogs (SMER18a, c-h, j, l) (e), or 47 μM SMER28 and its analogs (SMER28a-k) (f), were expressed as odds ratios. All the analogs were used in the cell culture media at 1:400 dilution of 5 mg ml-1 stock solution (in DMSO). Error bars: 95 % confidence interval. p=0.003 (SMER10a), p=0.004 (SMER10) (d); p<0.0001 (SMER18a, c, d, f, h), p=0.009 (SMER18e), p=0.001 (SMER18g), p=0.382 (SMER18j), p=0.067 (SMER18l), p=0.031 (SMER18) (e); p<0.0001 (SMER28a, c, e-g, i-k, SMER28), p=0.574 (SMER28b), p=0.041 (SMER28d), p=0.002 (SMER28h) (f).
***, p<0.001; **, p<0.01; *, p<0.05; NS, Non-significant.
SMER10 is an aminopyrimidone. The pyrimidone functionality of SMER10 is important for its autophagy-inducing activity, because substitution of a bulky phenyl group at the 2 position [SMER10b (41)], or creating a fused tetrazole [SMER10c (42)], nearly abolishes activity (Fig. 5a). However, removal of the amino group at the 3 position yielding hypoxanthine [SMER10a (40)] may slightly increase activity compared to the parent compound (Fig. 5a).
SMER18 is a vinylogous amide. The SMER18 analogs assess the tolerance of the two terminal aromatic rings to substitutions (Fig. 5b). For example, changing the hydroxyl group from the meta position either to the para [SMER18g (49)] or the ortho [SMER18f (48)] positions reduces but does not abolish activity; yet removing the hydroxyl group [SMER18i (51)] does abolish activity, suggesting its importance to activity (Fig. 5b).
SMER28 is a bromo-substituted quinazoline. A similar SAR pattern emerges here: the majority of substitutions are well tolerated individually, multiple concurrent substitutions fare worse, and none of the analogs are significantly more potent than the parent compound (Fig. 5c). Interestingly, two analogs of SMER28 [SMER28h (63) and i (64)] that have been functionalized in a way suitable for affinity chromatography are also active.
In conclusion, we have developed a high-throughput screening strategy for identifying small-molecule modulators of mammalian autophagy, as SMERs in yeast appear to act as autophagy inducers on their own in mammalian cells. Autophagy inducers may have value for a range of both neurodegenerative and infectious diseases. Autophagy inhibitors may also have use in the treatment of certain cancers.
Materials and methods
See Supplementary Methods online for details of yeast strains and media, primary screen, dose responses and selectivity profiling, characterisations of SMERs and SMIRs in yeast, plasmid constructs, mammalian cell culture and transfection, microscopy, detailed statistical analysis (for counting aggregation, cell death, EGFP-LC3 vesicles and densitometry on immunoblots), toxicity analyses and synthesis of SMER28 analogs.
Mammalian cell lines
Cell lines used were COS-7; HeLa; stable HeLa cells expressing EGFP-LC319 or UbG76V-EGFP degron25; wild-type ATG5+/+ or ATG5−/− MEFs15; inducible PC12 stable cell line expressing HA-tagged A53T α-synuclein mutant5. For details of cell culture and transfection, see Supplementary Methods online.
Quantification of aggregate formation and cell death
Approximately 200 EGFP-positive cells were counted by fluorescence microscope for the proportion of cells with EGFP-HDQ74 aggregates, as described previously3, 6, 8, 27. Nuclei were stained with DAPI and those showing apoptotic morphology were considered abnormal. Experiments were done in triplicate and with the scorer blinded to treatment.
Western Blot Analysis
Cell pellets were lysed on ice in Laemmli buffer (62.5 mM Tris-HCl pH 6.8, 5 % β-mercaptoethanol, 10 % glycerol, and 0.01 % bromophenol blue) for 30 min in presence of protease inhibitors (Roche Diagnostics). Primary antibodies include anti-EGFP (8362-1, Clontech), anti-HA (12CA5, Covance), anti-mTOR (2972), anti-Phospho-mTOR (Ser2448) (2971), anti-p70 S6 Kinase (9202), anti Phospho-p70 S6 Kinase (Thr389) (9206), anti-4E-BP1 (9452), anti-Phospho-4E-BP1 (Thr37/46) (9459) (all from Cell Signaling Technology), anti-Beclin-1 (3738, Cell Signaling), anti-Atg5 (ab19130, Abcam), anti-Atg7 (600-401-487, Rockland), anti-Atg12 (36-6400, Zymed Laboratories), anti-actin (A2066, Sigma). Blots were probed with anti-mouse or anti-rabbit IgG-HRP and visualised using ECL detection kit (Amersham).
Clearance of mutant huntingtin and α-synucleins
Stable inducible PC12 cell line expressing A53T α-synuclein mutant was induced with 1 μg/ml doxycycline (Sigma) for 48 h and the transgene expression was switched off by removing doxycycline from medium5. Cells were treated with or without compounds for time-points as indicated in experiments. Clearance of A53T α-synuclein was measured by immunoblotting with antibody against HA respectively and densitometry analysis relative to actin.
Drosophila methods
Fly culture and crosses were carried out at 25°C and at 70 % humidity, using Instant Fly Food (Philip Harris, Ashby de la Zouch, UK) unless otherwise stated. Flies were raised with a 12 h light: 12 h dark cycle. Aliquots of SMERs in DMSO, or DMSO alone, were added to the water that was used to prepare the instant fly food.
Virgin female flies of genotype y w; gmr-httNterm(1-171)Q120 (gmrQ120)22 were mated with isogenised w1118 males28 in food vials for 48 h. Flies were then transferred to vials containing instant fly food containing either SMERs in DMSO or DMSO alone. Progeny were collected 0-4 h after eclosion, kept on food of the same composition as they had been reared on, and scored for photoreceptor degeneration using the pseudopupil technique29 two days after eclosion.
Statistical analysis
Pooled estimates for the changes in aggregate formation or cell death, resulting from perturbations assessed in multiple experiments, were calculated as odds ratios with 95 % confidence intervals. Odds ratios and p values were determined by unconditional logistical regression analysis, using the general log-linear analysis option of SPSS 9 software (SPSS, Chicago). Densitometry analysis on the immunoblots was done by Scion Image Beta 4.02 software (Scion Corporation) from three independent experiments (n=3). Significance for the clearance of mutant proteins was determined by factorial ANOVA test using STATVIEW software, version 4.53 (Abacus Concepts), where the control condition was set to 100 %. The y-axis values are shown in percentage (%) and the error bars denote standard error of mean (S.E.M.). ***, p<0.001; **, p<0.01; *, p<0.05; NS, Non-significant. For further details, see Supplementary Methods online.
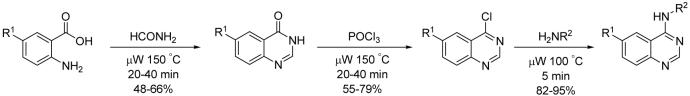
SMER 28 structural analog synthesis
The substituted quinazolinone was generated by reaction of an anthranilic acid with formamide in a microwave assisted Neimentowski reaction30. Treatment of the quinazolinone with phosphorus oxychloride gave the chloroquinazoline in high yield. The chloroquinazoline was then treated with a variety of primary amines to give the final aminoquinazolines. See Supplementary Methods online for further details.
Supplementary Material
Acknowledgements
We thank T. Yoshimori (National Institute of Genetics) for EGFP-LC3 construct, N. Mizushima (Tokyo Metropolitan Institute of Medical Science) for Atg5 and HA-Atg12 constructs, and wild-type and Atg5-deficient MEFs, A.M. Tolkovsky (University of Cambridge) for EGFP-LC3 HeLa stable cell line and N.P. Dantuma (Karolinska Institutet) for UbG76V-EGFP degron HeLa stable cell line. We thank the staff of the Broad Institute of Chemical Biology Program (formerly the Institute for Chemistry and Chemical Biology), B. Ravikumar, A. Williams, L. Jahreiss and R. Walker (University of Cambridge) for technical assistance; S. Haggarty for comments and discussion. This work was supported in part with federal funds from the NCI’s Initiative for Chemical Genetics, NIH, under Contract No. N01-CO-12400. We are grateful for Wellcome Trust Senior Fellowship in Clinical Science (D.C.R.), an MRC programme grant and E.U. Framework VI (EUROSCA) (D.C.R.) and the National Institute of General Medicine Sciences GM3862-7 (S.L.S.) for additional funding. S.L.S. is an Investigator at the Howard Hughes Medical Institute.
Footnotes
Competing financial interests
The authors declare no competing financial interests.
References
- 1.Klionsky DJ, Emr SD. Autophagy as a regulated pathway of cellular degradation. Science. 2000;290:1717–1721. doi: 10.1126/science.290.5497.1717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rubinsztein DC, Gestwicki JE, Murphy LO, Klionsky DJ. Potential therapeutic applications of autophagy. Nat. Rev. Drug Discov. 2007;6:304–312. doi: 10.1038/nrd2272. [DOI] [PubMed] [Google Scholar]
- 3.Ravikumar B, Duden R, Rubinsztein DC. Aggregate-prone proteins with polyglutamine and polyalanine expansions are degraded by autophagy. Hum. Mol. Genet. 2002;11:1107–1117. doi: 10.1093/hmg/11.9.1107. [DOI] [PubMed] [Google Scholar]
- 4.Ravikumar B, et al. Inhibition of mTOR induces autophagy and reduces toxicity of polyglutamine expansions in fly and mouse models of Huntington disease. Nat. Genet. 2004;36:585–595. doi: 10.1038/ng1362. [DOI] [PubMed] [Google Scholar]
- 5.Webb JL, Ravikumar B, Atkins J, Skepper JN, Rubinsztein DC. Alpha-Synuclein is degraded by both autophagy and the proteasome. J. Biol. Chem. 2003;278:25009–25013. doi: 10.1074/jbc.M300227200. [DOI] [PubMed] [Google Scholar]
- 6.Sarkar S, et al. Lithium induces autophagy by inhibiting inositol monophosphatase. J. Cell Biol. 2005;170:1101–1111. doi: 10.1083/jcb.200504035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rubinsztein DC. Lessons from animal models of Huntington’s disease. Trends Genet. 2002;18:202–209. doi: 10.1016/s0168-9525(01)02625-7. [DOI] [PubMed] [Google Scholar]
- 8.Sarkar S, Davies JE, Huang Z, Tunnacliffe A, Rubinsztein DC. Trehalose, a novel mTOR-independent autophagy enhancer,accelerates the clearance of mutant huntingtin and alpha-synuclein. J. Biol. Chem. 2007;282:5641–5652. doi: 10.1074/jbc.M609532200. [DOI] [PubMed] [Google Scholar]
- 9.Berger Z, et al. Rapamycin alleviates toxicity of different aggregate-prone proteins. Hum. Mol. Genet. 2006;15:433–442. doi: 10.1093/hmg/ddi458. [DOI] [PubMed] [Google Scholar]
- 10.Gutierrez MG, et al. Autophagy is a defense mechanism inhibiting BCG and Mycobacterium tuberculosis survival in infected macrophages. Cell. 2004;119:753–766. doi: 10.1016/j.cell.2004.11.038. [DOI] [PubMed] [Google Scholar]
- 11.Nakagawa I, et al. Autophagy defends cells against invading group A Streptococcus. Science. 2004;306:1037–1040. doi: 10.1126/science.1103966. [DOI] [PubMed] [Google Scholar]
- 12.Talloczy Z, Virgin HWT, Levine B. PKR-Dependent Autophagic Degradation of Herpes Simplex Virus Type 1. Autophagy. 2006;2:24–29. doi: 10.4161/auto.2176. [DOI] [PubMed] [Google Scholar]
- 13.Sarbassov DD, Ali SM, Sabatini DM. Growing roles for the mTOR pathway. Curr. Opin. Cell Biol. 2005;17:596–603. doi: 10.1016/j.ceb.2005.09.009. [DOI] [PubMed] [Google Scholar]
- 14.Huang J, et al. Finding new components of the target of rapamycin (TOR) signaling network through chemical genetics and proteome chips. Proc. Natl. Acad. Sci. USA. 2004;101:16594–16599. doi: 10.1073/pnas.0407117101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mizushima N, et al. Dissection of autophagosome formation using Apg5-deficient mouse embryonic stem cells. J. Cell Biol. 2001;152:657–668. doi: 10.1083/jcb.152.4.657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fenteany G, et al. Inhibition of proteasome activities and subunit-specific amino-terminal threonine modification by lactacystin. Science. 1995;268:726–731. doi: 10.1126/science.7732382. [DOI] [PubMed] [Google Scholar]
- 17.Kabeya Y, et al. LC3, a mammalian homologue of yeast Apg8p, is localized in autophagosome membranes after processing. EMBO J. 2000;19:5720–5728. doi: 10.1093/emboj/19.21.5720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mizushima N. Methods for monitoring autophagy. Int. J. Biochem. Cell Biol. 2004;36:2491–2502. doi: 10.1016/j.biocel.2004.02.005. [DOI] [PubMed] [Google Scholar]
- 19.Bampton ETW, Goemans CG, Niranjan D, Mizushima N, Tolkovsky AM. The dynamics of autophagy visualised in live cells: from autophagosome formation to fusion with endo/lysosomes. Autophagy. 2005;1:23–36. doi: 10.4161/auto.1.1.1495. [DOI] [PubMed] [Google Scholar]
- 20.Yamamoto A, et al. Bafilomycin A1 prevents maturation of autophagic vacuoles by inhibiting fusion between autophagosomes and lysosomes in rat hepatoma cell line, H-4-II-E cells. Cell Struct. Funct. 1998;23:33–42. doi: 10.1247/csf.23.33. [DOI] [PubMed] [Google Scholar]
- 21.Ekstrom P, Kanje M. Inhibition of fast axonal transport by erythro-9-[3-(2-hydroxynonyl)]adenine. J. Neurochem. 1984;43:1342–1345. doi: 10.1111/j.1471-4159.1984.tb05392.x. [DOI] [PubMed] [Google Scholar]
- 22.Jackson GR, et al. Polyglutamine-expanded human huntingtin transgenes induce degeneration of Drosophila photoreceptor neurons. Neuron. 1998;21:633–642. doi: 10.1016/s0896-6273(00)80573-5. [DOI] [PubMed] [Google Scholar]
- 23.Marsh JL, Thompson LM. Drosophila in the study of neurodegenerative disease. Neuron. 2006;52:169–178. doi: 10.1016/j.neuron.2006.09.025. [DOI] [PubMed] [Google Scholar]
- 24.Schmelzle T, Hall MN. TOR, a central controller of cell growth. Cell. 2000;103:253–262. doi: 10.1016/s0092-8674(00)00117-3. [DOI] [PubMed] [Google Scholar]
- 25.Dantuma NP, Lindsten K, Glas R, Jellne M, Masucci MG. Short-lived green fluorescent proteins for quantifying ubiquitin/proteasome-dependent proteolysis in living cells. Nat. Biotechnol. 2000;18:538–543. doi: 10.1038/75406. [DOI] [PubMed] [Google Scholar]
- 26.Iwata A, Riley BE, Johnston JA, Kopito RR. HDAC6 and microtubules are required for autophagic degradation of aggregated huntingtin. J. Biol. Chem. 2005;280:40282–40292. doi: 10.1074/jbc.M508786200. [DOI] [PubMed] [Google Scholar]
- 27.Narain Y, Wyttenbach A, Rankin J, Furlong RA, Rubinsztein DC. A molecular investigation of true dominance in Huntington’s disease. J. Med. Genet. 1999;36:739–746. doi: 10.1136/jmg.36.10.739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ryder E, et al. The DrosDel collection: a set of P-element insertions for generating custom chromosomal aberrations in Drosophila melanogaster. Genetics. 2004;167:797–813. doi: 10.1534/genetics.104.026658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Franceschini N. Pupil and pseudopupil in the compound eye of Drosophila. In: Wehner R, editor. Information processing in the visual system of Drosophila. Springer; Berlin: 1972. pp. 75–82. [Google Scholar]
- 30.Alexandre F-R, Berecibar A, Wrigglesworth R, Besson T. Novel series of 8H-quinazolino[4,3-b]quinazolin-8-ones via two Niementowski condensations. Tetrahedron. 2003;59:1413–1419. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.