Abstract
Autophagy is a major clearance route for intracellular aggregate-prone proteins causing diseases like Huntington’s disease. Autophagy induction with the mTOR inhibitor, rapamycin, accelerates clearance of these toxic substrates. As rapamycin has non-trivial side effects, we screened FDA-approved drugs to identify novel autophagy-inducing pathways. We found that L-type Ca2+ channel antagonists, the K+ATP channel opener minoxidil, and the Gi signaling activator clonidine, induce autophagy. These drugs revealed a cyclical mTOR-independent pathway regulating autophagy, where cAMP regulates IP3 levels, influencing calpain activity, which completes the cycle by cleaving and activating Gsα, which regulates cAMP levels. This pathway has numerous potential points where autophagy can be induced and we provide proof-of-principle for therapeutic relevance in Huntington’s disease using mammalian cell, fly and zebrafish models. Our data also suggest that insults that elevate intracytosolic Ca2+, like excitotoxicity, will inhibit autophagy, thus retarding clearance of aggregate-prone proteins.
Introduction
The autophagy-lysosomal and ubiquitin-proteasome pathways are major routes for protein and organelle clearance in eukaryotic cells. While the narrow pore of the proteasome barrel precludes clearance of large membrane proteins and protein complexes (including oligomers and aggregates), mammalian lysosomes can degrade protein complexes and organelles by macroautophagy, generally referred to as autophagy1. It involves the formation of double membrane structures called autophagosomes around a portion of cytosol. These fuse with lysosomes where their contents are degraded. Autophagy can be induced by several conditions, including starvation, and is regulated by a number of protein kinases, the best characterised being the mammalian target of rapamycin (mTOR)2.
Autophagy induction may represent a tractable therapeutic strategy for neurodegenerative disorders caused by aggregate-prone intracytosolic proteins, including Huntington’s disease (HD), an autosomal-dominant neurodegenerative disorder caused by a CAG trinucleotide repeat expansion (>35 repeats), which encodes an abnormally long polyglutamine (polyQ) tract in the N-terminus of the huntingtin protein1, 3. Mutant huntingtin toxicity is thought to be exposed after it is cleaved to form N-terminal fragments comprising the first 100-150 residues with the expanded polyQ tract, which are also the toxic species found in aggregates/inclusions3. Thus, HD pathogenesis is frequently modelled with exon 1 fragments containing expanded polyQ repeats which cause aggregate formation and toxicity in cell models and in vivo3 (Supplementary Fig. 1a online). The polyQ mutation in HD raises intracellular calcium (Ca2+) levels resulting in enhanced calpain activity, and this has been proposed as an important disease mechanism4. One possibility is that calpains enhance mutant huntingtin cleavage/processing5-7. However, the calpain cleavage sites are outside the exon 1 huntingtin fragment, so calpain cleavage of mutant huntingtin will not be a factor influencing the aggregation/toxicity in such N-terminal fragment disease models (Supplementary Fig. 1a online).
In addition to mutant huntingtin, autophagy also regulates the clearance of other aggregate-prone disease-causing proteins, like those causing spinocerebellar ataxias types 1 and 3, forms of tau (causing fronto-temporal dementias) and the A53T and A30P α-synuclein mutants [which cause familial Parkinson’s disease (PD)]8-15. Autophagy induction reduces both soluble mutant huntingtin levels and inclusion frequencies, and attenuates its toxicity in cell, Drosophila and mouse models of HD8-12. Autophagy induction may also be a valuable strategy in the treatment of infectious diseases, including tuberculosis and may protect against cell death in certain contexts16-18.
Currently, the only suitable pharmacological strategy for upregulating autophagy in mammalian brains is to use rapamycin (1), which inhibits mTOR9. Also, since rapamycin is an immunosuppressant, it is contra-indicated for use in diseases like tuberculosis. The mechanism by which mTOR regulates autophagy remains unclear and this kinase controls several cellular processes besides autophagy, probably contributing to the complications seen with its long-term use19. Thus, we sought to identify novel pathways and therapeutic agents that enhance autophagy. We found that L-type Ca2+ channel antagonists, a K+ATP channel opener, and Gi signaling activators, induce autophagy. These drugs revealed a cyclical mTOR-independent pathway regulating autophagy, where cAMP (2) regulates inositol 1,4,5-trisphosphate (IP3) (3) levels, influencing calpain activity, which completes the cycle by cleaving and activating Gsα, which regulates cAMP levels. This pathway has numerous potential points where autophagy can be induced and we provide proof-of-principle for therapeutic relevance in Huntington’s disease using cell, fly and zebrafish models.
Results
Screen for autophagy enhancers
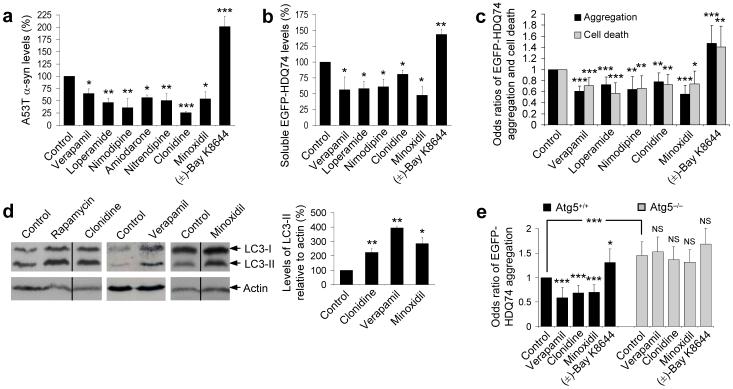
We screened for autophagy enhancers using a library of 253 compounds that had previously been into man without major toxic side effects, and pharmacological probes (see Materials and Methods). Our primary screen assayed clearance of A30P α-synuclein, a known autophagy substrate, in stable inducible PC12 cells14, 20. All compounds that visibly altered A30P α-synuclein clearance were retested in multiple experiments in similar PC12 cells lines expressing A53T α-synuclein and were successfully validated. A53T α-synuclein clearance was enhanced by compounds including known autophagy inducers like rapamycin and valproate11, 14 (4) (data not shown) and the following hits: 5 drugs that antagonise L-type Ca2+ channel activity [verapamil (5), loperamide (6), nimodipine (7), nitrendipine (8) and amiodarone (9)], minoxidil (10) (an ATP-sensitive K+ channel agonist) and clonidine (11) (binds to α2-adrenergic and type I imidazoline receptors and activates Gi-protein signalling pathways) (Fig. 1a and Supplementary Fig. 2a online). (±)-Bay K8644 (12) (an L-type Ca2+ channel agonist21) retarded A53T α-synuclein clearance (Fig. 1a and Supplementary Figs. 2a, b online). Supplementary Fig. 1b online summarises characteristics of screen hits and other compounds used in the paper.
Figure 1. Identification of autophagy-inducing drugs.
a, Densitometric analysis relative to actin of A53T α-synuclein clearance in stable inducible PC12 cell line expressing A53T α-synuclein. Transgene expression was induced with doxycycline for 48 h, and then switched off (by removing doxycycline) with drug (all 1 μM) or DMSO (vehicle control) treatment for 24 h. Control condition is set to 100%. Error bars: standard error of mean.
b, Densitometric analysis relative to actin of soluble EGFP-HDQ74 clearance in stable inducible PC12 cell line expressing EGFP-HDQ74. Transgene expression was induced with doxycycline for 8 h, and then switched off (by removing doxycycline) with drug (all 1 μM) or DMSO (vehicle control) treatment for 96 h. Control condition is set to 100%. Error bars: standard error of mean.
c, SK-N-SH cells transfected with EGFP-HDQ74 construct for 4 h were treated with drugs (all 1 μM) or DMSO (vehicle control) for 48 h post-transfection. The proportions of EGFP-positive cells with aggregates or cell death were expressed as odds ratios and the control was taken as 1. Error bars: 95% confidence interval.
d, PC12 cells were treated with clonidine, minoxidil and verapamil (all 1 μM) or DMSO (vehicle control) for 24 h. Endogenous LC3-II levels were detected with anti-LC3 antibody and quantified relative to actin. Rapamycin (0.2 μM) was positive control. Error bars: standard error of mean.
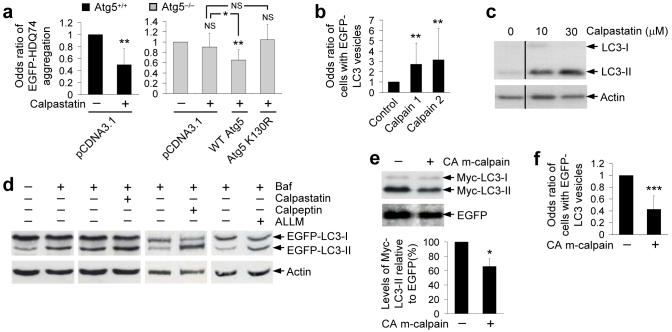
e, The proportions of EGFP-positive cells with EGFP-HDQ74 aggregates in wild-type (Atg5+/+) and knockout (ATG5−/−) Atg5 mouse embryonic fibroblasts, transfected with EGFP-HDQ74 for 4 h and then treated for 48 h with drugs (all 1 μM) or DMSO (vehicle control). Error bars: 95% confidence interval. ***, p<0.001; **, p<0.01; *, p<0.05; NS, Non-significant.
We prioritised our validation studies on three L-type Ca2+ channel antagonists that act at different sites on these channels (verapamil, loperamide and nimodipine22), minoxidil and clonidine. All of these compounds enhanced clearance of soluble mutant huntingtin exon 1 with 74Q (EGFP-HDQ74) in stable PC12 cells (which show no toxicity at these time points) and reduced its aggregation and toxicity in SK-N-SH (neuroblastoma) cells, whereas (±)-Bay K8644 had opposite effects (Figs. 1b, c and Supplementary Fig. 2c online).
Clonidine, minoxidil and verapamil increase autophagy
We assessed autophagosome numbers using the microtubule-associated protein 1 light chain 3 (LC3)23. LC3 is processed post-translationally into LC3-I, then converted to LC3-II, the only known protein that specifically associates with autophagosome membranes24. LC3-positive vesicle numbers or LC3-II levels (versus actin) correlate with autophagosome numbers23. LC3-II levels were increased by clonidine, minoxidil and a representative Ca2+ channel antagonist, verapamil, suggesting enhanced autophagy (Fig. 1d and Supplementary Fig. 2d online).
We confirmed that these drugs and (±)-Bay K8644 influenced huntingtin aggregation in an autophagy-dependent manner, as they had no effects in autophagy-deficient mouse embryonic fibroblasts (MEFs) that have a knockout of the key autophagy gene Atg5 (ATG5−/−), while their effects in wild-type MEFs (Atg5+/+) mirrored that in SK-N-SH cells (Fig. 1e).
Clonidine signals via the imidazoline type 1 receptor
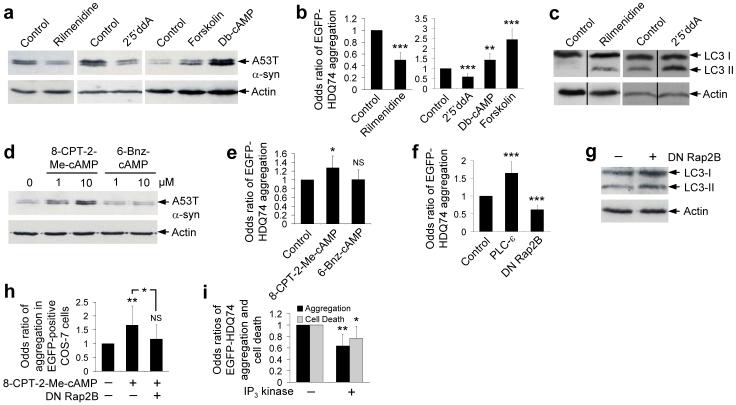
Clonidine binds both α2-adrenergic (α2-AR) and imidazoline-1 (I1R) receptors. When clonidine binds α2-AR, Gi signalling pathways are activated that reduce cAMP levels by inhibiting adenylyl cyclase25. I1R binding also reduces cAMP levels, although whether this is mediated by G-proteins is disputed26, 27 (Supplementary Fig. 3a online). As clonidine had beneficial effects in both PC12 cells (which do not have α2-AR but have I1R) and SK-N-SH cells (which have both these receptors), we confirmed that rilmenidine (13) (a clinically-approved centrally-acting drug which binds I1R at ∼30-fold higher affinity than the α2-AR) could also enhance A53T α-synuclein clearance and decrease EGFP-HDQ74 aggregation in PC12 and SK-N-SH cells, respectively (Fig. 2a, b and Supplementary Fig. 3b online). Rilmenidine, like clonidine and verapamil, also increased LC3-II levels in a dose-dependent manner (Fig. 2c and Supplementary Figs. 2d, 3c online).
Figure 2. Imidazoline-1 receptor agonists act through cAMP to Rap2B and PLC-ε to modulate autophagy.
a, A53T α-synuclein clearance in stable PC12 cells as in Fig. 1a, treated with or without 1 μM rilmenidine, 1 mM dibutyl-cAMP (db-cAMP), 24 μM forskolin or 500 μM 2′5′dideoxyadenosine (2′5′ddA) for 24 h. Note that control clearance levels for a given substrate may vary from figure to figure, due to different exposures of blots, which aid illustration of agents that either increase or retard clearance.
b, The proportions of EGFP-positive cells with EGFP-HDQ74 aggregates in SK-N-SH (for rilmenidine) and COS-7 (for db-cAMP, forskolin, 2′5′ddA) as in Fig. 1c, treated with concentrations used in Fig. 2a for 48 h post-transfection. Error bars: 95% confidence interval.
c, Endogenous LC3-II levels in PC12 cells treated with 1 μM rilmenidine or 500 μM 2′5′ddA for 24 h.
d, A53T α-synuclein clearance in stable PC12 cells as in Fig. 1a, treated with or without 8-CPT-2Me-cAMP or 6-Bnz-cAMP (1 μM or 10 μM).
e, The proportions of EGFP-positive COS-7 cells with aggregates as in Fig. 1c, treated with or without 10 μM 8-CPT-2Me-cAMP or 6-Bnz-cAMP for 48 h post-transfection. Error bars: 95% confidence interval.
f, COS-7 cells, transfected for 4 h with EGFP-HDQ74 along with empty vector (pcDNA3.1), dominant-negative S17N Rap2B (DN Rap2B) or wild-type PLC-ε (1:3 ratio), were assessed for the proportions of EGFP-positive cells with aggregates after 48 h. Error bars: 95% confidence interval.
g, Endogenous LC3-II levels in COS-7 cells transfected with a dominant-negative Rap2B (DN Rap2B) for 4 h were assessed after 48 h expression.
h, The proportions of EGFP-positive COS-7 cells with aggregates, transfected with EGFP-HDQ74 along with empty vector (pcDNA3.1) or dominant-negative Rap2B (1:3 ratio) for 4 h, were treated with or without 10 μM 8-CPT-2Me-cAMP for 48 h. Error bars: 95% confidence interval.
i, COS-7 cells, transfected with EGFP-HDQ74 and dsRed2 or cytosolic dsRed2-IP3 kinase A (1:3 ratio) for 4 h, were assessed at 48 h post-transfection for aggregation and cell death (as in Fig. 1c). Error bars: 95% confidence interval. ***, p<0.001; **, p<0.01; *, p<0.05; NS, Non-significant.
I1R agonists signal via cAMP/Epac/Rap2B/PLC
Clonidine and rilmenidine are likely to act by reducing cAMP levels, as the adenylyl cyclase inhibitor 2′5′-dideoxyadenosine (2′5′ddA) (14) also enhanced A53T α-synuclein clearance and decreased EGFP-HDQ74 aggregation, while the cAMP analogue (dibutyryl cAMP) (15) and the adenylyl cyclase activator (forskolin) (16) had opposite effects (Fig. 2a, b and Supplementary Figs. 3a, b online). Furthermore, LC3-II levels were increased by 2′5′ddA (Fig. 2c and Supplementary Fig. 3c online).
cAMP can signal to at least three different pathways; protein kinase A (PKA), Epac (a guanine nucleotide exchange factor) and through cyclic nucleotide activated ion channels (Supplementary Fig. 3a online)28. As the latter route does not operate at the cAMP levels expected by some of our treatments28, we tested the former options. The Epac-specific cAMP analogue, 8-CPT-2-Me-cAMP (17), slowed A53T α-synuclein clearance and enhanced mutant huntingtin aggregation, while the PKA-specific cAMP analogue, 6-Bnz-cAMP (18), had no effect (Figs. 2d, e and Supplementary Fig. 3d online). Likewise, KT 5720 (19), a PKA inhibitor, did not enhance A53T α-synuclein clearance - it had the reverse effect (Supplementary Fig. 3e online). Henceforth, we focused on the Epac pathway and tested its downstream components.
Epac activates the small GTPase Rap2B, which in turn activates the ubiquitously expressed phospholipase C (PLC)-ε isoform29, which hydrolyses phosphatidylinositol 4,5-bisphosphate (PIP2) (20) to form IP3 and diacylglycerol (DAG) (21) (Supplementary Fig. 3a online). This pathway is well conserved in many cell types including neurons, neuronal cells like PC12 cells, and COS-7 cells30-33. We have confirmed Rap2B activation in PC12 cells treated with forskolin (Supplementary Fig. 3f online). This established pathway from Gi-coupled receptors to PLC-ε was a strong candidate autophagy regulator, as IP3 negatively regulates autophagy in an mTOR-independent fashion and a recent study has shown induction of autophagy after IP3 receptor (IP3R) knockdown11, 34, 35. Consistent with this hypothesis, dominant-negative S17N Rap2B decreased EGFP-HDQ74 aggregation, while overexpression of PLC-ε enhanced EGFP-HDQ74 aggregation (Fig. 2f). The protective effects of dominant-negative Rap2B on EGFP-HDQ74 aggregation were associated with an increase in LC3-II levels (Fig. 2g and Supplementary Fig. 3g online). Consistent with a link between Epac and Rap2B, the deleterious effects of the Epac-specific cAMP analogue (8-CPT-2-Me-cAMP) on huntingtin aggregation, were abrogated by dominant-negative Rap2B (Fig. 2h and Supplementary Fig. 3h online). Furthermore, overexpression of cytosolic IP3 kinase A36, which catalyses the Ca2+-regulated phosphorylation of IP3, thereby inactivating the signal for ER Ca2+ release and generating inositol 1,3,4,5-tetrakisphosphate (IP4) (22), decreased EGFP-HDQ74 aggregation and toxicity (Fig. 2i).
PLC/IP3 and L-type Ca2+ channels regulate calpain activity
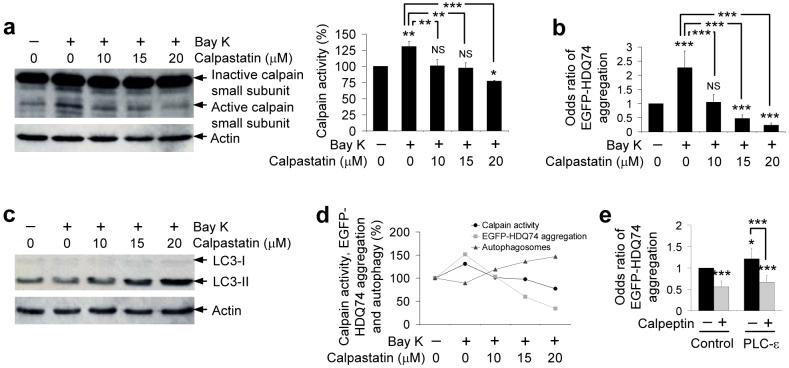
The pathway from cAMP to IP3 regulates intracytosolic Ca2+ levels, as IP3 binds to ER receptors facilitating Ca2+ release from this organelle. Likewise, L-type Ca2+ channel antagonists are known to decrease intracytosolic Ca2+ and L-type Ca2+ channel agonists increase intracytosolic Ca2+ concentrations (Supplementary Figs. 3a, i online). Intracytosolic Ca2+ activates calpains37. The two ubiquitously expressed mammalian calpains, calpain 1 (μ-calpain) and calpain 2 (m-calpain), are heterodimeric proteins comprising a distinct 80 kDa large catalytic subunit and a common 28 kDa small regulatory subunit, which is converted from the 28 kDa (inactive) form to a 21 kDa polypeptide (active) following activation by increased cytosolic Ca2+ [e.g., with thapsigargin (23) (Supplementary Fig. 3j online)]37. PLC-ε overexpression, and treatments with (±)-Bay K8644 and 8-CPT-2-Me-cAMP also increased calpain activity (Fig. 3a and Supplementary Figs. 3k, l online).
Figure 3. The effects of L-type Ca2+ channel agonist on autophagy and mutant huntingtin aggregation are abrogated by calpain inhibition.
a, SK-N-SH cells were pre-treated with or without 10, 15 or 20 μM calpastatin for 15 min followed by addition of 1 μM (±)-Bay K8644 for 4 h. Calpain activity was detected by immunoblotting with anti-calpain small subunit antibody. Densitometry analysis is relative to actin. Error bars: Standard error of mean.
b, SK-N-SH cells transfected with EGFP-HDQ74 for 4 h, then pre-treated with or without 10, 15 or 20 μM calpastatin for 15 min followed by addition of 1 μM (±)-Bay K8644 for 48 h post-transfection, were assessed for the proportion of EGFP-positive cells with EGFP-HDQ74 aggregates. Error bars: 95% confidence interval.
c, SK-N-SH cells were pre-treated with or without 10, 15 or 20 μM calpastatin for 15 min followed by addition of 1 μM (±)-Bay K8644 for 4 h. Endogenous LC3-II levels were detected by immunoblotting with anti-LC3 antibody.
d, Comparison between calpain activity, EGFP-HDQ74 aggregation and autophagosomes as seen in Fig. 3a-c. The control condition for all the assessments was set at 100%. Calpastatin lowered the increased calpain activity and EGFP-HDQ74 aggregation caused by (±)-Bay K8644 in a dose-dependent manner with a simultaneous increase in LC3-II levels (autophagosomes).
e, COS-7 cells transfected with empty vector (pcDNA3.1) or wild-type PLC-ε for 4 h and treated with or without 50 μM calpeptin for 48 h post-transfection were assessed for the proportion of EGFP-positive cells with EGFP-HDQ74 aggregates. Error bars: 95% confidence interval. ***, p<0.001; **, p<0.01; *, p<0.05; NS, Non-significant.
Calpain activity is enhanced in HD in vivo4 and we confirmed that this elevation could be normalised by calpastatin or verapamil in our stable inducible cell models (Supplementary Fig. 3m online). Very low calpain activity in the basal state makes reliable assessment of inhibitory effects even of specific calpain inhibitors difficult in whole cells with normal intracellular Ca2+ concentrations.
Minoxidil decreases whole-cell L-type Ca2+ channel currents in a concentration-dependent manner38. In contrast to minoxidil, K+ATP channel blockers, such as quinine sulphate (24) and tolazamide (25), slow clearance of autophagy substrates (Supplementary Figs. 2e, f online). Because minoxidil is likely to act via the same pathway as the L-type Ca2+ channel blockers, we have restricted further pathway analyses to the Ca2+ channel blockers.
Calpain inhibition reduces mutant huntingtin aggregation
We tested calpains as central to autophagy regulation as they are common targets of the two major pathways identified by our screen hits - the cAMP-Epac-PLC-IP3 (clonidine) and the L-type Ca2+ channel pathways. This possibility was consistent with the calpain inhibitor calpastatin inhibiting the levels of calpain activation mediated by (±)-Bay K8644 in a dose-dependent manner (Fig. 3a). Both calpastatin and verapamil (which also abrogates calpain activation) rescued the effects of (±)-Bay K8644 on EGFP-HDQ74 aggregation (Fig. 3b and Supplementary Figs. 3n, o online). Likewise, calpastatin enhanced LC3-II levels in the presence of (±)-Bay K8644 in a manner that correlated with the decrease in EGFP-HDQ74 aggregation and calpain activity (Figs. 3c, d). Furthermore, calpeptin (26), another calpain inhibitor, rescued the increased EGFP-HDQ74 aggregation induced by overexpression of PLC-ε (Fig. 3e).
It is important to point out that 10 μM calpastatin reduced calpain activation and EGFP-HDQ74 aggregation, and increased autophagic activity to around baseline levels (as in DMSO treated control condition) in (±)-Bay K8644-treated cells (Figs. 3a-d). However, 20 μM calpastatin resulted in lower calpain activity and EGFP-HDQ74 aggregation, and simultaneously increased autophagy in (±)-Bay K8644-treated cells, even compared to DMSO-treated (control) cells (Figs. 3a-d). Thus, this places calpain downstream of L-type Ca2+ channels in this pathway (Supplementary Fig. 3a online).
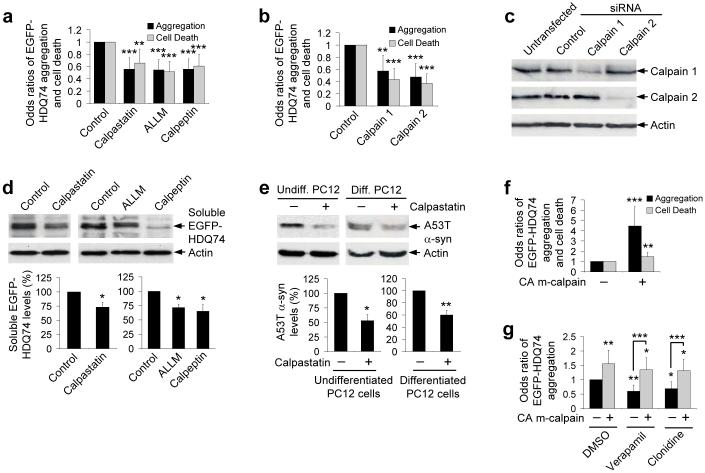
We confirmed the predictions from the above data that calpain inhibitors and siRNA knockdown of either calpain 1 or calpain 2 would reduce EGFP-HDQ74 aggregation/toxicity in cells not exposed to other drugs (Figs. 4a-c). Calpain inhibition enhanced the clearance of EGFP-HDQ74 and A53T α-synuclein (in undifferentiated and differentiated neuronal PC12 cells) (Figs. 4d, e).
Figure 4. Calpain inhibition increases aggregate-prone protein clearance.
a, The proportions of EGFP-positive SK-N-SH cells with EGFP-HDQ74 aggregates or cell death as in Fig. 1c, treated with or without 10 μM calpastatin, 50 μM ALLM or 50 μM calpeptin for 48 h post-transfection. DMSO was control for ALLM and calpeptin. Error bars: 95% confidence interval.
b, HeLa cells were transfected with EGFP-HDQ74 construct along with siGLO (control siRNA) in presence or absence of calpain 1 or calpain 2 siRNA (1:3 ratio) for 96 h. EGFP- and siGLO-positive cells were assessed for aggregation and cell death as in Fig. 1c. Error bars: 95% confidence interval.
c, HeLa cells, transfected with siGLO (control siRNA) or siRNAs for calpain 1 or calpain 2 for 96 h, were analysed with anti-calpain 1 or anti-calpain 2 domain III antibodies.
d, Clearance of soluble EGFP-HDQ74 in stable PC12 cells as in Fig. 1b, treated with drugs (concentrations in Fig. 4a) for 96 h. Densitometry is relative to actin. Error bars: Standard error of mean.
e, A53T α-synuclein clearance in undifferentiated or non-mitotic neuronally differentiated (with 100 ng/ml nerve growth factor for 5 d) stable PC12 cells as in Fig. 1a, treated with or without 10 μM calpastatin for 24 h. Densitometry is relative to actin. Error bars: Standard error of mean.
f, COS-7 cells, transfected with EGFP-HDQ74 along with empty vector (pcDNA3.1) or constitutive active (CA) m-calpain (1:3 ratio) for 48 h, were assessed for aggregation and cells death as in Fig. 1c. Error bars: 95% confidence interval.
g, SK-N-SH cells, transfected with EGFP-HDQ74 along with empty vector (pcDNA3.1) or constitutive active m-calpain (1:3 ratio) for 4 h, were subsequently treated with DMSO (control), 1 μM verapamil or 1 μM clonidine for 48 h. The proportions of EGFP-positive cells with aggregates were assessed as in Fig. 1c. Error bars: 95% confidence interval. ***, p<0.001; **, p<0.01; *, p<0.05.
Conversely, overexpression of constitutively active S50E human m-calpain increased EGFP-HDQ74 aggregation and toxicity (Fig. 4f). The effects of constitutively active m-calpain on EGFP-HDQ74 aggregation were not abrogated by either verapamil or clonidine (Fig. 4g), again consistent with calpain being downstream of the L-type Ca2+ channel (verapamil) and the cAMP-Epac-PLC-IP3 (clonidine) pathways.
Consistent with these data, thapsigargin increased EGFP-HDQ74 aggregates/toxicity and delayed A53T α-synuclein clearance; effects that were attenuated by calpain inhibitors (Supplementary Figs. 4a-j online).
Calpain inhibition induces autophagy
We next tested if the reduced EGFP-HDQ74 aggregation mediated by the calpain inhibitors was autophagy-dependent. Calpastatin could not reduce EGFP-HDQ74 aggregates in ATG5−/−, but reduced aggregates in Atg5+/+ MEFs, or when wild-type, but not conjugation-deficient Atg5 (K130R), was overexpressed in ATG5−/− cells (Fig. 5a).
Figure 5. Calpain inhibition induces autophagy.
a, The proportions of EGFP-positive MEFs with aggregates, transfected with EGFP-HDQ74 and pcDNA3.1 (empty vector) in Atg5+/+ cells, or transfected with EGFP-HDQ74 and pcDNA3.1, WT Atg5 or K130R Atg5 constructs in ATG5−/− cells for 4 h (1:3 ratios), were treated for next 48 h with or without 10 μM calpastatin. All data are from the same experiment, but odds ratios for reference conditions in specific experiments have been set to one, to facilitate comparisons. ATG5−/− cells had increased EGFP-HDQ74 aggregates compared to Atg5+/+ cells10, 12. Error bars: 95% confidence interval.
b, HeLa cells were transfected with siGLO (control) with or without calpain 1 siRNA or calpain 2 siRNA (1:3 ratio) for 96 h, followed by transfection with EGFP-LC3 construct for 4 h. Cells were fixed after a further 2 h. The proportions of transfected cells with >5 EGFP-LC3 vesicles were assessed. Error bars: 95% confidence interval.
c, Endogenous LC3-II levels in SK-N-SH cells treated with or without 10 μM or 30 μM calpastatin for 24 h.
d, HeLa cells stably expressing EGFP-LC3 were treated for 4 h with 400 nM bafilomycin A1 in presence or absence of 10 μM calpastatin, 50 μM ALLM or 50 μM calpeptin. Calpain inhibitors were treated for 24 h before bafilomycin A1 addition. EGFP-LC3-II was detected with anti-EGFP antibody. Data from different calpain inhibitors are from different blots.
e, COS-7 cells, transfected with myc-LC3 and EGFP-C1 with pcDNA3.1 (empty vector) or constitutive active (CA) m-calpain (1:1:3 ratio) for 4 h, were analysed after 24 h post-transfection for LC3-II levels by immunoblotting with anti-myc antibody. Error bars: Standard error of mean.
f, COS-7 cells, transfected with EGFP-LC3 and pcDNA3.1 (empty vector) or constitutive active m-calpain (1:3 ratio) for 4 h and fixed 24 h post-transfection, were assessed for proportions of EGFP-positive cells with >5 EGFP-LC3 vesicles. Error bars: 95% confidence interval. ***, p<0.001; **, p<0.01; *, p<0.05; NS, Non-significant.
Calpain 1 or calpain 2 knockdown by siRNA and calpain inhibitors increased LC3 vesicle/autophagosome numbers (Fig. 5b and Supplementary Figs. 5a-c online). Calpeptin also increased the number of autophagosome-like structures and decreased the numbers of mitochondria (which are endogenous autophagy substrates2) in COS-7 cells as evaluated by electron microscopy (Supplementary Figs. 5d, e online). Consistent with the data in Supplementary Figs. 4f-j online, thapsigargin-treated cells pre-treated with calpain inhibitors had more autophagosomes than cells treated with thapsigargin only (Supplementary Figs. 4k, l online).
LC3-II levels also accumulated in cells treated with calpastatin (Fig. 5c and Supplementary Fig. 5f online). LC3-II accumulation can occur due to increased autophagosome formation or impaired autophagosome-lysosome fusion. Thus, we assayed LC3-II in the presence of bafilomycin A1 (27), which blocks autophagosome-lysosome fusion, to assess autophagosome formation39. Bafilomycin A1 increases LC3-II (Fig. 5d and Supplementary Fig. 5g online) and the concentration used is saturating for LC3-II levels in this assay (data not shown). Further blockage of autophagosome-lysosome fusion via a bafilomycin A1-independent mechanism, using the dynein inhibitor erythro-9-[3-(2-hydroxynonyl)] adenine (EHNA)40 (28), along with this dose of bafilomycin A1, results in no increase in LC3-II compared to bafilomycin A1 alone (data not shown). We have previously demonstrated that autophagy-inducers can further increase LC3-II levels in bafilomycin A1-treated cells, indicating that such compounds increase autophagosome formation10, 12. To test if calpain inhibitors induce autophagosome formation, we pre-treated EGFP-LC3 HeLa cells with the inhibitors for 24 h and then added bafilomycin A1 to the cells for a further 4 h. We assumed that calpain inhibitors act by blocking clearance/cleavage of key autophagy regulators and thus some time is needed for these putative regulators to accumulate. Calpain inhibitors increased LC3-II levels in the presence of bafilomycin A1, compared to bafilomycin A1 alone, suggesting induction of autophagy (Fig. 5d and Supplementary Fig. 5g online). Likewise, L-type Ca2+ channel antagonist (verapamil) showed a similar trend (Supplementary Fig. 5h online). Conversely, overexpression of constitutively active m-calpain reduced Myc-LC3-II levels and EGFP-LC3 vesicle numbers (Figs. 5e, f).
While our data suggest that thapsigargin inhibits clearance of various autophagy dependent substrates by activating calpains (Supplementary Figs. 4a-j online), it appears to have additional effects on autophagy. In contrast to overexpression of m-calpain, which reduces autophagosome numbers (Figs. 5e, f), thapsigargin increased steady-state levels of LC3 vesicles and LC3-II (Supplementary Figs. 4m, n online). However, thapsigargin did not increase LC3-II levels in bafilomycin A1-treated cells, compared to the levels with bafilomycin A1 alone (Supplementary Fig. 4n online), which was in contrast to the effects of various autophagy inducers10, 12 (Fig. 5d and Supplementary Figs. 5g, h online). The phenomenon of an agent that decreases clearance of autophagy substrates, and that also increases steady-state autophagosome numbers but does not increase autophagosome levels in cells treated with bafilomycin A1, is characteristic of compounds/genes that block autophagosome-lysosome fusion10, 40. Such agents/genes also result in lower proportions of LC3 vesicles colocalising with the lysosomal marker lgp120 (Llamp1), which we observed with thapsigargin (Supplementary Fig. 4o online)10, 12. Thus, the most parsimonious explanation for our data is that thapsigargin decreases the clearance of autophagic substrates (Supplementary Figs. 4a-d online) via at least two mechanisms - a calpain-dependent process that reduces autophagosome synthesis (Figs. 5e, f and Supplementary Figs. 4f-l online), and a calpain-independent process that probably decreases autophagosome lysosome-fusion (Supplementary Figs. 4m-o online).
Autophagy induction by calpain inhibitors is not predominantly mediated by effects on Ca2+ levels in intracellular stores (like the ER)41. While the autophagy-inhibitory effect of thapsigargin was rescued by calpain inhibitors, there were no significant differences in the stored Ca2+ between thapsigargin-treated cells with or without calpain inhibitors (Supplementary Figs. 4p, q online).
Gsα is a calpain substrate regulating autophagy
In order to select candidate calpain substrates regulating autophagy, we considered which substrates would inhibit autophagy if subjected to low levels of cleavage. An appealing candidate was the alpha subunit of heterotrimeric G-proteins (Gsα) that increases its activity after calpain cleavage42, since Gsα enhances adenylyl cyclase activity. We confirmed that the hallmark 20 kDa Gsα cleavage product appeared in our cells overexpressing constitutive active m-calpain, or in cells treated with thapsigargin (Supplementary Fig. 6a online).
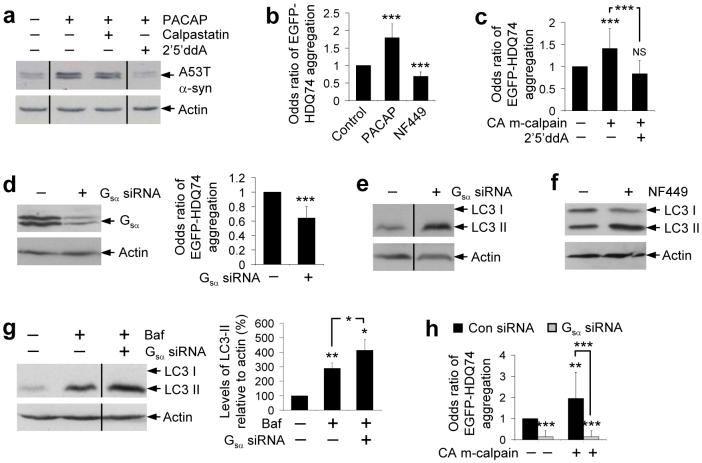
Compatible with our hypothesis, activation of Gsα with its natural ligand, pituitary adenylyl cyclase-activating polypeptide (PACAP), increased cAMP levels and retarded A53T α-synuclein clearance (Fig. 6a and Supplementary Figs. 6b-d online). This effect was not blocked by calpastatin (as PACAP activation can occur without Gsα cleavage) but was abolished by inhibition of its downstream target, adenyl cyclase (with 2′5′ddA) (Fig. 6a and Supplementary Fig. 6c online). PACAP also enhanced EGFP-HDQ74 aggregation (Fig. 6b). Consistent with the hypothesis that calpains activate Gsα, which mediates effects by enhancing adenylyl cyclase activity (confirmed in Supplementary Figs. 6d, e online, where constitutively active m-calpain increased cAMP levels), 2′5′ddA decreased the enhanced EGFP-HDQ74 aggregation caused by overexpression of constitutively active m-calpain (Fig. 6c), in a manner similar to what we observed with PACAP (Fig. 6a).
Figure 6. Calpain cleaves Gsα creating a link between the cAMP and Ca2+-calpain pathways.
a, A53T α-synuclein clearance in stable PC12 cells as in Fig. 1a, treated with or without 100 nM PACAP with or without 10 μM calpastatin or 500 μM 2′5′ddA for 24 h.
b, The proportion of EGFP-positive cells with aggregates in SK-N-SH cells as in Fig. 1c, treated for 48 h with 1 μM PACAP or 200 μM NF449. Error bars: 95% confidence interval.
c, Aggregation in COS-7 cells transfected with either pcDNA3:1 (empty vector) or constitutively active (CA) m-calpain and EGFP-HDQ74 (3:1 ratio) for 4 h and then treated with or without 500 μM 2′5′ddA for 48 h. Error bars: 95% confidence interval.
d, HeLa cells transfected with control or Gsα siRNA for 72 h were analysed for Gsα levels by immunoblotting with anti-Gsα antibody. Aggregation was assessed in HeLa cells transfected with control or Gsα siRNA and EGFP-HDQ74 for 72 h. Error bars: 95% confidence interval.
e, Endogenous LC3-II levels in HeLa cells transfected with control or Gsα siRNA for 72 h.
f, Endogenous LC3-II levels in PC12 cells treated with or without 200 μM NF449 for 24 h.
g, Endogenous LC3-II levels in HeLa cells transfected with control or Gsα siRNA for 72 h and treated with or without 400 nM bafilomycin A1 for the last 4 h. Densitometric analysis is relative to actin. Error bars: Standard error of mean.
h, The proportions of EGFP-positive HeLa cells with aggregates, transfected with control or Gsα siRNA for 48 h, and then re-transfected together with pcDNA 3.1 (empty vector) or constitutively active (CA) m-calpain and EGFP-HDQ74 (3:1 ratio) for a further 48 h. Error bars: 95% confidence interval. ***, p<0.001; **, p<0.01; *, p<0.05; NS, Non-significant.
Knockdown of Gsα with siRNA or treatment with its chemical inhibitor NF449 (29) decreased EGFP-HDQ74 aggregation and increased LC3-II levels (Figs. 6b, d-f and Supplementary Fig. 6f online). The increase in LC3-II levels with Gsα knockdown by siRNA was also seen in cells treated with bafilomycin A1, suggesting that knockdown of Gsα increases autophagosome synthesis (Fig. 6g). Gsα is likely to mediate many of the autophagy-related effects of calpains on EGFP-HDQ74 aggregation, as the increased aggregation mediated by overexpression of constitutively active m-calpain was abrogated by Gsα siRNA, and the beneficial effects of calpain inhibition are not further enhanced in calpastatin-treated cells with Gsα knockdown (Figs. 6h and Supplementary Fig. 6g online).
Another candidate we considered was Atg5, which may be cleaved by calpains in certain cell death contexts43. However, we observed no reduction in full-length Atg5 and no obvious cleavage products in cells treated with thapsigargin and no increase in Atg5 levels with calpastatin (Supplementary Fig. 6h online).
Calpain-regulated autophagy is mTOR-independent
Calpain-regulated autophagy appeared to be independent of, or downstream of, the known pathway that is negatively regulated by mTOR, as its kinase activity, inferred by the levels of phosphorylation of its substrates, ribosomal S6 protein kinase (S6K1, also known as p70S6K) and eukaryotic initiation factor 4E-binding protein 1 (4E-BP1)19, was not reduced by calpain inhibitors (Supplementary Figs. 6i, j online). Likewise, drugs that perturb intracellular Ca2+ like thapsigargin had no effect on mTOR signalling (Supplementary Fig. 4r online).
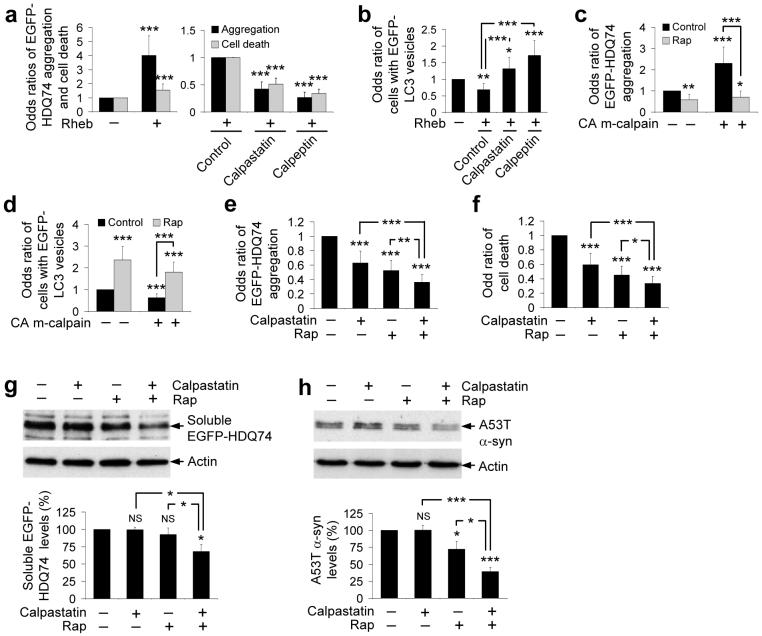
Overexpression of the small G-protein rheb, which greatly enhances mTOR signalling, markedly increased EGFP-HDQ74 aggregates and cell death in COS-7 cells9 (Fig. 7a). However, the calpain inhibitors reduced EGFP-HDQ74 aggregation/toxicity and increased EGFP-LC3 vesicle numbers in rheb-transfected cells (Figs. 7a, b), suggesting that induction of autophagy by calpain inhibition occurs even when mTOR is activated. Thus, calpain inhibitors do not act upstream of mTOR.
Figure 7. Regulation of autophagy by calpain is mTOR-independent.
a, The proportions of EGFP-positive COS-7 cells with aggregates or cell death, after transfection with EGFP-HDQ74 and rheb or pcDNA3.1 (empty vector) (1:3 ratio) for 4 h and treatment with or without 10 μM calpastatin or 50 μM calpeptin for 48 h post-transfection. The control condition for assessing the effect of calpain inhibitors in rheb-transfected cells was taken as 1. Error bars: 95% confidence interval.
b, The proportions of EGFP-positive COS-7 cells with >5 EGFP-LC3 vesicles, transfected with EGFP-LC3 and pcDNA3.1 (empty vector) or rheb (1:3 ratio) for 4 h and treated with or without 10 μM calpastatin or 50 μM calpeptin for 24 h post-transfection. Error bars: 95% confidence interval.
c, The proportions of EGFP-positive COS-7 cells with aggregates, transfected with EGFP-HDQ74 and constitutively active (CA) m-calpain or pcDNA3.1 (empty vector) (1:3 ratio) for 4 h and treated with or without 0.2 μM rapamycin for 48 h post-transfection. Error bars: 95% confidence interval.
d, The proportions of EGFP-positive COS-7 cells with >5 EGFP-LC3 vesicles, transfected with EGFP-LC3 and constitutively active (CA) m-calpain or pcDNA3.1 (empty vector) (1:3 ratio) for 4 h and treated with or without 0.2 μM rapamycin for 24 h post-transfection. Error bars: 95% confidence interval.
e, f, The proportions of EGFP-positive COS-7 cells with aggregates (e) and cell death (f) as in Fig. 1c, treated with or without 0.2 μM rapamycin, 10 μM calpastatin or both for 48 h. Error bars: 95% confidence interval.
g, Clearance of soluble EGFP-HDQ74 in stable PC12 cells as in Fig. 1b, treated with or without 0.2 μM rapamycin, 10 μM calpastatin or both for the 48 h switch-off period. Error bars: Standard error of mean.
h, Clearance of A53T α-synuclein in stable PC12 cells as in Fig. 1a, treated with or without 0.2 μM rapamycin, 10 μM calpastatin or both for the 8 h switch-off period. Error bars: Standard error of mean. ***, p<0.001; **, p<0.01; *, p<0.05; NS, Non-significant.
We next tested whether mTOR inhibition induced autophagy when calpains were activated. This is relevant to HD where calpains are activated, and where rapamycin is protective in in vivo models4, 5, 8, 9. In cells overexpressing constitutively active m-calpain, rapamycin reduced EGFP-HDQ74 aggregation and increased autophagosome numbers, further suggesting that mTOR and calpains regulate autophagy via independent pathways (Figs. 7c, d). Similarly, rapamycin reduced the increased EGFP-HDQ74 aggregation/toxicity induced by thapsigargin, and increased EGFP-LC3 vesicle numbers in thapsigargin-treated cells (Supplementary Figs. 4s-u online).
Simultaneous inhibition of calpain and mTOR
As calpain inhibition induces autophagy independently of mTOR, we confirmed that calpastatin and rapamycin have additive effects in reducing EGFP-HDQ74 aggregation/toxicity and enhancing the clearance of soluble EGFP-HDQ74 and A53T α-synuclein, compared to single treatments with calpastatin or rapamycin (Figs. 7e-h). The clearance effect was assessed at early time-points where no major reductions in the levels of mutant proteins were observed with single treatments. We have used a saturating dose of rapamycin11. However, co-treatment with calpastatin and verapamil (both acting through the same pathway) did not facilitate any further clearance of the aggregate-prone proteins at this early time-point (data not shown).
Therapeutic potential in HD models
In order to test whether compounds acting via the pathway identified in this screen were potentially relevant to therapy, we first tested them in a Drosophila model of HD (see Supplementary Methods online). Flies expressing a mutant huntingtin fragment with 120Q in the photoreceptors exhibit photoreceptor degeneration that is not observed in flies expressing the wild-type protein with 23Q44. The photoreceptor degeneration in 120Q flies is attenuated with drugs acting on L-type Ca2+ channels (verapamil) or on the cAMP arm of the pathway (clonidine) (Supplementary Figs. 7a, b online). Valproate (that induces autophagy by reducing IP3 levels11, among other activities that may be relevant to HD), also had protective effects (Supplementary Fig. 7c online).
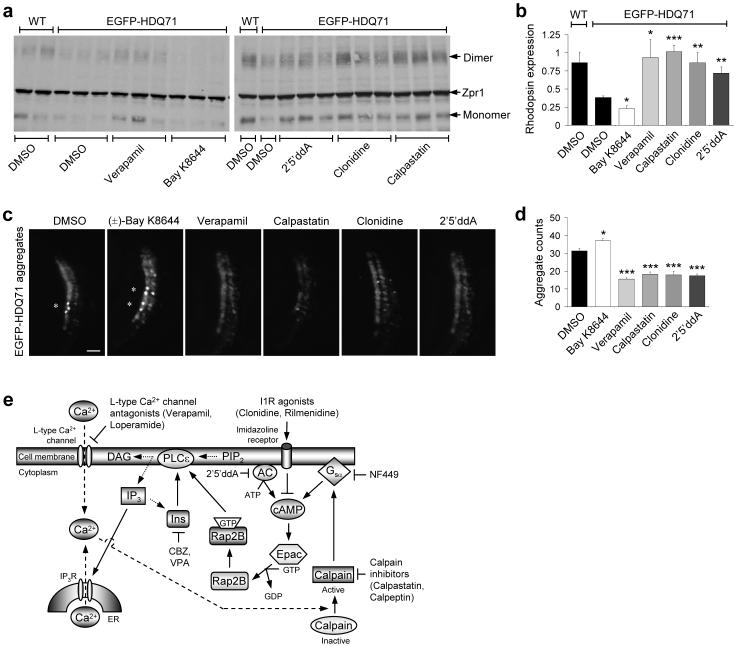
As verapamil, clonidine and valproate slowed this neurodegeneration phenotype to a similar extent as various histone deacetylase inhibitors, which are being extensively investigated in HD45, 46, we next tested representative compounds acting on our putative pathway in a zebrafish HD model we have generated that expresses EGFP-tagged huntingtin exon 1 with 71Q (EGFP-HDQ71) in the rod photoreceptors using the rhodopsin promoter. In this model, constructs with EGFP-HDQ71 form aggregates (which are not seen with EGFP-HDQ23 constructs), and induce a loss of rhodopsin expression which correlates with rod photoreceptor degeneration (Fig. 8a and Supplementary Figs. 8a-c online). Zebrafish EGFP-HDQ71 models treated with verapamil, calpastatin, clonidine and 2′5′ddA had significantly fewer aggregates (like with rapamycin) and significantly enhanced rhodospin expression compared to untreated fish, while aggregation was increased and rhodopsin expression was decreased with (±)-Bay K8644 (Figs. 8a-d).
Figure 8. L-type Ca2+ channel antagonists, imidazoline-1 receptor agonists, cAMP antagonists and calpain inhibitors rescue Huntington’s disease phenotypes in zebrafish.
a, Western blot showing EGFP-HDQ71-induced degeneration of rod photoreceptors at 9 d.p.f. as indicated by the lack of monomeric (∼36 kDa) and dimeric rhodopsin (∼72 kDa), that is further reduced by 3 μM (±)-Bay K8644, and recovered upon treatment with 3 μM verapamil, 100 μM 2′5′ ddA, 3 μM clonidine and 1 μM calpastatin. No general toxicity to the zebrafish at these concentrations was observed.
b, Expression of EGFP-HDQ71 significantly reduces the levels of rhodopsin expression, when compared to wild-type (WT), non-transgenic controls. This affect is further abrogated by 3 μM (±)-BayK8644, while 3 μM verapamil, 100 μM 2′5′ddA, 3 μM clonidine and 1 μM calpastatin all protected against rod photoreceptor degeneration. Error bars: Standard error of mean.
c, Magnified bright field and EGFP images of zebrafish eye sections, taken at the ventral marginal zone for each of the indicated treatments (with concentrations as in Fig. 8b), are shown. Bar, 10 μm.
d, The total number of aggregates across 8×12 μm sections were counted and compared to the EGFP-HDQ71 DMSO-treated control. 3 μM (±)-Bay K8644 was found to significantly increase the aggregate burden, while 3 μM verapamil, 1 μM calpastatin, 3 μM clonidine and 100 μM 2′5′ddA decreased the aggregate load. As in our cell models, these aggregates were insoluble in 4% triton/SDS. Error bars: Standard error of mean. ***, p<0.001; **, p<0.01; *, p<0.05.
e, Schematic representation of an mTOR-independent autophagy pathway involving cAMP-Ca2+-calpains-Gsα with multiple drug targets. Intracellular cAMP levels are increased by adenylyl cyclase (AC) activity, thereby activating Epac which subsequently activates Rap2B, a small G-protein that activates PLC-ε resulting in the production of IP3 and consequent Ca2+ release from the ER. Intracellular Ca2+ levels are also increased by L-type Ca2+ channel agonists. An increase in intracytosolic Ca2+ activates calpains, which cleave and activate Gsα that activates adenylyl cyclase to increase cAMP levels, thereby forming a loop. Activation of this pathway inhibits autophagy. Multiple drug targets acting at various places in this pathway induce autophagy, such as imidazoline-1 receptors (I1R) agonists (clonidine and rilmenidine) and adenylyl cyclase inhibitor (2′5′ddA) that decrease cAMP levels, agents lowering inositol and IP3 levels [carbamazepine (CBZ) and valproic acid (VPA)]11, L-type Ca2+ channel antagonists (verapamil and loperamide), calpain inhibitors (calpastatin and calpeptin) and Gsα inhibitor (NF449).
Discussion
Our compound screen has identified a pathway regulating autophagy that provides many possibilities for therapeutic intervention (Fig. 8e). Previous studies have shown that the clearance of mutant huntingtin exon 1 (both EGFP-tagged or HA-tagged), mutant full-length huntingtin and A53T or A30P α-synuclein is largely mediated by autophagy8, 13, 14, 20. Conversely, wild-type full-length or exon 1 huntingtin and wild-type α-synuclein have very low dependencies on autophagy for their clearance8, 14, 20. Consistent with these expectations, a range or autophagy-inducing drugs have no obvious effects on wild-type huntingtin exon 1 or wild-type α-synuclein clearance10, 14. Similarly, the drugs acting on the pathway we have identified in this study have no obvious effects on the clearance of wild-type huntingtin exon 1 (EGFP-HDQ23) or wild-type α-synuclein (Supplementary Figs. 2g, h online).
Our data suggest that raised cAMP levels act via Epac and PLC-ε to enhance calpain activity, which retards the clearance of autophagy substrates (Figs. 2a, b, d-f, 4f, 6a-c and Supplementary Figs. 3b, d, f, j-l, 4a-d, 6b-e online). The Gsα-Epac-PLC-ε-Ca2+ release pathway operates in neurons and neuronal cells32, 47. Importantly, this pathway can also be inhibited in neuronal cells by Gi stimulation (as would occur with clonidine)34. Likewise, L-type Ca2+ channel agonists slow the clearance of mutant proteins by blocking autophagy through calpain activation (Figs. 1a-c, 3a-d and Supplementary Figs. 2a-c, e, f online). Gsα provides a link between these two pathways as it is activated following calpain cleavage leading to increased cAMP levels (Fig. 8e and Supplementary Figs. 6a, d, e online). Interestingly, autophagosome synthesis can be induced and mutant huntingtin aggregation can be significantly inhibited by genetic knockdown of Gsα (Figs. 6d, e, g and Supplementary Fig. 6f online). This is the first demonstration of the importance of the Epac pathway in autophagy regulation. Previous studies in yeast have focused on PKA and many of the mammalian studies that implicated PKA were performed before the Epac pathway was discovered48, 49. The validity of the Epac pathway is consistent with the known activities of four additional compounds we identified in our screen that inhibit A53T α-synuclein clearance and enhance EGFP-HDQ74 aggregation: FPL61476 (30) has Ca2+ channel agonist activity, quinine sulphate and tolazamide are ATP-sensitive K+ channel antagonists, and rolipram (31) inhibits phophodiesterase 4 (leading to increased levels of cAMP) (Supplementary Figs. 2e, f online).
It is possible that changes in IP3 and Ca2+ levels may have additional effects on autophagy. These are considered in the context of previous literature in the Supplementary Discussion online, which also deals with issues of cell-type pathway specificity.
Our data provides a possible link between excitotoxicity, which results from increased Ca2+ entry into neurons via glutamate receptors, and enhanced intracytosolic protein aggregation due to impaired autophagy. Excitotoxicity is believed to contribute to a number of neurodegenerative diseases, including HD and Alzheimer’s disease (AD)6, 7, and it is notable that the proteins that aggregate in many of these diseases are autophagy substrates1, 8, 15. Calpain inhibition of autophagy may also contribute to the link between β-amyloid toxicity (which may elevate cytosolic Ca2+ levels) and tau accumulation in AD, as insoluble tau accumulates when autophagy is blocked15. Finally, it is tempting to speculate that our data may also go some way to explaining why apoptosis and autophagy generally do not coexist, as many apoptotic (and necrotic) cell death processes are associated with, or stimulated by, raised cytosolic Ca2+ levels50.
Calpain activation has been proposed to contribute to HD pathogenesis by promoting huntingtin cleavage - the toxicity of the HD mutation appears to be exposed or enhanced after cleavage4, 5. As the huntingtin fragments we used are N-terminal to the calpain cleavage sites at residues 469 and 536 (Supplementary Fig. 1a online), the cumulative data strongly argue that calpain inhibition will reduce mutant huntingtin fragment levels by impairing both cleavage (production) and by enhancing degradation (Figs. 4a-e). In addition to these effects that result in lowered levels of the toxic protein, calpain inhibition may have other benefits in neurodegenerative diseases, including protection against cell death (Figs. 4a, b).
Our data suggest that drugs acting on this mTOR-independent pathway may have added efficacy for neurodegenerative diseases in combination with rapamycin, providing a new direction for combinatorial treatment of disorders like HD by enhancing autophagy through two different routes (Figs. 7e-h and Supplementary Figs. 6i, j online). Combination therapy with more moderate calpain and mTOR inhibition may be safer for long-term treatment compared to using higher doses of either compound that result in more severe inhibition of a single pathway. This strategy may allow a larger safety window before toxic effects from non-autophagy-related effects of each drug are seen.
The discovery that autophagy induction can be mediated by Ca2+ channel blockers is particularly exciting, as these drugs are safer and better tolerated than rapamycin - for instance, verapamil has been used for decades to treat hypertension with minimal side effects. Our findings may have broad applicability, as autophagy appears to regulate the clearance of a range of intra-cytosolic aggregate-prone proteins associated with neurodegeneration and various intracellular bacteria1.
Materials and Methods
See Supplementary Information online for details of plasmids; compounds; mammalian cell lines, culture and transfection; quantification and analysis of aggregate formation and cell death; assessment of autophagy by LC3; analysis of autophagosome-lysosome fusion; immunocytochemistry; western blot analysis; measurement of stored intracellular Ca2+; RNA interference; assay for cAMP levels; Rap2B activation assay; electron microscopy; Drosophila analysis; zebrafish analysis.
Screen design
We screened for autophagy enhancers using a library of 253 compounds that had previously been into man without major toxic side effects, and pharmacological probes.
We first assayed clearance of A30P α-synuclein, a well characterized autophagy substrate14, in stable inducible PC12 (neuronal precursor) cells. A30P and A53T α-synuclein are not substrates for chaperone-mediated autophagy20. While these mutant are also dependent on the ubiquitin-proteasome system for their clearance, we have found these proteins to be excellent reporters for changes in autophagic activity through both mTOR-dependent and mTOR-independent pathways10-12, 14. Autophagy inducers enhance A30P and A53T α-synuclein clearance resulting in lower levels of the transgene which are readily detectable after 24 h switch-off period, compared to the control. Conversely, clearance is retarded when autophagy is blocked leading to higher levels of the transgene, compared to control after 24 h switch-off period. All compounds that visibly perturbed A30P α-synuclein clearance were retested and successfully validated in similar stable inducible PC12 cell lines expressing A53T α-synuclein.
Clearance of mutant huntingtin and A53T α-synuclein
Stable inducible PC12 cell lines expressing EGFP-HDQ74, or α-synuclein mutants (A53T or A30P) were induced with 1 μg ml-1 doxycycline (Sigma) for 8 h and 48 h, respectively8, 14. Transgene expression was switched off by removing doxycycline from medium. Cells were treated with or without compounds for time-points as indicated in experiments. If transgene levels are followed at various times after switching off expression after an initial induction period, the effect of different drugs on its clearance can be assessed, as its expression decays when synthesis is stopped. Compounds were replenished every 24 h for EGFP-HDQ74 clearance. Clearance of soluble mutant huntingtin or α-synuclein mutants was detected with anti-EGFP or anti-HA antibody, respectively.
Autophagy methods
Assessment of autophagy by EGFP-LC3 vesicles
The percentage of EGFP-positive cells with >5 EGFP-LC3 vesicles was counted, as previously described11.
Assessment of autophagy by LC3-II levels
LC3-II levels, which directly correlate with autophagosome numbers23, 24, is detected with anti-LC3 antibody (NB100-2220, Novus Biologicals) and densitometry analysis relative to actin or tubulin. To see if accumulation of LC3-II is due to increased autophagosome formation or impaired autophagosome-lysosome fusion, LC3-II is assessed in the presence of bafilomycin A1, that blocks autophagosome-lysosome fusion39, as shown previously10, 12.
Statistical analysis by odds ratio
Pooled estimates for the changes in aggregate formation, cell death or EGFP-LC3 vesicles, resulting from perturbations assessed in multiple experiments, were calculated as odds ratios with 95% confidence intervals8, 11, 12. Odds ratios and p values were determined by unconditional logistical regression analysis, using the general log-linear analysis option of SPSS 9 software (SPSS, Chicago). ***, p < 0.001; **, p < 0.01; *, p < 0.05; NS, Non-significant.
Statistical analysis on immunoblots
Densitometry analysis on the immunoblots was done by Scion Image Beta 4.02 software (Scion Corporation) from three independent experiments (n = 3). Significance for the clearance of mutant proteins was determined by factorial ANOVA test using STATVIEW software, version 4.53 (Abacus Concepts). The control condition was set to 100% and the error bars denote standard error of mean.
Supplementary Material
Discussion
Materials and Methods
References
Figures
Acknowledgements
We thank T. Yoshimori for LC3 antibody, myc-LC3 and EGFP-LC3 constructs, N. Mizushima for wild-type and knock-out Atg5 mouse embryonic fibroblasts, wild-type Atg5 and K130R Atg5 constructs, G. Jackson for the gmrQ120 flies, K.L. Guan for rheb construct, A. Wells for constitutive active S50E m-calpain construct, J. Lomasney for wild type phospholipase C ε construct, Jean de Gunzburg for dominant negative Rap2B construct, R.F. Irvine for cytosolic IP3 kinase A construct, M. Mizuguchi for human LC3B construct, P. Luzio for GFP-lgp20 construct, R. Tsien for mCherry construct, A.M. Tolkovsky for EGFP-LC3 HeLa stable cells, N.P. Dantuma for UbG76V-GFP HeLa stable cells, M. Mahaut-Smith for use of spectrophotometer, J.N. Skepper for E.M., A. Roach for critical comments and A. Cordenier for technical assistance. We are grateful for Gates Cambridge Scholarship and Hughes Hall Research Fellowship (Sov.S.), M.R.C. Studentships (A.W., E.T.), B.B.S.R.C. Career Development Award (C.J.O’K.), Eli Lilly Pergolide Fellowship (Shi.S.), Wellcome Grant GR077273MA (B.K.), A.M.S./M.R.C. Clinical Scientist Fellowship (R.A.F.), Wellcome Trust Senior Fellowship in Clinical Science (D.C.R.), KTP grant (Department of Trade and Industry) an M.R.C. Programme Grant, M.R.C. Link Grant, E.U. Framework VI (EUROSCA) and the NIHR Biomedical Research Centre at Addenbrooke’s Hospital for funding.
Footnotes
Conflict of interest: D.P. and A.F. are employees of Summit plc and have share options in this company. A.F. and P.G. are shareholders in Summit plc. Sov.S., A.W., D.C.R., E.T., C.O’K. and R.A.F. are inventors on patents relating to the use of autophagy activation in various diseases.
References
- 1.Rubinsztein DC, Gestwicki JE, Murphy LO, Klionsky DJ. Potential therapeutic applications of autophagy. Nat. Rev. Drug Discov. 2007;6:304–312. doi: 10.1038/nrd2272. [DOI] [PubMed] [Google Scholar]
- 2.Klionsky DJ, Emr SD. Autophagy as a regulated pathway of cellular degradation. Science. 2000;290:1717–1721. doi: 10.1126/science.290.5497.1717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rubinsztein DC. Lessons from animal models of Huntington’s disease. Trends Genet. 2002;18:202–209. doi: 10.1016/s0168-9525(01)02625-7. [DOI] [PubMed] [Google Scholar]
- 4.Gafni J, Ellerby LM. Calpain activation in Huntington’s disease. J. Neurosci. 2002;22:4842–4849. doi: 10.1523/JNEUROSCI.22-12-04842.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gafni J, et al. Inhibition of calpain cleavage of huntingtin reduces toxicity: accumulation of calpain/caspase fragments in the nucleus. J. Biol. Chem. 2004;279:20211–20220. doi: 10.1074/jbc.M401267200. [DOI] [PubMed] [Google Scholar]
- 6.Zeron MM, et al. Increased sensitivity to N-methyl-D-aspartate receptor-mediated excitotoxicity in a mouse model of Huntington’s disease. Neuron. 2002;33:849–860. doi: 10.1016/s0896-6273(02)00615-3. [DOI] [PubMed] [Google Scholar]
- 7.Tang TS, et al. Huntingtin and huntingtin-associated protein 1 influence neuronal calcium signaling mediated by inositol-(1,4,5) triphosphate receptor type 1. Neuron. 2003;39:227–239. doi: 10.1016/s0896-6273(03)00366-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ravikumar B, Duden R, Rubinsztein DC. Aggregate-prone proteins with polyglutamine and polyalanine expansions are degraded by autophagy. Hum. Mol. Genet. 2002;11:1107–1117. doi: 10.1093/hmg/11.9.1107. [DOI] [PubMed] [Google Scholar]
- 9.Ravikumar B, et al. Inhibition of mTOR induces autophagy and reduces toxicity of polyglutamine expansions in fly and mouse models of Huntington disease. Nat. Genet. 2004;36:585–595. doi: 10.1038/ng1362. [DOI] [PubMed] [Google Scholar]
- 10.Sarkar S, Davies JE, Huang Z, Tunnacliffe A, Rubinsztein DC. Trehalose, a novel mTOR-independent autophagy enhancer,accelerates the clearance of mutant huntingtin and alpha-synuclein. J. Biol. Chem. 2007;282:5641–5652. doi: 10.1074/jbc.M609532200. [DOI] [PubMed] [Google Scholar]
- 11.Sarkar S, et al. Lithium induces autophagy by inhibiting inositol monophosphatase. J. Cell Biol. 2005;170:1101–1111. doi: 10.1083/jcb.200504035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sarkar S, et al. Small molecules enhance autophagy and reduce toxicity in Huntington’s disease models. Nat. Chem. Biol. 2007;3:331–338. doi: 10.1038/nchembio883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shibata M, et al. Regulation of intracellular accumulation of mutant Huntingtin by Beclin 1. J. Biol. Chem. 2006;281:14474–14485. doi: 10.1074/jbc.M600364200. [DOI] [PubMed] [Google Scholar]
- 14.Webb JL, Ravikumar B, Atkins J, Skepper JN, Rubinsztein DC. Alpha-Synuclein is degraded by both autophagy and the proteasome. J. Biol. Chem. 2003;278:25009–25013. doi: 10.1074/jbc.M300227200. [DOI] [PubMed] [Google Scholar]
- 15.Berger Z, et al. Rapamycin alleviates toxicity of different aggregate-prone proteins. Hum. Mol. Genet. 2006;15:433–442. doi: 10.1093/hmg/ddi458. [DOI] [PubMed] [Google Scholar]
- 16.Gutierrez MG, et al. Autophagy is a defense mechanism inhibiting BCG and Mycobacterium tuberculosis survival in infected macrophages. Cell. 2004;119:753–766. doi: 10.1016/j.cell.2004.11.038. [DOI] [PubMed] [Google Scholar]
- 17.Nakagawa I, et al. Autophagy defends cells against invading group A Streptococcus. Science. 2004;306:1037–1040. doi: 10.1126/science.1103966. [DOI] [PubMed] [Google Scholar]
- 18.Ogawa M, et al. Escape of intracellular Shigella from autophagy. Science. 2005;307:727–731. doi: 10.1126/science.1106036. [DOI] [PubMed] [Google Scholar]
- 19.Sarbassov DD, Ali SM, Sabatini DM. Growing roles for the mTOR pathway. Curr. Opin. Cell Biol. 2005;17:596–603. doi: 10.1016/j.ceb.2005.09.009. [DOI] [PubMed] [Google Scholar]
- 20.Cuervo AM, Stefanis L, Fredenburg R, Lansbury PT, Sulzer D. Impaired degradation of mutant alpha-synuclein by chaperone-mediated autophagy. Science. 2004;305:1292–1295. doi: 10.1126/science.1101738. [DOI] [PubMed] [Google Scholar]
- 21.Greenberg DA, Cooper EC, Carpenter CL. Calcium channel ‘agonist’ BAY K 8644 inhibits calcium antagonist binding to brain and PC12 cell membranes. Brain Res. 1984;305:365–368. doi: 10.1016/0006-8993(84)90444-x. [DOI] [PubMed] [Google Scholar]
- 22.Hockerman GH, Peterson BZ, Johnson BD, Catterall WA. Molecular determinants of drug binding and action on L-type calcium channels. Annu. Rev. Pharmacol. Toxicol. 1997;37:361–396. doi: 10.1146/annurev.pharmtox.37.1.361. [DOI] [PubMed] [Google Scholar]
- 23.Mizushima N. Methods for monitoring autophagy. Int. J. Biochem. Cell Biol. 2004;36:2491–2502. doi: 10.1016/j.biocel.2004.02.005. [DOI] [PubMed] [Google Scholar]
- 24.Kabeya Y, et al. LC3, a mammalian homologue of yeast Apg8p, is localized in autophagosome membranes after processing. EMBO J. 2000;19:5720–5728. doi: 10.1093/emboj/19.21.5720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Osborne NN. Inhibition of cAMP production by alpha 2-adrenoceptor stimulation in rabbit retina. Brain Res. 1991;553:84–88. doi: 10.1016/0006-8993(91)90233-l. [DOI] [PubMed] [Google Scholar]
- 26.Felsen D, et al. Identification, localization and functional analysis of imidazoline and alpha adrenergic receptors in canine prostate. J. Pharmacol. Exp. Ther. 1994;268:1063–1071. [PubMed] [Google Scholar]
- 27.Greney H, et al. Coupling of I(1) imidazoline receptors to the cAMP pathway: studies with a highly selective ligand, benazoline. Mol. Pharmacol. 2000;57:1142–1151. [PubMed] [Google Scholar]
- 28.Kopperud R, Krakstad C, Selheim F, Doskeland SO. cAMP effector mechanisms. Novel twists for an ‘old’ signaling system. FEBS Lett. 2003;546:121–126. doi: 10.1016/s0014-5793(03)00563-5. [DOI] [PubMed] [Google Scholar]
- 29.Kelley GG, Reks SE, Ondrako JM, Smrcka AV. Phospholipase C(epsilon): a novel Ras effector. EMBO J. 2001;20:743–754. doi: 10.1093/emboj/20.4.743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Enserink JM, et al. A novel Epac-specific cAMP analogue demonstrates independent regulation of Rap1 and ERK. Nat. Cell Biol. 2002;4:901–906. doi: 10.1038/ncb874. [DOI] [PubMed] [Google Scholar]
- 31.Shi GX, Rehmann H, Andres DA. A novel cyclic AMP-dependent Epac-Rit signaling pathway contributes to PACAP38-mediated neuronal differentiation. Mol. Cell Biol. 2006;26:9136–9147. doi: 10.1128/MCB.00332-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ster J, et al. Exchange protein activated by cAMP (Epac) mediates cAMP activation of p38 MAPK and modulation of Ca2+-dependent K+ channels in cerebellar neurons. Proc. Natl. Acad. Sci. USA. 2007;104:2519–2524. doi: 10.1073/pnas.0611031104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Qiao J, Mei FC, Popov VL, Vergara LA, Cheng X. Cell cycle-dependent subcellular localization of exchange factor directly activated by cAMP. J. Biol. Chem. 2002;277:26581–26586. doi: 10.1074/jbc.M203571200. [DOI] [PubMed] [Google Scholar]
- 34.vom Dorp F, et al. Inhibition of phospholipase C-epsilon by Gi-coupled receptors. Cell Signal. 2004;16:921–928. doi: 10.1016/j.cellsig.2004.01.009. [DOI] [PubMed] [Google Scholar]
- 35.Criollo A, et al. Regulation of autophagy by the inositol trisphosphate receptor. Cell Death Differ. 2007;14:1029–1039. doi: 10.1038/sj.cdd.4402099. [DOI] [PubMed] [Google Scholar]
- 36.Schell MJ, Erneux C, Irvine RF. Inositol 1,4,5-trisphosphate 3-kinase A associates with F-actin and dendritic spines via its N terminus. J. Biol. Chem. 2001;276:37537–37546. doi: 10.1074/jbc.M104101200. [DOI] [PubMed] [Google Scholar]
- 37.Goll DE, Thompson VF, Li H, Wei W, Cong J. The calpain system. Physiol. Rev. 2003;83:731–801. doi: 10.1152/physrev.00029.2002. [DOI] [PubMed] [Google Scholar]
- 38.Hayashi S, Horie M, Okada Y. Ionic mechanism of minoxidil sulfate-induced shortening of action potential durations in guinea pig ventricular myocytes. J. Pharmacol. Exp. Ther. 1993;265:1527–1533. [PubMed] [Google Scholar]
- 39.Yamamoto A, et al. Bafilomycin A1 prevents maturation of autophagic vacuoles by inhibiting fusion between autophagosomes and lysosomes in rat hepatoma cell line, H-4-II-E cells. Cell Struct. Funct. 1998;23:33–42. doi: 10.1247/csf.23.33. [DOI] [PubMed] [Google Scholar]
- 40.Ravikumar B, et al. Dynein mutations impair autophagic clearance of aggregate-prone proteins. Nat. Genet. 2005;37:771–776. doi: 10.1038/ng1591. [DOI] [PubMed] [Google Scholar]
- 41.Gordon PB, Holen I, Fosse M, Rotnes JS, Seglen PO. Dependence of hepatocytic autophagy on intracellularly sequestered calcium. J. Biol. Chem. 1993;268:26107–26112. [PubMed] [Google Scholar]
- 42.Sato-Kusubata K, Yajima Y, Kawashima S. Persistent activation of Gsalpha through limited proteolysis by calpain. Biochem. J. 2000;347:733–740. [PMC free article] [PubMed] [Google Scholar]
- 43.Yousefi S, et al. Calpain-mediated cleavage of Atg5 switches autophagy to apoptosis. Nat. Cell Biol. 2006;8:1124–1132. doi: 10.1038/ncb1482. [DOI] [PubMed] [Google Scholar]
- 44.Jackson GR, et al. Polyglutamine-expanded human huntingtin transgenes induce degeneration of Drosophila photoreceptor neurons. Neuron. 1998;21:633–642. doi: 10.1016/s0896-6273(00)80573-5. [DOI] [PubMed] [Google Scholar]
- 45.Steffan JS, et al. Histone deacetylase inhibitors arrest polyglutamine-dependent neurodegeneration in Drosophila. Nature. 2001;413:739–743. doi: 10.1038/35099568. [DOI] [PubMed] [Google Scholar]
- 46.Butler R, Bates GP. Histone deacetylase inhibitors as therapeutics for polyglutamine disorders. Nat. Rev. Neurosci. 2006;7:784–796. doi: 10.1038/nrn1989. [DOI] [PubMed] [Google Scholar]
- 47.Hucho TB, Dina OA, Levine JD. Epac mediates a cAMP-to-PKC signaling in inflammatory pain: an isolectin B4(+) neuron-specific mechanism. J. Neurosci. 2005;25:6119–6126. doi: 10.1523/JNEUROSCI.0285-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Budovskaya YV, Stephan JS, Reggiori F, Klionsky DJ, Herman PK. The Ras/cAMP-dependent protein kinase signaling pathway regulates an early step of the autophagy process in Saccharomyces cerevisiae. J. Biol. Chem. 2004;279:20663–20671. doi: 10.1074/jbc.M400272200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Holen I, Gordon PB, Stromhaug PE, Seglen PO. Role of cAMP in the regulation of hepatocytic autophagy. Eur. J. Biochem. 1996;236:163–170. doi: 10.1111/j.1432-1033.1996.00163.x. [DOI] [PubMed] [Google Scholar]
- 50.Orrenius S, Zhivotovsky B, Nicotera P. Regulation of cell death: the calcium-apoptosis link. Nat. Rev. Mol. Cell Biol. 2003;4:552–565. doi: 10.1038/nrm1150. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Discussion
Materials and Methods
References
Figures