Abstract
Prior work has shown that heterozygosity G/C of single nucleotide polymorphism (SNP rs2910164) within the precursor of microRNA-146a predisposes to PTC (odds ratio = 1.62, P = 0.000007) although the mechanism was unclear. Here, we show that GC heterozygotes differ from both GG and CC homozygotes by producing 3 mature microRNAs: 1 from the leading strand (miR-146a), and 2 from the passenger strand (miR-146a*G and miR-146a*C), each with its distinct set of target genes. TaqMan analysis of paired tumor/normal samples revealed 1.5- to 2.6-fold overexpression of polymorphic miR-146a* in 7 of 8 tumors compared with the unaffected part of the same gland. The microarray data showed that widely different transcriptomes occurred in the tumors and in unaffected parts of the thyroid from GC and GG patients. The modulated genes are mainly involved in regulation of apoptosis leading to exaggerated DNA-damage response in heterozygotes potentially explaining the predisposition to cancer. We propose that contrary to previously held views transcripts from the passenger strand of miRs can profoundly affect the downstream effects. Heterozygosity for polymorphisms within the premiR sequence can cause epistasis through the production of additional mature miRs. We propose that mature miRs from the passenger strand may regulate many genetic processes.
Keywords: papillary thyroid carcinoma, polymorphism, PTC
MicroRNAs (miRs) are small noncoding RNA molecules that typically function as negative regulators of the expression of protein-encoding genes. MiRs are transcribed from endogenous DNA and processed from primary transcript (pri-microRNA) to a hairpin precursor (premicroRNA) comprising 2 strands: the leading strand used for production of the mature microRNA and the passenger strand that is believed to be degraded. Mature miRs target and bind to transcripts and interfere with their translation into protein. Watson–Crick complementarity between the target and the “seed” region comprising 2–8 nt of the mature miR is both necessary and sufficient for targeting and regulation of mRNAs by miRs. The sequence of this “seed” region is the basis of most genome-wide predictions of miR regulatory sites within miR-regulated genes.
The first evidence of a potential role for microRNAs in papillary thyroid carcinoma (PTC) was an overexpression of several miRs in tumor tissue compared with unaffected tissue of the thyroid gland (1). The list of highly up-regulated miRs (up to 19.3-fold) included miR-146, miR-221/222, miR-155, miR-34, and miR-181 and was soon confirmed (1, 2). We recently noted that some of these miRs, including miR-146a, are already overexpressed in the unaffected part of thyroid glands of patients with PTC compared with normal tissue from non-PTC individuals. This finding suggests that altered microRNAs might be early factors playing a role in the tumorigenesis of PTC. Further evidence supporting this argument came from our study showing a genetic association between PTC and a single nucleotide polymorphism (SNP, rs2910164) in the precursor of miR-146a. Individuals heterozygous for the SNP had an increased risk of acquiring PTC (OR-1.62, 95%CI 1.3–2.0, P = 0.000007) (3). We further demonstrated that the processing of the microRNA was affected by the SNP (3). Heterozygosity as a genetic risk rather than either homozygosity is a rare phenomenon (referred to as overdominance) and should be critically evaluated. It might be called a special form of genetic epistasis, where the phenotype of heterozygotes differs from the sum of the phenotypes of both alleles (4, 5).
We here propose that the phenomenon is caused by the existence of additional mature microRNAs generated from the passenger strand of the miR-146a precursor. That miRs produced from passenger strands can be functional was reported very recently and could be of great importance (6, 7). This realization should alter the previously held view that microRNA genes produce 1 functional molecule. In the case of miR-146a the presence of the SNP potentially generates 2 isoforms (marked *): miR-146a*G from the allele carrying G, and miR-146a*C from the C allele (Fig. 1A). Thus, GG and CC homozygotes each produce 2 mature molecules (miR-146a from the leading strand, and miR-146a*G or *C, respectively, from the passenger strand) whereas GC heterozygotes differ from both homozygotes by producing 3 mature miRs (miR-146a and both miR-146a*G and miR-146a*C).
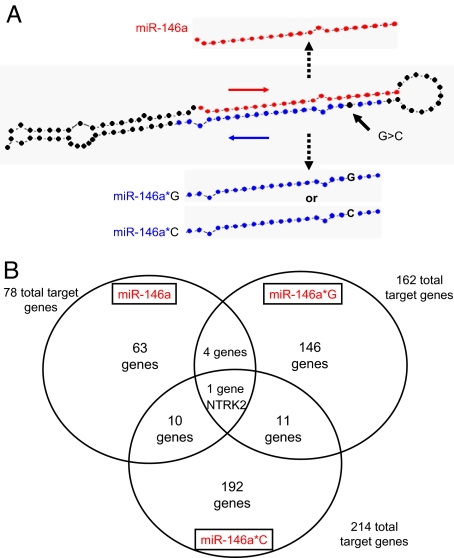
Fig. 1.
Structure and target genes of the 3 mature microRNAs produced from premiR-146a. (A) The predicted structure of premiR-146a. The mature miR-146a is shown by red dots. The location of passenger strand miR-146a* is shown by blue dots. The location of the G/C SNP is shown by an arrow. (B) Target genes of the 3 mature products of premiR-146a (Target Rank score >0.35).
Results and Discussion
Expression of miR-146a* in Transfected Cells and PTC Tumors.
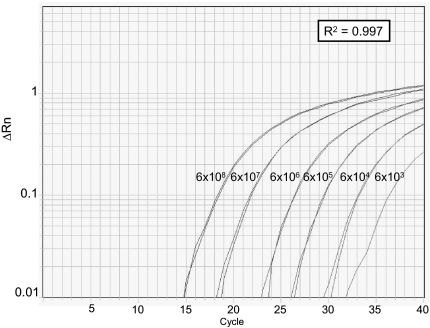
The expression of miR-146a* was detected by custom designed stem-loop TaqMan real-time RT-PCR assays. First, high sensitivity and specificity of the method was established using serial dilutions of synthesized miR-146a*G and miR-146a*C (Fig. 2). Then we evaluated the expression of both variants of miR-146a* in U2OS cells transfected with an expression vector carrying 1054nt of the G or C allele of pri-miR-146a. Both alleles were processed toward mature forms and expression of passenger strand miR-146a*G and miR-146a*C was detected in allele G and C, respectively. Finally, we confirmed the expression of miR-146a* in 16 samples of thyroid tissue derived from PTC patients (Table 1).
Fig. 2.
Quality control of custom designed stem loop real-time RT-PCR assay. Plots show the amplification of serial dilutions of synthesized mature miR-146a*G oligo. Similar results were obtained from the assay for miR-146a*C with serial dilutions of mature miR-146a*C oligo.
Table 1.
Expression of miR-146a*G and miR-146a*C determined by TaqMan
| samples | SNP | Number of molecules |
|
|---|---|---|---|
| miR-146a*G | miR-146a*C | ||
| PTC-01-Tumor | GC | 4,179 | 9,161 |
| PTC-01-Normal | GC | 6,106 | 5,746 |
| PTC-02-Tumor | GC | 5,392 | 9,108 |
| PTC-02-Normal | GC | 3,716 | 6,501 |
| PTC-03-Tumor | GC | 34,439 | 158,823 |
| PTC-03-Normal | GC | 23,473 | 417,861 |
| PTC-04-Tumor | GG | 20,253 | 0 |
| PTC-04-Normal | GG | 9,063 | 0 |
| PTC-05-Tumor | GG | 13,998 | 0 |
| PTC-05-Normal | GG | 5,552 | 0 |
| PTC-06-Tumor | GG | 38,737 | 0 |
| PTC-06-Normal | GG | 16,997 | 0 |
| PTC-07-Tumor | GG | 25,385 | 0 |
| PTC-07-Normal | GG | 57,739 | 0 |
| PTC-08-Tumor | GG | 6,820 | 0 |
| PTC-08-Normal | GG | 2,596 | 0 |
| pcDNA3-G | 491,698 | 0 | |
| pcDNA3-C | 0 | 149,017 | |
| both | 485,210 | 87,953 | |
| pcDNA3-empty | 0 | 0 | |
| mock | 0 | 0 | |
TaqMan ″stem-loop″ real-time reverse transcription (RT)-PCR assay in 8 PTC tumor/normal pairs, and the U2OS cell line transfected with an expression vector carrying 1054nt of the G or C allele or 50% of each allele of pri-miR-146a. We used total RNA in the amount of 200 ng (PTC cases) and 120 ng (transient transfection).
Impact of SNP on miR-146a* Target Genes.
The SNP is located in the crucial “seed” sequence of miR-146a*, so it determines its complementarity to potential target genes affecting the functionality of both isoforms. Using the TargetRank software (8) we found that the two alternative mature forms of the passenger strand miR* were predicted to have profoundly different target genes (162 genes for miR-146a*G and 214 genes for miR-146a*G, using a Target Rank score of >0.35) with only 12 genes being shared by the two isoforms (Fig. 1B and Table S1 and Table S2). Of note, one of the shared genes, neurotrophic tyrosine kinase receptor type 2 or NTRK2, is also targeted by miR-146a from the leading strand. The NTRK2 proto-oncogene encodes a kinase that, upon neurotrophin binding, phosphorylates members of the MAPK pathway leading to cell differentiation.
Because each mature miR binds to a distinct set of target genes, different target genes are affected by the miRs produced by GG or CC homozygotes (miR-146a and miR-146a*G or *C, respectively) and a third set is produced by GC heterozygotes (miR- 146a and both miR-146a*G and miR-146a*C). Thus, heterozygosity is the only state in which the set of target genes comprises all 3 subsets (totally 454 regulated genes). We propose that this may explain the demonstrated predisposition to PTC by the heterozygous state of the SNP.
Impact of SNP on Transcriptomes of Unaffected Thyroid Tissue.
To query whether homozygotes and heterozygotes display different transcriptomes we reanalyzed existing cDNA microarray data (HG-U133 array of Affymetrix) of thyroid tissue derived from 16 PTC patients. The data were generated and validated by us with the purpose of describing the gene expression signature of PTC tumors (9). In the present study we first determined the SNP status of available samples and then compared the transcriptomes of GC (n = 4) and GG (n = 6) samples derived from the unaffected part of the thyroid glands of PTC cases. CC homozygotes were not analyzed due to limited availability (n = 2). To identify genes expressed differentially between GC and GG samples, 2-sample t tests with random variance model was applied using BRB-Array Tools Version 3.7 (http://linus.nci.nih.gov/BRB-ArrayTools.html). The effect of miRs on target genes is mostly on the translational rather than transcriptional level. Thus, the expression signatures most probably reflect changes in genes downstream from the miR-targeted and translationally repressed mRNA. Transcriptomes of GC and GG samples differed by the expression of 358 genes (fold change >1.2, P < 0.025) (Table S3). To analyze the function of those genes, we used the DAVID software (10), which queries for biological activities according to the Gene Ontology (GO) annotations. The DAVID functional analysis (Table 2) showedthat the main biological processes altered in heterozygotes were apoptosis (27 genes, P = 0.011), localization (77 genes, P = 0.019), cell differentiation (47 genes, P = 0.037), and blood vessel development (9 genes, P = 0.032), which are often involved in tumorigenesis.
Table 2.
Gene ontology (GO) categories enriched in unaffected parts of the thyroidand and in tumors
| P | GO name | No. of genes |
GO identifiers | ||||
|---|---|---|---|---|---|---|---|
| Total | Down | Up | |||||
| Unaffected parts of thyroid | |||||||
| 0.011 | Apoptosis | 27 | 10 | 17 | 0006915 | 0012501 | 0016265 |
| 0.019 | Localization | 77 | 34 | 43 | 0051179 | 0006810 | 0051234 |
| 0.037 | Cell differentiation | 47 | 23 | 24 | 0030154 | 0048468 | 0048869 |
| 0.025 | Neurogenesis | 12 | 9 | 3 | 0022008 | 0030182 | 0048699 |
| 0.032 | Blood vessel development | 9 | 3 | 6 | 0001568 | 0001944 | 0048514 |
| 0.004 | Metanephros development | 6 | 4 | 2 | 0001656 | 0001655 | 0001822 |
| 0.044 | Phosphorylation | 29 | 11 | 18 | 0016310 | 0006796 | 0006793 |
| Tumors | |||||||
| 0.0001 | Apoptosis | 44 | 12 | 32 | 0006915 | 0042981 | 0012501 |
| 0.0087 | Positive regulation of NF-kappaB cascade | 12 | 1 | 11 | 0043123 | 0007249 | 0043122 |
| 0.0001 | Anatomical structure development | 108 | 55 | 53 | 0048856 | 0048731 | 0007275 |
| 0.0000 | Regulation of cellular process | 207 | 94 | 113 | 0050794 | 0065007 | 0050789 |
| 0.0003 | mRNA processing | 22 | 5 | 17 | 0006397 | 0016071 | 0008380 |
| 0.003 | Mitotic cell cycle | 22 | 7 | 15 | 0000278 | 0007067 | 0000087 |
| 0.006 | Posttranslational protein modification | 72 | 30 | 42 | 0043687 | 0006464 | 0043412 |
The numbers refer to genes down- or up-regulated in GC individuals compared to GG individuals.
Impact of SNP on Transcriptomes of PTC Tumors.
Finally we compared transcriptomes derived from tumor tissues of GC (n = 8) heterozygous and GG (n = 8) homozygous PTC cases and found very similar but more complex results. A total of 575 genes were differentially expressed between the GC and GG genotypes (fold change >1.2, P < 0.025) (Table S4). By DAVID software analysis, the main GO categories altered in heterozygotes (Table 2) were again regulation of apoptosis (44 genes, P = 0.0001) and notably, positive regulation of the NF-kappa-B cascade (12 genes, P = 0.009), and regulation of cellular processes (207 genes, P = 0.000001) with modulated mRNA processing (22 genes,P = 0.0003), mitotic cell cycle (22 genes, P = 0.003), and posttranslational protein modification (72 genes, P = 0.006). The main metabolic pathway significantly targeted in heterozygotes was the HIV-1 negative effector of the Fas and TNF pathway (9 genes, P = 0.0025), which is part of the GO category of apoptosis. As shown in Fig. S1 DNA-damage response leading to apoptosis seems to be exaggerated in GC heterozygotes compared with GG homozygotes. The DNA-damage response pathway is activated in early stage lesions of several human cancers (11, 12). Nevertheless, we also noticed that 11 genes of the anti-apoptotic NF-kappa B pathway were up-regulated and only one down-regulated in GC heterozygous tumors.
Induction of expression of miR-146a as a result of NF-kappa B activation is described in refs. 13–15. It is suggested that miR-146a by targeting IRAK1 and TRAF6 is part of a negative feedback loop and acts as an inhibitor of the activated NF-kappa B pathway (13). The level of that modulation may depend on the miR-146a G vs. C genotype because the inhibition of IRAK1 and TRAF6 is much lower in the presence of the C allele as shown in ref. 3.
Conclusions
We conclude that GC heterozygotes differ from both homozygotes by producing 3 mature microRNAs: 1 from the leading strand (miR-146a), and two from the passenger strand (miR-146a*G and miR-146a*C), each with its distinct set of target genes. We show widely different transcriptomes in GC and GG tumors and, notably, also in unaffected parts of the thyroid from PTC patients. We propose that the production of distinct miRs and regulation of different target genes by heterozygotes compared with homozygotes may explain the predisposition to PTC displayed by individuals who are heterozygous.
Materials and Methods
Thyroid Tissue Samples.
After approval of the Institutional Review Board and patient consent, fresh samples from PTC tumor tissue and unaffected thyroid tissue from the contralateral lobe were obtained from patients with sporadic PTC undergoing surgical resection; the samples were snap-frozen in liquid nitrogen and stored at −80 °C.
PremiR-146a Expression Vectors and U2OS Cell Transfection.
To create premiR-146a+60G and premiR-146a+60C expression vectors genomic fragments (1054bp) corresponding to the premiR and its flanking regions were amplified from human genomic DNA and cloned into the pcDNA3 vector (Invitrogen). For details, see ref. 3.
Real-time RT-PCR Detection of miR-146a*.
To evaluate the miR-146a*G and miR-146a*C expression levels, “stem-loop” real-time TaqMan reverse transcription (RT)-PCR was used. Total RNA was used for RT reactions that were performed using High Capacity cDNA Reverse Trancription Kit (Applied Biosystems) according to the manufacturer's protocol. Real-time PCR was performed on the ABI Prism 7900HT Sequence Detection System with U6 RNA as an endogenous loading control. The RT primer was the same for both miR-146a*G and mir-146a*C: 5′-GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACCTGAAG-3′. The qPCR primers for miR-146a*G were 5′-GTGCAGGGTCCGAGGT-3′ and 5′-GCCGCCCTGTGAAATTCAGTT-3′. The qPCR primers for miR-146a*C were 5′-GTGCAGGGTCCGAGGT-3′ and 5′-GCCGCCCTCTGAAATTCAGTT-3′. The TaqMan probe for both miRs* was 6-FAM-TGGATACGACCTGAAG(MGB). The cycle number at which the product level exceeded an arbitrarily chosen threshold (CT) was determined for each target sequence, and the amount of molecules was determined based on the standard curve. Standard curves were produced using serial dilutions of synthesized mature miR-146a*G and miR-146a*C (600 mln to 600 molecules).
Gene Expression Arrays.
Affymetrix HG-U133 chips (Affymetrix) were used to evaluate genomewide gene expression levels. For details, see ref. 9.
Statistical and Bioinformatics Analysis of Microarray Data.
Data from Affymetrix chips were analyzed by using the robust multichip average to calculate normalized expression values. To identify genes that are differentially expressed between two groups, Student's t test was applied.
Supplementary Material
Acknowledgments.
We thank Drs. Dorota Kula and Michal Jarzab for their assistance, and Dr. Denis Guttridge for critical reading of the manuscript. This work was supported by National Cancer Institute Grants CA16058 (to the Ohio State University Comprehensive Cancer Center) and CA124570 (to M.D.R.); an American Thyroid Association Thy-Ca Research Grant (to K.J), Polish Ministry of Science and Higher Education Grant 2P05A02230 (to B.J), and an Abbott Thyroid Research Clinical Fellowship Award presented by the Endocrine Society (to K.J.).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0812591106/DCSupplemental.
References
- 1.He H, et al. The role of microRNA genes in papillary thyroid carcinoma. Proc Natl Acad Sci USA. 2005;102:19075–19080. doi: 10.1073/pnas.0509603102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pallante P, et al. MicroRNA deregulation in human thyroid papillary carcinomas. Endocr Relat Cancer. 2006;13:497–508. doi: 10.1677/erc.1.01209. [DOI] [PubMed] [Google Scholar]
- 3.Jazdzewski K, et al. Common SNP in pre-miR-146a decreases mature miR expression and predisposes to papillary thyroid carcinoma. Proc Natl Acad Sci USA. 2008;105:7269–7274. doi: 10.1073/pnas.0802682105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Carlborg O, Haley CS. Epistasis: Too often neglected in complex trait studies? Nat Rev Genet. 2004;5:618–625. doi: 10.1038/nrg1407. [DOI] [PubMed] [Google Scholar]
- 5.Boone C, et al. Exploring genetic interactions and networks with yeast. Nat Rev Genet. 2007;8:437–449. doi: 10.1038/nrg2085. [DOI] [PubMed] [Google Scholar]
- 6.Okamura K, et al. The regulatory activity of microRNA* species has substantial influence on microRNA and 3′ UTR evolution. Nat Struct Mol Biol. 2008;15:354–363. doi: 10.1038/nsmb.1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kim S, et al. MicroRNA miR-199A* regulates the Met proto-oncogene and the downstream extracellular signal-regulated kinase 2 (ERK2) J Biol Chem. 2008;283:18158–18166. doi: 10.1074/jbc.M800186200. [DOI] [PubMed] [Google Scholar]
- 8.Nielsen CB, et al. Determinants of targeting by endogenous and exogenous microRNAs and siRNAs. RNA. 2007;13:1–18. doi: 10.1261/rna.768207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jarzab B, et al. Gene expression profile of papillary thyroid cancer: Sources of variability and diagnostic implications. Cancer Res. 2005;65:1587–1597. doi: 10.1158/0008-5472.CAN-04-3078. [DOI] [PubMed] [Google Scholar]
- 10.Dennis G, Jr, et al. DAVID: Database for annotation, visualization, and integrated discovery. Genome Biol. 2003;4:P3. [PubMed] [Google Scholar]
- 11.Gorgoulis VG, et al. Activation of the DNA damage checkpoint and genomic instability in human precancerous lesions. Nature. 2005;434:907–913. doi: 10.1038/nature03485. [DOI] [PubMed] [Google Scholar]
- 12.Bartkova J, et al. DNA damage response as a candidate anti-cancer barrier in early human tumorigenesis. Nature. 2005;434:864–870. doi: 10.1038/nature03482. [DOI] [PubMed] [Google Scholar]
- 13.Taganov KD, et al. NF-kB-dependent induction of microRNA miR-146, an inhibitor targeted to signaling proteins of innate immune responses. Proc Natl Acad Sci USA. 2006;103:12481–12486. doi: 10.1073/pnas.0605298103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Motsch N, et al. Epstein–Barr Virus-encoded latent membrane protein 1 (LMP1) induces the expression of the cellular microRNA miR-146a. RNA Biol. 2007;4:131–137. doi: 10.4161/rna.4.3.5206. [DOI] [PubMed] [Google Scholar]
- 15.Cameron JE, et al. Epstein–Barr virus latent membrane protein 1 induces cellular microRNA miR-146a, a modulator of lymphocyte signaling pathways. J Virol. 2008;82:1946–1958. doi: 10.1128/JVI.02136-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.