Abstract
Organisms have evolved elaborate systems that ensure the homeostasis of the thiol redox environment in their intracellular compartments. In Escherichia coli, the cytoplasm is kept under reducing conditions by the thioredoxins with the help of thioredoxin reductase and the glutaredoxins with the small molecule glutathione and glutathione reductase. As a result, disulfide bonds are constantly resolved in this compartment. In contrast to the cytoplasm, the periplasm of E. coli is maintained in an oxidized state by DsbA, which is recycled by DsbB. Thioredoxin 1, when exported to the periplasm turns from a disulfide bond reductase to an oxidase that, like DsbA, is dependent on DsbB. In this study we set out to investigate whether a subclass of the thioredoxin superfamily, the glutaredoxins, can become disulfide bond-formation catalysts when they are exported to the periplasm. We find that glutaredoxins can promote disulfide bond formation in the periplasm. However, contrary to the behavior of thioredoxin 1 in this environment, the glutaredoxins do so independently of DsbB. Furthermore, we show that glutaredoxin 3 requires the glutathione biosynthesis pathway for its function and can oxidize substrates with only a single active-site cysteine. Our data provides in vivo evidence suggesting that oxidized glutathione is present in the E. coli periplasm in biologically significant concentrations.
Keywords: protein oxidation, monothiol, SRP
Protein disulfide bond formation in the E. coli periplasm is catalyzed by DsbA, a member of the thioredoxin superfamily. Members of this superfamily use an active-site motif, usually consisting of Cys-X-X-Cys, to carry out either protein oxidation or reduction. In its active state in vivo, DsbA contains these 2 cysteines joined in a disulfide bond, which it transfers to substrate proteins. The resulting reduced cysteines of DsbA are reoxidized into the active form by the membrane protein DsbB (1).
An important determinant of whether thioredoxin superfamily members act to efficiently reduce protein disulfide bonds or promote their formation is their redox potential. For instance, in E. coli, DsbA with a redox potential of −120 mV acts as an oxidant, while thioredoxin 1 with a redox potential of −270 mV acts as reductant (2). However, another factor that can influence the role of such proteins is the environment of their subcellular localization. Thus, thioredoxin 1, normally a cytoplasmic reductant, can promote disulfide bond formation when exported to the periplasm (3). This is made possible by the ability of DsbB to oxidize thioredoxin 1 (4). DsbB is also able to oxidize other non-native thioredoxin family members from very distant species, including eukaryotic protein disulfide isomerase (4). There is, however, an engineered form of periplasmically localized thioredoxin 1 containing a catalytic [2Fe-2S] cluster that can promote disulfide bond formation independently of DsbB (5).
The glutaredoxins, a subclass of the thioredoxin superfamily, share the thioredoxin fold as well as a Cys-X-X-Cys/Ser active site motif. Glutaredoxins are cytoplasmic proteins that often share similar roles with the thioredoxins, but require for their function as reducing proteins the small molecule glutathione. While the cytoplasmic thioredoxins are kept in their reduced state by thioredoxin reductase, the glutaredoxins require both glutathione and glutathione reductase (6).
Initially, we were interested in the substrate specificity of DsbB and asked whether DsbB is generally able to accept members of the thioredoxin superfamily as substrates. To do this, we exported various glutaredoxins to the periplasm and determined (i) whether they could promote disulfide bond formation and (ii) whether this activity is dependent on the presence of DsbB. As a result of initial findings, our studies concentrated on glutaredoxin 3. Here we show that exported glutaredoxin 3 can promote disulfide bond formation in the periplasm. The exported enzyme was not dependent on DsbB for its function in vivo. However, the protein oxidase activity of exported glutaredoxin 3 requires only 1 active-site cysteine and is dependent on the glutathione biosynthetic pathway in the cytoplasm. The simplest explanation for these results is that oxidized glutathione is present in the periplasm and collaborates with glutaredoxin 3 to oxidize proteins. The existence of glutathione in the secretory compartments of bacteria had not been established because small molecules readily diffuse from the cytoplasm in the course of cell fractionation experiments used to isolate the contents of the periplasm.
Results
Glutaredoxins Exported to the Periplasm Catalyze Disulfide Bond Formation.
To export 3 E. coli glutaredoxins (glutaredoxin 1, 2, and 3) and the bacteriophage T4 glutaredoxin 1, we fused the SRP-dependent signal sequence of TorT to the N terminus of the respective proteins. In addition, we examined the effect of fusing the TorT signal sequence to thioredoxin 1 and thioredoxin 2. In the case of glutaredoxin 3 we also fused it to the TAT pathway signal sequence of TorA. The resulting constructs were assessed for export by fractionation experiments (data not shown). The TorT signal sequence allowed efficient export of thioredoxin 1 (TrxAP) and thioredoxin 2, glutaredoxin 3 (GrxCP), and bacteriophage T4 glutaredoxin 1 (NrdCP). Glutaredoxin 1 and glutaredoxin 2 were not efficiently exported, resulting in the accumulation of precursor protein in the spheroplast fraction even at very low expression levels. To avoid possible artifacts arising from poor export efficiency, we did not pursue further the glutaredoxin 2 construct.
We asked whether glutaredoxins, when localized to the periplasm, promote disulfide bond formation in cell envelope proteins, thus changing their activities from protein disulfide bond reductants to oxidants, as it is the case for thioredoxin 1 (3). We first determined whether the exported proteins could restore motility to strains that are deleted for dsbA. The P-ring protein FlgI of the E. coli flagella motor, which is required for motility, contains a disulfide bond. In dsbA strains, FlgI is missing the disulfide bond, resulting in a non-motile phenotype (7). To promote disulfide bond formation in the periplasm, TrxAP has to be oxidized by DsbB. Recent findings showed that the periplasmic reductive thiol redox pathway, composed of DsbC and DsbD, also affects TrxAP function (8). To circumvent any interference of the DsbC-DsbD system on our constructs, we used strains that lack besides dsbA also dsbC and dsbD.
Expression of periplasmically localized glutaredoxin 3 (GrxCP) in cells lacking dsbA dsbC dsbD led to increased motility on rich media, comparable to an isogenic strain expressing TrxAP. The inefficient export of glutaredoxin 1 (GrxAP) likely explains why this construct led to less motility (Table 1); GrxAP was not studied further. Expression of periplasmically localized thioredoxin 2 is as effective as TrxAP in restoring motility, whereas strains producing NrdCP did not show any increase in motility compared to the empty vector controls (Table 1).
Table 1.
Motility phenotype of strains expressing various thioredoxin or glutaredoxin constructs
| Construct on plasmid | Rich Medium |
Minimal Medium |
||
|---|---|---|---|---|
| dsbB+ | ΔdsbB | dsbB+ | ΔdsbB | |
| empty vector | − | − | − | − |
| TorTss-TrxA | ++ | − | ++ | − |
| TorAss-TrxA | ++ | − | ++ | − |
| TorTss-TrxA[CPHC] | ++++ | − | ++++ | − |
| TorTss-TrxC | ++ | − | ++ | − |
| TorTss-GrxA | + | + | ± | ± |
| TorTss-GrxC | ++ | ++ | ± | ± |
| TorAss-GrxC | ++ | ++ | ± | ± |
| TorTss-GrxC[CPHC] | ++ | ++ | ± | ± |
| TorTss-NrdC | − | − | − | − |
| TorTss-NrdC[CPHC] | − | − | − | − |
Motility phenotype of various strains. Strains (HK453 or MER144) expressing different constructs from plasmids were examined on NZ medium or M63 minimal medium. The motility was evaluated by comparing the motility to that of the negative control (empty vector) and that of the positive control (strain expressing TrxAP) on each media.
We also found that when thioredoxin 1 and glutaredoxin 3 are exported by the TAT pathway signal sequence of TorA they allow a comparable level of motility to that of the same enzymes exported by the SRP-dependent TorT signal sequence (Table 1). As judged from Western analysis, constructs with either signal sequence were expressed to similar levels (data not shown). These results suggest that the choice of the export pathway (either the SRP pathway or the TAT pathway) does not affect the oxidizing ability of these 2 proteins in the periplasm, in contrast to the thioredoxin 1 mutant containing an [2Fe-2S] cluster (5).
As judged by restoration of motility, the glutaredoxins tested are not as oxidizing as thioredoxin 1 when exported to the periplasm, especially in strains grown on minimal medium. The nature of the amino acid residues between the active site cysteines can greatly influence the redox potential of thioredoxin-like proteins. For example, thioredoxin 1 becomes much more oxidizing when its active site (Cys-Gly-Pro-Cys) is changed to that of DsbA (Cys-Pro-His-Cys) (4). We asked whether changing the amino acid sequence of the active site motif of the glutaredoxins to that of DsbA (GrxCP[CPHC] and NrdCP[CPHC], respectively) might enhance their ability to act as oxidases in the periplasm. While a control consisting of TrxAP with a DsbA active site (TrxAP[CPHC]) results in a large increase in motility of our test strains, expression of GrxCP[CPHC] or NrdCP[CPHC] gave no change in the motility phenotype from that seen with the wild-type sequence constructs (Table 1). The finding that, unlike thioredoxin 1, altering the active site of these glutaredoxins to that of the highly oxidizing protein DsbA does not improve the oxidizing ability of these enzymes, suggests that the glutaredoxins differ from TrxAP in the mechanism by which they promote disulfide bond formation in the periplasm (see the following sections).
Periplasmic Glutaredoxin 3 Does Not Require DsbB for Protein Oxidation.
To continuously promote oxidation of FlgI, periplasmic thioredoxins or glutaredoxins must themselves be reoxidized by transferring the electrons they receive from FlgI to an electron acceptor. Since DsbB reoxidizes not only its native partner DsbA but also non-native substrates such as thioredoxin 1 and the alpha domain of PDI, we asked whether glutaredoxin 3 is also reoxidized by DsbB (3, 4). We tested motility of strains expressing various periplasmic thioredoxins and glutaredoxins in the presence or absence of DsbB (Table 1). Surprisingly, GrxCP and GrxAP conferred motility independently of DsbB, indicating that they are reoxidized by a mechanism different from that used by DsbA and TrxAP.
We also noticed that motility of the strains expressing periplasmic glutaredoxin 1 or 3 is enhanced in rich media compared with motility of these strains in minimal media, suggesting that a component in the rich media contributed to oxidation of the glutaredoxins in the periplasm (Table 1). In contrast, NrdCP failed to restore the motility in the dsbA mutant on either of the 2 nutrient media.
Comparison of the Oxidative Efficiency of GrxCP with Other DsbA Substrates.
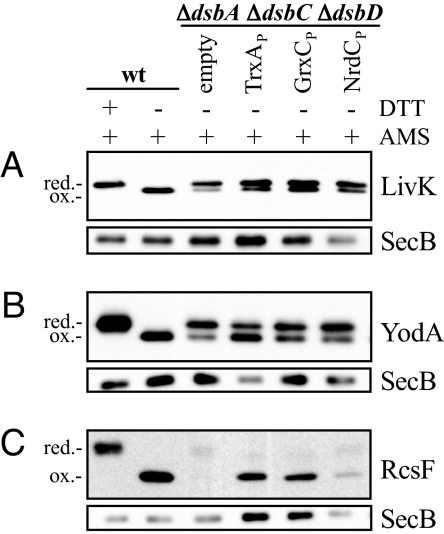
We tested the ability of the periplasmically localized glutaredoxins and thioredoxin to oxidize other DsbA substrates, including LivK, YodA, and RcsF (9). Plasmids, each encoding one of the DsbA substrates fused to the c-Myc epitope tag sequence, were expressed in the ΔdsbA ΔdsbC ΔdsbD strain together with either TrxAP, GrxCP, or NrdCP. As a control, the DsbA substrate constructs were also expressed in wild-type cells harboring an empty vector. Cells were grown at 30 °C in M63 minimal media supplemented with glycerol and arabinose. Samples were collected and subjected to alkylation with AMS. Alkylation of a free thiol with AMS adds a mass of approximately 0.5 kDa to the protein, which can be used to analyze the oxidation state of cysteines in a protein by Western analysis (10). To control for protein amounts we used the cytoplasmic protein SecB as a marker.
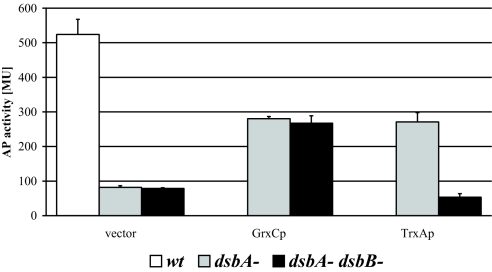
All tested substrate proteins were fully oxidized in the wild-type strain (Fig. 1). In a ΔdsbA ΔdsbC ΔdsbD strain background, they were predominantly in their reduced forms or degraded (Fig. 1), as seen before (9). Co-expression of TrxAP resulted in increased accumulation of oxidized protein, especially in the case of RcsF (Fig. 1C). Coexpression of GrxCP and NrdCP gave different results. While GrxCP increased the amounts of oxidized RcsF and LivK, it was inefficient in the oxidation of YodA when compared with TrxAP. NrdCP also oxidized RcsF, albeit at a reduced efficiency; some reduced RcsF remained. We quantitated the efficiency of disulfide bond formation of exported glutaredoxins by measuring the level of enzymatic activity of the DsbA substrate alkaline phosphatase (AP). AP activity depends on the formation of its disulfide bonds (11). We constructed dsbA or dsbA dsbB strains that carried a phoR mutation allowing constitutive expression of AP (12), and transformed them with a plasmid expressing GrxCP or TrxAP (Fig. 2). In strains lacking the Dsb proteins, expression of GrxCP increased AP activity 3-fold over a control carrying an empty plasmid, reaching approximately 60% of the wild-type level. As with the motility assay, the increased AP activity was independent of DsbB (Fig. 2). Expression of TrxAP in dsbA strains lead to a DsbB-dependent 3-fold increase of AP activity. GrxCP expression in strains with deletions of dsbA, dsbC, and dsbD caused the same 3-fold increase in AP activity (data not shown). Overall, GrxCP is a rather effective oxidant of AP, RcsF, and LivK and less so of YodA and FlgI. NrdCP did not measurably oxidize AP (data not shown). Thus, RcsF was the only protein that NrdCP was able to oxidize among the DsbA substrate proteins tested. In the remaining experiments, we have focused on GrxCP.
Fig. 1.
Redox state of various DsbA substrates. LivK-c-Myc (A), YodA-c-Myc (B), or RcsF-c-Myc (C) were expressed from plasmids (pMER154, pMER155, and pMER156) and detected by Western blot using anti-c-Myc antibody. Strains used were HK295 (wt) and HK453 (ΔdsbA ΔdsbC ΔdsbD) harboring empty vector (pMER79) or plasmids encoding TrxAP, GrxCP, or NrdCP (pMER90, pMER94, and pMER96). Cultures were grown and samples were prepared as described in materials and methods. Where indicated, samples were treated with 100 mM DTT (DTT) before AMS alkylation. The mobility of DTT-treated proteins indicates the position of the reduced form. As a loading control, the same sample volumes were subjected to an immunoblot detecting SecB.
Fig. 2.
Restoration of AP activity by GrxCP or TrxAp. Wild-type (MER360), ΔdsbA (MER390), or ΔdsbA ΔdsbB (MER392) strains expressing GrxCP or TrxAp from plasmids (pTrc99a-ssTorA-GrxC or pTrc99a-ssTorA-TrxA) were grown in M63 minimal medium at 30 °C, aliquots were taken at early log-phase and AP activity measured as described in materials and methods.
Oxidative Activity of GrxCP Depends on the Cytoplasmic Glutathione Biosynthetic Pathway.
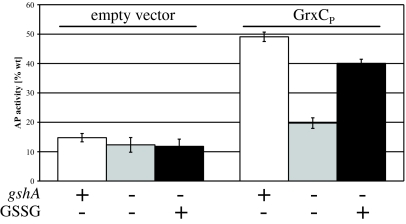
If GrxCP does not require DsbB for its oxidative activity, what is its source of oxidative power? In the cytoplasm of E. coli, glutaredoxins are maintained in the reduced state by reduced glutathione (GSH) (6). We considered the possibility that the periplasmic version of glutaredoxin 3, GrxCP, is oxidized through a reaction with oxidized glutathione. GrxCP-dependent motility was enhanced on rich media compared to minimal media (Table 1) perhaps due to the presence of small oxidizing molecules, such as glutathione, present in rich media (1). In minimal media, there may be sufficient export of glutathione from the cytoplasm (13, 14) to allow the oxidation reaction to occur in minimal medium. If the oxidative activity of GrxCP is dependent on oxidized glutathione, then mutations that eliminate glutathione biosynthesis should prevent the oxidation of substrates from taking place. We constructed strains lacking the gene for the first enzyme in the glutathione biosynthesis pathway (gshA) and analyzed its effect on GrxCP-promoted alkaline phosphatase activity. Strains were grown in minimal media to avoid any interfering effects of glutathione present in rich media.
We found that the elimination of glutathione biosynthesis resulted in a dramatic drop in AP activity, nearly eliminating the effect of GrxCP on disulfide bond formation in AP (Fig. 3). We also showed that adding to a culture lacking the glutathione biosynthetic pathway (ΔgshA) 1 μM oxidized glutathione (GSSG), a concentration that corresponds to physiological concentrations of this molecule reported to accumulate in culture supernatants during exponential phase (14), restores AP activity, but only when GrxCP was present. These experiments are consistent with the proposal that GrxCP functions as an oxidant using periplasmic oxidized glutathione.
Fig. 3.
Influence of glutathione on GrxCP-dependent restoration of AP activity. A ΔdsbA ΔdsbB strain (MER392) harboring an empty vector (pMER79) or the same strain expressing GrxCP from plasmid pTrc99a-ssTorA-GrxC or isogenic strains with a deletion in gshA (MER396, MER382) were grown to log-phase at 30 °C in M63 minimal medium containing 0 μM or 1 μM oxidized glutathione (GSSG). AP activity is expressed as % of wild-type activity.
We also compared the effect of adding varying amounts of oxidized glutathione (GSSG) or reduced glutathione (GSH) with cultures lacking GshA (data not shown). In the absence of GrxCP, AP activity remained unchanged at all tested concentrations of GSH or GSSG (0, 0.1, 1, 10, or 100 μM final concentration), indicating that glutathione at these concentrations cannot promote the oxidation of the substrate on its own. In contrast, in the presence of GrxCP and either GSSG or GSH at concentrations greater than or equal to 1 μM, AP activity was enhanced. Addition of 10 μM GSH or GSSG conferred 75% AP activity compared with wild-type cells. Since we added glutathione to the cultures at the point of culture inoculation, we suspect that GSH is readily converted to GSSG by air oxidation (15).
GrxCP Can Act as an Oxidant with Only One of Its Redox-active Cysteines.
In the cytoplasm, glutaredoxins resolve disulfide bonds in substrate proteins by either a dithiol or monothiol mechanism (16). The dithiol mechanism involves a disulfide bond-exchange reaction between reduced glutaredoxin and oxidized substrate such that the glutaredoxin becomes oxidized and the substrate reduced. This mechanism requires 2 cysteines in the glutaredoxin active site. The monothiol pathway functions first via attack of GSH on an oxidized substrate resulting in a substrate-glutathione mixed disulfide complex. This complex is then resolved by a glutaredoxin using only 1 cysteine yielding a reduced substrate and a glutathionylated glutaredoxin. The latter is resolved by the attack of a second glutathione molecule (16, 17).
Up until this point, we have implicitly considered mechanisms for GrxCP oxidative activity in the periplasm that were similar to that of DsbA in which the 2 cysteines of the active site are joined in a disulfide bond that is directly transferred to substrates. However, given the existence of a monothiol mechanism for reduction in the cytoplasm, we wondered whether a monothiol mechanism might be used for oxidation in the periplasm. Thus, we asked whether the oxidation reaction carried out by GrxCP required both of its active site cysteines.
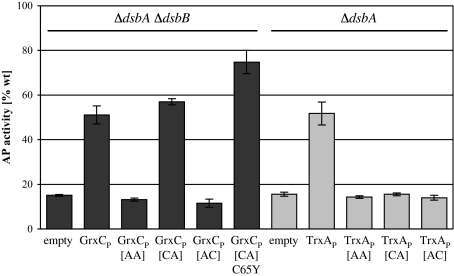
First, as a control, we showed that TrxAP requires both active site cysteines for its activity as an oxidant (Fig. 4). Similarly to TrxAP, GrxCP does not oxidize AP when its first cysteine (Cys-11) or both cysteines (Cys-11 and Cys-14) are changed to alanine. However, in contrast to thioredoxin 1, GrxCP is fully active in oxidizing AP as a monothiol glutaredoxin when the second active site cysteine (Cys-14) is changed to alanine. The third, non-active site cysteine in GrxCP is not involved in AP oxidation since a GrxCP variant having Cys-14 mutated to alanine and Cys-65 mutated to tyrosine, the latter being a well characterized mutation that has no effect on the glutaredoxin's in vitro reductase activity (2), was also able to oxidize AP, albeit at somewhat higher activity than wild type GrxCP. Previous studies suggested that mutating Cys-65 might prevent non-productive side reactions with glutathione (2), which could explain the observed increase in activity of this mutant in the context of the periplasm. That a monothiol glutaredoxin can act as a protein oxidant had been shown in vitro with yeast (18) and E. coli glutaredoxin 1 (19). In both studies, the authors outline mechanisms for how oxidized glutathione behaves as the final electron acceptor in this process.
Fig. 4.
Restoration of AP activity by active site mutants of GrxCP or TrxAP. AP activity of strains lacking dsbA (MER390) or dsbA and dsbB (MER392) expressing GrxCP or TrxAP active site variants from mutated plasmids derived from pTrc99a-ssTorA-GrxC or pTrc99a-ssTorA-TrxA, respectively. TrxAP[AA] or GrxCP[AA] have both active-site cysteines mutated to alanines, GrxCP[CA] has a Cys-14→Ala mutation, GrxCP[AC] has a Cys-11→Ala mutation, and GrxCP[CA]C65Y has Cys-14→Ala and Cys-65→Tyr mutations. TrxAP[CA] has a Cys-35→Ala mutation and TrxAP[AC] has a Cys-32→Ala mutation. Strains were grown as in previous experiments.
Discussion
In this article, we describe the conversion of the E. coli cytoplasmic reductant glutaredoxin 3 to an oxidant by promoting its export to the periplasmic space. The mechanism by which GrxCP carries out disulfide bond formation in the periplasm differs from the normal process of disulfide bond formation in bacteria. In particular, our results indicate that the source of oxidizing power for GrxCP is oxidized glutathione. This hypothesis is based on the findings that (i) GrxCP can function with only 1 active site cysteine, and (ii) mutations in the glutathione biosynthesis pathway abolish GrxCP activity. That glutathione could play such a role in the bacterial periplasm in the strains we have generated raises the possibility that glutathione in bacteria may be used in other extracytoplasmic pathways as a source of oxidizing or perhaps even as reducing power.
Bacteria vary in their ability to make disulfide bonds in proteins. Some make a high proportion of their proteins with disulfide bonds, others a low proportion and some may make none (20). The pathways for disulfide bond formation in those bacteria that do make disulfide bonds share common mechanistic features. These features include (i) the use of DsbA or, more generally, thioredoxin homologues, for the direct oxidization of a substrate protein's cysteines (4, 21); (ii) dependence on both cysteines of the active site Cys-X-X-Cys motif of DsbA for the generation of disulfide bonds in substrate proteins; and (iii) restoration of the oxidizing power of DsbA using membrane-bound quinones and a membrane protein, DsbB, in many bacteria or a homologue of eukaryotic vitamin K epoxide reductase in others (1, 20, 22).
Dithiol glutaredoxins, including glutaredoxin 3, share the protein fold and the Cys-X-X-Cys motif found in all thioredoxin family members, including DsbA (23). Despite these similarities, the oxidizing activity of GrxCP when exported to the periplasm is not dependent on DsbB. Furthermore, in contrast to the properties of TrxAP and DsbA, GrxCP only requires the first active-site cysteine for its oxidizing activity. Finally, the DsbB-independent oxidizing activity of GrxCP requires the presence of the cytoplasmic glutathione biosynthetic pathway. In a mutant that eliminates glutathione biosynthesis (gshA), the functioning of GrxCP as an oxidant is lost but can be restored by the addition of exogenously added glutathione.
DsbA or TrxAP catalyze disulfide bond formation via a disulfide bond-exchange mechanism, requiring both active site cysteines (24). The active site cysteines are connected by a disulfide bond, which is transferred to a substrate protein. GrxCP can oxidize proteins using only its N-terminal cysteine, excluding a simple dithiol disulfide bond-exchange reaction. The observed activities of GrxCP and its active site mutant, GrxCP[CA], are almost equal, suggesting that the wild-type GrxCP uses only its first active site cysteine for disulfide bond formation. Xiao and coworkers have previously shown in vitro that, in the presence of glutathione, glutaredoxins can catalyze disulfide bond formation requiring only the amino-terminal cysteine of the Cys-X-X-Cys motif (19). They presented a model (Fig. 5) explaining how glutaredoxins can catalyze disulfide bond formation by a monothiol mechanism. This model requires the activation of the glutaredoxin active site cysteine by glutathionylation, generating a highly reactive mixed disulfide that consequently is transferred to the substrate protein. The analogous behavior of GrxCP in our in vivo experiments makes it highly likely that it functions through a similar reactive mixed disulfide with a thiol based redox active molecule. Our findings that the gshA mutation eliminates the oxidative activity of GrxCP and that the activity is restored by the addition of exogenous glutathione suggests that glutathione is that thiol-based redox active molecule. Our results provide support for the hypothesis that glutathione is present in the periplasm at biologically significant concentrations. Consistent with our interpretation of the monothiol active version of GrxCP is our finding that the poorly oxidizing NrdCP can be converted to a more active oxidant by mutational elimination of the second cysteine of its Cys-X-X-Cys motif (unpublished results), reminiscent of in vitro oxidase activity of a yeast glutaredoxin 1 monothiol mutant (18).
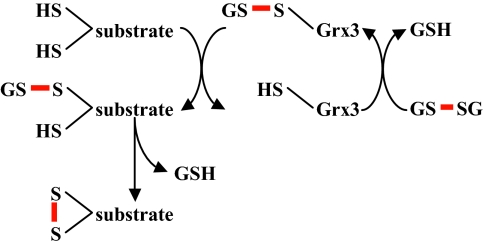
Fig. 5.
Model for disulfide bond formation by a monothiol glutaredoxin, adopted from (19). A glutathionylated glutaredoxin is attacked by a substrate cysteine, producing reduced glutaredoxin and glutathionylated substrate. In a second step, a second cysteine within the substrate attacks the glutathione-substrate disulfide, resulting in oxidized substrate.
High concentrations of glutathione (combined oxidized and reduced) are known to accumulate in supernatants of logarithmically growing E. coli culture up to 5 μM (14). The outer membrane porins allow the diffusion of molecules ≤600 Da from the extracellular fluid to the periplasm (25). For this reason it is reasonable to assume that extracellular glutathione (307 Da reduced; 612 Da oxidized) is at equilibrium with the periplasmic space. Our finding that we can restore GrxCP activity in gshA strains when either oxidized or reduced glutathione is added to the culture medium indicates that both forms can enter the periplasm. Our results also argue that the inability of E. coli to use oxidized glutathione as a sulfur source, reported elsewhere, is unlikely to be due to a permeability barrier for the molecule (26).
The E. coli cytoplasm of cells grown under standard conditions is reducing with a GSH/GSSG ratio between 200 and 300. By contrast, in the medium of growing cells, the GSH/GSSG ratio is 16.8 (27, 28). Approximately 30% of the entire glutathione pool is found outside the cell (14). These results indicate either that more oxidized than reduced glutathione is exported from cells or that exported glutathione is more liable for oxidation. If the lower GSH/GSSG ratio of the cell exterior reflects that of the periplasm, GrxCP may be presented with much higher levels of oxidized glutathione than it would be in the cytoplasm, thus allowing the activity we observe. Experiments probing the redox-potential of periplasmic E. coli extracts suggest that glutathione may take part in the periplasmic redox balance (29). While these arguments are consistent with the proposal that oxidized glutathione is the source of the oxidizing potential for GrxCP, we cannot rule out the possibility that the properties of the gshA mutant are due to an indirect effect of the absence of cytoplasmic glutathione on some process of electron-transfer across the cytoplasmic membrane.
We have attempted to manipulate the concentration of glutathione in the periplasm by employing a gshA mutant or external addition of glutathione. We have also examined the effects of mutations in several genes, including cydD (encoding a proposed glutathione exporter), ggt (encoding periplasmic γ-glutamyl transpeptidase), and mdlA (encoding a multidrug resistant-like ABC transporter) (13, 30, 31), which might influence the concentration of periplasmic glutathione. However, none of these mutations affected the ability of GrxCP to promote disulfide bond formation in AP (data not shown).
Our work and that of others indicate that glutaredoxins are capable of promoting protein disulfide bond formation. However, do glutaredoxins carry out oxidizing reactions under any conditions in vivo, for instance in the cytoplasm? Østergaard and coworkers describe an in vivo situation in which glutaredoxins behave in such a way: They found that the oxidation rate of a redox sensor yellow fluorescent protein was increased by yeast glutaredoxin 1 and 2 when the cytosolic GSH/GSSG ratio was altered (32). This result suggests that these glutaredoxins are not only taking part in the reduction of the redox probe but also in its oxidation in vivo, which could occur when the GSH/GSSG ratio is shifted toward more oxidizing conditions. Furthermore, during the course of this work, Rouhier and coworkers showed that the highly unusual poplar glutaredoxin 1, containing an [2Fe-2S] center, can oxidize proteins when exported to the E. coli periplasm in the absence of DsbB (33). However, the source of oxidative power, allowing the exported poplar glutaredoxin 1 to oxidize proteins in the E. coli periplasm, is unknown.
While the glutaredoxin 3 we have studied was artificially localized to the periplasm, we wondered whether there were any examples of periplasmically localized glutaredoxins in bacteria. We found that there exist several glutaredoxin homologues in various organisms that have predicted signal sequences (e.g., gi: 3963154 of Rhodoferax ferrireducens). Some of these organisms also have homologues of the glutathione biosynthesis pathway proteins GshA and GshB, suggesting that these organisms produce glutathione. These predicted periplasmic glutaredoxins might be considered candidates for periplasmic disulfide bond formation catalysts in these species.
The finding of an apparent extracytoplasmic activity of glutathione via our forced export of glutaredoxin to the periplasm raises the possibility that glutathione plays a physiological role in this compartment. If so, it is likely that this role of glutathione is related to its function as a regulator of redox reactions. One approach to assessing potential roles of glutathione in the periplasm is to obtain mutants that no longer accumulate glutathione in the cell envelope and examine the properties of such strains. The oxidative activity of glutaredoxin that we have reported here provides phenotypes (the accumulation of disulfide bonds in specific proteins), which should allow selections or screens for mutants defective in glutathione export to the periplasm.
Materials and Methods
Bacterial Strains, Plasmids, and Growth Conditions.
All E. coli strains and plasmids used in this work are listed in Table S1 and oligonucleotides are listed in Table S2. Standard techniques were used for cloning and DNA analysis, PCR, transformation, and P1 transduction (34). Strains were grown routinely in NZ-amine medium (35) at 37 °C, when necessary in the presence of the appropriate antibiotic. Antibiotics used to select for plasmids or chromosomal markers were present in following final concentrations: ampicillin (200 μg/ml), kanamycin (40 μg/ml), chloramphenicol (10 μg/ml), and spectinomycin (30 μg/ml). D-glucose (0.2%) or glycerol (0.2%) and L-arabinose (0.2%) were used for growth in M63 minimal medium (36), the latter condition to induce genes on pBAD42-derived plasmids. Genes on pDSW204-derived plasmids were induced by 10 μM IPTG (Shelton Scientific, Inc.) in liquid culture or 20 μM in low agar-motility plates. M63 minimal medium contained 18 aa (all except cysteine and methionine). When indicated, medium was supplemented with reduced or oxidized glutathione as stated. Motility plates were either NZ or M63 medium containing 0.35% agar, inoculated by stabbing a single colony into the medium and incubated at 30 °C for 24 to 40 h.
Plasmid and Strain Constructions.
Standard techniques (34) were used for the construction of strains and plasmids and are described in detail in SI Text.
Western Blots.
Protein samples were separated by Tris-Glycine SDS/PAGE and transferred by semidry electrotransfer according to the manufacturers protocol (Bio-Rad) onto an Immobilon-P membrane (Millipore). Anti-FLAG (M2) (Sigma), anti-c-Myc (A-14) (Santa Cruz Biotechnology, Inc.), anti-β-lactamase (5′→3′, Inc.), anti-thioredoxin 1 (Sigma), anti-SecB (laboratory collection) were used as previously stated (37, 38). Immunodetection was performed using anti-mouse and anti-rabbit conjugates to horseradish peroxidase in combination with ECL detection reagents, used according to the manufacturer (Amersham).
Determination of Redox States.
The redox state of DsbA substrates was determined by alkylating free protein thiols with AMS and subsequent protein separation by SDS/PAGE and visualization by Western blot as described in ref. 39 with the following changes. Cells were grown in M63 minimal medium containing 0.2% glycerol, 0.2% L-arabinose, and 10 μM IPTG. After growth at 30 °C to log phase, samples were immediately TCA precipitated (10% final concentration) and free protein thiols were alkylated using AMS. Proteins were separated by Tris-Glycine SDS/PAGE (10%, 12%, 20% acrylamide for LivK, YodA, and RcsF respectively) and subsequently subjected to Western analysis using antibodies as indicated.
Alkaline Phosphatase Activity Assays.
Cultures to be assayed were grown at 30 °C in M63 minimal medium to log phase. Alkaline phosphatase activity assays were carried out as published (40) and activities were calculated in Miller units [MU] and when applicable expressed as percent of wild-type strain (MER360) activity. AP activity was calculated from at least 3 independent experiments.
Chemicals and Reagents.
If not stated otherwise, chemicals and reagents were obtained from Sigma-Aldrich, Mallinckrodt, and Baker, Inc. Restriction enzymes and DNA modifying enzymes were purchased from New England Biolabs. Oligonucleotides were synthesized by Invitrogen or IDT Inc.
Supplementary Material
Acknowledgments.
We thank Dana H. Boyd (Harvard Medical School, Boston), Melinda Faulkner (Harvard Medical School, Boston), Linda Benson (Oregon State University, Corvallis, OR), and Chris Mathews (Oregon State University, Corvallis, OR) for generously providing us with strains and plasmids and present and former members of the J.B. and G.G. laboratories for helpful critical discussions. This work was funded by grants from the National Institute of General Medical Sciences Grants GMO41883 and GMO550905 and New England Biolabs.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0812596106/DCSupplemental.
References
- 1.Bardwell JC, Lee JO, Jander G, Martin N, Belin D, Beckwith J. A pathway for disulfide bond formation in vivo. Proc Natl Acad Sci USA. 1993;90:1038–1042. doi: 10.1073/pnas.90.3.1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aslund F, Berndt KD, Holmgren A. Redox potentials of glutaredoxins and other thiol-disulfide oxidoreductases of the thioredoxin superfamily determined by direct protein–protein redox equilibria. J Biol Chem. 1997;272:30780–30786. doi: 10.1074/jbc.272.49.30780. [DOI] [PubMed] [Google Scholar]
- 3.Debarbieux L, Beckwith J. The reductive enzyme thioredoxin 1 acts as an oxidant when it is exported to the Escherichia coli periplasm. Proc Natl Acad Sci USA. 1998;95:10751–10756. doi: 10.1073/pnas.95.18.10751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jonda S, Huber-Wunderlich M, Glockshuber R, Mossner E. Complementation of DsbA deficiency with secreted thioredoxin variants reveals the crucial role of an efficient dithiol oxidant for catalyzed protein folding in the bacterial periplasm. EMBO J. 1999;18:3271–3281. doi: 10.1093/emboj/18.12.3271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Masip L, et al. An engineered pathway for the formation of protein disulfide bonds. Science. 2004;303:1185–1189. doi: 10.1126/science.1092612. [DOI] [PubMed] [Google Scholar]
- 6.Holmgren A. Hydrogen donor system for Escherichia coli ribonucleoside-diphosphate reductase dependent upon glutathione. Proc Natl Acad Sci USA. 1976;73:2275–2279. doi: 10.1073/pnas.73.7.2275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dailey FE, Berg HC. Mutants in disulfide bond formation that disrupt flagellar assembly in Escherichia coli. Proc Natl Acad Sci USA. 1993;90:1043–1047. doi: 10.1073/pnas.90.3.1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Masip L, Klein-Marcuschamer D, Quan S, Bardwell JC, Georgiou G. Laboratory evolution of Escherichia coli thioredoxin for enhanced catalysis of protein oxidation in the periplasm reveals a phylogenetically conserved substrate specificity determinant. J Biol Chem. 2008;283:840–848. doi: 10.1074/jbc.M705147200. [DOI] [PubMed] [Google Scholar]
- 9.Kadokura H, Tian H, Zander T, Bardwell JC, Beckwith J. Snapshots of DsbA in action: Detection of proteins in the process of oxidative folding. Science. 2004;303:534–537. doi: 10.1126/science.1091724. [DOI] [PubMed] [Google Scholar]
- 10.Kobayashi T, et al. Respiratory chain is required to maintain oxidized states of the DsbA-DsbB disulfide bond formation system in aerobically growing Escherichia coli cells. Proc Natl Acad Sci USA. 1997;94:11857–11862. doi: 10.1073/pnas.94.22.11857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Akiyama Y, Ito K. Folding and assembly of bacterial alkaline phosphatase in vitro and in vivo. J Biol Chem. 1993;268:8146–8150. [PubMed] [Google Scholar]
- 12.Brickman E, Beckwith J. Analysis of the regulation of Escherichia coli alkaline phosphatase synthesis using deletions and phi80 transducing phages. J Mol Biol. 1975;96:307–316. doi: 10.1016/0022-2836(75)90350-2. [DOI] [PubMed] [Google Scholar]
- 13.Pittman MS, Robinson HC, Poole RK. A bacterial glutathione transporter (Escherichia coli CydDC) exports reductant to the periplasm. J Biol Chem. 2005;280:32254–32261. doi: 10.1074/jbc.M503075200. [DOI] [PubMed] [Google Scholar]
- 14.Owens RA, Hartman PE. Export of glutathione by some widely used Salmonella typhimurium and Escherichia coli strains. J Bacteriol. 1986;168:109–114. doi: 10.1128/jb.168.1.109-114.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Heath RL, Tappel AL. A new sensitive assay for the measurement of hydroperoxides. Anal Biochem. 1976;76:184–191. doi: 10.1016/0003-2697(76)90277-3. [DOI] [PubMed] [Google Scholar]
- 16.Bushweller JH, Aslund F, Wuthrich K, Holmgren A. Structural and functional characterization of the mutant Escherichia coli glutaredoxin (C14—S) and its mixed disulfide with glutathione. Biochemistry. 1992;31:9288–9293. doi: 10.1021/bi00153a023. [DOI] [PubMed] [Google Scholar]
- 17.Holmgren A, Aslund F. Glutaredoxin. Methods Enzymol. 1995;252:283–292. doi: 10.1016/0076-6879(95)52031-7. [DOI] [PubMed] [Google Scholar]
- 18.Bjornberg O, Ostergaard H, Winther JR. Mechanistic insight provided by glutaredoxin within a fusion to redox-sensitive yellow fluorescent protein. Biochemistry. 2006;45:2362–2371. doi: 10.1021/bi0522495. [DOI] [PubMed] [Google Scholar]
- 19.Xiao R, Lundstrom-Ljung J, Holmgren A, Gilbert HF. Catalysis of thiol/disulfide exchange: Glutaredoxin 1 and protein disulfide isomerase use different mechanisms to enhance oxidase and reductase activities. J Biol Chem. 2005;280:21099–21106. doi: 10.1074/jbc.M411476200. [DOI] [PubMed] [Google Scholar]
- 20.Dutton RJ, Boyd D, Berkmen M, Beckwith J. Bacterial species exhibit diversity in their mechanisms and capacity for protein disulfide bond formation. Proc Natl Acad Sci USA. 2008;105:11933–11938. doi: 10.1073/pnas.0804621105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bardwell JC, McGovern K, Beckwith J. Identification of a protein required for disulfide bond formation in vivo. Cell. 1991;67:581–589. doi: 10.1016/0092-8674(91)90532-4. [DOI] [PubMed] [Google Scholar]
- 22.Singh AK, Bhattacharyya-Pakrasi M, Pakrasi HB. Identification of an atypical membrane protein involved in the formation of protein disulfide bonds in oxygenic photosynthetic organisms. J Biol Chem. 2008;283:15762–15770. doi: 10.1074/jbc.M800982200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Aslund F, et al. Glutaredoxin-3 from Escherichia coli. Amino acid sequence, 1H AND 15N NMR assignments, and structural analysis. J Biol Chem. 1996;271:6736–6745. doi: 10.1074/jbc.271.12.6736. [DOI] [PubMed] [Google Scholar]
- 24.Yu J, McLaughlin S, Freedman RB, Hirst TR. Cloning and active site mutagenesis of Vibrio cholerae DsbA, a periplasmic enzyme that catalyzes disulfide bond formation. J Biol Chem. 1993;268:4326–4330. [PubMed] [Google Scholar]
- 25.Decad GM, Nikaido H. Outer membrane of gram-negative bacteria. XII. Molecular-sieving function of cell wall. J Bacteriol. 1976;128:325–336. doi: 10.1128/jb.128.1.325-336.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Roberts RB. Studies of biosynthesis in Escherichia coli. WA: Carnegie Institution of Washington; 1955. [Google Scholar]
- 27.Aslund F, Zheng M, Beckwith J, Storz G. Regulation of the OxyR transcription factor by hydrogen peroxide and the cellular thiol-disulfide status. Proc Natl Acad Sci USA. 1999;96:6161–6165. doi: 10.1073/pnas.96.11.6161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Smirnova GV, Muzyka NG, Oktyabrsky ON. Effects of cystine and hydrogen peroxide on glutathione status and expression of antioxidant genes in Escherichia coli. Biochemistry (Mosc) 2005;70:926–934. doi: 10.1007/s10541-005-0204-2. [DOI] [PubMed] [Google Scholar]
- 29.Messens J, Collet JF, Van Belle K, Brosens E, Loris R, Wyns L. The oxidase DsbA folds a protein with a nonconsecutive disulfide. J Biol Chem. 2007;282:31302–31307. doi: 10.1074/jbc.M705236200. [DOI] [PubMed] [Google Scholar]
- 30.Suzuki H, Kumagai H, Tochikura T. gamma-Glutamyltranspeptidase from Escherichia coli K-12: Purification and properties. J Bacteriol. 1986;168:1325–1331. doi: 10.1128/jb.168.3.1325-1331.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hirrlinger J, Moeller H, Kirchhoff F, Dringen R. Expression of multidrug resistance proteins (Mrps) in astrocytes of the mouse brain: A single cell RT-PCR study. Neurochem Res. 2005;30:1237–1244. doi: 10.1007/s11064-005-8795-y. [DOI] [PubMed] [Google Scholar]
- 32.Ostergaard H, Tachibana C, Winther JR. Monitoring disulfide bond formation in the eukaryotic cytosol. J Cell Biol. 2004;166:337–345. doi: 10.1083/jcb.200402120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rouhier N, et al. Functional, structural, and spectroscopic characterization of a glutathione-ligated [2Fe-2S] cluster in poplar glutaredoxin C1. Proc Natl Acad Sci USA. 2007;104:7379–7384. doi: 10.1073/pnas.0702268104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sambrook J, Fritsch EF. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 35.Guzman LM, Barondess JJ, Beckwith J. FtsL, an essential cytoplasmic membrane protein involved in cell division in Escherichia coli. J Bacteriol. 1992;174:7716–7728. [PMC free article] [PubMed] [Google Scholar]
- 36.Miller J. A Short Course in Bacterial Genetics. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1992. [Google Scholar]
- 37.Kadokura H, Beckwith J. Four cysteines of the membrane protein DsbB act in concert to oxidize its substrate DsbA. EMBO J. 2002;21:2354–2363. doi: 10.1093/emboj/21.10.2354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Huber D, Boyd D, Xia Y, Olma MH, Gerstein M, Beckwith J. Use of thioredoxin as a reporter to identify a subset of Escherichia coli signal sequences that promote signal recognition particle-dependent translocation. J Bacteriol. 2005;187:2983–2991. doi: 10.1128/JB.187.9.2983-2991.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kobayashi T, Ito K. Respiratory chain strongly oxidizes the CXXC motif of DsbB in the Escherichia coli disulfide bond formation pathway. EMBO J. 1999;18:1192–1198. doi: 10.1093/emboj/18.5.1192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rietsch A, Belin D, Martin N, Beckwith J. An in vivo pathway for disulfide bond isomerization in Escherichia coli. Proc Natl Acad Sci USA. 1996;93:13048–13053. doi: 10.1073/pnas.93.23.13048. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.