Abstract
Processing of amyloid precursor protein (APP) by γ-secretase is the last step in the formation of the Aβ peptides associated Alzheimer's disease. Solid-state NMR spectroscopy is used to establish the structural features of the transmembrane (TM) and juxtamembrane (JM) domains of APP that facilitate proteolysis. Using peptides corresponding to the APP TM and JM regions (residues 618–660), we show that the TM domain forms an α-helical homodimer mediated by consecutive GxxxG motifs. We find that the APP TM helix is disrupted at the intracellular membrane boundary near the ε-cleavage site. This helix-to-coil transition is required for γ-secretase processing; mutations that extend the TM α-helix inhibit ε cleavage, leading to a low production of Aβ peptides and an accumulation of the α- and β-C-terminal fragments. Our data support a progressive cleavage mechanism for APP proteolysis that depends on the helix-to-coil transition at the TM-JM boundary and unraveling of the TM α-helix.
Keywords: Alzheimer's disease, GxxxG motifs, progressive cleavage, solid-state-NMR spectroscopy
Amyloid precursor protein (APP) is an integral membrane protein with a single transmembrane (TM) domain that is expressed in a wide number of different cell types including neurons (1). The processing of the protein occurs by the action of several proteases; the α- and β-secretases cleave between the extracellular domain and the TM domain to generate a membrane-anchored C-terminal fragment (α- or β-CTF) and the γ-secretase cleaves the CTFs within their TM domain. The cleavage of the β-CTF generates a 38–43-residue Aβ peptide. The cleavage of both α- and β-CTFs releases a 50-residue APP intracellular domain (AICD) peptide. The soluble AICD peptide may be involved in the regulation of gene transcription (2).
The most unusual feature of APP proteolysis is the intramembraneous cleavage by the γ-secretase complex. The mechanism of proteolysis is of considerable interest because of its role in (i) generating the Aβ peptides associated with Alzheimer's disease and (ii) releasing the AICD involved in APP-dependent gene transcription. Several cleavage sites have been identified that generate different length Aβ peptides. The γ-cleavage site cuts the APP sequence in the middle of the TM domain to predominantly produce the Aβ40 peptide, and to a lesser extent the Aβ42 peptide. However, Aβ42 has a higher propensity to form aggregates than the shorter isoforms and is the most toxic peptide generated by γ cleavage (3). There is another cleavage site (4), referred to as the ε-cleavage site, a few residues downstream between Leu-645 and Val-646 that has been identified by N-terminal sequencing of the AICD peptide (4). An open question has been whether the same enzyme activity is responsible for both the γ- and ε-cleavage sites.
The γ-secretase complex has a diverse set of type I membrane protein substrates. Notch, Cd44, ErbB4, and E-cadherin are cleaved by the γ-secretase in vivo. For each of these substrates, truncation of the extracellular domain to just a few amino acids is required to bind to the γ-secretase complex. These substrates are all cleaved near the intracellular TM-JM boundary. However, like APP, Notch, and Cd44 are also cleaved in the middle of the TM domain, although their sequences are not conserved (see SI).
To address the mechanism of intramembrane proteolysis, we focus on the structure of the TM domain of APP in membrane bilayers. Proteolysis requires local unraveling of the helical secondary structure of the TM domain to expose backbone carbonyl carbons for nucleophilic attack by polarized water in the enzyme active site. This requirement raises the question of whether there are sequence motifs in the TM domain of APP that destabilize the helical structure in cell membranes in a fashion similar to that proposed for the conserved Asn-Pro sequence in the sterol regulatory element binding protein (SREBP), the substrate of the site-2 protease (5).
There are 2 unusual features of the TM sequence of APP that have the potential to distort or destabilize local helical secondary structure. The first is the high density of β-branched amino acids surrounding the Aβ40- and Aβ42-cleavage sites. The β-branched amino acids (Val, Thr, and Ile) are known to have a high propensity for forming extended β-structure. Sequential β-branched residues can have a destabilizing effect on the secondary structure of TM helices (6, 7).
The second unusual feature in the TM domain of APP is the high occurrence of glycines upstream of the Aβ40- and Aβ42-cleavage sites. Li et al. (6) have shown that in the aqueous environment of SDS micelles, glycines in TM domains can promote extended secondary structure. In the APP TM helix, glycines may play a role in helix destabilization, particularly within the context of the γ-secretase complex.
Several of the glycines in the APP TM domain occur in GxxxG motifs. However, rather than destabilizing helical secondary structure, GxxxG motifs within TM sequences are known to mediate helix dimerization (8). Mutational studies of APP (9, 10) indicate that these GxxxG glycines are important in both dimerization and APP processing. Whereas the mutation of the GxxxG motifs has been shown to significantly decrease the generation of Aβ40 (9, 10), the influence on dimerization is less clear. For example, mutation of Gly-625 and Gly-629 to isoleucine diminishes the ability of APP to dimerize (9), whereas mutation of these same residues to leucine leads to SDS-resistant dimers that apparently adopt an interface nonproductive for γ-processing (10). In contrast, recent studies by Gorman et al. (11) have shown using fluorescence energy transfer that peptides corresponding to the TM domain of APP dimerize and proposed that a GxxxA motif, rather than the GxxxG sequences, mediate dimerization. They found that dimerization influences the ratio of Aβ40 to Aβ42 produced by the γ-secretase complex. Consequently, to address the role of the TM sequence in APP processing, not only is it important to establish the secondary structure of APP, but also to determine the helix interface that mediates dimerization.
Solid-state NMR spectroscopy is used to probe the secondary structure and dimerization of the TM domain of APP. Using peptides corresponding to the TM-JM sequence (residues 618–660, APP695 numbering), we target the glycines within the TM domain, and residues at the γ- and ε-cleavage sites. We find that GxxxG motifs involving Gly-625, Gly-629, and Gly-633 mediate TM helix homodimerization, and that the TM helix breaks at the transition point near the ε-cut site. Finally, we show that the insertion of 3 consecutive leucines at the transition point in APP695 inhibits ε cleavage leading to a low production of Aβ peptides and an accumulation of the α- and β-CTFs. The leucine insertion extends the TM domain by 1 helical turn, whereas an insertion of 3 glycines does not, demonstrating that the helix-to-coil transition is required for γ-secretase processing.
Results
TM Region of APP is Helical in Membrane Bilayers.
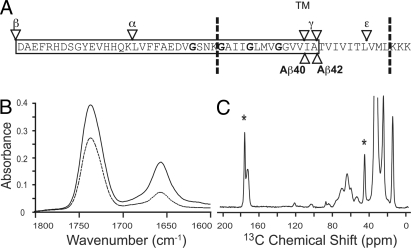
Glycines and β-branched amino acids both contribute to helix destabilization in soluble proteins. As a result, the abundance of these residues within the TM region of APP raises the question of whether the secondary structure is locally unraveled at either the γ- or ε-cut sites (Fig. 1A). Polarized IR spectroscopy can be used to establish the global secondary structure of peptides reconstituted into membrane bilayers. Polarized IR spectra of APP (618–660) exhibit an amide I vibration at 1,657 cm−1, a frequency characteristic of α-helical structure (Fig. 1B). Deconvolution of the amide I band of APP (618–660) reveals only a small shoulder at lower frequency (1,630 cm−1). The dichroic ratio of the amide I band is sensitive to the orientation of the TM helix relative to the plane of membrane. The observed dichroic ratio of 3.46 corresponds to a helix orientation of 15.5° relative to the membrane normal (12). The high dichroic ratio, which provides a way to assess our ability to reconstitute TM peptides in a homogeneous fashion (12), indicates that the APP (618–660) peptide can be reconstituted into DMPC: DMPG (10:3) bilayers in a stable (nonaggregated) helical TM orientation.
Fig. 1.
Sequence of the APP TM and JM regions along with polarized IR and 13C MAS NMR of APP (618–660) in membrane bilayers. (A) Schematic representation of the TM and JM domains of human APP. The positions of the glycines in the 3 consecutive GxxxG are in bold. The cleavage sites of α-, β-, and γ- (γ and ε) secretase activities are indicated by arrows. (B) Polarized IR spectra of APP (618–660) were obtained using parallel (solid line) and perpendicular polarized light (dashed line). (C) 13C MAS NMR spectra of of APP (618–660). The 13C-labeled α-carbon of Gly-634 and the carbonyl carbon of Ala-638 are marked by asterisks. Other peaks in the spectrum correspond to natural abundance 13C of the lipid.
Solid-state NMR spectroscopy can be used to probe local secondary structure. The 13C magic angle spinning (MAS) NMR spectrum of membrane-reconstituted APP (618–660) containing 1-13C-Ala-638 and 2-13C-Gly-634 is shown in Fig. 1C. The observed 13C NMR resonances correspond predominately to the natural abundance 13C carbons of the lipids. The 1-13C resonance of Ala-638 is observed at 175.1 ppm (asterisk), indicative of α-helical secondary structure. The Ala-638-Thr-639 peptide bond corresponds to the γ-cut site for the production of the Aβ42 peptide. β-structure yields lower chemical shifts in the range of 168–173 ppm (13). The Val-636-Ile-637 bond corresponds to the γ-cut site for the Aβ40 peptide. The 13C chemical shifts of the carbonyl carbons at Val-636 (174.8 ppm) and Ile-637 (174.8 ppm) are also characteristic of α-helical structure (data not shown).
For glycines, the carbonyl chemical shifts are approximately 172 ppm for helices and 168 ppm for extended β-structure (13). In APP, the carbonyl chemical shifts of the TM glycines are observed at 172 ppm or greater. These results, together with the data presented in Fig. 1, show that the structure in the region of γ cleavage is helical in membrane bilayers and that the TM glycines are not locally unwinding the APP TM helix (see also SI).
Computational Searches of APP-APP Interactions.
The ability of APP to dimerize has been suggested by cross-linking and gel filtration (14), and more recently by FRET (11) and TOXCAT (15) measurements. On the basis of the helical secondary structure observed for the TM domain of APP, we undertook computational searches of low-energy dimer structures involving the TM domain of APP. The strategy behind these searches is described in the SI.
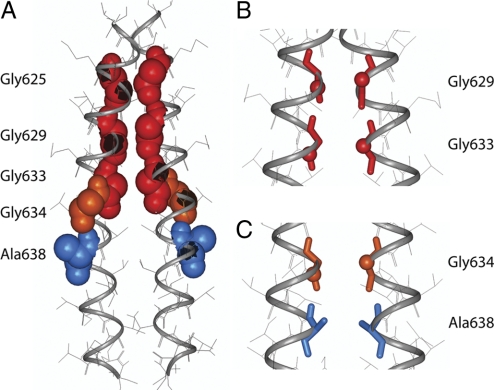
Two clusters of low energy dimer structures are typically found in the computational searches of APP dimers. The structures in these clusters have helices with right-handed crossing angles. In the first cluster, the GxxxG motifs mediate dimerization (Figs. 2A and B). In the second cluster (Fig. 2C), the Gly634xxxAla638 sequence mediates dimerization.
Fig. 2.
Computational searches of APP-APP interactions. (A) Low-energy structure of the APP TM dimer based on computational searches of APP helix–helix interactions. The glycines in the GxxxG motifs (red van der Waals spheres) line the dimer interface. The Gly-634 (orange) and Ala-638 (blue) residues in the GxxxA motif are oriented away from the dimer interface. (B) View of Gly-629…Gly-629 and Gly-633…Gly-633 interactions in the APP homodimer shown in (A). (C) Contact region involving the GxxxA sequence in the second low energy cluster identified in APP TM dimer searches. See SI for details.
APP TM Helix Forms Homodimers through Sequential GxxxG Motifs.
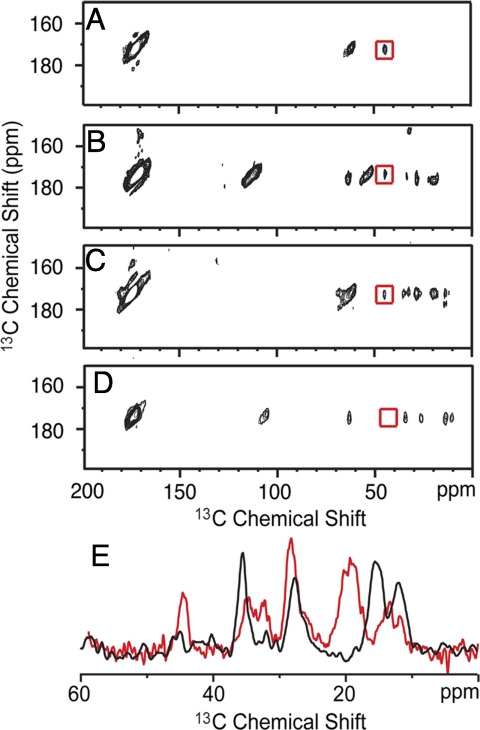
To test whether any of the low energy dimer structures seen in the computational studies actually occur in membrane bilayers, namely whether the GxxxG or the GxxxA motifs contact one another in the APP TM dimer, solid-state NMR experiments were undertaken of the membrane-reconstituted APP TM domain. Fig. 3A presents the results of NMR measurements between APP TM peptides labeled at Gly-625, Gly-629, and Gly-633. Two peptides were synthesized for these experiments; one with 1-13C-Gly and the second with 2-13C-Gly at each of the 3 glycine positions. These resonances have distinct chemical shifts. The 2 peptides were reconstituted in a 1:1 molar ratio. The observation of a 13Cα…13C′ cross-peak (see boxed cross-peak in Fig. 3A) in the 2D dipolar-assisted rotational resonance (DARR) NMR experiment indicates that these carbons are in contact. The observation of a contact is highly significant because it indicates that stable and well-defined APP homodimers are present in our reconstituted samples. The dimers must be in a head-to-head orientation and tight association (i.e., little or no monomer). On the basis of the intensity of the interhelical 13Cα…13C′ contacts observed, we can estimate that >80% of the dimers are associated in a stable uniform structure (at least in the region of glycine contacts).
Fig. 3.
2D 13C DARR NMR of APP-APP interactions. (A) Region from the 2D DARR spectrum showing a cross-peak (red box) between 2-13C glycine on APP TM peptide 1 and 1-13C glycine on APP TM peptide 2. Gly-625, 629, and 633 are 13C-labeled on each peptide. (B) Region of the 2D DARR spectrum showing a cross-peak (red box) between 2-13C Gly-629 and 1-13C Gly-629. (C) Region of the 2D DARR spectrum showing a cross-peak (red box) between 2-13C Gly-633 and 1-13C Gly-633. (D) Same experiment as in (B and C) with 1-13C- and 2-13C-labeled glycine at Gly-634. The absence of a Gly-634-Gly-634 cross-peak is consistent with the GxxxA motif having a lipid-facing orientation. (E) 1D rows through the glycine cross-peak from (C) (red) and (D) (black). The Gly-633 cross-peak at 44 ppm (red spectrum) is approximately the same intensity as the cross-peaks corresponding to the intramolecular correlations of U-13C-labeled Val-636 and Ile-641. In contrast, the Gly-634 cross-peak at 44 ppm is much weaker in intensity than the nearby intramolecular cross-peaks from Ile-637. The resonance at 175 ppm corresponds to the diagonal C = O resonance in the 2D spectrum. Spectra in (A) and (C) were obtain at carbon frequency of 90 MHz and the peaks at 65 ppm correspond to a spinning sidebands. Spectra in (B) and (D) were obtained at a carbon frequency of 150 MHz and the peaks at 105 ppm correspond to spinning sidebands. The remaining peaks in (B–D) correspond to Ile and Val intramolecular correlations.
To verify the translational position of the glycine residues in the dimer and to address the possibility that the GxxxA sequence mediates dimerization, we undertook DARR NMR experiments on APP (618–660) labeled at individual glycines: Gly-629, Gly-633, and Gly-634. Fig. 3B presents the 2D NMR spectrum obtained using APP peptides reconstituted as before and separately labeled at position 629 with 1-13C-Gly (peptide 1) and 2-13C Gly (peptide 2). We observe an interhelical cross-peak between the Gly-629 residues consistent with the packing shown in Figs. 2A and B. In a similar fashion, we observe a cross-peak in the 2D DARR NMR spectrum of an equimolar mixture of APP peptides containing either 1-13C- and 2-13C-Gly at position 633 (Fig. 3C). In contrast, DARR NMR spectra (Fig. 4D) obtained in a parallel experiment using APP peptides separately labeled at position 634 with 1-13C-Gly and 2-13C-Gly did not exhibit a Gly-Gly peak.
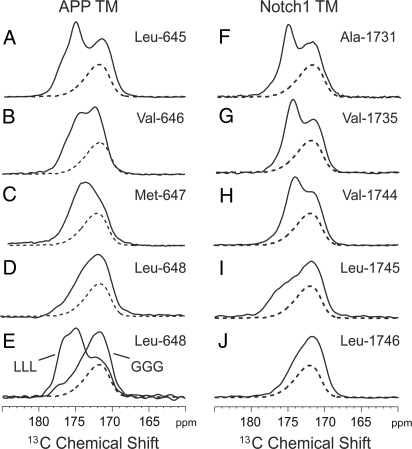
Fig. 4.
13C MAS NMR of the region containing carbonyl carbon resonances at the TM-JM boundary of APP and Notch1. Four APP peptides were specifically 13C-labeled at Leu-645 (A), Val-646 (B), Met-647 (C), and Leu-648 (D), and 5 Notch1 peptides were specifically 13C-labeled at Ala-1,731 (F), Val-1,735 (G), Val-1,744 (H), Leu-1,745 (I), and Leu-1,746 (J). We also obtained 13C MAS NMR data on APP 3L and 3G peptides containing 1-13C-labeled Leu-648 (E). In each case, the peptides were reconstituted into DMPC:DMPG membranes at a 1:50 molar ratio of peptide-to-lipid. The dashed line indicates the MAS spectrum of lipid alone.
In the 3 experiments above using APP peptides containing individually 13C-labeled glycines, the peptides also contained uniformly 13C-labeled Val-640 (Fig. 3B), Val-636 and Ile-641 (Fig. 3C), and Ile-637 (Fig. 3D) for solution NMR measurements (not discussed here). The intraresidue 13C-13C cross-peaks from these residues serve as internal controls for the DARR NMR measurements. Fig. 3E presents a comparison of the rows through the carbonyl diagonal resonance in Figs. 3C and D. The intermolecular Gly-Gly cross-peak between the Gly-633 13C labels has an intensity that is comparable with the intraresidue cross-peaks. The strong intensity indicates the Gly-633 CH2 and C = O groups are in van der Waals contact across the dimer interface, suggesting the presence of a Cα-H…O hydrogen bond (16).
TM Helix of APP Breaks at the Cytoplasmic Membrane Surface.
The sensitivity of the 13C chemical shifts to secondary structure allows us to determine whether there is a change in the local structure around the ε-cut site (Leu-645-Val-646). In Figs. 4A–D, we compare the 13C MAS NMR spectra of APP (618–660) specifically 13C-labeled at Leu-645, Val-646, Met-647, and Leu-648. The bold lines correspond to the lipid-reconstituted peptide; dotted lines correspond to lipid alone. The 13C MAS NMR spectrum of Leu-645 exhibits 2 resonances at 175.0 and 172.1 ppm (Fig. 4A), assigned to the backbone 13C = O of Leu-645 and the lipid C = O, respectively. The 13C chemical shift indicates that Leu-645 is in α-helical secondary structure. Comparison of the Leu-645 spectrum with the spectra of Val-646-Leu-648 shows that the peptide carbonyl resonances shift to lower frequency, characteristic of random coil structure.
The above data indicate that there is a break in helical secondary structure near the ε-cut site at the TM-JM boundary. Nicholson and coworkers have previously shown that the N-terminal residues of the isolated JM domain of APP are unstructured in solution and suggested that the transient local structure and prolyl cis/trans isomerization induced by phosphorylation at Thr-668 may function as a regulatory switch (17, 18).
Finally, we obtained the carbonyl chemical shifts at the γ- and ε-cut sites of Notch1 to show that a helix-to-random coil transition at the position of the ε-cut sites is also observed in this γ-secretase substrate (Figs. 4F–J). Notch1 (TM residues 1,720–1,760) was membrane reconstituted in a fashion parallel to that for APP (618–660), and exhibited a helical amide I vibration at 1,654 cm−1 with a dichroic ratio of 3.3. The NMR data indicate that the local structure of the TM domain at the γ- or S4-cut site (Ala-1,731) is helical and there is a helix break in the region of the ε- or S3 cut site (Leu-1,744). Previous studies have shown that the Notch TM domain is dimeric (15), indicating that both the secondary structure and oligomerization state are similar to that in APP.
Extension of the APP TM Helix Leads to an Accumulation of CTFs.
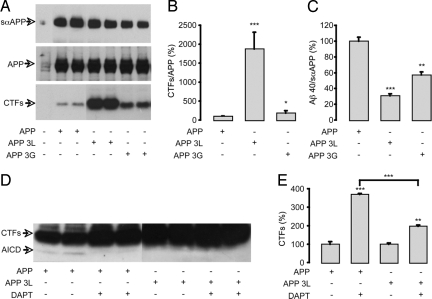
To test the proposal that the secondary structure at the TM-JM boundary is critical for APP processing, we present data on APP with 2 insertions at the intracellular TM-JM boundary. The insertion of 3 leucines before the intracellular KKK sequence (APP 3L) is designed to extend the length of the TM helix. The insertion of 3 glycines (APP 3G) should maintain the helix break point as in the wild-type protein (Fig. 4A). To verify that the 3L insertion extended the α-helical structure at the TM-JM boundary, whereas the 3G extension retained the helix-to-coil transition, 13C MAS NMR spectra were obtained of APP TM peptides containing the 3L and 3G inserts, and labeled at position 648 with 1-13C leucine. The carbonyl chemical shifts of the 3L and 3G peptides observed in Fig. 4E clearly indicate that the TM helix has been extended only with the 3L insertion.
The APP mutants were expressed in Chinese Hamster Ovary (CHO) cells. The insertion of 3G or 3L did not alter APP expression. The production of sαAPP, an indicator of nonamyloidogenic processing, was not impaired in cells expressing APP 3L or APP 3G (Fig. 5A). Strikingly, significant accumulation of APP C-terminal fragments, mainly αCTF, occurred in cells expressing APP 3L and to a lesser extent in cells expressing APP 3G (Fig. 5B). In the context of unmodified α cleavage, the accumulation of αCTF is likely a result of an impairment of the γ cleavage of C-terminal fragments. This result was confirmed by the quantification of extracellular Aβ40. The release of Aβ40 (normalized to sαAPP) decreased by 70% in cells expressing APP 3L (Fig. 5C). We next analyzed AICD production in these cells (Fig. 5D). AICD results from the cleavage at the ε position performed by the γ-secretase complex. Treatment with DAPT, a γ-secretase inhibitor (19), led as expected to the inhibition of AICD production and the strong accumulation of CTFs in cells expressing APP (Fig. 5E). Importantly, there was no AICD detected in cells expressing APP 3L and the DAPT treatment had only weak effects on the accumulation of CTFs as compared with wild type APP (Fig. 5D and E, respectively).
Fig. 5.
Influence of the 3L and 3G insertions in the intracellular TM-JM boundary of APP. (A) Schematic representation of the TM-JM domains of human APP, APP 3L, and APP 3G. (B) Expression of cellular APP and soluble αAPP (sαAPP) were analyzed by Western blot analysis using the WO-2 antibody on cell lysates and cell supernatants, respectively. The CTFs (α and βCTF) were detected in cell lysates by the C17 antibody. (C) The CTF-to-APP ratio was calculated and represented as a percentage of the CTF/APP level in nonmutated APP controls. (D) The Aβ40-to-sαAPP ratio was calculated and represented as a percentage of Aβ40/sαAPP production in nonmutated controls. AICD was detected by Western blot analysis using the C17 antibody (E), and the effects of DAPT on CTF accumulation were quantified in the same experiments and given as a percentage of CTF levels in APP or APP 3L nontreated controls (F). Values are means ± SEM, n = 4; *P < 0.05, ** P < 0.01, *** P < 0.001, compared with control or as indicated (C, D, F).
Discussion
The molecular mechanism of intramembrane proteolysis is not well understood. Proteolysis by aspartyl proteases is mediated by water and requires local unraveling of helical TM secondary structure to expose a backbone carbonyl. Here, we have characterized the structure of the TM-JM regions of the APP protein in membrane bilayers to gain insight into how γ-secretase catalyzes proteolysis.
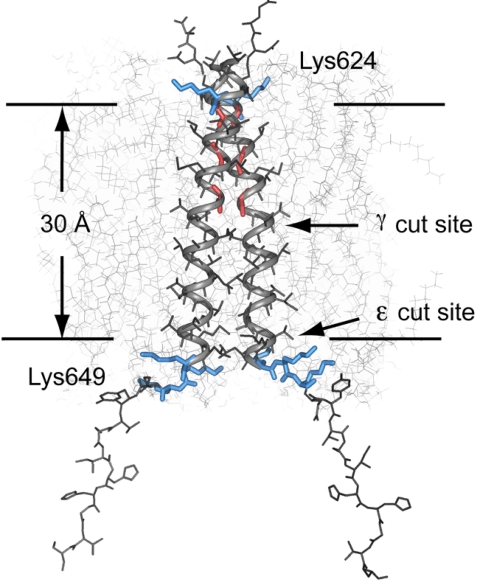
Fig. 6 presents the structure of the APP TM dimer. The ε-cut site lies at the boundary between the hydrophobic core and the polar head group region of the bilayer, whereas the γ-cut site is at the bilayer center. The TM dimer structure is consistent with the ε cleavage site being the sole active site in the γ-secretase complex and with proposals for progressive cleavage from the ε- to the γ-cut site (20) (see SI).
Fig. 6.
Structural model of the APP TM domain. Proposed structure of the APP (618–660) TM dimer in relation to a DOPC membrane bilayer (downloaded from http://persweb.wabash.edu/facstaff/fellers/). The structural studies of the APP TM domain indicate that the ε-cut site is approximately 32 Å from Lys-624. Cleavage at a single site would lead to local unraveling of the helix and a shift of amino acids into the binding site. To place the Aβ42-cleavage site at the same position would result in unraveling of the TM helix to the Gly-633-Gly-634 sequence.
The progressive cleavage model is consistent with 1) the detection of Aβ peptides with lengths intermediate between Aβ49 (ε cleavage) and Aβ42 (γ cleavage) (21, 22), 2) measurements of equimolar production of Aβ and AICD (23), and 3) the fact that it has not been possible to detect longer AICD peptides that would be expected if γ cleavage occurs before ε cleavage (23, 24). Indirect support for the progressive cleavage model comes from sequence comparisons of γ-secretase substrates, where the only common motif that these substrates share is the sequence of basic and hydrophobic amino acids at the TM-JM boundary. Alternative models have been proposed on the basis of FAD and other mutations, where the γ- and ε-secretase activities may be associated with different presenilin complexes (25, 26). In this case, one must imagine that presenilin unravels the TM α-helix in a sequence independent manner within the middle of the TM domain to generate the γ cleavage.
There is not yet a high-resolution crystal structure for the γ-secretase complex. However, the structures of 2 intramembrane proteases provide insights into how the γ-secretase may function. In both the rhomboid protease (27) and site-2 protease (28), the substrate cleavage site is not in the middle of TM helix, but just 3–4 amino acids from the TM-JM boundary as in APP. The cut site on the SREBP protein is found 3 residues into the TM domain, immediately before a required charged cytoplasmic Asp-Arg-Ser-Arg sequence (29). To accommodate cleavage, the substrate has an extended structure at the TM-JM boundary and the catalytic residues are accessible to water. Moreover, the SREBP substrate contains a conserved Asn-Pro sequence (5) within the middle of the TM helix that may allow the peptide to unwind in a fashion during proteolysis, as suggested for the TM glycines in APP.
In addition to the break in helical secondary structure at the TM-JM boundary, dimerization of the TM domain may be a common feature of γ-secretase substrates (9–11, 15, 18). The role of dimerization of APP and other γ-secretase substrates in proteolysis, however, remains controversial. Based on mutational studies, Munter et al. (9) concluded that dimerization facilitates Aβ42 production. In contrast, several studies have indicated the opposite. Gorman et al. (11) using FAD mutants and our studies (10) using TM glycine mutants found that increased dimerization can reduce the Aβ42/Aβ40 ratio. One possibility is that helix orientation within the TM dimer, and the strength of dimerization, controls APP processing. We have determined the orientation of the TM helices in the wild-type APP TM dimer (Figs. 2A and B, and 6), where GxxxG motifs (rather than the GxxxA sequence) mediate dimerization. However, it will be necessary to correlate the detailed structures of the TM and JM regions of APP mutants with the generation of different length Aβ peptides to fully address the influence of structure on processing.
The structural model in Fig. 6 shows the APP TM dimer with a break in helical secondary structure at the TM-JM boundary. To show that the helix-to-coil transition is required for APP processing of full length APP by the γ-secretase complex, we inserted 3 additional leucines at the TM-JM boundary to extend the TM helix without changing the TM region mediating dimerization. The 3L insertion was found to block APP processing by γ-secretase, whereas the processing of APP with the GGG insertion was similar to the WT protein. The lack of γ and ε cleavage of the 3L mutant of APP is consistent with the requirement that the TM domain must be locally unstructured for proteolysis and that there is only a single cleavage site at the TM-JM boundary.
Materials and Methods
Materials.
13C-labeled amino acids were purchased from Cambridge Isotope Laboratories. Other amino acids and octyl-ß-glucoside were obtained from Sigma Chemical. DMPC and DMPG were obtained from Avanti Polar Lipids as lyophilized powders and used without further purification.
Peptide Synthesis and Purification.
Peptides corresponding to the TM and JM regions of APP (Glu-618-Val-660) and Notch1 (Pro-1,720-Pro-1,760) were synthesized by solid-phase methods. The C terminus was amidated and we added an N-terminal lysine to prevent head-to-tail dimerization. The synthetic peptides were purified by reverse phase HPLC on a C4 column with a gradient of formic acid/1- propanol (4:1) over formic acid/water (2:3). The purity was confirmed with MALDI mass spectrometry and analytical reverse phase HPLC.
Reconstitution of Peptides into Membrane Bilayers.
The APP and Notch1 peptides were cosolubilized in DMPC, DMPG and octyl-β-glucoside in hexafluoroisopropanol. The peptide:lipid molar ratio was 1:50; the molar ratio between DMPC:DMPG was 10:3. The solution was incubated for 90 min at 37 °C, after which the solvents were removed under a stream of argon gas and then under vacuum. Mes buffer (50 mM Mes, 50 mM NaCl, and 5 mM DTT, pH 6.2) was added to the solid from the previous step and mixed at 37 °C for 6 h. The octyl-ß-glucoside was removed by dialysis (12). The reconstituted membranes were pelleted and loaded into NMR rotors.
ATR-FTIR Spectroscopy.
Polarized attenuated total reflection (ATR) FTIR spectra were obtained as previously described (12). We use a value of α = 41.8° based on parallel measurements on bacteriorhodopsin (12).
Solid-State NMR Spectroscopy.
NMR experiments were performed on either a 360 or 600 MHz Bruker AVANCE spectrometer. The MAS spinning rate was set to 9–11 KHz. The ramped amplitude cross polarization contact time was 2 ms. Two-pulse phase-modulated decoupling was used during the evolution and acquisition periods with a field strength of 80 kHz. Internuclear 13C…13C distance constraints were obtained from 2D DARR NMR experiments (30) using a mixing time of 600 ms. The sample temperature was maintained at 205 K.
Cell Culture, Transfection, and APP Expression.
Plasmids expressing APP and the APP 3L and 3G mutants were generated by PCR overlap extension as previously described (10). The cells used were CHO cells lines. For transient transfection, cells were seeded at a density of 3.105 cells per cm2 24 h before transfection (2 μg plasmid per well) with Lipofectamine™ according to manufacturer's instructions (Invitrogen). Cells were harvested 48 h after transfection and Western blot analysis was performed on cell culture media or cell lysates (10, 31). Primary antibodies used were the human-specific WO-2 antibody (The Genetics Company) and the C17 antibody directed against the APP C terminus (32). Quantification of APP and the C-terminal fragment were performed by direct acquisition of chemiluminescence on a ChemiDoc device (Bio-Rad). Human Aβ40 was quantified in the culture media by fluorescent sandwich ELISA (BioSource).
Statistical Analysis.
The number of samples (n) in each experimental condition is indicated in the Fig. 5 legend. When two experimental conditions were compared, statistical analysis was performed using an unpaired t test. Otherwise, statistical analyses were performed by 1-way ANOVA followed by Bonferroni's multiple comparison posttest.
Supplementary Material
Acknowledgments.
This work was supported by grants from the National Institutes of Health (AG-27317 and GM-46732) to S.O.S., and by grants from the Fonds National de la Recherche Scientifique (F.N.R.S.), the Fédération Belge contre le Cancer and the de Hovre Foundation to S.N.C, the Interuniversity Attraction Poles Program-Belgian Sate-Belgian Science Policy to J.N.O and S.N.C. and the Foundation for Research on Alzheimer Disease (FRMA) to P.K.C. Excellent technical support from Mingli Li, Joanne Van Hees and Martine Ziliox is acknowledged. S.N.C. is a Research Associate of the F.N.R.S. Belgium.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0812261106/DCSupplemental.
References
- 1.Selkoe DJ. Alzheimer disease: Mechanistic understanding predicts novel therapies. Ann Intern Med. 2004;140:627–638. doi: 10.7326/0003-4819-140-8-200404200-00047. [DOI] [PubMed] [Google Scholar]
- 2.Cao XW, Südhof TC. A transcriptively active complex of APP with Fe65 and histone acetyltransferase Tip60. Science. 2001;293:115–120. doi: 10.1126/science.1058783. [DOI] [PubMed] [Google Scholar]
- 3.Burdick D, et al. Assembly and aggregation properties of synthetic Alzheimer's A4/β amyloid peptide analogs. J Biol Chem. 1992;267:546–554. [PubMed] [Google Scholar]
- 4.Weidemann A, et al. A novel epsilon-cleavage within the transmembrane domain of the Alzheimer amyloid precursor protein demonstrates homology with notch processing. Biochemistry. 2002;41:2825–2835. doi: 10.1021/bi015794o. [DOI] [PubMed] [Google Scholar]
- 5.Ye J, Dave UP, Grishin NV, Goldstein JL, Brown MS. Asparagine-proline sequence within membrane-spanning segment of SREBP triggers intramembrane cleavage by Site-2 protease. Proc Natl Acad Sci USA. 2000;97:5123–5128. doi: 10.1073/pnas.97.10.5123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Li SC, Deber CM. Glycine and β-branched residues support and modulate peptide helicity in membrane environments. FEBS Lett. 1992;311:217–220. doi: 10.1016/0014-5793(92)81106-v. [DOI] [PubMed] [Google Scholar]
- 7.Deber CM, et al. Val–>Ala mutations selectively alter helix-helix packing in the transmembrane segment of phage M13 coat protein. Proc Natl Acad Sci USA. 1993;90:11648–11652. doi: 10.1073/pnas.90.24.11648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Russ WP, Engelman DM. The GxxxG motif: A framework for transmembrane helix-helix association. J Mol Biol. 2000;296:911–919. doi: 10.1006/jmbi.1999.3489. [DOI] [PubMed] [Google Scholar]
- 9.Munter LM, et al. GxxxG motifs within the amyloid precursor protein transmembrane sequence are critical for the etiology of Aβ42. EMBO J. 2007;26:1702–1712. doi: 10.1038/sj.emboj.7601616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kienlen-Campard P, et al. Amyloidogenic processing but not amyloid precursor protein (APP) intracellular C-terminal domain production requires a precisely oriented APP dimer assembled by transmembrane GXXXG motifs. J Biol Chem. 2008;283:7733–7744. doi: 10.1074/jbc.M707142200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gorman PM, et al. Dimerization of the transmembrane domain of amyloid precursor proteins and familial Alzheimer's disease mutants. BMC Neuroscience. 2008;9:17. doi: 10.1186/1471-2202-9-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Smith SO, et al. Implications of threonine hydrogen bonding in the glycophorin A transmembrane helix dimer. Biophys J. 2002;82:2476–2486. doi: 10.1016/S0006-3495(02)75590-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Saito H, Tuzi S, Naito A. Empirical versus nonempirical evaluation of secondary structure of fibrous and membrane proteins by solid-state NMR: A practical approach. Annu Rep NMR Spectrosc. 1998;36:79–121. [Google Scholar]
- 14.Scheuermann S, et al. Homodimerization of amyloid precursor protein and its implication in the amyloidogenic pathway of Alzheimer's disease. J Biol Chem. 2001;276:33923–33929. doi: 10.1074/jbc.M105410200. [DOI] [PubMed] [Google Scholar]
- 15.Vooijs M, Schroeter EH, Pan YH, Blandford M, Kopan R. Ectodomain shedding and intramembrane cleavage of mammalian notch proteins is not regulated through oligomerization. J Biol Chem. 2004;279:50864–50873. doi: 10.1074/jbc.M409430200. [DOI] [PubMed] [Google Scholar]
- 16.Senes A, Ubarretxena-Belandia I, Engelman DM. The Cα-H….O hydrogen bond: A determinant of stability and specificity in transmembrane helix interactions. Proc Natl Acad Sci USA. 2001;98:9056–9061. doi: 10.1073/pnas.161280798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ramelot TA, Gentile LN, Nicholson LK. Transient structure of the amyloid precursor protein cytoplasmic tail indicates preordering of structure for binding to cytosolic factors. Biochemistry. 2000;39:2714–2725. doi: 10.1021/bi992580m. [DOI] [PubMed] [Google Scholar]
- 18.Theresa A, Ramelot TA, Nicholson LK. Phosphorylation-induced structural changes in the amyloid precursor protein cytoplasmic tail detected by NMR. J Mol Biol. 2001;307:871–884. doi: 10.1006/jmbi.2001.4535. [DOI] [PubMed] [Google Scholar]
- 19.Dovey HF, et al. Functional gamma-secretase inhibitors reduce beta-amyloid peptide levels in brain. J Neurochem. 2001;76:173–181. doi: 10.1046/j.1471-4159.2001.00012.x. [DOI] [PubMed] [Google Scholar]
- 20.De Strooper B. Loss-of-function presenilin mutations in Alzheimer disease - Talking point on the role of presenilin mutations in Alzheimer disease. EMBO Rep. 2007;8:141–146. doi: 10.1038/sj.embor.7400897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yu CJ, et al. Characterization of a presenilin-mediated amyloid precursor protein carboxyl-terminal fragment gamma - Evidence for distinct mechanisms involved in gamma-secretase processing or the APP and Notch1 transmembrane domains. J Biol Chem. 2001;276:43756–43760. doi: 10.1074/jbc.C100410200. [DOI] [PubMed] [Google Scholar]
- 22.Qi-Takahara Y, et al. Longer forms of amyloid β protein: Implications for the mechanism of intramembrane cleavage by gamma-secretase. J Neurosci. 2005;25:436–445. doi: 10.1523/JNEUROSCI.1575-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kakuda N, et al. Equimolar production of amyloid β-protein and amyloid precursor protein intracellular domain from β-carboxyl-terminal fragment by γ-secretase. J Biol Chem. 2006;281:14776–14786. doi: 10.1074/jbc.M513453200. [DOI] [PubMed] [Google Scholar]
- 24.Chandu D, Huppert SS, Kopan R. Analysis of transmembrane domain mutants is consistent with sequential cleavage of Notch by γ-secretase. J Neurochem. 2006;96:228–235. doi: 10.1111/j.1471-4159.2005.03547.x. [DOI] [PubMed] [Google Scholar]
- 25.Wiley JC, Hudson M, Kanning KC, Schecterson LC, Bothwell M. Familial Alzheimer's disease mutations inhibit γ-secretase-mediated liberation of β-amyloid precursor protein carboxy-terminal fragment. J Neurochem. 2005;94:1189–1201. doi: 10.1111/j.1471-4159.2005.03266.x. [DOI] [PubMed] [Google Scholar]
- 26.Hecimovic S, et al. Mutations in APP have independent effects on Aβ and CTFγ generation. Neurobiol Dis. 2004;17:205–218. doi: 10.1016/j.nbd.2004.04.018. [DOI] [PubMed] [Google Scholar]
- 27.Wang YC, Zhang YJ, Ha Y. Crystal structure of a rhomboid family intramembrane protease. Nature. 2006;444:179–183. doi: 10.1038/nature05255. [DOI] [PubMed] [Google Scholar]
- 28.Feng L, et al. Structure of a site-2 protease family intramembrane metalloprotease. Science. 2007;318:1608–1612. doi: 10.1126/science.1150755. [DOI] [PubMed] [Google Scholar]
- 29.Duncan EA, Dave UP, Sakai J, Goldstein JL, Brown MS. Second-site cleavage in sterol regulatory element-binding protein occurs at transmembrane junction as determined by cysteine panning. J Biol Chem. 1998;273:17801–17809. doi: 10.1074/jbc.273.28.17801. [DOI] [PubMed] [Google Scholar]
- 30.Takegoshi K, Nakamura S, Terao T. 13C-1H dipolar-assisted rotational resonance in magic-angle spinning NMR. Chem Phys Lett. 2001;344:631–637. [Google Scholar]
- 31.Huysseune S, Kienlen-Campard P, Octave JN. Fe65 does not stabilize AICD during activation of transcription in a luciferase assay. Biochem Biophys Res Commun. 2007;361:317–322. doi: 10.1016/j.bbrc.2007.06.186. [DOI] [PubMed] [Google Scholar]
- 32.Sergeant N, et al. Progressive decrease of amyloid precursor protein carboxy terminal fragments (APP-CTFs), associated with tau pathology stages, in Alzheimer's disease. J Neurochem. 2002;81:663–672. doi: 10.1046/j.1471-4159.2002.00901.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.