Abstract
Reliable structure-prediction methods for membrane proteins are important because the experimental determination of high-resolution membrane protein structures remains very difficult, especially for eukaryotic proteins. However, membrane proteins are typically longer than 200 aa and represent a formidable challenge for structure prediction. We have developed a method for predicting the structures of large membrane proteins by constraining helix–helix packing arrangements at particular positions predicted from sequence or identified by experiments. We tested the method on 12 membrane proteins of diverse topologies and functions with lengths ranging between 190 and 300 residues. Enforcing a single constraint during the folding simulations enriched the population of near-native models for 9 proteins. In 4 of the cases in which the constraint was predicted from the sequence, 1 of the 5 lowest energy models was superimposable within 4 Å on the native structure. Near-native structures could also be selected for heme-binding and pore-forming domains from simulations in which pairs of conserved histidine-chelating hemes and one experimentally determined salt bridge were constrained, respectively. These results suggest that models within 4 Å of the native structure can be achieved for complex membrane proteins if even limited information on residue-residue interactions can be obtained from protein structure databases or experiments.
Keywords: de novo protein structure prediction, ROSETTA
Membrane proteins constitute ≈30% of all proteins and perform crucial functions that range from cell–cell communication to energy transduction to the transport of small key molecules. Despite recent progress, experimental high-resolution structural determination for membrane proteins is still difficult, making structure prediction an important alternative approach.
Membrane proteins can be classified into 2 groups: transmembrane helical (TMH) bundles and beta-barrels. For TMH proteins, the physical constraints imposed by the anisotropic environment of the lipid bilayer lead to characteristic distributions of amino acids that depend on their depth in the membrane. These observations have enabled the development of topology prediction schemes that have become quite sophisticated and powerful over recent years (1). In principle, 3-dimensional (3D) structure modeling based on an existing structure of a close homolog can provide atomic-level structural detail (2–4). However, with few structures known, homology modeling cannot yet be universally applied to membrane protein structures. Previous studies have shown that de novo structure prediction can be successful for small membrane protein domains (5) and can generate models that can be refined to higher resolution (6). However, structure prediction of full-length membrane proteins is hindered by the considerable size of these polypeptides and represents a formidable unsolved challenge. Fortunately, the conformational space sampled by the majority of TMH pairs can be described by a limited number of TMH orientations (7) and recurrent sequence motifs such as the well-studied GXXXG motif (8) appear to favor one particular TMH pair configuration. A significant fraction of membrane proteins bind cofactors with well-defined coordination geometries, therefore imposing further constraints on the structure of TMH assemblies. To take advantage of these conformational restrictions in structure-prediction approaches, we have developed a method to generate models of membrane proteins from sequence in which TMH orientations are constrained using residue-residue interactions either predicted from sequence/structure correlations or derived from experiments. In this study, we describe the validation of the method on a set of membrane proteins with diverse size, topologies, and functions.
Results
Folding with Constraints.
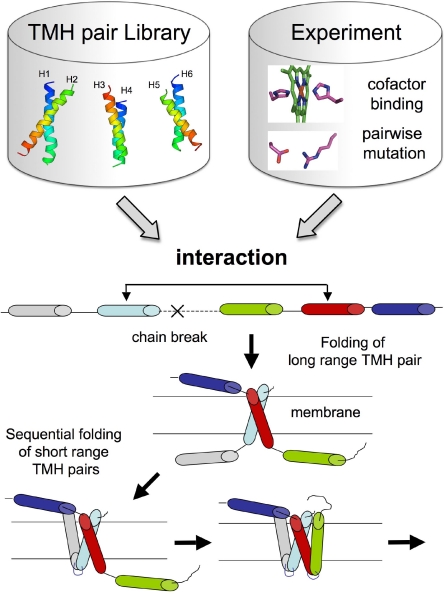
We adapted a technique recently developed for sampling nonlocal beta-sheet topologies (9) to fold membrane proteins from sequence in which the relative orientation of TMH pairs is fixed at two particular positions during folding by long-range pairwise constraints. Briefly, for each long-range constraint between two helices, a “fold tree” is constructed for the polypeptide chain in which two Cα positions from the two helices are connected and fixed in space during folding (9). To allow for this non-local connection in the tree, the peptide chain is cut between the two connected positions (see Materials and Methods and Fig. 1). The cut is randomly selected within predicted loop regions of the proteins with a bias toward long loops. This avoids disrupting subdomains composed of few TMHs connected by short loops, which can be folded properly using continuous chain fragment insertion methods that we have developed previously (5). In a typical run, we generate models using many independent trajectories in which a single randomly selected interaction from a set of predicted TM helix–helix constraints is enforced (Figs. S1–S3). TM helix–helix interactions enriched in low energy models are identified and then used to seed a subsequent round of model generation (Fig. S4). During this process, the average fraction of trajectories constrained with a near-native interaction increased from 16% at iteration 1 to 24% at iteration 2 and to 29% at iteration 3. However, in most cases, the coarse-grained models with the lowest rmsd to the native structure cannot be identified by energy alone. The final coarse-grained models are therefore clustered in structurally related families and refined at the all-atom level (see Materials and Methods).
Fig. 1.
Ab initio folding protocol with long-range interactions. Interactions can be predicted from sequence information using a database of TMH pairs of known structure (Fig. S1) or can be inferred from experiments (see Materials and Methods). Once an interaction is selected, the two helices connected through space by that interaction are inserted and folded in the membrane. Adjacent individual TMHs are then randomly selected and folded in the membrane by Monte-Carlo fragment insertion sampling. After all TMHs are assembled in the membrane, the initial chain break is closed.
Structure Generation Using Predicted Constraints.
Construction of TM helix–helix constraint library.
To predict structural constraints from sequence information, we developed a method that extracts the configuration of TMHs at interacting positions from a database of TMH pairs of known structures (see Materials and Methods, Fig. S1). This database of interacting TMH pairs is searched for local sequence matches with all possible pairs of predicted TMHs in the query sequence using a sliding window (see Materials and Methods, SI Text). This scanning produces for each pair of predicted helices in the query a library of possible interaction geometries defined by the interacting positions and the backbone conformations of the TMH pair from the database. In each folding trajectory, a single randomly selected predicted interaction in the library is used to constrain a particular helix pair to the helix–helix arrangement of the structural template (see above). Ten predicted interactions are included for each helix pair, which allows correct models to be generated despite the low overall accuracy of the interaction library since only one of the 10 need be correct (Figs. S2 and S3).
Validation of the Method.
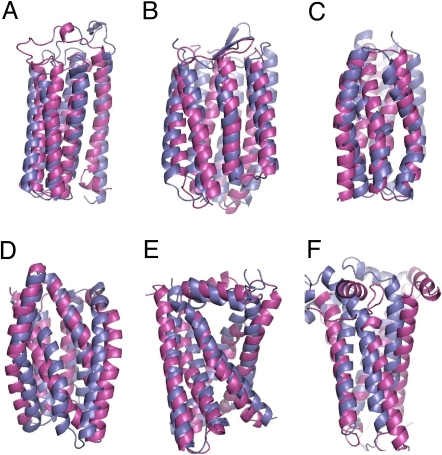
To test the ability of the method to select relevant contacts from the structure database of TMH pairs and use these constraints to generate near-native membrane protein structures, we generated structures for membrane proteins with different sizes and topological complexities. The 4 TMH subdomain of bacteriorhodopsin and the 4 TMH subunit of V-type Na+ ATPase have simple topologies, are limited in size (<150 residues), and can be folded correctly to near-native structure without any long-range constraint (5, 6). We carried out multiple folding trajectories for these polypeptides, each enforcing a single randomly selected interaction from the library of constraints and generated similar near-native structures albeit in lower proportion. The 5 TMH subdomain of cytochrome c has a more complex topology with a long loop connecting the first and second helix. The lowest rmsd models generated without constraints did not recapitulate completely the native topology and had 70% of the residues superimposable on the native structure to within 4 Å (Table 1). When models were generated with a single randomly selected predicted constraint, however, a 40-fold enrichment in low rmsd structures was observed and the lowest rmsd structure was native-like with 93% of the residues superimposable on the native structure (Table 1). After all-atom refinement, 1 of the top 5 lowest energy models was native-like and had 100% of the residues superimposable on the TMH region of the native structure (Fig. 2A). The lowest rmsd model of the full-length 7 TMHs bacteriorhodopsin generated without constraint was not entirely native-like with 68% of residues superimposable on the native structure. By contrast, when bacteriorhodopsin was folded constraining a single randomly selected constraint, a 9-fold enrichment in low rmsd structures was observed and the lowest rmsd model had 93% of the residues superimposable on the native structure (Table 1). After all-atom refinement, 1 of the top 5 lowest energy models was native-like and had 99% of the residues superimposable on the TMH region of the native structure (Fig. 2B).
Table 1.
Structure prediction of membrane proteins.
| No. of TMHs/no. of residues | Constraint type | Highest maxsub in 5,000 simulations (old) | Highest maxsub in 5,000 simulations (new) | Enrichment in high maxsub models (new versus old) | Highest maxsub (full/TMH) | Highest maxsub among 5 lowest energy models (full/TMH) | |
|---|---|---|---|---|---|---|---|
| Bacteriorhodopsin subdomain | 4/123 | One predicted | 1.0 | 1.0 | 0.1 | 1.0/1.0 | 1.0/1.0 |
| V-type ATPase subunit | 4/145 | One predicted | 1.0 | 1.0 | 0.5 | 1.0/1.0 | 0.99/1.0 |
| Cytochrome c | 5/191 | One predicted | 0.70 | 0.88 | 40.3 | 0.93/0.96 | 0.91/1.0 |
| Lac permease Nterminal subunit | 6/190 | One predicted | 0.71 | 0.65 | 0.1 | 0.74/0.82 | — |
| Lac permease Cterminal subunit | 6/185 | One predicted | 0.82 | 0.91 | 1.4 | 0.98/0.99 | — |
| Bacteriorhodopsin | 7/227 | One predicted | 0.68 | 0.83 | 8.7 | 0.93/0.96 | 0.89/0.99 |
| Sensory rhodopsin | 7/217 | One predicted | 0.71 | 0.81 | 3.2 | 0.93/0.95 | 0.52/0.54 |
| Halorhodopsin | 7/239 | One predicted | 0.64 | 0.73 | 4.5 | 0.79/0.89 | 0.48/0.57 |
| Bovine rhodopsin | 7/278 | One predicted | 0.56 | 0.52 | 0.2 | 0.58/0.69 | 0.40/0.53 |
| Beta2 adrenergic receptor | 7/282 | One predicted | 0.46 | 0.55 | 6.3 | 0.61/0.74 | 0.36/0.46 |
| Fumarate reductase | 5/216 | Two pairs of histidine binding hemes | 0.59 | 0.73 | 3.2 | 0.86/0.92 | 0.84/1.0 |
| Cytochrome bc1 | 5/222 | Two pairs of histidine binding hemes | 0.43 | 0.62 | 197.2 | 0.66/0.82 | 0.55/0.80 |
| Lac permease Cterminal subunit | 6/185 | One experimentally-determined salt bridge | 0.82 | 0.99 | 47.6 | 1.0/1.0 | — |
The highest maxsub in 5,000 simulations generated without constraints [using the ″old″ version of RosettaMembrane (5)] and with constraints (“new”, current version of RosettaMembrane) is reported in columns 4 and 5, respectively. Maxsub is the fraction of residues in the model that are superimposable within 4 Å to the X-ray structure of the target (18). The increase in the frequency of models with maxsub greater than or equal to that in column 4 in the constrained runs is reported in column 6. The model closest to the native structure generated by the current version of RosettaMembrane using the protocol described in Materials and Methods is reported in column 7 with maxsub given for both the full-length (″full″) and the transmembrane helical core regions [″TMH″, as predicted by Octopus (15)]. The most accurate (highest maxsub) model among the 5 lowest all-atom energy models is reported in the last column for each target except the isolated subunits of the Lac permease.
Fig. 2.
Prediction of membrane protein structures. Superposition between the most accurate (highest maxsub) models of the 5 lowest all-atom energy models (magenta) and X-ray structure of: chain A of cytochrome c (A), Bacteriorhodopsin (B), chain H of fumarate reductase (E), and chain D of cytochrome bc1 (F). Because individual subunits of the Lactose permease virtually expose pore-lining polar residues to the lipids, near-native structures cannot be selected by energy alone. The cluster size was used as an initial filter for the selection of the models. Superposition between the most accurate (highest maxsub) of the lowest all-atom energy model in the 2 largest clusters (magenta) and X-ray structure of: N-terminal subunit of lactose permease (C, view from the channel), C-terminal subunit of lactose permease (D, view from the channel).
We also tested the method on proteins with complex topologies composed of 6 and 7 TMHs: Lac permease N- and C- terminal domains, sensory rhodopsin, halorhodopsin, bovine rhodopsin, and the beta2 adrenergic receptor. Both subunits of lactose permease have a complicated topology with each of the TMHs making little contact with the next or previous TMH in the sequence. Constraining the chain with a single randomly selected constraint during folding slightly increased the fraction of near-native models for the C-terminal subunit compared with the same simulations performed without constraints (Table 1). The lowest rmsd models had 82% and 99% of the residues superimposable on the TMH region of the native structure for the N- and C- terminal domains, respectively (Table 1). When refined at all-atom, native-like models clustered in one of the largest family of structures but were not the lowest in energy. Polar residues present in the pore region of the transporter become exposed to the hydrophobic region of the lipid bilayer in each isolated subunit, therefore penalizing energetically near-native models. Sensory rhodopsin and halorhodopsin show little sequence identity (<30%) but are structurally similar to bacteriorhodopsin. The structure of these two targets was modeled de novo with a single randomly selected constraint selected from the structure database of TMH pairs (Table 1). A 3- to 5-fold enrichment in low rmsd structures compared to simulations performed without constraints was observed. The lowest rmsd model generated for sensory rhodopsin has 93% of the residues superimposable on the native structure. Except for the long distorted beta hairpin connecting the second and the third TMH, the lowest rmsd model of halorhodopsin is native-like with 89% of the residues from the TMH region superimposable on the native structure (Table 1). Due to the absence of the chromophore and to the particular constraints between TMH pairs for these targets, the near-native models were too tightly packed in the region binding the chromophore. Consequently, these models could not be recovered and refined at all-atom and less accurate models were selected by energy (Table 1). Bovine rhodopsin and the beta2 adrenergic receptor have nearly 300 residues, a complex topology characterized by distorted helices, a significant number of contacts between helices not adjacent in sequence, and a long loop buried in the core of the TMH bundle connecting the second and third helix. Models were generated de novo using a single randomly selected constraint from the structure database of TMH pairs. No enrichment in low rmsd models was observed for bovine rhodopsin. A 6-fold enrichment in low rmsd models was observed for the beta2 adrenergic receptor and the lowest rmsd model had 74% of the residues from the TMH region superimposable on the native structure (Table 1). These models were not close enough to the native structure to be selected by energy alone.
Structure Modeling with Positions of Contacts Inferred from Experiments.
Construction of libraries of experimentally derived structural constraints.
Experimental data were incorporated by restricting the sequence profile search described above to database of template helix pairs selected using the constraint information (see Materials and Methods). This approach generated libraries of interactions sampling the conformational diversity consistent with the chemical interactions identified from experiments.
Modeling with constraints from cofactor binding.
The presence of a cofactor imposes stringent constraints on the protein structure that can be judiciously exploited in structure prediction. One such cofactor is the heme, a FeIII atom chelated by a porphyrin cycle and bound to the protein by 2 histidines providing the 2 axial nitrogen atom ligands of the iron. We modeled the distribution of orientations between the histidines using a library of non-homologous pairs of helices binding hemes extracted from the protein structure database (see Materials and Methods). The ability of our method to generate near-native structures of heme-binding membrane proteins using such constraints was tested on the heme-binding subunits of fumarate reductase and cytochrome bc1. A 3- and 197-fold enrichment in low rmsd models was observed for fumarate reductase and cytochrome bc1, respectively. While the exact positions of interfacial helices and unconstrained loops were not well predicted, in the TMH core regions, the lowest rmsd models had 92% and 82% of the residues superimposable on the native structure for fumarate reductase and cytochrome bc1, respectively (Table 1). After all-atom refinement, the lowest energy model among the two largest clusters was native-like and had 100% and 80% of the residues superimposable on the TMH region of the native structure for fumarate reductase and cytochrome bc1, respectively (Table 1 and Fig. 2 E and F).
Modeling with constraints from compensatory mutations.
The C-terminal domain of lactose permease was folded by constraining a single randomly selected interaction from the structure database compatible with the salt bridge between Asp 237 and Lys 358 inferred from mutagenesis data [(10), see Materials and Methods]. A 48-fold enrichment in low rmsd models was observed compared with simulations performed without constraint (Table 1). The lowest rmsd model had 100% of the residues superimposable on the native structure (Table 1). After all-atom refinement, one of the near-native models, which belong to the second largest cluster was 1 of the top5 lowest energy model and had a Calpha rmsd of 4.2 Å to the native structure (Fig. 2D).
Discussion
Despite the crucial functions performed by membrane proteins in living cells, few high-resolution structures of these proteins have been solved to date. Reliable methods to predict their structures are therefore of high interest but creating such method is a formidable challenge given the size and the complexity of membrane proteins. We provide in this study a step toward a solution to the sampling problem for TMH assemblies, which is conceptually similar to that proposed recently for beta-sheet proteins (9). We developed a method that folds membrane proteins by constraining helix–helix packing arrangements at particular positions predicted from sequence or suggested from experiments to mediate the interaction between the TMHs. We validated the method by generating models for 12 membrane proteins of diverse size, topologies and functions (Table 1). By enforcing a single constraint during the folding simulations, the population of near-native models was enriched for 9 of the targets with more than 4 TM helices (Table 1). Using a single randomly-selected constraint predicted from sequence information alone, near-native structures were generated for the 5 TMH domain of cytochrome c, full-length bacteriorhodopsin, sensory rhodopsin, the C- terminal domain of the lactose permease, and for the TMH core domain of halorhodopsin. Using experimentally derived constraints, native-like structures were obtained for the C-terminal domain of the lactose permease and for the heme-binding TMH regions of the fumarate reductase and cytochrome bc1. For 7 of these 12 proteins, the most accurate models were close enough to the native structure to be selected based their very low energies.
Our extraction by sequence profile matches of plausible interactions between TMHs from the structure database shows relatively low accuracy, but since 10 possibilities are considered for each pair during folding, high accuracy is not necessary. This is analogous to the selection of short peptide fragments based on local sequence in soluble protein structure prediction using ROSETTA. The libraries of local structures and TM helix–helix interactions represent the ensemble of states consistent with local sequence, which is frequently quite ambiguous. Successful prediction requires only that at least one of the helix–helix interactions in the library selected for a given helix pair is correct. Our results suggest also that non-native interactions generating high-energy models can be filtered out from the initial library by a simple iterative refinement protocol, therefore enriching the library from 16% to nearly 30% of native-like interactions. In the future, information from the analysis of coevolving residues (i.e., contact predictor) may be used to improve the prediction of pairs of interacting residues at TMH interfaces. A recent study performed on membrane proteins suggests that sparse residue-residue contacts can now be predicted with high specificity from coevolution information (11).
While high-resolution structures are difficult to obtain for membrane proteins, many experiments can be performed to probe residue-residue interactions and derive effective constraints to feed into our structure prediction method. We have used 2 classes of experimental data from which structural information with different level of accuracy can be extracted. The binding of cofactors provides many structural constraints providing the ligand residues are known as illustrated by our results for heme-binding proteins. More sophisticated spectroscopic data could be used in the future to further constrain the orientation of the cofactor with regard to the membrane bilayer. Interactions between non-covalently linked residues inferred from compensatory mutations provide structural information of lower resolution that can still be useful as illustrated by our results with the C-terminal subunit of the lactose permease. Disulfide bonds between cysteines and chemical cross-links are widely used to probe residue-residue interactions in membrane protein (12) and such constraints can be readily input into our structure calculation procedure.
Using one constraint, our method generated near-native structures of membrane proteins with up to 6 TMHs and on larger but topologically rather simple prokaryotic GPCR-like proteins. The lower accuracy models obtained for the topologically more complex eukaryotic GPCRs clearly point to several directions for improvement. First, these results suggest that for such proteins multiple constraints may be necessary to obtain accurate models. Second, as in bovine rhodopsin and the beta2 adrenergic receptor, long partially buried loops can make substantial contacts with the core of the TMH domain and may partially dictate the precise topology of the TMH bundle. Therefore, it could be advantageous to fold large loops in the early stages of the folding process. The precise conformation of long loops is often difficult to predict by the insertion of short peptide fragments. As suggested by the work of Zhang and Skolnick (4), the identification and sampling of longer peptide fragments may better capture sequence/structure signals governing the conformation of long loops. Third, many membrane proteins covalently or reversibly bind ligand or cofactors in specific cavities of their structures. If the cofactors/ligands are not modeled explicitly in the structure prediction calculations, models that are too tightly packed at these particular binding sites are generated (e.g., for sensory rhodopsin). A solution would be to model explicitly at the coarse-grained level the ligand during the folding of the polypeptide chain, providing constraints can be derived for binding the ligand. Finally, while the all-atom refinement of the coarse-grained models was in many cases able to discriminate by energy near-native from non-native structures, it is very sensitive to small inaccuracies in the constraints enforced during coarse-grained folding. More effective refinement strategies may involve the sampling of rigid-body degrees of freedom of the TMHs to overcome the inaccuracies in the predicted TM helix–helix interaction templates.
While the method has not been tested yet in a blind prediction experiment, our results suggest that it can be used to predict near-native structures of membrane-embedded single polypeptide chains, providing TM helix–helix interactions can be predicted from sequence or extracted from experiments. In this study we experimented with the use of single constraints and could identify near native models using energy based selection for membrane proteins with up to 230 residues; for larger proteins, however, multiple constraints are likely to be necessary to obtain accurate models. Such models should prove useful to guide and rationalize future experimental investigations on the many systems for which no high-resolution structural information is yet available.
Materials and Methods
Selection of Long-Range Pairwise Interactions from a Library of TMH Pairs with Known Structure.
A library of 621 interacting transmembrane helical pairs was constructed from 79 high-resolution membrane proteins chains with <90% pairwise sequence identity, taken from the protein database as of April 2007. The boundaries for TM helical segments were taken from the MPtopo database (13). Two helices were considered to interact if 5 or more pairs of Cα atoms were within 8 Å.
Sequence profiles were constructed for all helical pairs in the database by PSI-BLAST (14) with the -j 2 option using the BLOSUM62 substitution with E-value cutoff 10−3 against Uniref90 (uniref) for the whole protein chain sequence and parsing out the specific regions corresponding to the TM helical pairs. To ensure that no templates from homologs were present in the final library, all hits to templates from proteins with a BLAST hit better than E-value 5E−2 to the query sequence were filtered out.
To search the library, a sequence profile is constructed for the query sequence as described above and the specific regions corresponding to transmembrane regions predicted by Octopus (15) are parsed out. In the next step each possible pair of predicted transmembrane helices is compared with the profiles for each pair in the library using a gapless log average profile-profile scoring (16) over a 14-residue sliding window, other window sizes were tried but 14 performed best (data not shown). To compare a helix pair in the library (H1,H2) with a helix pair from the query (h1, h2), H1 is compared with h1 by sliding one window over H1 and one over h1 and calculating the log average profile-profile score for each position of the two windows; the same is done for H2 and h2. Only registers in which all residues in the two 14 residue windows are aligned are considered. The final score for a match is the sum of the best scores from the H1-h1 and H2-h2 comparisons. This procedure gives a score for each possible position of the 4 windows (Fig. S1). Overlaps between the windows were avoided by requiring that the center of the window on h1 be separated by at least 20 residues from the center of the window on h2. Once a match is found, the backbone orientation (i.e., coordinates of the N, Cα and C positions) for the closest point of interaction (closest distance) in the matching windows for the template helices H1 and H2 is copied to the equivalent positions in query helices h1 and h2. By taking the closest point of interaction instead of the residues in the center of the windows, potential helix–helix interaction motifs do not need to be in the middle of the window to be captured.
Long-Range Pairwise Interactions Extracted from Experiments.
The sequence matching technique described above was applied to subsets of template helix pairs preselected based on the experimental data. If experimental data suggest that 2 helices interact via a particular pairwise interaction, the template library of interacting TMH pairs with known structure is searched for local sequences matching that particular interaction. As for the pure “sequence only” search (see above), each selected template is used to constrain the configuration of the 2 helices during folding by fixing the backbone coordinates of the 2 interacting positions to those found in the template. In our study, 2 different experimentally derived pairwise constraints were considered: (i) constraints for pairs of histidines that chelate hemes by providing the 2 axial nitrogens coordinating the FeIII were selected from a library of high-resolution heme-binding protein structures; (ii) constraints for pairs of polar residues involved in salt bridges were derived from a library of interacting TMH pairs interacting via the salt bridge.
Ab Initio Folding Protocol with Long-Range Constraints.
Once a long-range constraint between 2 helices is identified, a fold tree is constructed for the polypeptide chain in which 2 Cα positions from these 2 helices are connected and fixed in space during folding (9). To allow for this non-local connection in the tree, the peptide chain is cut at a randomly selected position within predicted loop regions of the proteins with a bias toward long loops. The folding process involves the following steps (Fig. 1): (i) 2 helices connected through space by the long-range interaction are inserted in the membrane; (ii) individual adjacent TMHs are randomly selected and inserted in the membrane by Monte-Carlo fragment insertion sampling as described in ref. 5. This process is repeated until all TMHs are folded in the membrane; and (iii) once all TMHs and connecting loops are folded, a final cycle of fragment insertions is performed to close the chain break created by the initial cut in the polypeptide chain. For each protein, a total of two hundred thousand coarse-grained models were generated in several steps using an iterative approach to select the most promising set of TM helix–helix interactions (SI Text and Fig. S4). In most cases, the coarse-grained models with the lowest rmsd to the native structure could not be identified from energy alone, which prompted us to refine them at the all-atom level. The “old” protocol used as the control in Table 1 carries out the stage 2 continuous chain fragment insertion for the whole trajectory (5).
All-Atom Refinement of Coarse-Grained Models.
The full-atom structure relaxation of the coarse-grained models is performed using an all-atom potential developed recently for membrane proteins (6). Instead of relaxing all coarse-grained models, we implemented a more efficient refinement protocol that aims at selecting rapidly which models are likely to occupy energy minima in the all-atom energy landscape. Coarse-grained models were first clustered into structurally related families. The clusters with energies below the median energy were selected for all-atom refinement. For each of these selected clusters, both the center and the 10 lowest energy structures were refined using a stochastic Monte Carlo minimization protocol. Each move in this landscape involves a random perturbation of backbone torsion angles followed by discrete optimization of side-chain rotamers and then by gradient-based local minimization on all conformational degrees of freedom (17). A faster version of the original refinement protocol for membrane proteins (6) was used that consists of 3 iterative cycles of side-chain rotamer repacking and gradient-based minimization on backbone and side-chain degrees of freedom. In the initial cycle, the repulsive component of the Lennard-Jones potential is heavily damped. The damping factor is then iteratively decreased in the next cycles. This procedure eases the transition from centroid to atomic structures by accommodating and iteratively relaxing structural inaccuracies present in the centroid models. The lowest energy all-atom structure was selected as the final refined structure for each starting centroid model (SI Text and Fig. S5).
Choice of the Benchmark Test.
The membrane proteins used to validate the method were selected based on several criteria: first, a structure determined experimentally by X-ray crystallography at a resolution <3.5 Å; second, a protein length between 100 and 300 residues with 4 to 7 TMHs and third, a range of topologies with different level of complexities and structural irregularities such as TMH kinks, coils and interfacial regions. To generate a dataset of proteins for which contacts could be also deduced from experiments, we incorporated a number of proteins with residue-residue contacts identified by different experimental techniques (Table 1).
Metric for Assessing the Structural Quality of the Models.
The quality of a structural model is usually measured by the root mean square deviations over a given set of atoms between the model and the experimentally determined structure. For larger proteins, however, large deviations from the native structure in localized regions often lead to large rmsd values, which can mask the quality of the prediction in the other regions of the protein. For the large proteins studied in this work, the proportion of residues superimposable within 4 Å on the native structure [as measured by maxsub (18)] was found to be a more suitable metric of the quality of the predictions.
Acknowledgments.
This work was supported by the Howard Hughes Medical Institute, the National Institutes of Health and the European Union 6th Framework Program Rosetta-Membrane Project Contract MOIF-CT-2006-40496 (to B.W.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0808323106/DCSupplemental.
References
- 1.Elofsson A, von Heijne G. Membrane protein structure: Prediction versus reality. Annu Rev Biochem. 2007;76:125–140. doi: 10.1146/annurev.biochem.76.052705.163539. [DOI] [PubMed] [Google Scholar]
- 2.Qian B, et al. High-resolution structure prediction and the crystallographic phase problem. Nature. 2007;450:259–264. doi: 10.1038/nature06249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Forrest LR, Tang CL, Honig B. On the accuracy of homology modeling and sequence alignment methods applied to membrane proteins. Biophys J. 2006;91:508–517. doi: 10.1529/biophysj.106.082313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhang Y, Devries ME, Skolnick J. Structure modeling of all identified G protein-coupled receptors in the human genome. PLoS Comput Biol. 2006;2:e13. doi: 10.1371/journal.pcbi.0020013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yarov-Yarovoy V, Schonbrun J, Baker D. Multipass membrane protein structure prediction using Rosetta. Proteins. 2006;62:1010–1025. doi: 10.1002/prot.20817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Barth P, Schonbrun J, Baker D. Toward high-resolution prediction and design of transmembrane helical protein structures. Proc Natl Acad Sci USA. 2007;104:15682–15687. doi: 10.1073/pnas.0702515104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Walters RF, DeGrado WF. Helix-packing motifs in membrane proteins. Proc Natl Acad Sci USA. 2006;103:13658–13663. doi: 10.1073/pnas.0605878103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Senes A, Engel DE, DeGrado WF. Folding of helical membrane proteins: The role of polar, GxxxG-like and proline motifs. Curr Opin Struct Biol. 2004;14:465–479. doi: 10.1016/j.sbi.2004.07.007. [DOI] [PubMed] [Google Scholar]
- 9.Bradley P, Baker D. Improved beta-protein structure prediction by multilevel optimization of nonlocal strand pairings and local backbone conformation. Proteins. 2006;65:922–929. doi: 10.1002/prot.21133. [DOI] [PubMed] [Google Scholar]
- 10.Zhao M, Zen KC, Hubbell WL, Kaback HR. Proximity between Glu126 and Arg144 in the lactose permease of Escherichia coli. Biochemistry. 1999;38:7407–7412. doi: 10.1021/bi9906524. [DOI] [PubMed] [Google Scholar]
- 11.Fuchs A, et al. Co-evolving residues in membrane proteins. Bioinformatics. 2007;23:3312–3319. doi: 10.1093/bioinformatics/btm515. [DOI] [PubMed] [Google Scholar]
- 12.Wu J, Kaback HR. A general method for determining helix packing in membrane proteins in situ: Helices I and II are close to helix VII in the lactose permease of Escherichia coli. Proc Natl Acad Sci USA. 1996;93:14498–14502. doi: 10.1073/pnas.93.25.14498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jayasinghe S, Hristova K, White SH. MPtopo: A database of membrane protein topology. Protein Sci. 2001;10:455–458. doi: 10.1110/ps.43501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Altschul SF, et al. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Viklund H, Elofsson A. OCTOPUS: Improving topology prediction by two-track ANN-based preference scores and an extended topological grammar. Bioinformatics. 2008;24:1662–1668. doi: 10.1093/bioinformatics/btn221. [DOI] [PubMed] [Google Scholar]
- 16.von Ohsen N, Sommer I, Zimmer R. Profile-profile alignment: A powerful tool for protein structure prediction. Pac Symp Biocomput. 2003:252–263. [PubMed] [Google Scholar]
- 17.Schueler-Furman O, Wang C, Bradley P, Misura K, Baker D. Progress in modeling of protein structures and interactions. Science. 2005;310:638–642. doi: 10.1126/science.1112160. [DOI] [PubMed] [Google Scholar]
- 18.Siew N, Elofsson A, Rychlewski L, Fischer D. MaxSub: An automated measure for the assessment of protein structure prediction quality. Bioinformatics. 2000;16:776–785. doi: 10.1093/bioinformatics/16.9.776. [DOI] [PubMed] [Google Scholar]
- 19.Adamian L, Liang J. Prediction of transmembrane helix orientation in polytopic membrane proteins. BMC Struct Biol. 2006;6:13. doi: 10.1186/1472-6807-6-13. [DOI] [PMC free article] [PubMed] [Google Scholar]