Abstract
gC1qR is one of the C1q receptors implicated in the regulation of innate and adaptive immunity. We found that gC1qR inhibits RIG-I and MDA5-dependent antiviral signaling. Double stranded RNA and virus trigger the translocation of gC1qR to the mitochondrial outer membrane leading to the interaction of gC1qR with the RIG-I and MDA5 adaptor, VISA/MAVS/IPS-1/Cardif. The interaction of gC1qR with VISA/MAVS/IPS-1/Cardif at mitochondria results in the disruption of RIG-I and MDA5 signaling and the promotion of virus replication. Knockdown of endogenous gC1qR enhances RIG-I-dependent antiviral signaling, and augments the inhibition of virus proliferation. Therefore, gC1qR is a physiological inhibitor of the RIG-I and MDA5-mediated antiviral signaling pathway. These data uncover a new viral mechanism used to negatively control antiviral signaling in host cells.
Keywords: innate immunity, RIG-I signaling, virus
Upon viral infection, the host cells trigger innate immune signaling cascades critical for effective antiviral immune responses through both Toll-like receptor (TLR)-dependent and -independent pathways, resulting in the induction of type I IFN, including IFN-β and the family of IFN-α (1–3). TLRs recognize extracellular viral components such as viral DNA (sensed by TLR9), dsRNA (sensed by TLR3), and ssRNA (sensed by TLR7/8) (4). In contrast, RIG-I and MDA5 are RNA helicases, which function as cytoplasmic viral RNA sensors to mediate TLR-independent antiviral responses (5, 6).
RIG-I and MDA5 belong to the family of DExD/H box RNA helicases that contain 2 caspase-recruiting domains (CARD) at their N termini and a single DExD/H box RNA helicase domain at their C termini. The helicase domain is required for the recognition of intracellular viral RNA. The CARD domain is responsible for transducing signals to the downstream adaptor VISA (virus-induced signaling adapter) [also identified as IPS-1, Cardif, or MAVS (7–10)] through a CARD-CARD interaction. VISA contains an N-terminal CARD-like domain and a C-terminal transmembrane domain, both of which are essential for signaling. The transmembrane domain guides VISA to the mitochondria, where it functions as an adaptor for RIG-I and MDA5 (8). These findings demonstrate a new role for mitochondria in the innate immune response.
Although the induction of type I IFN plays a key role in the control of viral infection, limiting signaling initiated by type I IFN is essential to prevent this protective response from causing injury to the host. Recently, negative regulators of RIG-I and MDA5 signaling have been identified, including A20 (11) and NLRX1 (12). However, the relevance of these negative regulators to physiological functions is not clear.
Human gC1qR (receptor for globular head domain of complement component C1q), a ubiquitously expressed protein, has been implicated in various ligand-mediated cellular responses. It is found in the mitochondria (13–15), nucleus (14), cytoplasm (16, 17) and at the cell surface (18–20). The N terminus of the immature gC1qR includes a mitochondrial targeting sequence. We here identified human gC1qR as a physiological inhibitor of RIG-I and MDA5 signaling. It translocates to the mitochondrial outer membrane upon viral infection, which leads to the inhibition of RIG-I and MDA5 mediated antiviral response through the interaction with VISA at mitochondria.
Results
gC1qR Inhibits RIG-I and MDA5 Signaling.
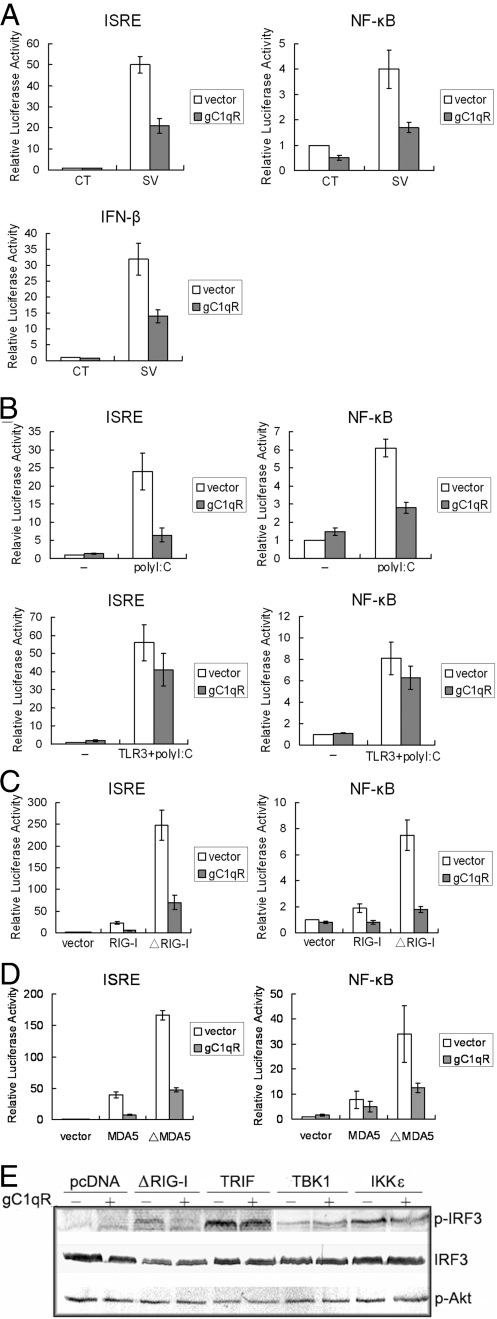
Recent reports suggest that gC1qR, a ubiquitously expressed protein (Fig. S1), might be important for virus replication (21, 22), prompting us to investigate whether gC1qR participates in viral infection. We first addressed the effect of gC1qR on virus activated signaling. We transfected gC1qR with IFN stimulated response element (ISRE), NF-κB, or IFN-β reporter plasmids into 293T cells, and then infected the cells with Sendai virus, a RNA virus. As shown in Fig. 1A, virus infection activated ISRE, NF-κB and IFN-β promoters. The activation was significantly suppressed in cells overexpressing exogenous gC1qR.
Fig. 1.
gC1qR inhibits RIG-I and MDA5-mediated signaling. (A) gC1qR inhibits Sendai virus-induced activation of ISRE, NF-κB and IFN-β promoter. The 293T cells (1 × 105) were transfected with the indicated luciferase reporter plasmids (0.1 μg), control vector or gC1qR expression plasmid (0.5 μg). 12 h after transfection, cells were infected with Sendai virus or left uninfected for 12 h before luciferase assays were performed. (B) gC1qR inhibits poly(I:C) activated intracellular signaling. (Lower) The 293T cells (1 × 105) were transfected with the indicated luciferase reporter plasmids (0.1 μg) and expression plasmids (0.5 μg of each), and were treated with poly(I:C) (20 μg/ml) for 6 h before luciferase assays were performed. (Upper) Cells were transfected with gC1qR expression plasmids and poly(I:C) (0.5 μg of each) followed by luciferase assays. (C and D) gC1qR inhibits activation of ISRE and NF-κB mediated by RIG-I, ΔRIG-I, MDA5 and ΔMDA5. The 293T cells (1 × 105) were transfected with the indicated luciferase reporter plasmids (0.1 μg) and expression plasmids (0.5 μg of each). Eighteen hours after transfection, luciferase assays were performed. (E) gC1qR blocks ΔRIG-I-mediated IRF-3 activation. The 293T cells were transfected with the indicated expression plasmids (0.5 μg). Whole cell extracts were analyzed by immunoblot assays.
Virus activated signaling may be mediated by TLR3-dependent or -independent pathways. To determine which pathway(s) gC1qR inhibits, we exposed cells overexpressing a control vector or gC1qR to the viral dsRNA homologue polyinosinic:polycytidylic acid [poly(I:C)] either extracellularly (to activate TLR3) or intracellularly (to activate RIG-I and MDA5). Poly(I:C) cannot activate an ISRE reporter in TLR3-deficient 293T cells or primary HUVECs when added in the medium. However, when poly(I:C) was transfected into cells, the ISRE promoter was activated (Fig. S2). Moreover, Overexpression of gC1qR significantly suppressed NF-κB and ISRE promoter activation by transfected poly(I:C), but not by poly(I:C) added to the culture medium in 293T transfected with TLR3 (Fig. 1B). Transfection of poly(I:C) has been shown to activate RIG-I and MDA5-dependent signaling (5, 23, 24). Therefore, gC1qR might suppress RIG-I and MDA5-dependent signaling. To test this notion, we cotransfected gC1qR and RIG-I, and then measured ISRE and NF-κB reporter activity. Overexpression of RIG-I or its CARD domain (ΔRIG-I) activated ISRE or NF-κB reporters (Fig. 1C). ΔRIG-I exhibited more potent effect on the activation of promoters than full-length RIG-I did. The activation driven by RIG-I and ΔRIG-I was significantly blocked in cells overexpressing exogenous gC1qR. MDA5 is a RIG-I homologue and plays the similar role as RIG-I in antiviral signaling (25). It also contains CARD motifs. Therefore, we evaluated the effect of gC1qR on MDA5 mediated signaling. gC1qR also suppressed the activation of ISRE and NF-κB by full length MDA5 and its CARD (ΔMDA5) (Fig. 1D). These results indicate that gC1qR specifically inhibits both RIG-I and MDA5 signaling pathway.
IFN regulatory factor 3 (IRF-3) is a transcription factor that activates ISRE. It is phosphorylated upon viral infection, and binds to the ISRE to activate the transcription of a set of genes including IFN-β. Several pathways can mediate the activation of IRF3, including the RIG-I and TLR pathways. We evaluated the effect of gC1qR on RIG-I and TLR mediated IRF3 activation. gC1qR was cotransfected into 293T cells with ΔRIG-I, or the TLR pathway components TRIF, TBK1 or IKKε and immunoblot assay was performed. The result indicated that gC1qR almost completely blocked ΔRIG-I mediated IRF3 phosphorylation (Fig. 1E), but only weakly affected TRIF mediated IRF3 phosphorylation, and did not affect TBK1 and IKKε mediated phosphorylation of IRF3, consistent with the data from Fig. 1B. We also tested whether overexpression of gC1qR has an effect on the activation of PI3K signaling by examining the phosphorylation of Akt, a downstream transducer of PI3K. No change in Akt phosphorylation was observed (Fig. 1E), suggesting that the involvement of gC1qR in RIG-I signaling is not related to PI3K pathway.
Virus Infection Induces the Translocation of gC1qR into Mitochondria.
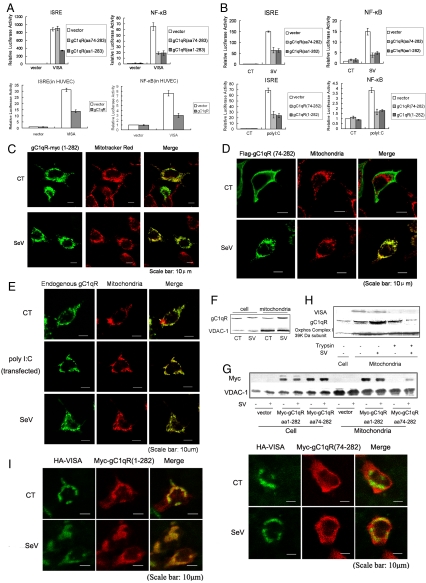
Because both gC1qR and VISA contain a mitochondrial targeting sequence required for their mitochondrial localization (8, 15), and mitochondrial localization is required for VISA signaling, we addressed whether the mitochondrial targeting sequence is required for the inhibition of RIG-I and MDA5 signaling by gC1qR. One to 73 aa of the gC1qR mitochondrial targeting sequence was deleted to form the truncated gC1qR 74–282, also considered a mature form, because amino acids 1–73 are excised to generate the mature form of gC1qR (15). Interestingly, we found that the full length and mature forms of gC1qR behaved differently. Mature gC1qR suppressed the activation of the NF-κB promoter, but not the ISRE promoter, in cells overexpressing VISA (Fig. 2A). Mature gC1qR was also able to suppress activation of ISRE and NF-κB reporters in cells infected by virus or transfected with poly(I:C) (Fig. 2B). In contrast, full length gC1qR suppressed activation of both ISRE and NF-κB reporters induced by either VISA overexpression, viral infection, or transfection with poly(I:C) (Fig. 2 A and B). The suppression occurred not only in 293T cells (Fig. 2A Upper), but also in human umbilical vein endothelial cells (HUVECs) (Fig. 2A Lower). These data demonstrate that mitochondrial localization is required for the suppression of ISRE activation by gC1qR, and suggest that viral infection, especially double stranded RNA, the product of virus replication, induces the translocation of gC1qR to mitochondria.
Fig. 2.
Virus infection induces the translocation of gC1qR into mitochondria. (A) gC1qR inhibits VISA-mediated activation of ISRE and NF-κB in both 293T cells (Upper) and primary HUVECs (Lower). The experiments were performed as in Fig. 1C. (B) (Upper) Virus induces the suppression activity of gC1qR on ISRE activation. The experiments were performed as in Fig. 1A. (Lower) PolyI:C induces the suppression activity of gC1qR on ISRE activation. The experiments were performed as in 1B. (C and D) Translocation of ectopic full length gC1qR and gC1qR74–282 aa. The 293T cells were transfected with indicated expression plasmids followed by viral infection. Cells were subjected to confocal assay probed with Mitotracker Red and antibody against the tag. (E and F) Translocation of endogenous gC1qR. 293T cells were infected with Sendai virus or treated with poly(I:C). (E) Cells were subjected to confocal assay probed with antibody against gC1qR monoclonal antibody and Mitotracker red after treatment. (F) Mitochondrial protein was extracted and analyzed by immunoblot assay with antibodies against gC1qR and VDAC-1, a mitochondria marker. (G) Identification of translocation of ectopic gC1qR74–282 aa and full length gC1qR by immunoblot assay. The 293T cells were transfected with indicated expression plasmids followed by viral infection. Cellular protein and mitochondrial protein were analyzed by immunoblot assay with specific antibody. (H) Endogenous gC1qR localizes at outer membrane of mitochondria after translocation induced by virus. Cells were infected with Sendai virus. Mitochondria were isolated according to the method described in ref. 33. Mitochondria were then incubated with trypsin (1 mg/ml) for 20 min at room temperature. Cellular protein and mitochondrial protein were respectively extracted after digestion, followed by immunoblot assay using antibody against VISA, gC1qR and Oxphos Complex I 39K Da subunit. (I) Colocalization of gC1qR and VISA. The 293T cells were transfected with indicated expression plasmids followed by viral infection. Cells after treatment were subjected to confocal assay probed with antibody against the tag.
To examine gC1qR localization, we expressed a Myc-tagged version of gC1qR in 293T cells and visualized it using fluorescent confocal microscopy. Full length gC1qR labeling overlapped with mitochondrial staining, and viral infection enhanced this overlap (Fig. 2C). Mature gC1qR did not show mitochondrial localization when overexpressed in cells (Fig. 2D Upper), and it translocated to mitochondria only in cells infected by virus (Lower). To determine whether the virus-induced translocation of gC1qR is a physiological response rather than a result of overexpression, we used a gC1qR monoclonal antibody to monitor endogenous gC1qR in cells infected with Sendai virus or transfected with poly(I:C). As shown in Fig. 2E, endogenous gC1qR was observed in both mitochondria and cytoplasm in uninfected cells (Fig. 2E Top). When cells were infected by virus (Fig. 2E Middle) or transfected with poly(I:C) (Fig. 2E Bottom), the cytoplasmic staining for gC1qR was decreased, but mitochondrial staining was increased. It demonstrated that dsRNA is the viral component that induces the translocation of endogenous gC1qR to mitochondria upon infection. Thus, the translocation of gC1qR to mitochondria is a physiological host response to viral infection.
We then performed biochemical experiments to confirm the translocation of exogenous and endogenous gC1qR into mitochondria upon viral infection. Cellular and mitochondrial proteins were isolated from cells infected with Sendai virus and immunoblotted, using a monoclonal antibody for gC1qR or a Myc antibody. Endogenous gC1qR levels clearly increased in the mitochondrial fraction upon viral infection, whereas total cellular levels of gC1qR remained the same (Fig. 2F). Cells expressing gC1qR 74–282 aa also showed the same results after viral infection, whereas full length gC1qR was enriched at mitochondria without infection (Fig. 2G). Thus, the subcellular localization of endogenous and exogenous gC1qR after viral infection consistently supported the conclusion that viral infection triggers the translocation of gC1qR into mitochondria.
Although VISA has been identified to be in the outer membrane (8), the exact localization of gC1qR at mitochondria is not well defined. To determine the localization of translocated gC1qR, we isolated mitochondria from virally infected or uninfected cells, incubated them with trypsin, and then immunoblotted the lysates with gC1qR antibody (Fig. 2H). After trypsin digestion the outer membrane protein VISA could no longer be found in mitochondria in either uninfected or infected cells (Fig. 2H), whereas the inner membrane protein Oxphos Complex I 39K Da subunit, remained unchanged. In contrast, trypsin digestion slightly reduced gC1qR level in mitochondria from uninfected cells, but largely reduced gC1qR level in mitochondria from virus infected cells (Fig. 2H). These results suggested that basal levels of gC1qR may be found in mitochondrial matrix and in cytoplasm, and that viral infection induces the translocation of gC1qR to the mitochondrial outer membrane where it is retained by its interaction with VISA, because the total gC1qR was stable upon viral infection in both mRNA and protein level (Fig. S3). Confocal microscopic visualization of exogenous VISA and gC1qR confirmed their colocalization in the same cellular compartment (Fig. 2I), which could be observed both in uninfected and virally infected cells. Mature gC1qR was colocalized with VISA only in cells infected with virus. Finally, we determined whether gC1qR interacts with VISA within cells by FRET (fluorescence resonance energy transfer). Results clearly showed that full length gC1qR interacted with VISA within cells, whereas the truncated form gC1qR1–167 aa failed to interact with VISA (Fig. S4). Taken together, these data demonstrate that virus-induced translocation to mitochondria is required for gC1qR inhibition of VISA-mediated ISRE activation.
gC1qR Interacts with VISA upon Viral Infection.
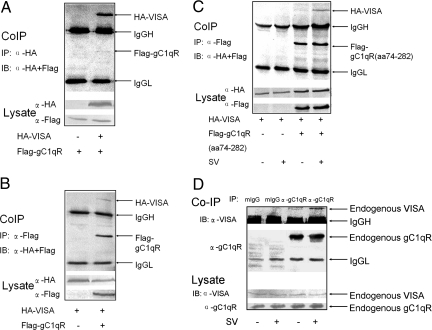
VISA is a signaling component in both RIG-I and MDA5 signaling pathways (10). gC1qR suppresses both RIG-I- and MDA5-mediated (Fig. 1 C and D) and VISA-mediated (Fig. 2A) signaling, suggesting that VISA could be a target of gC1qR. We therefore examined whether the suppression of ISRE or NF-κB activation is mediated by the interaction between gC1qR and VISA. When both tagged proteins are overexpressed, VISA could be coimmunoprecipitated with gC1qR and vice versa, demonstrating that these two proteins interact in cells (Fig. 3 A and B). Intriguingly, gC1qR 74–282 showed only a weak interaction with VISA in uninfected cells. The interaction was dramatically enhanced in cells infected by Sendai virus (Fig. 3C). As shown in Fig. 3D, endogenous gC1qR also weakly associated with endogenous VISA in uninfected cells, and this association was significantly enhanced in cells infected with virus. These results demonstrate that viral infection plays an important role in the association of gC1qR with VISA, and such an association is a physiological event in response to viral infection. Furthermore, it has been shown that the interaction between VISA and ΔRIG-I was disrupted by overexpression of gC1qR (Fig. S5). It suggests that the disruption of the interaction between VISA and RIG-I by gC1qR results in the inhibition of RIG-I signaling.
Fig. 3.
gC1qR interacts with VISA upon viral infection. (A and B) Interaction of ectopic full length gC1qR with ectopic VISA. The 293T cells were transfected as indicated. Protein interaction was analyzed by immunoprecipitated with anti-HA antibody (A) and anti-Flag antibody (B), followed by immunoblot assay, using anti-Flag and anti-HA antibody, respectively. (C) Interaction of ectopic gC1qR74–282 aa with ectopic VISA. The 293T cells were transfected as indicated. 12 h after transfection, cells were infected with Sendai virus or left uninfected for 12 h. The protein association was analyzed as in B. (D) Interaction of endogenous gC1qR and VISA. 293T cells were infected with Sendai virus or left uninfected. Total cellular protein was extracted followed by pull down with gC1qR monoclonal antibody. The following steps were performed as in B.
To map the inhibition region on gC1qR, we made a series of deletion constructs as illustrated in Fig. S6. Co-IP experiments were performed to examine the interaction between the gC1qR mutants and VISA. The interaction of gC1qR with VISA was mainly mediated by the region between 168 and 213 aa. The mitochondrial targeting sequence of gC1qR facilitated the interaction, because gC1qR without the mitochondrial targeting sequence showed only a weak interaction (Fig. S6). These results are consistent with the data in Fig. 2I that full length gC1qR was colocalized with VISA in uninfected cells whereas mature gC1qR was not. We next performed ISRE and NF-κB reporter assays to evaluate the effects of gC1qR deletion mutants on virus- and VISA-mediated signaling. Again, 168–213 aa was the main region required for the inhibition of NF-κB promoter activation. This region without mitochondrial targeting sequence inhibited both VISA-mediated and viral infection-induced activation of NF-κB (Fig. S7). For the inhibition of ISRE activation, either the mitochondrial targeting sequence or virus infection was required (Fig. S8). These functional results are consistent with the results from the coimmunoprecipitation assays.
gC1qR siRNA Results in Enhanced Antiviral Response.
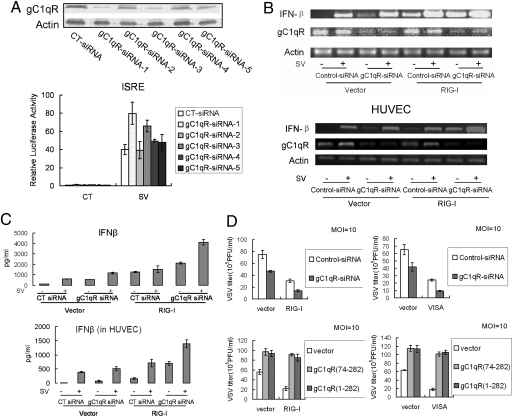
We wanted to determine whether endogenous gC1qR also suppresses IFN-β production and leads to the reduction of antiviral response. Although it has been reported that double stranded RNA can induce the immune response (26), gene silencing and immunostimulation are two independent functional characteristics of RNA oligonucleotides. Therefore, we used siRNA to knock down gC1qR and measured IFN-β levels induced by RIG-I expression or viral infection. Five different siRNA oligonucleotides for gC1qR were designed and used for the knockdown assay. It showed that siRNA-1 had the strongest knocked down effect on endogenous gC1qR (Fig. 4A Upper) and it significantly affected activation of ISRE by virus (Lower). Therefore, siRNA-1 was used for the following experiments. As expected, IFN-β mRNA and protein were potently induced by RIG-I expression or viral infection, and the induction was significantly enhanced in cells where endogenous gC1qR was knocked down in both 293T cells and primary HUVECs by siRNA-1 compared with control siRNA (Fig. 4 B and C).
Fig. 4.
gC1qR siRNA results in enhanced antiviral response. (A) gC1qR siRNA knocks down endogenous gC1qR. Cells were transfected with 5 different siRNAs as indicated. Forty-eight hours after transfection, total cellular protein was analyzed by immunoblot assay (Upper). For reporter assay, cells were transfected with the ISRE reporter plasmid and different siRNAs, respectively. Twelve hours after transfection, cells were infected with Sendai virus or left uninfected for 12 h before luciferase assays (Lower). (B and C) gC1qR siRNA enhances virus-triggered and RIG-I-mediated expression of IFN-β at the levels of mRNA and protein in both 293T cells (Upper) and HUVECs (Lower). Cells were transfected with the indicated siRNA and expression plasmids. Thirty-six hours after transfection, cells were infected with Sendai virus or left uninfected for 12 h. RT-PCR was performed with specific primers (B). For secreted IFN-β analysis, supernatants were collected after the treatment and subjected to ELISA (C). (D) Effects of gC1qR on RIG-I and VISA mediated antiviral response. Cells were transfected with the indicated expression plasmids or siRNA. Thirty-six (Upper) or 24 (Lower) hours after transfection, cells were infected with VSV. Twelve hours after infection, supernatants were harvested and analyzed for VSV titer by standard plaque assays.
RIG-I signaling is required for establishing the antiviral state independent of TLR3. The results described above demonstrate that gC1qR suppresses RIG-I-mediated activation of ISRE and NF-κB, and the induction of IFN-β. We wanted to determine whether gC1qR has an effect on the RIG-I-mediated antiviral response. VSV assay was used to evaluate the effect of gC1qR on virus proliferation. Knockdown of endogenous gC1qR significantly inhibited viral proliferation, and RIG-I- and VISA-mediated suppression of viral proliferation was enhanced in gC1qR silenced cells (Fig. 4D Upper). These data are consistent with the conclusion that silencing endogenous gC1qR augments the induction of IFN-β (Fig. 4 B and C). Conversely, viral titers were increased in cells overexpressing either full length gC1qR or gC1qR 74–282 (Fig. 4D Lower). Suppression of viral proliferation by RIG-I and VISA expression was also prevented by overexpression of either full length gC1qR or gC1qR 74–282 (mature gC1qR) (Fig. 4D Lower). Thus, gC1qR is a physiological inhibitor of the RIG-I-mediated antiviral response.
Discussion
gC1qR is a multicompartmental and multibinding protein, and thus, a multifunctional protein. Although recent evidence suggests that gC1qR may be involved in the regulation of innate immunity and adaptive immunity (27–29), its precise physiological functions and their underlying molecular mechanisms, especially in virus infection, remain to be determined. In this study, we report that gC1qR is a physiological inhibitor of RIG-I and MDA5-dependent antiviral signaling through the interaction with VISA at mitochondria.
Pathogen invasion triggers innate immune host responses required to contain and eradicate foreign microbes. However, the innate immune system must also limit its response to pathogens, because excessive and prolonged activation of innate immunity is harmful to the host and, in some cases fatal, because of severe tissue damage and circulatory failure. Some factors such as A20 (11), NLRX1 (12), SIKE (30), and DAK (31), which serve as native negative regulators in RIG-I and MDA5 and TLR3 antiviral signaling, may contribute to limiting the antiviral host response. For example, as an inhibitor of steady-state, SIKE was associated with TBK1 in uninfected cells to block RIG-I and TRIF mediated signaling pathway. Upon viral infection or TLR3 engagement, SIKE was dissociated from the TBK1 complex. Similarly, DAK bound to MDA5 in uninfected cells and their interaction was disrupted by viral infection.
However, gC1qR may act differently from these negative regulators. It seems to function similarly to PI3K during bacterial infection. LPS activates the TLR4 signaling pathway in host cells. In parallel, it also activates a feedback PI3K pathway that negatively regulates TLR4 signaling to balance the immune response (32). Interestingly, viruses also evolved the ability to use host components for proliferation and survival. It seems that gC1qR might be the primary target of some viruses as a negative feedback regulator, although gC1qR is stable upon viral infection in both mRNA and protein level (Fig. S3). Viral infection activates the translocation of gC1qR to mitochondria and then enhances the association between gC1qR and VISA (Fig. 2 and 3), which disrupts the interaction between VISA and RIG-I (Fig. S5) or downstream molecules and leads to the inhibition of RIG-I- and MDA5-mediated antiviral responses. The inhibition of RIG-I- and MDA5-mediated signaling by gC1qR was not limited to cultured 293T cell lines, and was also observed in primary HUVECs (Figs. 2A and 4 B and C). These results indicate that gC1qR plays a critical role in suppressing RIG-I- and MDA5-mediated signaling in physiologically immunocompetent cells. Taken together, our data identify a new role for gC1qR in RIG-I- and MDA5-mediated antiviral responses, but there are still many issues remaining to be investigated. For example, how does gC1qR translocate to mitochondria?
Our findings provide evidence that viruses use the host protein gC1qR to inhibit host antiviral responses and promote viral proliferation by activating a suppressive pathway to negatively regulate antiviral signaling. Inhibition of gC1qR translocation to mitochondria could be a therapeutic approach to enhance the antiviral response of the host, and may be an alternative means to treat virus associated diseases.
Materials and Methods
Antibodies, Plasmids, and Reagents.
All reagents are listed in SI Materials and Methods.
RT-PCR.
Total RNA was isolated from 293T or HUVECs, using TRIzol reagent (Invitrogen) according to the manufacturer's instructions. Two micrograms of RNA was then reverse transcribed using SuperScript III Reverse Transcriptase (Invitrogen) to generate first strand cDNA. It was then subjected to PCR with specific primers (Table S1).
ELISA.
The supernatants of 293T cells or HUVECs were collected after treatment as indicated. IFN-β level was measured by ELISA kit (Biosource).
Other Methods.
Other methods are described in SI Materials and Methods, including the construction of gC1qR mutants, transfection and reporter gene assays, coimmunoprecipitation and western blot analysis, vesicular stomatitis virus (VSV) plaque assay, immunofluorescence assay, and preparation of cellular and mitochondrial proteins.
Supplementary Material
Acknowledgments.
We thank Dr. Hongbing Shu (Peking University, Beijing) for ISRE, NF-κB, SV40, IFN-β promoter luciferase reporter plasmids, mammalian expression plasmids for human RIG-I and VISA. We thank Dr. Zhijian Chen (University of Texas Southwestern Medical Center, Dallas, TX) and Dr. Bruce Beutler (The Scripps Research Institute, La Jolla, CA) for proofreading of the article. We also thank Dr. Eva Marie Moresco (The Scripps Research Institute) for revising the manuscript. This work was supported by National Science Foundation (China) Grant 30671905 and Ministry of Science and Technology (China), National Basic Research Program “973” project Grant 2006CB503802.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0811029106/DCSupplemental.
References
- 1.Theofilopoulos AN, Baccala R, Beutler B, Kono DH. Type I interferons (alpha/beta) in immunity and autoimmunity. Annu Rev Immunol. 2005;23:307–336. doi: 10.1146/annurev.immunol.23.021704.115843. [DOI] [PubMed] [Google Scholar]
- 2.Beutler B, et al. Genetic analysis of resistance to viral infection. Nat Rev Immunol. 2007;7:753–766. doi: 10.1038/nri2174. [DOI] [PubMed] [Google Scholar]
- 3.Akira S, Uematsu S, Takeuchi O. Pathogen recognition and innate immunity. Cell. 2006;124:783–801. doi: 10.1016/j.cell.2006.02.015. [DOI] [PubMed] [Google Scholar]
- 4.Akira S, Takeda K. Toll-like receptor signalling. Nat Rev Immunol. 2004;4:499–511. doi: 10.1038/nri1391. [DOI] [PubMed] [Google Scholar]
- 5.Yoneyama M, et al. The RNA helicase RIG-I has an essential function in double-stranded RNA-induced innate antiviral responses. Nat Immunol. 2004;5:730–737. doi: 10.1038/ni1087. [DOI] [PubMed] [Google Scholar]
- 6.Andrejeva J, et al. The V proteins of paramyxoviruses bind the IFN-inducible RNA helicase, mda-5, and inhibit its activation of the IFN-beta promoter. Proc Natl Acad Sci USA. 2004;101:17264–17269. doi: 10.1073/pnas.0407639101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Xu LG, et al. VISA is an adapter protein required for virus-triggered IFN-beta signaling. Mol Cell. 2005;19:727–740. doi: 10.1016/j.molcel.2005.08.014. [DOI] [PubMed] [Google Scholar]
- 8.Seth RB, Sun L, Ea CK, Chen ZJ. Identification and characterization of MAVS, a mitochondrial antiviral signaling protein that activates NF-kappaB and IRF 3. Cell. 2005;122:669–682. doi: 10.1016/j.cell.2005.08.012. [DOI] [PubMed] [Google Scholar]
- 9.Meylan E, et al. Cardif is an adaptor protein in the RIG-I antiviral pathway and is targeted by hepatitis C virus. Nature. 2005;437:1167–1172. doi: 10.1038/nature04193. [DOI] [PubMed] [Google Scholar]
- 10.Kawai T, et al. IPS-1, an adaptor triggering RIG-I- and Mda5-mediated type I interferon induction. Nat Immunol. 2005;6:981–988. doi: 10.1038/ni1243. [DOI] [PubMed] [Google Scholar]
- 11.Lin R, et al. Negative regulation of the retinoic acid-inducible gene I-induced antiviral state by the ubiquitin-editing protein A20. J Biol Chem. 2006;281:2095–2103. doi: 10.1074/jbc.M510326200. [DOI] [PubMed] [Google Scholar]
- 12.Moore CB, et al. NLRX1 is a regulator of mitochondrial antiviral immunity. Nature. 2008;451:573–577. doi: 10.1038/nature06501. [DOI] [PubMed] [Google Scholar]
- 13.van Leeuwen HC, O'Hare P. Retargeting of the mitochondrial protein p32/gC1Qr to a cytoplasmic compartment and the cell surface. J Cell Sci. 2001;114:2115–2123. doi: 10.1242/jcs.114.11.2115. [DOI] [PubMed] [Google Scholar]
- 14.Matthews DA, Russell WC. Adenovirus core protein V interacts with p32—a protein which is associated with both the mitochondria and the nucleus. J Gen Virol. 1998;79:1677–1685. doi: 10.1099/0022-1317-79-7-1677. [DOI] [PubMed] [Google Scholar]
- 15.Dedio J, Jahnen-Dechent W, Bachmann M, Muller-Esterl W. The multiligand-binding protein gC1qR, putative C1q receptor, is a mitochondrial protein. J Immunol. 1998;160:3534–3542. [PubMed] [Google Scholar]
- 16.Petersen-Mahrt SK, et al. The splicing factor-associated protein, p32, regulates RNA splicing by inhibiting ASF/SF2 RNA binding and phosphorylation. EMBO J. 1999;18:1014–1024. doi: 10.1093/emboj/18.4.1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Krainer AR, Mayeda A, Kozak D, Binns G. Functional expression of cloned human splicing factor SF2: Homology to RNA-binding proteins, U1 70K, and Drosophila splicing regulators. Cell. 1991;66:383–394. doi: 10.1016/0092-8674(91)90627-b. [DOI] [PubMed] [Google Scholar]
- 18.Ghebrehiwet B, Peerschke EI. Structure and function of gC1q-R: A multiligand binding cellular protein. Immunobiology. 1998;199:225–238. doi: 10.1016/S0171-2985(98)80029-6. [DOI] [PubMed] [Google Scholar]
- 19.Ghebrehiwet B, et al. Evidence that the two C1q binding membrane proteins, gC1q-R and cC1q-R, associate to form a complex. J Immunol. 1997;159:1429–1436. [PubMed] [Google Scholar]
- 20.Joseph K, Ghebrehiwet B, Peerschke EI, Reid KB, Kaplan AP. Identification of the zinc-dependent endothelial cell binding protein for high molecular weight kininogen and factor XII: Identity with the receptor that binds to the globular “heads” of C1q (gC1q-R) Proc Natl Acad Sci USA. 1996;93:8552–8557. doi: 10.1073/pnas.93.16.8552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Beatch MD, Everitt JC, Law LJ, Hobman TC. Interactions between rubella virus capsid and host protein p32 are important for virus replication. J Virol. 2005;79:10807–10820. doi: 10.1128/JVI.79.16.10807-10820.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Marschall M, et al. Cellular p32 recruits cytomegalovirus kinase pUL97 to redistribute the nuclear lamina. J Biol Chem. 2005;280:33357–33367. doi: 10.1074/jbc.M502672200. [DOI] [PubMed] [Google Scholar]
- 23.Kato H, et al. Cell type-specific involvement of RIG-I in antiviral response. Immunity. 2005;23:19–28. doi: 10.1016/j.immuni.2005.04.010. [DOI] [PubMed] [Google Scholar]
- 24.Saito T, Gale M., Jr Differential recognition of double-stranded RNA by RIG-I-like receptors in antiviral immunity. J Exp Med. 2008;205:1523–1527. doi: 10.1084/jem.20081210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yoneyama M, et al. Shared and unique functions of the DExD/H-box helicases RIG-I, MDA5, and LGP2 in antiviral innate immunity. J Immunol. 2005;175:2851–2858. doi: 10.4049/jimmunol.175.5.2851. [DOI] [PubMed] [Google Scholar]
- 26.Schlee M, Hornung V, Hartmann G. siRNA and isRNA: Two edges of one sword. Mol Ther. 2006;14:463–470. doi: 10.1016/j.ymthe.2006.06.001. [DOI] [PubMed] [Google Scholar]
- 27.Waggoner SN, Cruise MW, Kassel R, Hahn YS. gC1q receptor ligation selectively down-regulates human IL-12 production through activation of the phosphoinositide 3-kinase pathway. J Immunol. 2005;175:4706–4714. doi: 10.4049/jimmunol.175.7.4706. [DOI] [PubMed] [Google Scholar]
- 28.Eisen-Vandervelde AL, et al. Hepatitis C virus core selectively suppresses interleukin-12 synthesis in human macrophages by interfering with AP-1 activation. J Biol Chem. 2004;279:43479–43486. doi: 10.1074/jbc.M407640200. [DOI] [PubMed] [Google Scholar]
- 29.Kittlesen DJ, Chianese-Bullock KA, Yao ZQ, Braciale TJ, Hahn YS. Interaction between complement receptor gC1qR and hepatitis C virus core protein inhibits T-lymphocyte proliferation. J Clin Invest. 2000;106:1239–1249. doi: 10.1172/JCI10323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Huang J, et al. SIKE is an IKK epsilon/TBK1-associated suppressor of TLR3- and virus-triggered IRF-3 activation pathways. EMBO J. 2005;24:4018–4028. doi: 10.1038/sj.emboj.7600863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Diao F, et al. Negative regulation of MDA5- but not RIG-I-mediated innate antiviral signaling by the dihydroxyacetone kinase. Proc Natl Acad Sci USA. 2007;104:11706–11711. doi: 10.1073/pnas.0700544104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fukao T, Koyasu S. PI3K and negative regulation of TLR signaling. Trends Immunol. 2003;24:358–363. doi: 10.1016/s1471-4906(03)00139-x. [DOI] [PubMed] [Google Scholar]
- 33.Bonifacino JS. New York: John Wiley; 1998. Current protocols in cell biology; p. v. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.