Abstract
Protein dynamics are inextricably linked to protein function but there are few techniques that allow protein dynamics to be conveniently interrogated. For example, mutations and translocations give rise to aberrant proteins such as Bcr-Abl where changes in protein conformation and dynamics are believed to result in deregulated kinase activity that provides the oncogenic signal in chronic myelogeous leukemia. Although crystal structures of the down-regulated c-Abl kinase core have been reported, the conformational impact of mutations that render Abl resistant to small-molecule kinase inhibitors are largely unknown as is the allosteric interplay of the various regulatory elements of the protein. Hydrogen exchange mass spectrometry (HX MS) was used to compare the conformations of wild-type Abl with a nonmyristoylated form and with 3 clinically relevant imatinib resistance mutants (T315I, Y253H and E255V). A HX-resistant core localized to the interface between the SH2 and kinase domains, a region known to be important for maintaining the down-regulated state. Conformational differences upon demyristoylation were consistent with the SH2 domain moving to the top of the small lobe of the kinase domain as a function of activation. There were conformational changes in the T315I mutant but, surprisingly, no major changes in conformation were detected in either the Y253H or the E255V mutants. Taken together, these results provide evidence that allosteric interactions and conformational changes play a major role in Abl kinase regulation in solution. Similar analyses could be performed on any protein to provide mechanistic details about conformational changes and protein function.
Keywords: allosteric interactions, chronic mylogenous leukemia, hydrogen exchange, mass spectrometry
The mammalian Abl family of nonreceptor tyrosine kinases plays a significant role in human leukemia (reviewed in ref. 1). The family includes 2 homologous proteins: c-Abl and the Abl-related gene product, Arg. In 95% of patients with chronic myelogenous leukemia (CML), chromosomal translocation fuses the c-abl and bcr loci leading to expression of the constitutively active oncoprotein, Bcr-Abl. Other leukemias such as acute lymphocytic leukemia (ALL) also express Bcr-Abl fusion proteins. The unregulated tyrosine kinase activity of Bcr-Abl drives several proliferative and antiapoptotic pathways. Bcr-Abl causes cytokine-independent proliferation of hematopoietic progenitor cells and is sufficient to induce CML- and ALL-like syndromes in mice, providing strong evidence that it is the driving force behind these diseases. Imatinib, nilotinib, and dasatinib are ATP-competitive inhibitors of Bcr-Abl catalytic activity that have demonstrated remarkable efficacy in chronic-phase CML (reviewed in ref. 2). Patients in the blast crisis phase of CML often relapse after imatinib therapy because of emergence of imatinib resistant cells expressing point mutations in Bcr-Abl. All of the mutant clones with the exception of the “gatekeeper” mutation T315I can be inhibited by the second generation drugs nilotinib and dasatinib.
Multiple intramolecular interactions involving the SH3/SH2 regulatory domains and the kinase domain have been observed in the crystal structures of the down-regulated Abl core (3, 4), see also Fig. S1. The SH3 domain binds the SH2-kinase linker, an interaction necessary to suppress kinase activity (reviewed in ref. 1). Unlike the related Src-family kinases, which have a C-terminal tail phosphotyrosine involved in regulatory binding to the SH2 domain, Abl SH2 does not engage a C-terminal phosphorylated tail. Rather, other stabilizing forces including interactions between the SH2 domain and the kinase domain (5), and increased SH3:linker binding propensity (6) are thought to compensate for the lack of a phosphorylated C-terminal tail in Abl. The NCap also contributes to the stability of the down-regulated conformation (5, 7). Last, a deep pocket in the C-lobe of the kinase domain was shown to create a binding site for myristic acid. The crystal structures and biological data strongly suggest that the myristoylated N terminus of c-Abl binds within this pocket and thereby positions the NCap and associated sequences to help hold SH2 and SH3 in a down-regulatory position (reviewed in ref. 1). Interestingly, small molecules that mimic the interaction of the NCap myristoyl group with the kinase domain are also effective c-Abl kinase inhibitors (N. S. Gray, unpublished results), suggesting that binding to the myristic acid binding pocket is capable of inducing a conformation with down-regulated kinase activity.
Mutations in the Abl portion of Bcr-Abl can contribute to drug resistance and disease progression. The majority of resistance mutations for ATP-competitive inhibitors such as imatinib are located in and around the ATP binding site of the kinase domain (8), but other mutations in regions distant to the active site have also been observed experimentally (9). The most challenging Bcr-Abl mutation responsible for clinical drug resistance is T315I, located at the “gatekeeper” position in the center of the ATP-binding cleft. [Note that residues responsible for imatinib drug resistance are most commonly numbered according to the Abl 1a sequence (10)]. The T315I mutation is the most recalcitrant to inhibition because of a combination of several factors including steric hindrance of drug binding, loss of a key hydrogen-bonding interaction with the T315 side-chain hydroxyl group exploited by imatinib, nilotinib and dasatinib and potentially through increasing the intrinsic kinase activity (for example, see refs. 9 and 11).
In this work, we explored the overall solution conformational properties of the Abl core protein with hydrogen/deuterium exchange (HX) mass spectrometry (MS). These investigations represent, to our knowledge, the first HX MS analyses of a near full-length nonreceptor tyrosine kinase. HX MS is a well established method for the study of conformational changes and protein folding (12) and there are multiple advantages to using this methodology. Specific to Abl, the advantages of this technique are: (i) the studies are not limited to forms of Abl that will crystallize; (ii) the analyses are done in physiological solution conditions without the need for 15N of 13C labeling; (iii) very small quantities (picomoles) at low concentrations (typically 10 μM) are used relative to other methods such as NMR (13); (iv) the whole Abl core can be investigated rather than just the isolated domains; and (v) no inhibitors or ATP/Mg2+ were used during purification or data acquisition and therefore Abl sampled the greatest number of possible conformations of any version of Abl structurally probed in solution to date.
Results
Abl Proteins.
The wild-type Abl core was expressed in Sf9 insect cells and purified by affinity chromatography. This protein was N-terminally myristoylated as verified by intact mass analysis, trypsin mapping and tandem mass spectrometry (MS/MS; data not shown). This version of Abl core is referred to as Abl(Myr) (see also Fig. S1). Mass spectrometry also verified that Abl(Myr) was phosphorylated at Ser-69, as found in ref. 3. Abl core proteins bearing the imatinib-resistance mutations T315I, E255V and Y253H were prepared in a similar fashion and similarly characterized. [Note we are using the designations of these mutants according to established clinical conventions (10), but that we have used Abl 1b numbering (3) elsewhere in this work; for example, Abl position T315 is actually T334 in Abl 1b]. The wild-type Abl core protein was also expressed in Escherichia coli and purified. This version of Abl, termed Abl(NonMyr), is not N-terminally myristoylated and the NCap is slightly shorter (see Fig. S1). All of the recombinant c-Abl proteins used in this study were coexpressed with the YopH phosphatase to prevent autophosphorylation of the activation loop and other sites (4, 5, 14). MS analysis showed that all of the myristoylated proteins were devoid of phosphotyrosine, whereas ≈25% of the Abl(NonMyr) molecules were phosphorylated on Y412 (MS/MS data not shown; see also Fig. S2). The conformational and dynamic properties of all 5 of these purified Abl proteins were then compared by HX MS.
General Description of Abl(Myr) by HX MS.
We first measured hydrogen exchange in wild-type Abl(Myr). A review of hydrogen/deuterium exchange MS can be found in ref. 12. Exchange into Abl(Myr) provided baseline exchange for the protein in solution, which was used later in comparisons of exchange in the nonmyristoylated and mutant forms. Abl(Myr) was exposed to 2H2O for various periods of time ranging from 10 seconds to 4 h and the reaction quenched by adjusting the pH to 2.6 and the temperature to 0 °C. There was no ATP or Mg2+ in any of the buffers used for measurements described here. The quenched protein was digested with pepsin and deuterium incorporation into each pepsin fragment (>70 in total) (see Fig. S3) was analyzed. Chromatographic separation before MS analysis was accomplished with a newly-described UPLC system that provided very high-resolution separations under the constraints of an HX MS experiment (15) and only required ≈20 pmols (2 μg) for each time point. The quality of the data, examples of the chromatographic separation of each pepsin digest, representative mass spectra and the processing steps to convert the raw spectra into a deuterium uptake curves are shown in Fig. S4. The deuterium incorporation with time for each peptic peptide was determined, in duplicate experiments. The uptake curves for 45 peptides that cover 94% of the sequence of Abl(Myr) are provided in Fig. S5 (redundant and overlapping peptides not shown).
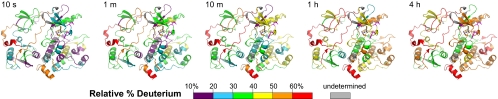
Hydrogen exchange data for all parts of Abl(Myr) are summarized in graphical form in Fig. 1 and in Movies S1 and S2. Although the HX MS data for the most part are consistent with the structure observed by crystallography, they provide the first ever insight into the dynamics of the c-Abl core. The data show that in myristoylated Abl, the NCap was rapidly deuterated, as were the linker and the α-helix at the C terminus. Within the first minute of deuterium labeling, these regions exchanged >50% of the possible backbone amide hydrogen sites. Other regions remained more protected during this time, including parts of the kinase and regulatory domains. However, after 1 h of labeling, many of the mobile or dynamic elements in Abl(Myr) were further deuterated. A protected core emerged that includes α-helices in the large lobe of the kinase domain and the SH2 domain. This “core” remained protected from deuterium incorporation even after 4 h of labeling, revealing that it is highly solvent protected and/or heavily hydrogen bonded. Disruption of this core, by mutation at the interface between the SH2 domain and the large lobe of the kinase domain, has been shown to cause catalytic activation (5).
Fig. 1.
Summary of all HX MS data for Abl(Myr) mapped onto the crystal structure of Abl [PDB entry 2FO0 (3)]. The relative deuterium levels for all residues of Abl(Myr) that were probed are mapped with color for each exchange point; the color code is explained at the bottom of the figure. This figure represents each frame of Movie S1. Regions colored gray after exchange began represent residues where deuterium levels were not determined.
Exchange into functionally important regions of Abl was revealing of the behavior of the protein in solution. Among them it is noteworthy to mention several peptides (see Fig. S6). Data for a peptide from the NCap showed that the NCap was mostly unstructured as it was able to incorporate a large amount of deuterium at the earliest time points (11 of 15 potential backbone amide hydrogens). This result was consistent with previous analysis of the conformation of NCap in smaller constructs of Abl (16). Regions within the SH3 domain, such as residues 100–118, appeared protected yet somewhat flexible, as revealed by the ability to incorporate deuterium over time. The SH2-kinase linker (residues 246–256) was rapidly deuterated and appeared mostly exposed to solvent as it was quickly deuterated and the deuterium level remained constant throughout the time course of exchange. The P-loop was somewhat protected from exchange and partially dynamic during the exchange time course.
The activation loop appeared very flexible and was rapidly deuterated even by the first time point. However, it was not totally deuterated by the end of the experiment and had a small degree of protection from exchange. [Note that Abl(Myr) was not phosphorylated on the activation loop tyrosine during these studies.] Peptic peptides 402–406 and 409–420 span the activation loop. As seen in the deuterium incorporation graphs (Fig. S6) there was slight protection from exchange, where it seems that in peptide 402–406 one amide hydrogen was protected from exchange and, in peptide 409–420, four amide hydrogens were protected from exchange. These results are more consistent with the conformation observed in the Abl core when bound to the inhibitor PD166326 (2FO0) and less consistent with the conformation of the Abl kinase domain only when bound to imatinib (1FPU) (see Fig. S6). In 1FPU, more hydrogen bonded amides were found in the region 402–420 therefore more protection from exchange would have been anticipated in both of the peptides covering this area had the protein adopted this conformation in solution. Recall, however, that the HX MS data discussed here do not involve inhibitor binding, but rather the protein in the naturally down-regulated conformation. The results further indicate that the activation loop is mobile and not in a highly structured conformation. This conclusion is supported by HX MS results with near-full length Lck and Hck tyrosine kinases in which the activation loop displayed similar rapid deuteration (J. Hochrein, T.E.S., and J.R.E., unpublished data). In addition, early crystal structures of c-Src and Hck (17, 18) were not able to resolve the activation loop, which was likely because of this inherent flexibility. When inhibitors were added for later crystal structures (19, 20), the activation loop was stabilized and its position could be resolved. Our HX MS results for Abl are entirely consistent with a mobile activation loop even in the myristoylated and down-regulated conformation that Abl(Myr) assumes.
Comparison of Kinase Activity of the Forms of Abl.
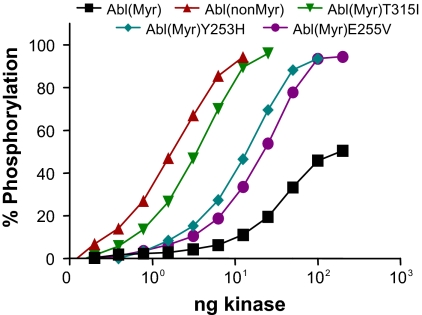
We next wished to compare the influence of myristoylation on the conformational properties of the Abl core, and to evaluate differences in HX MS of common imatinib-resistant mutants, some of which have been reported to have altered signaling properties in the context of Bcr-Abl in cells (21, 22). Before initiating the HX MS analyses, we compared the specific activity of each recombinant protein, using an in vitro kinase assay with a peptide substrate. Each kinase protein was titrated into the assay in increasing amounts to provide a comparative view of their relative activities in solution. As shown in Fig. 2, wild-type Abl(Myr) was the least active of the kinases, because it required the largest amount of input kinase to trigger activation, and failed to reach a maximal activity level in the assay. This finding is consistent with its down-regulated conformation. In contrast, Abl(NonMyr) was the most active kinase, consistent with the idea that the lack of myristoylation at the N terminus frees the SH3/SH2 regulatory machinery from its down-regulatory position at the back of the kinase domain (1). Interestingly, the imatinib-resistant mutant T315I showed almost equivalent activity to Abl(NonMyr), despite the confirmed presence of N-terminal myristoylation. The other two imatinib-resistance mutants, Y253H and E255V, also showed enhanced activity relative to wild-type Abl(Myr) but intermediate to that of T315I and Abl(NonMyr). These results show that kinase domain mutations causing imatinib resistance also disturb negative regulatory interactions within the kinase core in vitro with recombinant proteins. Very recently, Azam et al. showed that the T315I mutant was far more active in transfected mammalian cells than a mutant lacking the N-terminal glycine required for myristoylation (23), in agreement with our in vitro results. To better understand the impact of these mutations and myristoylation on Abl regulation, we probed each protein with HX MS.
Fig. 2.
In vitro kinase assay of recombinant Abl core proteins. Abl(Myr) and Abl(Myr) mutants T315I, Y253H, and E255V were coexpressed with the YopH phosphatase in Sf9 insect cells. Abl(NonMyr) was coexpressed with YopH in E. coli. These proteins were purified in their nonphosphorylated forms as described in Methods and Fig. S2. Kinase activity was determined using a FRET-based tyrosine kinase assay with a peptide substrate and increasing amounts of each recombinant Abl protein. Each condition was repeated in quadruplicate, and the extent of phosphorylation is expressed as mean percentage phosphorylation relative to a control phosphopeptide ± SD. The overall experiment was repeated three times with comparable results.
Comparison of Myristoylated Versus Nonmyristoylated Abl Core.
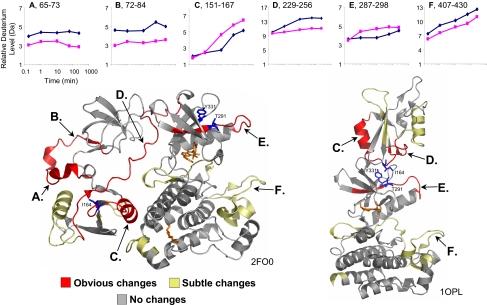
The deuterium exchange of Abl(NonMyr) was compared with that of Abl(Myr). The experiment was as described for Abl(Myr) except that both proteins were labeled and analyzed on the same day so that comparison between the two datasets could be made with the least error possible (see also ref. 12). The entire experiment was done in duplicate, and the results are summarized in Fig. 3(the complete dataset is shown Fig. S7). For much of Abl, there were no detectable differences in deuterium incorporation between the myristoylated versus the nonmyristoylated form. The cumulative error of measuring deuterium uptake in these assays is approximately ±0.2 Da. Anything larger than that was considered significant for the purposes of comparing the two datasets. The changes were grouped according to obvious changes (>1.0 Da separating the deuterium incorporation curves, after 2 replicates were averaged) and subtle changes (0.4–1.0 Da difference). Mapping the changes onto the crystal structure of the down-regulated form (PDB entry 2FO0) shows that there were small but significant changes to exchange in the NCap and the linker, and in the SH2 helix that interacts with the large lobe of the kinase domain in the down-regulated structure (Fig. 3 Lower Left). In addition, there were small but detectable changes to exchange in the small lobe of the kinase domain near the active site, likely reflective of kinase domain activation. The changes in deuterium incorporation were mostly a decrease in deuterium levels in Abl(NonMyr), with the exception of peptides 151–167 and 287–298 where it was the opposite. These results did not make sense in light of a model in which the protein adopts a more open and solvent exposed conformation when myristic acid is removed and engagement of the NCap with the pocket on the kinase domain does not exist. However, interpretation of the data in light of the structure determined by small angle X-ray scattering (SAXS) and a lower resolution crystal structure for activated Abl (3) made much more sense. In that structure (PDB entry 1OPL; Fig. 3 Lower Right), the SH2 domain moves to occupy a position on top of the small lobe of the kinase domain. Amino acids I164 from the SH2 domain interact with residues T291 and Y331 in the kinase domain (blue in Fig. 3). Differences in HX MS as a result of removal of the myristoyl group lie exactly at the interface of SH2 and the kinase domain in the structure of Abl where SH2 sits on top of the kinase domain (Fig. 3 Lower Right). The HX MS data showed that the differences were decreases in deuterium level in Abl(NonMyr), a result consistent with additional protection of some regions in this “top-hat” conformation (Fig. 3 Lower Right). Two peptides showed the opposite trend, that is more deuterium in Abl(NonMyr). These peptides were the regions in SH2 that include the critical I164 residue (peptide 151–167) and the part of the kinase domain that makes contact with SH2 (peptide 287–298). Slightly more deuterium in these peptides in Abl(NonMyr) argues that there is a loosening of the structure or subtle exposure to more solvent in some part of these peptide (keep in mind that it was a difference of a few daltons in peptides that contain >10 residues). Taken together, the data comparing Abl(Myr) with Abl(NonMyr) support the conformation in which the SH2 domain occupies a position on top of the small lobe of the kinase domain when the kinase is in the active state. This conformation has been shown by crystallography to exist in other kinases with SH2-kinase domain arrangements, such as the c-Fes tyrosine kinase (24). Mutagenesis data show that formation of this conformation is essential for maintenance of kinase activity (3, 24). This “top-hat” conformation is perhaps a general feature of the activated conformation of some tyrosine kinases, an idea supported by our HX MS measurements in solution.
Fig. 3.
Comparison of deuterium exchange in Abl(Myr) and Abl(NonMyr). The deuterium uptake curves for 6 representative peptides (the complete dataset is in Fig. S7) are shown in Upper [blue: Abl(Myr); pink: Abl(NonMyr)]. The location of each peptide, according to the labels A–F, is shown on the crystal structures (Left, PDB entry 2FO0; Right, PDB entry 1OPL). Obvious changes (colored red) were defined as a difference between deuterium exchange-in curves of 1.0 Da or more. Subtle changes (colored yellow) were 0.4–1.0 Da. No changes were differences of 0.0–0.4 Da. Residues I164, T291, and Y331 are colored blue and rendered as sticks.
Conformational Effects of Imatinib-Resistance Mutations.
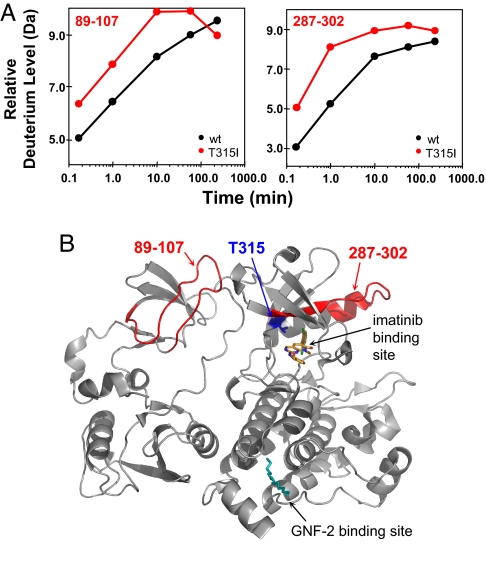
HX MS data comparing Abl(Myr) with Abl(Myr)T315I were obtained using the same methodology described above. Again, the changes that were detected in HX were not extremely large, suggesting that only small conformational disturbances accompany significant changes in activity (see Fig. 2) for the T315I mutant protein. From >40 peptic peptides, only 2 peptides showed differences in the amount of deuterium incorporation as a result of the T315I mutation: peptic peptide 89–107 (triply charged ion at m/z 686.7) and peptic peptide 287–302 (quadruply charged at m/z 467.8). Residues 287–302 are located in the kinase domain in the proximity of the drug binding site (Fig. 4). Deuterium incorporation data for this peptide indicated that in the T315I mutant, more deuterium was incorporated, implying less protection/structural organization in this region in the mutant form. We speculate that mutation from threonine to isoleucine destabilizes the backbone amide hydrogens in this peptide such that one more position can become deuterated. Concomitant disruption of the hydrophobic spine (23, 25) upon T315I mutation could also lead to a conformational destabilization of the region including residues 287–302, making the HX MS results consistent with the hydrophobic spine model. Surprisingly, the second region of significant change (residues 89–107) was located in the SH3 domain, specifically within the RT-loop. This part of Abl incorporates slightly more deuterium in Abl(Myr)T315I than in Abl(Myr) indicating conformational disruptions in this region. These results suggest that the T315I mutation not only influences conformation in the immediate region, but also alters hydrogen exchange in distant regions including the SH3 domain. This result is highly significant, because alterations in the dynamics of the SH3 domain RT loop could impact its negative regulatory influence on the kinase domain, contribute to the observed increase in kinase activity, and affect the ability of Abl to interact with signaling partners (see Discussion).
Fig. 4.
Summary of the HX MS differences between Abl(Myr) and Abl(Myr)T315I. (A) Two peptides that exhibit differences in the deuterium incorporation between wt Abl(Myr) and the T315I(Myr) mutant were 89–107 and 287–302. The location of these 2 peptides is shown in B. Gray, no changes; red, obvious changes as defined in Fig. 3.
Two other clinically relevant drug resistance mutants (Y253H and E255V) were also investigated with HX MS in the same way as described above. There were no significant differences in deuterium exchange for either mutant at any time point in any peptide in either replicate analysis (Fig. S8). Although Abl(Myr)Y253H and Abl(Myr)E255V were more active than wild-type Abl(Myr) (Fig. 2), they did not undergo conformational changes detectable by HX MS. One conclusion from these results may be that these mutations confer only subtle local changes to the shape and character of the ATP binding site, whereas the T315I mutation alters more than just its local environment. This reasoning could explain an additional feature that makes the T315I mutation so resistant to treatment with small molecule inhibitors in that other allosteric changes in conformation are driven by T315I that cannot be overcome by current FDA-approved ATP-competitive inhibitors.
Discussion
Conformational changes in solution are not necessarily obvious from crystal structures. Having such information is, however, extremely important especially if conformational changes play a part in regulation. This is the case for many proteins, not the least of which is the Abl kinase. Our results using HX MS have uncovered information about Abl that is extremely difficult to obtain by other methods and indicate overall that in Abl, conformational changes may contribute to the inability of small-molecules to block aberrant kinase activity.
In the HX MS analysis of the myristoylated form, as expected, the NCap, the linker, the activation loop and some parts of the regulatory SH2 and SH3 domains seem fairly dynamic and less protected from exchange (Fig. 1). Although Chen et al. (6, 16) have used HX MS to investigate several other mechanistic facets of the Abl kinase the results reported here were for the entire Abl kinase core in solution in the absence of inhibitors. A protected core consisting of the large lobe of the kinase domain and its interface with the SH2 domain was the most resistant to deuteration indicating that this region is the most stable and structured part of the molecule. As shown by Hantschel et al. (5), mutation within this core at the SH2-kinase interface leads to kinase activation. Rearrangement of the orientation of the domains of Abl as a result of removal of the myristic acid was suggested by previous data that showed the SH2 domain occupies a position on top of the small lobe of the kinase domain in nonmyristoylated forms (3). Our HX MS data support this model as changes in deuterium incorporation were found at the new interfaces created by the reorientation of the domains. It is therefore possible to distinguish and validate various crystallographic models in solution using HX MS, another broad benefit illustrated by these results.
In the case of the T315I imatinib-resistance mutant, conformational disturbances distant to the site of mutation were detected. There was an unexpected alteration to deuterium exchange in the RT-loop of the SH3 domain in T315I whereas no changes were detected in any location in the other 2 mutants. These results suggest that allosteric conformational changes may accompany the T315I mutation, the most difficult Bcr-Abl imatinib-resistance mutation to overcome therapeutically (26), whereas changes as a result of the other mutations are more likely local effects. The surprising up-regulation of kinase activity accompanying the T315I mutation may also relate to this allosteric change located distantly in the SH3 domain. Previous work has established that some c-Abl mutations far away from the active site are able to cause kinase activation (9). Activation by distant mutation may involve conformational changes that are communicated allosterically to the active site. We speculate that stabilization of the conformation in distant regions could assist inhibition of the T315I mutant specifically, and perhaps in other mutants that cause allosteric conformational changes. Comparative screening of such mutants with HX MS should be revealing for further drug design and development.
These results illustrate that important conformational data can be obtained for large proteins that are critical to understand for drug-development purposes. The location of changes can be determined and interpreted in light of complementary structural studies. Here, we used HX MS to investigate how clinically relevant mutations in Abl affect protein dynamics. However, HX MS analysis can also be used to investigate how protein conformation is influenced by other perturbations such a posttranslational modification, binding of regulatory proteins and small molecules.
Methods
Protein Expression and Activity Measurements.
Wild-type Abl core, T315I, E255V and Y253H mutants were overexpressed in Sf9 insect cells coexpressing YopH, as described for wild-type Abl in ref. 6. The sequence of wild-type Abl was identical in sequence to the down-regulated crystal structure of Nagar et al. (3) (Fig. S1). Proteins made in Sf9 cells [referred to as Abl(Myr) throughout the manuscript] were myristoylated at Gly-2 and phosphorylated at Ser-69, as confirmed by mass spectrometry (Fig. S2). Preparation of Abl(NonMyr) followed methods published in ref. 14, as described in Fig. S2. The purity and mass of all proteins were verified by mass spectrometry. Tyrosine kinase assays were performed using the FRET-based Z′-Lyte kinase assay system (Invitrogen) as described in ref, 27 and SI Materials and Methods.
Deuterium Exchange Reactions.
A stock solution of Abl(Myr) at 20 pmol/μL in 20 mM Tris, 100 mM NaCl, 3 mM DTT (pH 8.3), H2O was prepared. Deuterium exchange was initiated by dilution of the stock solution 15-fold with 20 mM Tris, 100 mM NaCl, 3 mM DTT buffer (p2H 8.3), 2H2O, 21 °C. At each deuterium exchange time point (from 10 s to 4 h) an aliquot from the exchange reaction was removed and labeling was quenched by adjusting the pH to 2.6 with an equal volume of quench buffer (50 mM potassium phosphate, pH 2.6, H2O). Quenched samples were immediately frozen on dry ice and stored at −80 °C until analysis. The same procedure was used for the labeling of the mutants and Abl(NonMyr).
Mass Analysis.
Each frozen sample was thawed rapidly to 0 °C and incubated with pepsin at a ratio of 1:1 (weight:weight) for 5 min at 0 °C. The resulting peptides were injected into a custom Waters nanoACQUITY UPLC system and analyzed as described in ref. 15 (see also Fig. S4). All comparison experiments were done under identical experimental conditions thus negating the need for back-exchange correction (12). Each experiment was performed in duplicate. The error of determining the deuterium levels was ±0.20 Da in the experimental setup used. Mass spectra were processed with the software HX-Express (28) by centroiding an isotopic distribution corresponding to the +2, +3, or +4 charge state of each peptide. The resulting relative deuterium levels were plotted versus the exchange-in time (see also Figs. S4–S7).
Supplementary Material
Acknowledgments.
We thank T.E. Wales and D.J. Houde for technical assistance. This work was supported by National Institutes of Health Grants GM070590 (to J.R.E.) and CA101828 (to T.E.S.), the Novartis Institute of Biomedical Research (N.S.G.), and Waters Corporation (J.R.E.). This is Barnett Institute contribution no. 931.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0811912106/DCSupplemental.
References
- 1.Hantschel O, Superti-Furga G Regulation of the c-Abl and Bcr-Abl tyrosine kinases. Nat Rev Mol Cell Biol. 2004;5:33–44. doi: 10.1038/nrm1280. [DOI] [PubMed] [Google Scholar]
- 2.Jabbour E, Cortes J, Kantarjian H. Novel tyrosine kinase inhibitors in chronic myelogenous leukemia. Curr Opin Oncol. 2006;18:578–583. doi: 10.1097/01.cco.0000245314.97638.d3. [DOI] [PubMed] [Google Scholar]
- 3.Nagar B, et al. Organization of the SH3-SH2 unit in active and inactive forms of the c-Abl tyrosine kinase. Mol Cell. 2006;21:787–798. doi: 10.1016/j.molcel.2006.01.035. [DOI] [PubMed] [Google Scholar]
- 4.Nagar B, et al. Structural Basis for the Autoinhibition of c-Abl tyrosine kinase. Cell. 2003;112:859–871. doi: 10.1016/s0092-8674(03)00194-6. [DOI] [PubMed] [Google Scholar]
- 5.Hantschel O, et al. A myristoyl/phosphotyrosine switch regulates c-Abl. Cell. 2003;112:845–857. doi: 10.1016/s0092-8674(03)00191-0. [DOI] [PubMed] [Google Scholar]
- 6.Chen S, Brier S, Smithgall TE, Engen JR. The Abl SH2-kinase linker naturally adopts a conformation competent for SH3 domain binding. Protein Sci. 2007;16:572–581. doi: 10.1110/ps.062631007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pluk H, Dorey K, Superti-Furga G. Autoinhibition of c-Abl. Cell. 2002;108:247–259. doi: 10.1016/s0092-8674(02)00623-2. [DOI] [PubMed] [Google Scholar]
- 8.Branford S, et al. High frequency of point mutations clustered within the adenosine triphosphate-binding region of BCR/ABL in patients with chronic myeloid leukemia or Ph-positive acute lymphoblastic leukemia who develop imatinib (STI571) resistance. Blood. 2002;99:3472–3475. doi: 10.1182/blood.v99.9.3472. [DOI] [PubMed] [Google Scholar]
- 9.Azam M, Latek RR, Daley GQ. Mechanisms of autoinhibition and STI-571/imatinib resistance revealed by mutagenesis of BCR-ABL. Cell. 2003;112:831–843. doi: 10.1016/s0092-8674(03)00190-9. [DOI] [PubMed] [Google Scholar]
- 10.Gorre ME, et al. Clinical resistance to STI-571 cancer therapy caused by BCR-ABL gene mutation or amplification. Science. 2001;293:876–880. doi: 10.1126/science.1062538. [DOI] [PubMed] [Google Scholar]
- 11.Modugno M, et al. Crystal structure of the T315I Abl mutant in complex with the aurora kinases inhibitor PHA-739358. Cancer Res. 2007;67:7987–7990. doi: 10.1158/0008-5472.CAN-07-1825. [DOI] [PubMed] [Google Scholar]
- 12.Wales TE, Engen JR. Hydrogen exchange mass spectrometry for the analysis of protein dynamics. Mass Spectrom Rev. 2006;25:158–170. doi: 10.1002/mas.20064. [DOI] [PubMed] [Google Scholar]
- 13.Vajpai N, et al. Solution conformations and dynamics of ABL kinase-inhibitor complexes determined by NMR substantiate the different binding modes of imatinib/nilotinib and dasatinib. J Biol Chem. 2008;283:18292–18302. doi: 10.1074/jbc.M801337200. [DOI] [PubMed] [Google Scholar]
- 14.Seeliger MA, et al. High yield bacterial expression of active c-Abl and c-Src tyrosine kinases. Protein Sci. 2005;14:3135–3139. doi: 10.1110/ps.051750905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wales TE, Fadgen KE, Gerhardt GC, Engen JR. High-speed and high-resolution UPLC separation at zero degrees celsius. Anal Chem. 2008;80:6815–6820. doi: 10.1021/ac8008862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen S, Dumitrescu TP, Smithgall TE, Engen JR. Abl N-terminal cap stabilization of SH3 domain dynamics. Biochemistry. 2008;47:5795–5803. doi: 10.1021/bi800446b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xu W, Harrison SC, Eck MJ. Three-dimensional structure of the tyrosine kinase c-Src. Nature. 1997;385:595–602. doi: 10.1038/385595a0. [DOI] [PubMed] [Google Scholar]
- 18.Sicheri F, Moarefi I, Kuriyan J. Crystal structure of the Src family tyrosine kinase Hck. Nature. 1997;385:602–609. doi: 10.1038/385602a0. [DOI] [PubMed] [Google Scholar]
- 19.Schindler T, et al. Crystal structure of Hck in complex with a Src family-selective tyrosine kinase inhibitor. Mol Cell. 1999;3:639–648. doi: 10.1016/s1097-2765(00)80357-3. [DOI] [PubMed] [Google Scholar]
- 20.Xu W, et al. Crystal structures of c-Src reveal features of its autoinhibitory mechanism. Mol Cell. 1999;3:629–638. doi: 10.1016/s1097-2765(00)80356-1. [DOI] [PubMed] [Google Scholar]
- 21.Griswold IJ, et al. Kinase domain mutants of Bcr-Abl exhibit altered transformation potency, kinase activity, and substrate utilization, irrespective of sensitivity to imatinib. Mol Cell Biol. 2006;26:6082–6093. doi: 10.1128/MCB.02202-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Skaggs BJ, et al. Phosphorylation of the ATP-binding loop directs oncogenicity of drug-resistant BCR-ABL mutants. Proc Natl Acad Sci USA. 2006;103:19466–19471. doi: 10.1073/pnas.0609239103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Azam M, et al. Activation of tyrosine kinases by mutation of the gatekeeper threonine. Nat Struct Mol Biol. 2008;15:1109–1118. doi: 10.1038/nsmb.1486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Filippakopoulos P, et al. Structural coupling of SH2-kinase domains links Fes and Abl substrate recognition and kinase activation. Cell. 2008;134:793–803. doi: 10.1016/j.cell.2008.07.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kornev AP, Haste NM, Taylor SS, Eyck LF. Surface comparison of active and inactive protein kinases identifies a conserved activation mechanism. Proc Natl Acad Sci USA. 2006;103:17783–17788. doi: 10.1073/pnas.0607656103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Deininger M, Buchdunger E, Druker BJ. The development of imatinib as a therapeutic agent for chronic myeloid leukemia. Blood. 2005;105:2640–2653. doi: 10.1182/blood-2004-08-3097. [DOI] [PubMed] [Google Scholar]
- 27.Trible RP, Emert-Sedlak L, Smithgall TE. HIV-1 Nef selectively activates Src family kinases Hck, Lyn, and c-Src through direct SH3 domain interaction. J Biol Chem. 2006;281:27029–27038. doi: 10.1074/jbc.M601128200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Weis DD, Engen JR, Kass IJ. Semi-automated data processing of hydrogen exchange mass spectra using HX-express. J Am Soc Mass Spectrom. 2006;17:1700–1703. doi: 10.1016/j.jasms.2006.07.025. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.