Abstract
All α-subunits of vertebrate heterotrimeric G proteins have been classified into 4 major classes, Gs, Gi, Gq, and G12, which possess orthologs already in sponges, one of the earliest animal phyla to evolve. Here we report the discovery of the fifth class of Gα protein, Gv, ancient like the other 4 classes, with members already in sponges, and encoded by 1–2 gnav genes per species. Gv is conserved across the animal kingdom including vertebrates, arthropods, mollusks, and annelids, but has been lost in many lineages such as nematodes, fruit fly, jawless fish, and tetrapods, concordant with a birth-and-death mode of evolution. All Gv proteins contain 5 G-box motifs characteristic of GTP-binding proteins and the expected acylation consensus sites in the N-terminal region. Sixty amino acid residues are conserved only among Gv, suggesting that they may constitute interaction sites for Gv-specific partner molecules. Overall Gv homology is high, on average 70% amino acid identity among vertebrate family members. The dN/dS analysis of teleost gnav genes reveals evolution under stringent negative selection. Genomic structure of vertebrate gnav genes is well conserved and different from those of the other 4 classes. The predicted full ORF of zebrafish gnav1 was confirmed by isolation from cDNA. RT-PCR analysis showed broad expression of gnav1 in adult zebrafish and in situ hybridization demonstrated a more restricted expression in larval tissues including the developing inner ear. The discovery of this fifth class of Gα proteins changes our understanding of G protein evolution.
Keywords: Danio rerio, evolution, metazoan, heterotrimeric G protein, birth-and-death mode
Heterotrimeric G proteins have a central role in cell biology. They transduce a broad range of extracellular signals received by G protein-coupled receptors (GPCRs) by coupling to many different intracellular signaling cascades (1). Disruption in human genes encoding G proteins has been shown to result in various diseases (2, 3). Among the 3 subunits α, β, and γ, the α-subunits interact with GPCRs directly (4). Compared to the large number of multigene families for GPCRs, the number of gna genes encoding Gα proteins is very small, only 16 functional gna loci in humans (5, 6). All of them, and in fact all vertebrate Gα proteins described so far, belong to 4 major classes (Gs, Gi, Gq, and G12) on the basis of their sequence homologies (2, 7). Each class can be subdivided into 2–4 families; the Gs class contains Gαs and Gαolf; Gi comprises Gαt, Gαo, Gαi, and Gαz; Gq encompasses Gαq, Gα11, Gα14, and Gα15/16; and G12 contains Gα12 and Gα13 (2). Each Gα protein family possesses a particular set of interaction partners, with respect to both GPCRs and effector proteins, but there is considerable overlap and also crosstalk between different pathways (1, 8).
In contrast to the well-investigated mammalian Gα proteins, our knowledge about the Gα protein family in lower vertebrates (and many invertebrate phyla) is still very fragmentary. In light of the fact that teleost species rapidly are becoming important animal models for human health and disease, we analyzed the gna gene family in zebrafish and found a unique Gα protein that cannot be grouped into any of the 4 established classes. Orthologous genes are broadly distributed across the animal kingdom and constitute a fifth class of Gα proteins, Gv, at the level of the other 4 classes. Such a discovery, years after the genomes became available, is a fundamental advance in the understanding of G protein evolution and also completely unexpected, as nearly 2 decades have passed since the fourth class of G proteins became known (7). We describe here the ancient evolutionary origin, frequent gene loss in many lineages, genomic properties, and expression pattern of this unique class of Gα proteins.
Results
Identification of Gv, a Fifth Major Class of Gα Proteins.
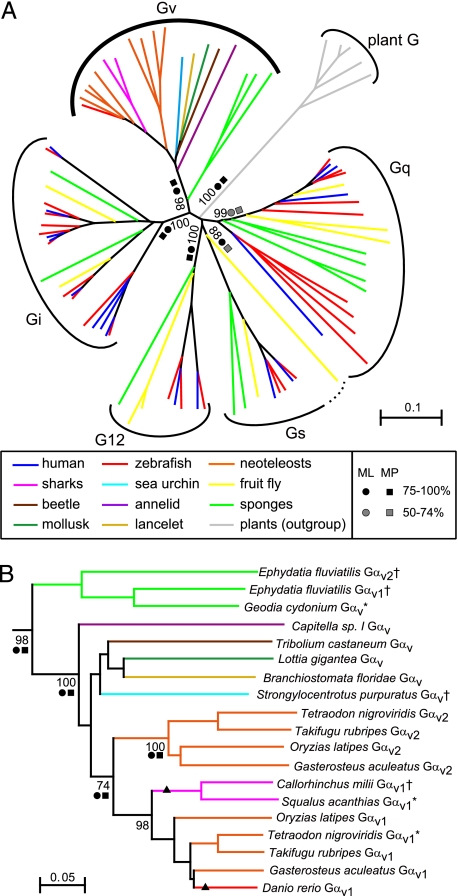
A recursive search in the Ensembl and NCBI genomic databases led to the identification of 26 gna genes in zebrafish, Danio rerio. In the phylogenetic analysis, all but one of these genes fall clearly into the 4 known Gα classes (Fig. 1A). However, the final gene did not conform to this pattern. To characterize this gene, we searched for related genes in many animals from different phyla and found 1–2 homologous sequences from 4 neoteleost species (medaka, three-spined stickleback, fugu, and tetraodon), 2 sharks, a cephalochordate (lancelet), a sea urchin, a beetle, an annelid (polychaete worm), a mollusk (limpet), and 2 sponges (supporting information (SI) Fig. S1 and Table S1). All these genes clearly belong to the animal Gα protein clade, using plant Gα proteins as an outgroup (Fig. 1A). Inside the animal Gα clade these genes form a monophyletic subclade that separates clearly from 4 other monophyletic subclades representing the other 4 classes of Gα proteins, here collected from human, fruit fly, and sponges. This branching is supported by near to maximal bootstrap values using 3 different algorithms (NJ, neighbor joining; MP, maximum parsimony; and ML, maximum likelihood; Fig. 1A). On the basis of these findings, we propose “class Gv” as nomenclature of this group, where “v” stands for the Roman numeral v. As this class includes members in sponges that are among the earliest diverging phyla of the animal kingdom, Gv has an ancient evolutionary origin at a very early stage of metazoan evolution. Thus, it appears to be as old as the other 4 classes of Gα proteins. Surprisingly, we failed to find orthologs in many other species including tetrapods, jawless fish, ascidians (sea squirt), fruit flies, leeches, nematodes, and cnidarians (sea anemone). Although some of the databases we used are still not complete, these results suggest that Gv has been lost in many lineages. On the other hand, at least two independent gene duplications appear to have occurred in the sponge and vertebrate lineages, respectively (Fig. 1B).
Fig. 1.
Phylogenetic analysis of Gv proteins: the phylogenetic tree (NJ algorithm) of 16 human (blue), 26 zebrafish (red), 9 fruit fly (yellow), 2 shark (pink), 8 neoteleost (orange), 1 cephalochordate (lancelet, ochre), 1 sea urchin (light blue), 1 annelid (polychaete worm, purple), 1 mollusk (limpet, dark green), 1 beetle (brown), and 12 sponge (green) Gα proteins. Gα proteins of 5 plants are included as an outgroup (gray). Dotted curve, fruit fly Gfα provisionally assigned as Gs (29). (A) Full tree including all 5 classes. All G proteins in the whole animal kingdom fall in these 5 classes except a nematode-specific family (30) not clustering with Gv (not shown). (B) Gαv clade as determined in A. Bootstrap values at major branches are shown as percentages. Strong (>75%, black circle, black box) and moderate (50–74%, gray circle, gray box) support for each branching derived from ML (circles) and MP (boxes) analyses is also indicated. Branches that change positions in ML and/or MP trees are marked by triangles. Note that all three seawater sponge Gαv proteins are excluded from MP analysis due to significant sequence gaps (cf. Fig. S1). Asterisks and daggers indicate corrected and partial sequences, respectively (see Fig. S1 for details). Scale bar shows amino acid substitution rate for the NJ tree.
Gv is more closely related to Gi than to the other classes, with the divergence between Gv and Gi comparable to that between Gs and G12 (Fig. 1A). Within the Gv clade, sponge proteins branch first, followed by the nonvertebrate Gv proteins, in rough accordance with the order of lineage separation during animal evolution. Vertebrate Gv proteins segregate reliably into 2 families, Gαv1 and Gαv2 (Fig. 1B), whose divergence is comparable with, e.g., that between the Gα12 and Gα13 proteins in class G12 (data not shown). Neoteleosts possess both paralogous genes, whereas the earlier-diverging species shark and zebrafish have only 1 gnav gene. Both appear to be orthologs of gnav1 (Fig. 1B and Fig. S1), consistent with a duplication event early in vertebrate evolution and subsequent losses of gnav2 in the shark and zebrafish lineages. Alternatively a duplication event in a neoteleost ancestor is conceivable. In the neoteleost lineage the genus Tetraodon may exemplify a currently ongoing gene loss, as the gnav1 gene could well be a pseudogene (3 nucleotide insertions, of which 2 generate stop codons in a functional domain; see Fig. S1), in contrast to the apparently intact fugu gnav genes.
Gv Proteins Possess Motifs Conserved Among GTP-Binding Proteins and Class-Specific Conserved Sequences.
Gα proteins typically contain a helical domain, whose six α-helices are inserted between the α1 helix and the β2 sheet of the Ras-like nucleotide-binding domain, and 3 switch regions (SWs) that undergo the conformational change upon GTP binding (4). All these elements are predicted in the Gαv proteins at the appropriate positions in the sequence (Fig. 2 and Fig. S1).
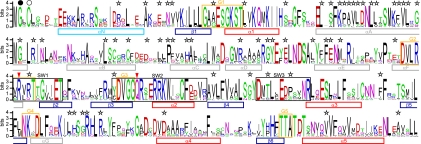
Fig. 2.
Conserved sequence features in Gαv proteins: the degree of conservation of 19 Gαv protein sequences shown as a sequence logo. Secondary structures are indicated below the logo with bars (light blue, N-terminal helix; gray, helices within helical domain; red, helices within GTPase domain; dark blue, β-sheet). G-boxes and switch regions are indicated with orange and black boxes, respectively. Black and white circles above the logo indicate putative sites for N-linked myristoylation and thio-palmitoylation, respectively. Gv-specific motifs (conservation >60%) are marked with stars. Red arrowheads indicate residues critical for GTPase activity.
All Gαv proteins, for which full-length sequence information is available, contain all 5 characteristic G-box motifs (Fig. 2 and Fig. S1) that are a hallmark of guanidine-nucleotide binding proteins and play a direct role in their nucleotide binding (9). Two residues that are critical for GTPase activity, the arginine in SW1 and the glutamine in SW2 (10), are also conserved among all Gαv proteins (Fig. 2 and Fig. S1). Co- and posttranslational acylation in the N-terminal region has been shown to affect the plasma membrane localization of Gα proteins (11, 12). The corresponding acylation sites are predicted in the N-terminal region of all Gαv, for which full-length sequence information is available (Fig. 2 and Fig. S1). In summary, all elements characteristic for Gα proteins are present in the Gv proteins, confirming their identification as Gα proteins.
A detailed comparison of Gv proteins with those from the other 4 classes reveals class-specific conservation of 60 residues (Fig. 2 and Fig. S1). They are located mainly in the helical domain, but also in SWs and may thus be involved in interaction with GPCRs, β/γ heterodimers, regulators, and effectors (4, 13–18), suggesting that the Gαv proteins have their specific interaction partners and consequently regulate distinct pathways. Furthermore, both N- and C-terminal regions, which are assumed to interact with GPCRs and β/γ heterodimers (4), contain several Gv-specific amino acids as well (Fig. 2 and Fig. S1).
In particular, Gv proteins differ from the phylogenetically most related Gi class (Fig. 1A) by the complete absence of two functionally relevant residues, a cysteine residue at the fourth position from the C-terminus, which is ADP-ribosylated by Pertussis toxin (4) in all Gi but Gαz, which exhibits a conserved serine at position 27, a substrate of PKC (19). None of the Gv proteins possesses either residue (Fig. S1), clearly separating the class Gv from the class Gi.
Strong Negative Selection in the Teleost Gαv Family.
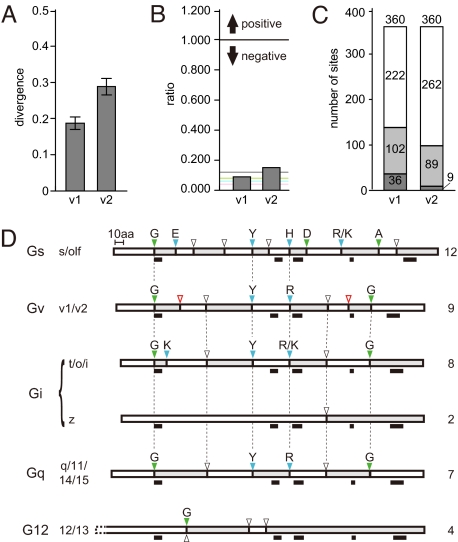
Vertebrate Gαv proteins are highly conserved with overall amino acid identities ranging from 59 to 89% (70% on average, values for full-length proteins). The average sequence divergence in the teleost families of Gαv1 and Gαv2 is 0.19 and 0.29, respectively (Fig. 3A), showing that Gαv2 is more divergent than Gαv1.
Fig. 3.
Selective pressure and class-specific splicing pattern of gnav genes. (A) Divergence calculated for teleost gnav1 and gnav2 families. Values given represent means ± SE of all possible pairwise comparisons. (B) Nonsynonymous vs. synonymous substitution ratio (dN/dS), global values calculated for each gnav family. Horizontal lines, average values for each class: gray, Gv; green, Gq; blue, G12; pink, Gs, Gi. (C) Numbers of neutral and negatively selected sites (codons) counted for each gnav family. White and gray bars are for neutral and negatively selected sites, respectively. Two different significance levels were used for detecting selected sites (dark gray, P < 0.1; light gray, P < 0.2). Total amino acid numbers analyzed for each family are shown on the top of the bars. (D) Schematic representation of the coding regions of gna genes. Genomic structures for all of 16 human and 26 zebrafish gna genes and 8 neoteleost gnav genes are summarized. Exons are indicated as alternating white and gray boxes. The N-terminus is to the left. Dashed line in G12, gna12 extends beyond gna13. Intron phases (0, 1, 2) at each splice junction are shown as white, green, and blue triangles, respectively. Residues at splice junctions of phases 1 and 2 are shown above. Gv-specific splice sites are shown in red. Black bars beneath indicate positions of the G-boxes (G1–5, from left to right). Vertical dotted lines indicate splice sites conserved among different classes. Classes and families are shown to the left and numbers of exons for each class (or family) are shown to the right. The human gnas gene has an extra exon between exons 2 and 3 (not shown). Note that all classes except Gq exhibit unique splice sites not shared with any other class. (Scale bar, 10 aa.)
As an indicator for the selective pressures acting on these genes, we calculated ratios of nonsynonymous vs. synonymous nucleotide substitutions (dN/dS) among teleost gnav genes. Both gnav families have very low global dN/dS values, 0.09 for gnav1 and 0.15 for gnav2 (Fig. 3B), showing that they are under strong negative selection (neutral selection corresponds to a ratio of 1). At the individual codon level we find 138 and 98 residues under negative selection in gnav1 and gnav2 genes, respectively, without a single positively selected residue (Fig. 3C and Fig. S2). Negatively selected residues in the proteins are not restricted to particular motifs (Fig. S2), indicating stringent constraint on the entire protein coding region.
However, compared to the other four classes, Gv proteins show a somewhat relaxed selection pressure, since dN/dS values for the other classes are even lower than those found for Gv (average values 0.04 for Gi and Gs, 0.06 for G12, and 0.08 for Gq, compared to 0.12 for Gv, Fig. 3B). This implies a certain acceleration of evolution in teleost gnav genes and might suggest a somewhat higher divergence of interaction partners and functions in the Gv class.
The Exon/Intron Structure of Teleost gnav Genes Is Strictly Conserved and Characteristically Different From That of the Other 4 Classes.
The 4 known classes of gna genes each exhibit a specific splicing pattern (20). We determined the genomic structure for all teleost gnav genes and compared it to all known gna genes of human and zebrafish. All teleost gnav genes consist of 9 exons, and the positions of exon/intron boundaries are well conserved (Fig. 3D and Fig. S1). The boundaries at exon 1/2, exon 4/5, and exon 5/6 of Gv are shared with Gs, Gi (except gnaz), and Gq both in position and in phase, suggesting that these junctions are older than the evolutionary separation of these classes. Three other junctions (exon 3/4, exon 6/7, and exon 8/9) are shared just with Gi (except gnaz) and Gq. These results place Gv closer to Gi and Gq than to Gs and furthest from G12. Most importantly, 2 further junctions in Gv (exon 2/3 and exon 7/8) are not present in any of the other classes. These Gv-specific junctions further support the designation of Gv as a class in its own right, independent from the other 4 classes.
Gnav Transcripts Are Expressed in Many Adult Zebrafish Tissues.
We explored the EST databases of five teleost species and dogfish shark and found 1 to several ESTs for zebrafish gnav1, medaka gnav1 and gnav2, stickleback gnav1, and fugu gnav2 (9, 12, 1, 2, and 1 clones, respectively; for a list of EST clones see Table S2; for shark see Fig. S1). Considering the incompleteness of EST databases, the most plausible interpretation is that gnav genes generally are expressed and presumably give rise to functional proteins.
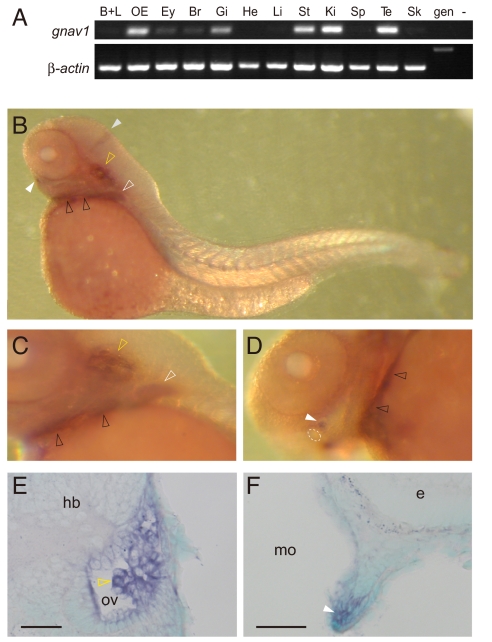
As a further test we isolated cDNA for gnav1 containing the full-length ORF by RT-PCR from zebrafish olfactory epithelium and determined the complete nucleotide sequence. This confirmed that the transcript predicted from the genomic database is correct and is transcribed in vivo. We then checked the mRNA distribution of zebrafish gnav1 in adult tissues by semiquantitative RT-PCR, using an intron-spanning primer pair. A band of the expected size was found in many tissues, with the highest band intensities observed for gill, kidney, olfactory epithelium, stomach, and testis at 35 cycles (Fig. 4A). At 40 cycles, weak to moderate expression was detected in barbels and lips, eye, brain, liver, spleen, and skin, whereas expression in heart could hardly be detected at all (data not shown).
Fig. 4.
Expression pattern of the zebrafish gnav1 gene. (A) Transcripts for zebrafish gnav1 were detected by RT-PCR using intron-spanning primer pairs. β-Actin was used as a positive control. B+L, barbels and lips; OE, olfactory epithelium; Ey, eye; Br, brain; Gi, gill; He, heart; Li, liver; St, stomach; Ki, kidney; Sp, spleen; Te, testis; Sk, skin; gen, genomic DNA; −, negative control without template DNA. (B–F) Whole-mount in situ hybridization with gnav1 probe. (B and C) Lateral views of the whole larva and posterior part of the head region, respectively. (D) Ventral view of the head region. Dotted circle, mouth. (E and F) Cross-sections after hybridization show expression in the developing inner ear (E) and lower lip (F). Sections were counterstained with methyl green. Dorsal is to the top. White and gray solid arrowheads indicate the cell clusters next to the lower lip and the midbrain–hindbrain boundary, respectively. White, yellow, and black open arrowheads point to labeled cells within pectoral fins, otic vesicle (ov), and branchial arches, respectively. e, eye; hb, hindbrain; mo, mouth cavity. (Scale bars: 50 μm.)
Specific Expression of gnav1 in Larval Zebrafish.
Finally, we performed whole-mount in situ hybridization of 3-day-old zebrafish larvae using two different, nonoverlapping gnav1 probes. Specific expression was evident in the inner ear and in bilateral cell clusters near the lower lip (Fig. 4 and Fig. S3). Expression was also observed in the branchial arches, the pectoral fins, and the midbrain-hindbrain boundary region. Signals in these regions were reproducible with both probes (data not shown) and absent with sense-strand controls (Fig. S3). All other regions did not contain detectable levels of gnav1 transcripts. This expression pattern is characteristically different from that of gna genes of the other 4 classes (Fig. S4). While we cannot exclude that the broader distribution observed in adult tissues may be explained by the higher sensitivity of the RT-PCR, it is conceivable that fully differentiated cells and tissues exhibit higher expression levels. In any case, we have shown that gnav genes are expressed in vivo and thus presumably give rise to functional proteins.
Discussion
In this study, we have identified a fifth class of Gα protein in metazoans. Gv orthologs occur already in sponges, members of one of the earliest diverged phyla in the animal kingdom, suggesting that Gv is as ancient as the other 4 classes. Gv proteins possess all domain structures, sequence motifs, and modification sites expected of Gα proteins. Their monophyletic origin together with their sequence motifs and exon/intron borders unique to Gv unambiguously delineate this new class. Gnav genes generally appear to encode functional Gαv proteins, whose expression is shown by EST analysis, RT-PCR, and in situ hybridization data.
As Gα proteins are an extensively characterized protein family, it was completely unexpected to find a new class of Gα protein at the level of the canonical 4 classes. The absence of Gv in human, mouse, fruit fly, and nematode, the most studied model organisms, seems to have hampered the identification of Gv. This may explain why Gv members from fresh water and marine sponge, sea urchin, and red flour beetle had been misassigned to other classes (Fig. S1). All these proteins both share the Gv-specific motifs and form a single clade with the vertebrate Gv proteins, and thus constitute invertebrate representatives of the Gv class.
One of the most striking features of this gene family is a considerable gene loss throughout animal evolution, resulting in the absence of the Gv class in many lineages. Despite our extensive search, we failed to find orthologous sequences in genomic or EST databases of mammals, chicken, reptiles, amphibians, jawless fish, ascidians, fruit fly, mosquitoes, bee, moth, several nematodes, leech, and cnidarians. However, Gv genes were detected in several neoteleosts, zebrafish, cartilaginous fish, a lancelet, a sea urchin, a polychaete worm, a limpet, a beetle, and 2 sponge species. Thus, gene loss events seem to have occurred at a basal level in the nematode phylum, but several independent losses are required to explain Gv occurrence in the phylum chordata, one of them in the ascidian lineage (urochordata), another one in sea lamprey (agnatha), and a third one in the ancestor of tetrapods, resulting in complete absence of Gv in all classes of tetrapods. The genus Tetraodon may exemplify a currently ongoing gene loss, as gnav1 appears to be a pseudogene in tetraodon but not in fugu. Similarly, in the phylum of arthropods the presence of Gv ortholog in red flour beetle, but not in fruit fly, bee, moth, or mosquito suggests that independent gene losses seem to have taken place after separation of class insecta. These recurrent losses of the Gv class in so many lineages are different from the more commonly observed pseudogenization events after gene duplication in larger protein families. To the best of our knowledge a similar pattern of gene losses has not been seen in any other gene family so far.
On the other hand, two independent gene gains are observed in the Gv class, one in a sponge and the other in jawed fish. Such a pattern of recurrent gene gains and losses suggests that the Gv class conforms to a birth-and-death mode of evolution (21).
So what could be the function of the Gv genes? The broad, but not ubiquitous expression pattern of zebrafish gnav1 revealed by RT-PCR suggests that Gαv is not involved in ubiquitous housekeeping processes. All but one species that possess Gv orthologs are living in an aqueous environment. Consistent with the gnav1 expression in zebrafish kidney, this raises the possibility that Gαv proteins might be involved in the regulation of cell osmolality in these species. Larval expression of gnav1 is quite different from that of other gna genes (Fig. S4) and suggests an involvement in cellular differentiation processes. The expression in the inner ear might indicate a role in sensory cell differentiation, and a role in taste bud differentiation could be conjectured from the expression in branchial arches, which are among the earliest sites for taste bud primordia to appear (22). The bilateral cell clusters near the lips expressing gnav1 might constitute barbel primordia. Taken together, larval expression may be linked to a subset of sensory tissues.
Previous studies with mammalian Gα proteins have implicated that both N- and C-terminal regions determine the coupling specificity to GPCRs (4). We found that N- and C-termini are uniquely conserved among Gv, including a characteristic length for the N-terminus (see Fig. S1). This is consistent with the concept that Gv may interact with a distinct set of GPCRs. In species that lost Gv one might expect either a loss of corresponding GPCRs or compensation by G proteins of other classes (see refs. 1 and 8).
In an attempt to identify potentially interacting regulators and/or effectors we have analyzed conserved G protein motifs in Gv proteins (Fig. S1). Mutagenesis studies and crystal structures in mammalian G proteins have identified single residues essential for interaction with regulators of G protein signaling-4, -9, and -16, all of them conserved in Gv proteins, and a larger, partially conserved motif interacting with phosphodiesterase γ (Fig. S1). Moreover, a set of Gv-specific interaction partners may be inferred from the presence of several extended Gv-specific motifs in the helical domain. The helical domain is a divergent region of Gα proteins in general, but seems to be conserved within a class or a family. Although the functions of this domain have not been fully understood so far, several studies have shown its effect on GTPase activity and involvement in the interaction with GPCRs, regulator and effector proteins, and possibly β/γ heterodimers (13–18).
In conclusion, we identified a fifth class of metazoan Gα protein, Gv, with an ancient evolutionary origin like the other 4 classes. The Gv class has been evolving under strong purifying selection. A striking and unexpected feature of Gv is its loss in many lineages during animal evolution, leading to its absence in several commonly used model organisms. However, Gv is retained in other lineages across the animal kingdom. Our discovery of a fifth class of Gα proteins should provide a unique opportunity for studying both the evolution of the Gα protein family and cell signaling mechanisms through heterotrimeric G proteins.
Materials and Methods
Identification of gnav Genes in Silico.
Annotated zebrafish Gα protein sequences (www.ncbi.nlm.nih.gov/) and automatic paralog predictions [www.ensembl.org/index.html, assembly version 7 (Zv7), release 48, December 2007], together with 16 human Gα protein sequences, served as queries for TBLASTN algorithm in the Ensembl zebrafish genomic DNA database. An expectation cutoff value of 10−10 was used to identify candidate Gα protein coding sequences. GenWise (www.ebi.ac.uk/Wise2/) was applied to find all exons of each gene by matching to orthologous human Gα protein sequences.
The Gαv orthologs in other species were identified through TBLASTN search in Ensembl genome databases (release 48, December 2007) for medaka Oryzias latipes, three-spined stickleback Gasterosteus aculeatus, fugu Takifugu rubripes, and tetraodon Tetraodon nigroviridis; in the NCBI EST database for dogfish shark Squalus acanthias, red flour beetle Tribolium castaneum, fresh water sponge Ephydatia fluviatilis, and marine sponge Geodia cydonium; in the NCBI whole genome shotgun database for elephant shark Callorhinchus milii; in the HGSC genome database (www.hgsc.bcm.tmc.edu/projects/) for sea urchin Strongylocentrotus purpuratus; and in the JGI genome database (http://genome.jgi-psf.org/euk_cur1.html) for lancelet Branchiostoma floridae, polychaete worm annelid Capitella sp. I, and limpet Lottia gigantea.
Phylogenetic Analysis.
Gα protein sequences were aligned with MAFFT 4.0. Sequence alignment was manually edited with MEGA4 (23) and gap positions present in >85% of sequences were removed. NJ, MP, and ML algorithms were used to construct trees with Clustal X (NJ), Protpars (MP), and Proml (ML) from the PHYLIP package (http://evolution.genetics.washington.edu/phylip.html). Bootstrapping was performed for each algorithm, 1,000, 100, and 100 times, respectively, using either Clustal X or Seqboot from the PHYLIP package. Horizontal and radial trees were visualized with Njplot and Unrooted, respectively.
Sequence Logo, Secondary Structure Prediction, and dN/dS Analysis.
A sequence logo was generated using WebLogo (24). Sequence alignment with 9 teleost, 2 cartilaginous fish, 1 lancelet, 1 sea urchin, 1 beetle, 1 annelid, 1 limpet, and 3 sponge Gαv proteins was manually edited with MEGA4 and gap positions present in >50% of sequences were removed. The secondary structure of each full-length Gαv protein was predicted with Geno3D (25), using default parameter settings and 3 structure templates in the protein data bank found by the program. The dN/dS analysis on overall proteins and single codons was performed as described (26).
RT-PCR and Whole-Mount in Situ Hybridization.
Total RNA samples were prepared from adult zebrafish tissues of a wild-type Ab/Tübingen strain with the RNeasy kit (QIAGEN). After digestion with DNaseI, 100 ng RNA for each tissue were subjected to the first-strand cDNA synthesis with RevertAid MmLV reverse transcriptase (Fermentas), using oligo(dT)15 primer. Subsequent PCR was performed using Red Taq mix (Bioline) with gene-specific primers listed in Table S3.
Two nonoverlapping digoxigenin-labeled RNA probes (gnav1-N and gnav1-M) were used. Whole-mount in situ hybridization with 3-day-old larvae was done as described (27, 28). For details see Fig. S3.
Supplementary Material
Acknowledgments.
We thank Mehmet Saltürk for taking good care of the zebrafish. This work was supported by a Deutsche Forschungsgemeinschaft grant (S.I.K.) and by the International Graduate School in Genetics and Functional Genomics, University of Cologne (L.R.S. and Y.Y.K.). Y.O. was partially supported by Yoshida scholarship foundation.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0809420106/DCSupplemental.
References
- 1.Wettschureck N, Offermanns S. Mammalian G proteins and their cell type specific functions. Physiol Rev. 2005;85:1159–1204. doi: 10.1152/physrev.00003.2005. [DOI] [PubMed] [Google Scholar]
- 2.Downes GB, Gautam N. The G protein subunit gene families. Genomics. 1999;62:544–552. doi: 10.1006/geno.1999.5992. [DOI] [PubMed] [Google Scholar]
- 3.Melien O. Heterotrimeric G proteins and disease. Methods Mol Biol. 2007;361:119–144. doi: 10.1385/1-59745-208-4:119. [DOI] [PubMed] [Google Scholar]
- 4.Oldham WM, Hamm HE. Heterotrimeric G protein activation by G-protein-coupled receptors. Nat Rev Mol Cell Biol. 2008;9:60–71. doi: 10.1038/nrm2299. [DOI] [PubMed] [Google Scholar]
- 5.Hurowitz EH, et al. Genomic characterization of the human heterotrimeric G protein α, β, and γ subunit genes. DNA Res. 2000;7:111–120. doi: 10.1093/dnares/7.2.111. [DOI] [PubMed] [Google Scholar]
- 6.Birnbaumer L. Expansion of signal transduction by G proteins. The second 15 years or so: from 3 to 16 α subunits plus βγ dimers. Biochim Biophys Acta. 2007;1768:772–793. doi: 10.1016/j.bbamem.2006.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Strathmann MP, Simon MI. Gα12 and Gα13 subunits define a fourth class of G protein α subunits. Proc Natl Acad Sci USA. 1991;88:5582–5586. doi: 10.1073/pnas.88.13.5582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Albert PR, Robillard L. G protein specificity: traffic direction required. Cell Signal. 2002;14:407–418. doi: 10.1016/s0898-6568(01)00259-5. [DOI] [PubMed] [Google Scholar]
- 9.Sprang SR. G protein mechanisms: insights from structural analysis. Annu Rev Biochem. 1997;66:639–678. doi: 10.1146/annurev.biochem.66.1.639. [DOI] [PubMed] [Google Scholar]
- 10.Majumdar S, Ramachandran S, Cerione RA. New insights into the role of conserved, essential residues in the GTP binding/GTP hydrolytic cycle of large G proteins. J Biol Chem. 2006;281:9219–9226. doi: 10.1074/jbc.M513837200. [DOI] [PubMed] [Google Scholar]
- 11.Marrari Y, Crouthamel M, Irannejad R, Wedegaertner PB. Assembly and trafficking of heterotrimeric G proteins. Biochemistry. 2007;46:7665–7677. doi: 10.1021/bi700338m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Milligan G, Kostenis E. Heterotrimeric G-proteins: a short history. Br J Pharmacol. 2006;147(Suppl 1):S46–55. doi: 10.1038/sj.bjp.0706405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Krieger-Brauer HI, Medda PK, Hebling U, Kather H. An antibody directed against residues 100–119 within the α-helical domain of Gα(s) defines a novel contact site for β-adrenergic receptors. J Biol Chem. 1999;274:28308–28313. doi: 10.1074/jbc.274.40.28308. [DOI] [PubMed] [Google Scholar]
- 14.Cherfils J, Chabre M. Activation of G-protein Gα subunits by receptors through Gα-Gβ and Gα-Gγ interactions. Trends Biochem Sci. 2003;28:13–17. doi: 10.1016/s0968-0004(02)00006-3. [DOI] [PubMed] [Google Scholar]
- 15.Skiba NP. The α-helical domain of Gαt determines specific interaction with regulator of G protein signaling 9. J Biol Chem. 1999;274:8770–8778. doi: 10.1074/jbc.274.13.8770. [DOI] [PubMed] [Google Scholar]
- 16.Soundararajan M, et al. Structural diversity in the RGS domain and its interaction with heterotrimeric G protein α-subunits. Proc Natl Acad Sci USA. 2008;105:6457–6462. doi: 10.1073/pnas.0801508105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu W, Northup JK. The helical domain of a G protein α subunit is a regulator of its effector. Proc Natl Acad Sci USA. 1998;95:12878–12883. doi: 10.1073/pnas.95.22.12878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Day PW, et al. Characterization of the GRK2 binding site of Gαq. J Biol Chem. 2004;279:53643–53652. doi: 10.1074/jbc.M401438200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cabrera-Vera TM, et al. Insights into G protein structure, function, and regulation. Endocr Rev. 2003;24:765–781. doi: 10.1210/er.2000-0026. [DOI] [PubMed] [Google Scholar]
- 20.Sarwal MM, Sontag JM, Hoang L, Brenner S, Wilkie TM. G protein α subunit multigene family in the Japanese puffer fish Fugu rubripes: PCR from a compact vertebrate genome. Genome Res. 1996;6:1207–1215. doi: 10.1101/gr.6.12.1207. [DOI] [PubMed] [Google Scholar]
- 21.Nei M, Rooney AP. Concerted and birth-and-death evolution of multigene families. Annu Rev Genet. 2005;39:121–152. doi: 10.1146/annurev.genet.39.073003.112240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hansen A, Reutter K, Zeiske E. Taste bud development in the zebrafish, Danio rerio. Dev Dyn. 2002;223:483–496. doi: 10.1002/dvdy.10074. [DOI] [PubMed] [Google Scholar]
- 23.Tamura K, Dudley J, Nei M, Kumar S. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol Biol Evol. 2007;24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- 24.Crooks GE, Hon G, Chandonia JM, Brenner SE. WebLogo: a sequence logo generator. Genome Res. 2004;14:1188–1190. doi: 10.1101/gr.849004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Combet C, Jambon M, Deleage G, Geourjon C. Geno3D: automatic comparative molecular modelling of protein. Bioinformatics. 2002;18:213–214. doi: 10.1093/bioinformatics/18.1.213. [DOI] [PubMed] [Google Scholar]
- 26.Saraiva LR, Korsching SI. A novel olfactory receptor gene family in teleost fish. Genome Res. 2007;17:1448–1457. doi: 10.1101/gr.6553207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Thisse C, Thisse B, Schilling TF, Postlethwait JH. Structure of the zebrafish snail1 gene and its expression in wild-type, spadetail and no tail mutant embryos. Development. 1993;119:1203–1215. doi: 10.1242/dev.119.4.1203. [DOI] [PubMed] [Google Scholar]
- 28.Kraemer AM, Saraiva LR, Korsching SI. Structural and functional diversification in the teleost S100 family of calcium-binding proteins. BMC Evol Biol. 2008;8:48. doi: 10.1186/1471-2148-8-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Quan F, Wolfgang WJ, Forte M. A Drosophila G-protein α subunit, Gfα, expressed in a spatially and temporally restricted pattern during Drosophila development. Proc Natl Acad Sci USA. 1993;90:4236–4240. doi: 10.1073/pnas.90.9.4236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bastiani C, Mendel J. Heterotrimeric G proteins in C. elegans. WormBook. 2006;1:25. doi: 10.1895/wormbook.1.75.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.