Abstract
Purpose
A linkage study on autosomal recessive high myopia (arHM) has not been reported, although several loci for autosomal dominant high myopia (adHM) have been mapped. Data from a consanguineous Chinese family with arHM were collected to map the genetic locus associated with this condition.
Methods
Phenotypic information and DNA samples were collected from family members. A genome-wide linkage scan combined with homozygosity mapping was performed by using 382 microsatellite DNA markers from the entire genome spaced at intervals of about 10 cM.
Results
The pedigree and clinical data of the family indicate that the high myopia is autosomal recessive. A genome-wide scan of chromosomes 1–22 gave a LOD score greater than 1.0 for 22 markers. Linkage to most of these markers was not supported by closely flanking markers except for three possible loci on chromosomes 11, 14, and 17. Fine mapping and haplotype analysis provide evidence for a locus at 14q22.1-q24.2 in a 25.23 Mb region between markers D14S984 and D14S999 with a maximum LOD score of 2.19. All 11 microsatellite markers inside the linkage interval as well as haplotype construction point to a gene at this locus. Linkage elsewhere on chromosome 11 and chromosome 17 could not be excluded due to the small size of the family.
Conclusions
Pedigree and clinical data suggest that an autosomal recessive gene is responsible for high myopia in a consanguineous Chinese family. Genome-wide linkage analysis was used to map the gene for high myopia to a few limited loci. The resultant information should help future studies identify the gene for arHM. To our knowledge, this report is the first clinical and linkage study on a consanguineous family with arHM.
Introduction
Myopia is a leading cause of visual impairment worldwide, and Chinese people living in industrialized or urban/suburban areas have a significantly higher incidence of myopia than other populations [1-4]. High myopia, its extreme form, is the fourth most common cause of irreversible blindness [5,6]. Evidence has shown that genetic factors play an important role in the development of high myopia [5,7-14].
High myopia may be inherited as an autosomal dominant, autosomal recessive, or X-linked recessive trait. Some high myopia could also be transmitted as a complex trait. Eight loci for autosomal dominant high myopia (adHM) and two loci for X-linked recessive high myopia (xlHM) have been mapped including myopia 1 (MYP1 [Xq28]) [12,15], MYP2 (18p11.31) [14,16], MYP3 (12q21-q23) [13,17], MYP4 (7q36) [18], MYP5 (17q21-q22) [5], MYP11 (4q22-q27) [10], MYP12 (2q37.1) [11], MYP13 (Xq23-q25) [9,19], and two others on 10q21.1 [8] and 5p15 [7] in which formal gene names have not yet been assigned. These loci, however, are estimated to be responsible for only a small portion of high myopia [20]. Although several candidate genes have been analyzed [21-26], the genes at these loci still remain unknown [21,27-29]. Furthermore, before this study, no genetic loci for autosomal recessive high myopia (arHM) have been reported. Identification of additional loci as well as of the causative genes is the first step toward understanding the molecular basis of myopia and, subsequently, toward the prevention and treatment of this sight threatening problem.
Here, we report a clinical and linkage study on a consanguineous Chinese family with arHM. A genome-wide linkage analysis combined with homozygosity mapping provides suggestive linkage of high myopia to a few loci including a novel locus in the q21-q24 region of chromosome 14 between markers D14S288 and D14S74.
Methods
Family and clinical data
The consanguineous Chinese family with arHM was one of the 628 families with high myopia ascertained as part of a project to identify the genetic causes of high myopia in China. This family was from the Guangdong province of China. It includes one consanguineous marriage with three affected individuals (Figure 1). Three affected and three unaffected individuals from the family participated in this study. Informed consent conforming to the tenets of the Declaration of Helsinki and following the Guidance of Sample Collection of Human Genetic Diseases (863-Plan) by the Ministry of Public Health of China was obtained from the participating individuals before the study. Ophthalmological examination was performed at the Eye Hospital, Zhongshan Ophthalmic Center. Refractive error was measured by retinoscopy after mydriasis with compound tropicamide (Mydrin®-P; Santen Pharmaceutical Co. Ltd., Osaka, Japan). A subject was considered to have high myopia if he or she met the following criteria: 1) the myopia was noted before school age; 2a) cycloplegic refraction of −6.00 diopters (D) or lower (spherical equivalent) in individuals under 30 years of age or 2b) manifest refraction of −6.00 D or lower (spherical equivalent) in individuals 30 years old or older as defined previously [9]; and 3) exclusion of other known ocular or systemic diseases. Electroretinogram (ERG) responses were recorded in the proband consistent with the International Society for Clinical Electrophysiology of Vision (ISCEV) standards [30]. Genomic DNA was prepared from venous blood.
Figure 1.
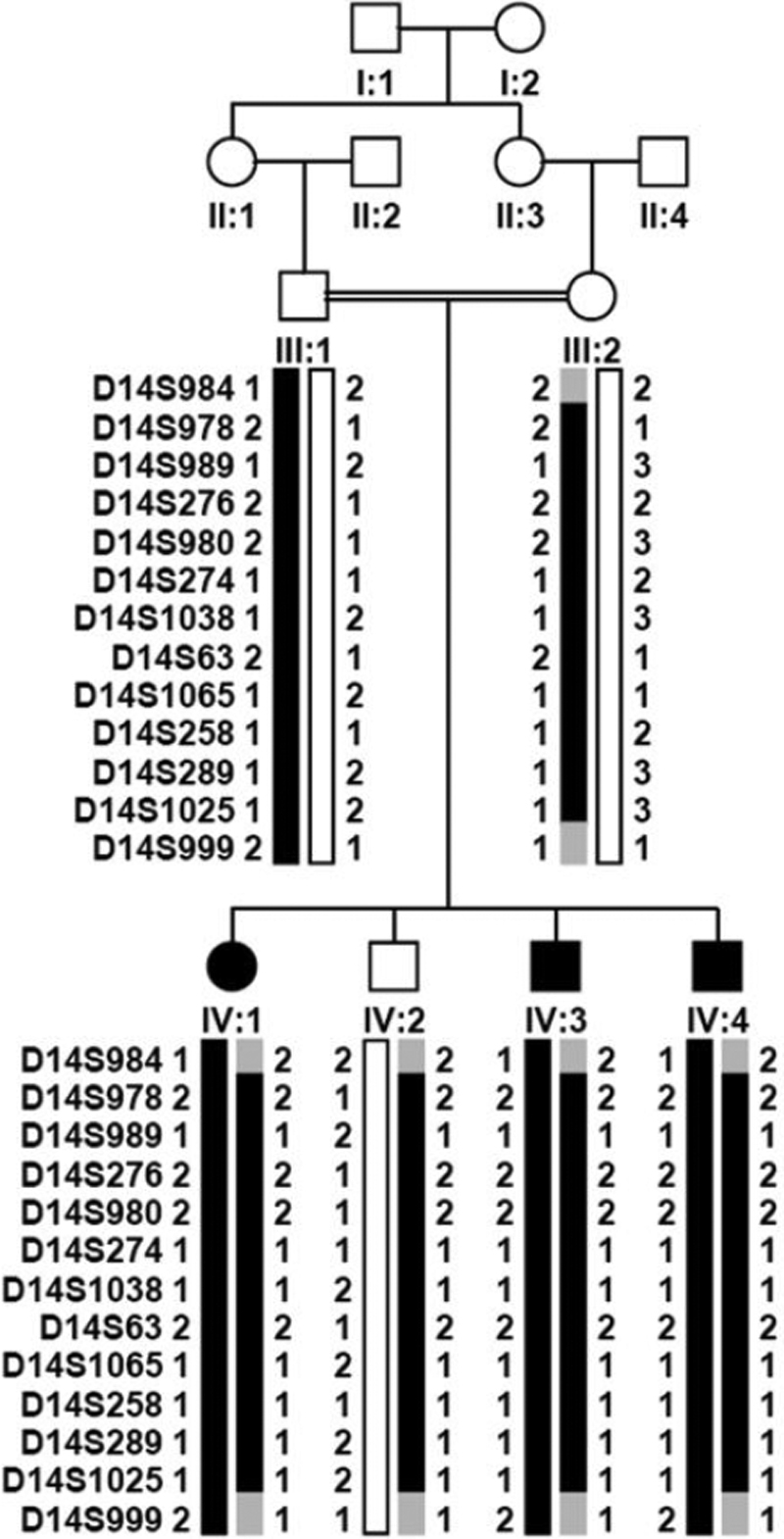
Pedigree and haplotype diagram of the family. Filled squares (male) or circles (female) represent individuals affected with high myopia. Blackened filled bars indicate the chromosomal regions that are derived from the ancestral disease-associated haplotype.
Genotyping, linkage analysis, and candidate gene screening
Genotyping for all participating family members was performed using 5′-fluorescently labeled microsatellite markers as previously described [10], except that the amplicons were separated on an ABI 3100 Genetic Analyzer (Applied Biosystems, Foster City, CA). Genotyping data were analyzed using the Gene Mapper version 3.5 software package (Applied Biosystems). Two-point linkage analysis was performed by using the MLINK program of the FASTLINK implementation of the LINKAGE program package [31,32]. The myopia in the family was analyzed as an autosomal recessive trait with full penetrance and with a disease-gene allele frequency of 0.0001. Homozygosity mapping was used to exclude those markers with positive LOD scores but without homozygosity for alleles [33,34]. For fine mapping around the possible region, additional markers were selected according to the National Center for Biotechnology Information (NCBI) map. Haplotypes were generated using the Cyrillic 2.1 program (CyrillicSoftware, Wallingford, Oxfordshire OX10 8BA) and confirmed by inspection. The criteria for establishing linkage have been previously described [35].
Results
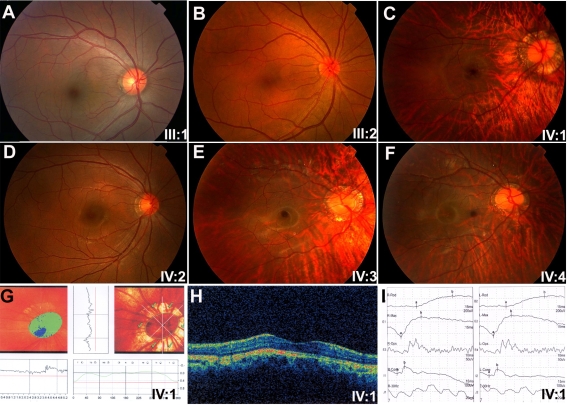
The consanguineous family originates from a small town in northeast Guangdong province, which is about 400 km away from Guangzhou, China (Figure 1). Three of the four children have been affected with high myopia since early childhood. None of the affected siblings had night blindness or photophobia. Neither the parents nor the grandparents were affected. Ophthalmological examination revealed that all three affected siblings had normal corneas, irises, and lenses. All three affected children had extreme high myopia with excessive extension of the axial length (Figure 2, Table 1). Fundus observation of the three affected siblings demonstrated similar changes including tigroid fundus and a circular choroidal defect around the optic disc (Figure 2C,E,F). Optical coherence tomography (OCT) and a Heidelberg Retina Tomograph (HRT) examination revealed normal optic discs and normal thickness of the retinal layers (Figure 2G,H). The electroretinogram (ERG) records of two patients showed a mild reduced amplitude of the cone response (Figure 2I). All patients had normal color vision based on Ishihara plate screening. No systemic abnormalities were noted in the affected individuals.
Figure 2.
Clinical phenotypes of arHM. A-F: Fundus photos from parents (A, B) and affected (C, E, F) and unaffected (D) offspring. The individual numbers at the lower right corner of each photo are the same as those in Figure 1. All affected patients had a tigroid fundus change and circular choroidal defects around the optic discs. G-I: Results of HRT (G), OCT (H), and ERG (I) from affected individual IV:1 are shown. Normal HRT and OCT and mild reduced cone responses were shown.
Table 1. Clinical data for the family with autosomal recessive high myopia.
| ID | Age at onset (years) | Age at exam (years) | Sex |
Unaided best visual acuity (corrected) |
Refraction |
Axial length (mm) |
Fundus | |||
|---|---|---|---|---|---|---|---|---|---|---|
| OD | OS | OD | OS | OD | OS | |||||
| III:1 |
|
47 |
M |
1.5 |
1.2 |
plano |
-0.25DCx45 |
23.17 |
23.12 |
Normal |
| III:2 |
|
46 |
F |
0.8 |
0.7 |
-1.50DS+2.25DCx170 |
-1.25DCx90 |
22.59 |
22.50 |
Normal |
| VI:1 |
Early childhood |
20 |
F |
0.03 (0.8) |
0.05 (0.8) |
-12.00DS-2.50DCx180 |
-10.50DS-2.50DCx10 |
27.29 |
26.86 |
Myopic |
| VI:2 |
16 |
19 |
M |
0.1 (1.0) |
1.0 |
-2.00DS |
plano |
23.96 |
22.85 |
Normal |
| VI:3 |
Early childhood |
17 |
M |
0.1 (1.0) |
0.2 (1.0) |
-15.50DS-2.50DCx15 |
-12.50DS-3.50DCx170 |
29.4 |
28.24 |
Myopic |
| VI:4 | Early childhood | 16 | M | 0.02 (0.8) | 0.02 (0.8) | -13.00DS-4.00DCx10 | -14.00DS-5.00DCx170 | 28.9 | 29.16 | Myopic |
All three affected individuals had high myopia since early childhood. They had extreme high myopia at examination with apparent elongation of the axial length of both eyes. In the table, “plano” indicates no refractive error or zero.
Linkage to all known loci for high myopia was initially excluded. A genome-wide scan excluded linkage to 313 of the 382 panel markers for chromosomes 1−22 by a LOD score equal to or lower than −2.0. Twenty markers yielding LOD scores between -2.0 and 0 were further excluded from linkage due to heterozygous alleles by homozygosity mapping. For the remaining 49 markers with LOD scores greater than −2.0, markers D7S513 and D12S310 were unsuccessful for genotyping, and 25 of these markers with positive LOD scores of less than 1.0 were not informative where the neighboring markers gave LOD scores of less than −2.0.
Only 22 markers yielded two-point LOD scores greater than 1.0 in a genome-wide scan of chromosomes 1−22 (Table 2). Linkage to most of these markers was not supported by closely flanking markers except for three loci on chromosomes 11, 14, and 17. Fine mapping and haplotype analysis provide strong evidence for a locus on chromosome 14q22.1-q24.2 (Figure 1, Table 3). This locus maps to a 25.23 Mb region between D14S984 and D14S999 with maximum LOD scores of 2.19 at θ=0 for D14S989, D14S980, D14S1038, D14S289, and D14S1025, reaching the theoretical maximum LOD score that could be generated in this type of family. All 11 microsatellite markers examined inside the linkage interval and the haplotype construction of these markers support this locus. However, linkage to regions of chromosomes 11 and 17 could not be excluded due to the small size of the family.
Table 2. Markers yielding a two-point LOD score over 1.0 from a genome-wide scan.
| Markers |
LOD score at θ= |
||||||
|---|---|---|---|---|---|---|---|
| 0.00 | 0.01 | 0.05 | 0.10 | 0.20 | 0.30 | 0.40 | |
| D1S199 |
1.43 |
1.39 |
1.27 |
1.11 |
0.80 |
0.49 |
0.22 |
| D3S1580 |
2.03 |
1.99 |
1.82 |
1.60 |
1.14 |
0.67 |
0.23 |
| D4S406 |
1.12 |
1.10 |
1.00 |
0.88 |
0.62 |
0.38 |
0.17 |
| D4S1597 |
1.43 |
1.39 |
1.27 |
1.11 |
0.80 |
0.49 |
0.22 |
| D7S657 |
2.19 |
2.14 |
1.96 |
1.72 |
1.24 |
0.77 |
0.33 |
| D9S286 |
1.43 |
1.39 |
1.27 |
1.11 |
0.80 |
0.49 |
0.22 |
| D10S249 |
1.12 |
1.10 |
1.00 |
0.88 |
0.62 |
0.38 |
0.17 |
| D10S189 |
2.03 |
1.98 |
1.8 |
1.56 |
1.08 |
0.61 |
0.19 |
|
D11S901 |
1.12 |
1.10 |
1.00 |
0.88 |
0.62 |
0.38 |
0.17 |
|
D11S898 |
1.43 |
1.39 |
1.27 |
1.11 |
0.80 |
0.49 |
0.22 |
|
D11S1320 |
1.43 |
1.39 |
1.27 |
1.11 |
0.80 |
0.49 |
0.22 |
|
D11S968 |
1.43 |
1.39 |
1.27 |
1.11 |
0.80 |
0.49 |
0.22 |
| D13S265 |
1.12 |
1.10 |
1.00 |
0.88 |
0.62 |
0.38 |
0.17 |
|
D14S276 |
1.43 |
1.39 |
1.27 |
1.11 |
0.80 |
0.49 |
0.22 |
|
D14S63 |
2.03 |
1.98 |
1.8 |
1.56 |
1.08 |
0.61 |
0.19 |
|
D14S258 |
1.12 |
1.10 |
1.00 |
0.88 |
0.62 |
0.38 |
0.17 |
| D16S404 |
1.12 |
1.10 |
1.00 |
0.88 |
0.62 |
0.38 |
0.17 |
| D16S3046 |
1.12 |
1.10 |
1.00 |
0.88 |
0.62 |
0.38 |
0.17 |
|
D17S1868 |
1.12 |
1.10 |
1.00 |
0.88 |
0.62 |
0.38 |
0.17 |
|
D17S787 |
2.19 |
2.15 |
1.98 |
1.76 |
1.30 |
0.83 |
0.37 |
| D18S452 |
1.43 |
1.39 |
1.27 |
1.11 |
0.80 |
0.49 |
0.22 |
| D19S571 | 1.43 | 1.39 | 1.27 | 1.11 | 0.80 | 0.49 | 0.22 |
It is common to have several dozen of markers with positive LOD scores in a genome-wide scan in a family of moderate size. A single marker with a LOD score over 1.0 is usually not an indication of suggestive linkage if it is not supported by closely flanking markers. To report all markers with positive LOD scores greater than 1.0 may provide useful clues for subsequent linkage study of other families with arHM. Markers on relevant regions of chromosomes 11, 14, and 17 are highlighted in bold.
Table 3. Two-point linkage results for markers at 14q22.1-q24.2.
| Markers |
Position |
LOD score at θ= |
|||||||
|---|---|---|---|---|---|---|---|---|---|
| cM* | Mb# | 0.00 | 0.01 | 0.05 | 0.10 | 0.20 | 0.30 | 0.40 | |
| D14S984 |
43.60 |
49.17 |
−2.28 |
−0.97 |
−0.39 |
−0.2 |
−0.13 |
−0.15 |
−0.13 |
| D14S978 |
44.20 |
50.98 |
2.03 |
1.98 |
1.80 |
1.56 |
1.08 |
0.61 |
0.19 |
| D14S989 |
46.20 |
52.77 |
2.19 |
2.14 |
1.96 |
1.72 |
1.24 |
0.77 |
0.33 |
| D14S276 |
47.00 |
54.75 |
1.43 |
1.39 |
1.27 |
1.11 |
0.80 |
0.49 |
0.22 |
| D14S980 |
50.90 |
56.22 |
2.19 |
2.14 |
1.96 |
1.72 |
1.24 |
0.77 |
0.33 |
| D14S274 |
53.80 |
56.73 |
1.12 |
1.10 |
1.00 |
0.88 |
0.62 |
0.38 |
0.17 |
| D14S1038 |
56.70 |
58.69 |
2.19 |
2.14 |
1.96 |
1.72 |
1.24 |
0.77 |
0.33 |
| D14S63 |
59.00 |
63.72 |
2.03 |
1.98 |
1.80 |
1.56 |
1.08 |
0.61 |
0.19 |
| D14S1065 |
63.00 |
67.98 |
1.43 |
1.39 |
1.27 |
1.11 |
0.80 |
0.49 |
0.22 |
| D14S258 |
65.80 |
69.65 |
1.12 |
1.10 |
1.00 |
0.88 |
0.62 |
0.38 |
0.17 |
| D14S289 |
67.10 |
70.63 |
2.19 |
2.14 |
1.96 |
1.72 |
1.24 |
0.77 |
0.33 |
| D14S1025 |
71.60 |
73.31 |
2.19 |
2.14 |
1.96 |
1.72 |
1.24 |
0.77 |
0.33 |
| D14S999 | 73.70 | 74.40 | −2.28 | −0.97 | −0.39 | −0.2 | −0.13 | −0.15 | −0.13 |
Fine mapping demonstrates strong evidence of suggestive linkage for a novel arHM locus on chromosome 14q22.1-q24.2. This locus encompasses a 25.23 Mb region between D14S984 and D14S999. All 11 microsatellite markers examined inside the linkage interval support this locus by yielding positive LOD scores, with maximum LOD scores of 2.19 at θ=0 for D14S989, D14S980, D14S1038, D14S289, and D14S1025. The asterisk indicates the genetic position (cM, centimorgan) of each marker based on data from Genethon; The sharp (hash mark) indicates the physical position (Mb, mega base pairs) of each marker according to the database of Homosapiens Build 36.3.
Discussion
In this study, we described a consanguineous Chinese family with three offspring affected with high myopia. All three affected family members presented with extreme high myopia in early childhood, which is characterized by a typical and similar myopic fundus and an obvious elongation of ocular axial length. Other ocular and systemic diseases were excluded. These similar characteristics together with pedigree and family information such as the comparatively normal refraction and fundus in parents suggest that the high myopia in this family is inherited as an autosomal recessive trait. Although arHM has been suggested by population and segregation analysis [36-38], arHM in any consanguineous family has not been previously reported with accompanying clinical and linkage data.
It has been suggested that arHM represents a common form of hereditary high myopia [36-39]. Unfortunately, linkage studies on this type of high myopia have been lacking. Of the 10 loci that have been previously mapped for high myopia, eight are for adHM and the two others for xlHM. None were for arHM. Two major problems may contribute to the difficulty in identifying the exact loci and genes for high myopia [10]. First, are we really able to differentiate hereditary high myopia from acquired high myopia? That is, do we determine the affected and unaffected status correctly in linkage analyses? Second, as high myopia is very common, the introduction of another myopia-related gene into a family such as by marriage can confound the identification of loci and genes. Therefore, a high myopia family with clearly defined phenotypes and minimum environmental impact would be ideal for finding the responsible locus or gene. The family reported in this study has typical hereditary high myopia and a clearly defined phenotype without any overlapping signs with unaffected individuals. This study, to our knowledge, is the first genome-wide linkage study performed for arHM. We have mapped the gene for arHM to a few limited loci, especially a 25.23 Mb region between D14S984 and D14S999 at chromosome 14q22.1-q24.2. Potential candidate genes in this linkage interval may include but are not limited to guanine nucleotide binding protein (G protein), gamma 2 (GNG2), G protein-coupled receptor 135 (GPR135), SIX homeobox 4 (SIX4), and regulator of G-protein signaling 6 (RGS6). Even though the exact locus and gene have not been defined, our study does provide useful clues for identifying the gene for arHM in further studies. Recruitment of additional families could possibly help to define the exact locus, which would lead to the cloning of the causative gene.
Acknowledgments
The authors thank all patients and family members for their participation. This study was supported in part by grants 30572006 and 30772390 from the National Natural Science Foundation of China and grant 7001571 from the Guangdong Natural Science Foundation. Professor Qingjiong Zhang is a recipient of the National Science Fund for Distinguished Young Scholars (30725044).
References
- 1.Feldkamper M, Schaeffel F. Interactions of genes and environment in myopia. Dev Ophthalmol. 2003;37:34–49. doi: 10.1159/000072037. [DOI] [PubMed] [Google Scholar]
- 2.Choo V. A look at slowing progression of myopia. Lancet. 2003;361:1622–3. doi: 10.1016/S0140-6736(03)13315-6. [DOI] [PubMed] [Google Scholar]
- 3.McCarty CA, Taylor HR. Myopia and vision 2020. Am J Ophthalmol. 2000;129:525–7. doi: 10.1016/s0002-9394(99)00444-4. [DOI] [PubMed] [Google Scholar]
- 4.Fan DS, Lam DS, Lam RF, Lau JT, Chong KS, Cheung EY, Lai RY, Chew SJ. Prevalence, incidence, and progression of myopia of school children in Hong Kong. Invest Ophthalmol Vis Sci. 2004;45:1071–5. doi: 10.1167/iovs.03-1151. [DOI] [PubMed] [Google Scholar]
- 5.Paluru P, Ronan SM, Heon E, Devoto M, Wildenberg SC, Scavello G, Holleschau A, Makitie O, Cole WG, King RA, Young TL. New locus for autosomal dominant high myopia maps to the long arm of chromosome 17. Invest Ophthalmol Vis Sci. 2003;44:1830–6. doi: 10.1167/iovs.02-0697. [DOI] [PubMed] [Google Scholar]
- 6.Pararajasegaram R. VISION 2020-the right to sight: from strategies to action. Am J Ophthalmol. 1999;128:359–60. doi: 10.1016/s0002-9394(99)00251-2. [DOI] [PubMed] [Google Scholar]
- 7.Lam CY, Tam PO, Fan DS, Fan BJ, Wang DY, Lee CW, Pang CP, Lam DSA. Genome-wide Scan Maps a Novel High Myopia Locus to 5p15. Invest Ophthalmol Vis Sci. 2008;49:3768–78. doi: 10.1167/iovs.07-1126. [DOI] [PubMed] [Google Scholar]
- 8.Nallasamy S, Paluru PC, Devoto M, Wasserman NF, Zhou J, Young TL. Genetic linkage study of high-grade myopia in a Hutterite population from South Dakota. Mol Vis. 2007;13:229–36. [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang Q, Guo X, Xiao X, Jia X, Li S, Hejtmancik JF. Novel locus for X linked recessive high myopia maps to Xq23-q25 but outside MYP1. J Med Genet. 2006;43:e20. doi: 10.1136/jmg.2005.037853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang Q, Guo X, Xiao X, Jia X, Li S, Hejtmancik JF. A new locus for autosomal dominant high myopia maps to 4q22-q27 between D4S1578 and D4S1612. Mol Vis. 2005;11:554–60. [PubMed] [Google Scholar]
- 11.Paluru PC, Nallasamy S, Devoto M, Rappaport EF, Young TL. Identification of a novel locus on 2q for autosomal dominant high-grade myopia. Invest Ophthalmol Vis Sci. 2005;46:2300–7. doi: 10.1167/iovs.04-1423. [DOI] [PubMed] [Google Scholar]
- 12.Young TL, Deeb SS, Ronan SM, Dewan AT, Alvear AB, Scavello GS, Paluru PC, Brott MS, Hayashi T, Holleschau AM, Benegas N, Schwartz M, Atwood LD, Oetting WS, Rosenberg T, Motulsky AG, King RA. X-linked high myopia associated with cone dysfunction. Arch Ophthalmol. 2004;122:897–908. doi: 10.1001/archopht.122.6.897. [DOI] [PubMed] [Google Scholar]
- 13.Young TL, Ronan SM, Alvear AB, Wildenberg SC, Oetting WS, Atwood LD, Wilkin DJ, King RA. A second locus for familial high myopia maps to chromosome 12q. Am J Hum Genet. 1998;63:1419–24. doi: 10.1086/302111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Young TL, Ronan SM, Drahozal LA, Wildenberg SC, Alvear AB, Oetting WS, Atwood LD, Wilkin DJ, King RA. Evidence that a locus for familial high myopia maps to chromosome 18p. Am J Hum Genet. 1998;63:109–19. doi: 10.1086/301907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schwartz M, Haim M, Skarsholm D. X-linked myopia: Bornholm eye disease. Linkage to DNA markers on the distal part of Xq. Clin Genet. 1990;38:281–6. [PubMed] [Google Scholar]
- 16.Young TL, Atwood LD, Ronan SM, Dewan AT, Alvear AB, Peterson J, Holleschau A, King RA. Further refinement of the MYP2 locus for autosomal dominant high myopia by linkage disequilibrium analysis. Ophthalmic Genet. 2001;22:69–75. doi: 10.1076/opge.22.2.69.2233. [DOI] [PubMed] [Google Scholar]
- 17.Nurnberg G, Jacobi FK, Broghammer M, Becker C, Blin N, Nurnberg P, Stephani U, Pusch CM. Refinement of the MYP3 locus on human chromosome 12 in a German family with Mendelian autosomal dominant high-grade myopia by SNP array mapping. Int J Mol Med. 2008;21:429–38. [PubMed] [Google Scholar]
- 18.Naiglin L, Gazagne C, Dallongeville F, Thalamas C, Idder A, Rascol O, Malecaze F, Calvas P. A genome wide scan for familial high myopia suggests a novel locus on chromosome 7q36. J Med Genet. 2002;39:118–24. doi: 10.1136/jmg.39.2.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang Q, Li S, Xiao X, Jia X, Guo X. Confirmation of a genetic locus for X-linked recessive high myopia outside MYP1. J Hum Genet. 2007;52:469–72. doi: 10.1007/s10038-007-0130-9. [DOI] [PubMed] [Google Scholar]
- 20.Farbrother JE, Kirov G, Owen MJ, Pong-Wong R, Haley CS, Guggenheim JA. Linkage analysis of the genetic loci for high myopia on 18p, 12q, and 17q in 51 U.K. families. Invest Ophthalmol Vis Sci. 2004;45:2879–85. doi: 10.1167/iovs.03-1156. [DOI] [PubMed] [Google Scholar]
- 21.Li T, Xiao X, Li S, Xing Y, Guo X, Zhang Q. Evaluation of EGR1 as a candidate gene for high myopia. Mol Vis. 2008;14:1309–12. [PMC free article] [PubMed] [Google Scholar]
- 22.Hayashi T, Inoko H, Nishizaki R, Ohno S, Mizuki N. Exclusion of transforming growth factor-beta1 as a candidate gene for myopia in the Japanese. Jpn J Ophthalmol. 2007;51:96–9. doi: 10.1007/s10384-006-0417-y. [DOI] [PubMed] [Google Scholar]
- 23.Scavello GS, Paluru PC, Ganter WR, Young TL. Sequence variants in the transforming growth beta-induced factor (TGIF) gene are not associated with high myopia. Invest Ophthalmol Vis Sci. 2004;45:2091–7. doi: 10.1167/iovs.03-0933. [DOI] [PubMed] [Google Scholar]
- 24.Scavello GS, Jr, Paluru PC, Zhou J, White PS, Rappaport EF, Young TL. Genomic structure and organization of the high grade Myopia-2 locus (MYP2) critical region: mutation screening of 9 positional candidate genes. Mol Vis. 2005;11:97–110. [PubMed] [Google Scholar]
- 25.Zhou J, Young TL. Evaluation of Lipin 2 as a candidate gene for autosomal dominant 1 high-grade myopia. Gene. 2005;352:10–9. doi: 10.1016/j.gene.2005.02.019. [DOI] [PubMed] [Google Scholar]
- 26.Zhang Q, Xiao X, Li S, Jia X, Yang Z, Huang S, Caruso RC, Guan T, Sergeev Y, Guo X, Hejtmancik JF. Mutations in NYX of individuals with high myopia, but without night blindness. Mol Vis. 2007;13:330–6. [PMC free article] [PubMed] [Google Scholar]
- 27.Tang WC, Yap MK, Yip SP. A review of current approaches to identifying human genes involved in myopia. Clin Exp Optom. 2008;91:4–22. doi: 10.1111/j.1444-0938.2007.00181.x. [DOI] [PubMed] [Google Scholar]
- 28.Young TL, Metlapally R, Shay AE. Complex trait genetics of refractive error. Arch Ophthalmol. 2007;125:38–48. doi: 10.1001/archopht.125.1.38. [DOI] [PubMed] [Google Scholar]
- 29.Morgan I, Rose K. How genetic is school myopia? Prog Retin Eye Res. 2005;24:1–38. doi: 10.1016/j.preteyeres.2004.06.004. [DOI] [PubMed] [Google Scholar]
- 30.Marmor MF, Holder GE, Seeliger MW, Yamamoto S. Standard for clinical electroretinography (2004 update). Doc Ophthalmol. 2004;108:107–14. doi: 10.1023/b:doop.0000036793.44912.45. [DOI] [PubMed] [Google Scholar]
- 31.Lathrop GM, Lalouel JM. Easy calculations of lod scores and genetic risks on small computers. Am J Hum Genet. 1984;36:460–5. [PMC free article] [PubMed] [Google Scholar]
- 32.Schaffer AA, Gupta SK, Shriram K, Cottingham RW., Jr Avoiding recomputation in linkage analysis. Hum Hered. 1994;44:225–37. doi: 10.1159/000154222. [DOI] [PubMed] [Google Scholar]
- 33.Faivre L, Le Merrer M, Al-Gazali LI, Ausems MG, Bitoun P, Bacq D, Maroteaux P, Munnich A, Cormier-Daire V. Homozygosity mapping of a Desbuquois dysplasia locus to chromosome 17q25.3. J Med Genet. 2003;40:282–4. doi: 10.1136/jmg.40.4.282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Weigell-Weber M, Sarra GM, Kotzot D, Sandkuijl L, Messmer E, Hergersberg M. Genomewide homozygosity mapping and molecular analysis of a candidate gene located on 22q13 (fibulin-1) in a previously undescribed vitreoretinal dystrophy. Arch Ophthalmol. 2003;121:1184–8. doi: 10.1001/archopht.121.8.1184. [DOI] [PubMed] [Google Scholar]
- 35.Lander E, Kruglyak L. Genetic dissection of complex traits: guidelines for interpreting and reporting linkage results. Nat Genet. 1995;11:241–7. doi: 10.1038/ng1195-241. [DOI] [PubMed] [Google Scholar]
- 36.Edwards M, Lewis WH. Autosomal recessive inheritance of myopia in Hong Kong Chinese infants. Ophthalmic Physiol Opt. 1991;11:227–31. [PubMed] [Google Scholar]
- 37.Karlsson JL. Influence of the myopia gene on brain development. Clin Genet. 1975;8:314–8. doi: 10.1111/j.1399-0004.1975.tb01508.x. [DOI] [PubMed] [Google Scholar]
- 38.Yu ZQ, Fu CW, Shen FM, Chu RY. Pedigree study of pathological myopia. Yi Chuan Xue Bao. 2005;32:130–5. [PubMed] [Google Scholar]
- 39.Chu R, Ni P, Ni M, Shen F. Genetic epidemiology study of pathological myopia. Zhonghua Yi Xue Yi Chuan Xue Za Zhi. 2000;17:178–80. [PubMed] [Google Scholar]