Native and selenomethionine-labeled crystals of Ebola VP35 interferon inhibitory domain were obtained by the hanging-drop vapor-diffusion method.
Keywords: Ebola VP35, interferon inhibitory domain, antiviral antagonists
Abstract
Ebola VP35 is a multifunctional protein that is important for host immune suppression and pathogenesis. VP35 contains an N-terminal oligomerization domain and a C-terminal interferon inhibitory domain (IID). Mutations within the VP35 IID result in loss of host immune suppression. Here, efforts to crystallize recombinantly overexpressed VP35 IID that was purified from Escherichia coli are described. Native and selenomethionine-labeled crystals belonging to the orthorhombic space group P212121 were obtained by the hanging-drop vapor-diffusion method and diffraction data were collected at the ALS synchrotron.
1. Introduction
Rare but deadly outbreaks of Ebola virus (EBOV) can cause an infection that leads to severe hemorrhagic fever owing to simultaneous subversion of the host immune system and enhancement of viral replication (Feldmann et al., 1993 ▶). EBOV is a nonsegmented negative-strand RNA virus (NNSV) that can strike both the host innate and adaptive immune systems like other NNSV viruses such as influenza, rabies and measles. Owing to the relatively small viral genome size, many virally encoded proteins perform multiple functions, often at different stages of the viral replication cycle.
Ebola viral protein 35 (VP35) is a multifunctional protein that is required for efficient host innate immune inhibition and viral pathogenesis. Decreased VP35 activity leads to lower viral amplification rates and reduced lethality in mice (Basler et al., 2003 ▶; Cardenas et al., 2006 ▶; Feng et al., 2007 ▶; Hartman, Bird et al., 2008 ▶; Enterlein et al., 2006 ▶). Recent studies have shown that the VP35 N-terminus is required for oligomerization (Moller et al., 2005 ▶; Reid et al., 2005 ▶), whereas the C-terminus is involved in the suppression of interferon (IFN) activity (Cardenas et al., 2006 ▶; Feng et al., 2007 ▶; Hartman et al., 2004 ▶). Furthermore, oligomerization-defective mutants fail to interact with the viral polymerase (L) protein and display reduced IFN inhibition, suggesting that the VP35 N-terminus is necessary for viral replication (Cardenas et al., 2006 ▶; Hartman et al., 2006 ▶). Interestingly, diminished IFN inhibition can largely be compensated by overexpression of the isolated VP35 C-terminus or by artificial tethering of the C-terminus to a heterologous oligomerization module (Reid et al., 2005 ▶). These results suggest that the N-terminus provides a critical oligomerization function, which facilitates efficient IFN inhibition through elements located at the C-terminus of VP35. Studies thus far show that the VP35 C-terminus can also bind dsRNA and that VP35-mediated IFN antagonism correlates with dsRNA-binding activity. Consistent with these observations, the mutation of a highly conserved basic residue, Arg312, leads to loss of VP35 dsRNA binding and IFN suppression. Moreover, viruses containing the Arg312Ala mutation activate IRF-3 and induce a stronger immune response compared with wild-type viruses. Interestingly, the same mutation does not significantly affect VP35 function as part of the viral polymerase complex, suggesting that the IFN inhibition and viral replication functions of VP35 may be uncoupled (Cardenas et al., 2006 ▶; Hartman, Bird et al., 2008 ▶; Hartman, Ling et al., 2008 ▶; Hartman et al., 2004 ▶).
The importance of VP35 in performing multiple functions, including immune suppression through its C-terminus, has been well documented (Basler et al., 2003 ▶; Hartman, Ling et al., 2008 ▶). However, the mechanism(s) by which VP35 inhibits IFN activity and a description of the structural properties of VP35, are currently lacking. In order to address these issues, we have initiated efforts to characterize the C-terminal region of Ebola VP35. Here, we report the expression, purification, crystallization and preliminary X-ray diffraction analysis of the VP35 interferon inhibitory domain (IID). The Ebola VP35 IID sequence is highly conserved among various isolates from the Filoviridae family and the structural information gathered here has the potential to provide information on related viral proteins from Ebola and Marburg viruses.
2. Materials and methods
2.1. Cloning and expression
We generated PCR products for subcloning the VP35 IID coding region (residues 215–340) into a modified pET15b vector (Novagen) by using a coding region for VP35 from the Ebola Zaire strain Mayinga (Gene ID 911827; Basler et al., 2000 ▶) as a template. The VP35 IID sequence was cloned immediately 3′ to the maltose-binding protein (MBP residues 1–496; GenBank accession No. AAB87675) fusion tag sequence and the tobacco etch virus protease recognition sequence. The resulting vector was verified by sequencing prior to transformation of Escherichia coli BL21(DE3) cells (Novagen). Protein expression for native crystals was carried out in cells grown in LB media and protein for MAD data collection was generated from cells grown in minimal media with appropriately labeled metabolites introduced following established protocols. Bacterial cells were cultured at 310 K. Protein expression was induced at an optical density of 0.8 at 600 nm with 0.5 mM IPTG and cells were grown overnight at 291 K.
2.2. Protein purification
Cells were harvested and resuspended in lysis buffer (25 mM sodium phosphate pH 7.0, 1 M NaCl, 20 mM imidazole and 5 mM β-mercaptoethanol). Resuspended cells were flash-frozen in liquid nitrogen, stored at 193 K for at least 2 h and subsequently thawed in an ice–water bath. Thawed cells were lysed using an EmulsiFlex-C5 homogenizer (Avestin) and clarified by centrifugation at 30 000g at 277 K for 30 min. The resulting supernatant was applied onto a 15 ml amylose column (XK 26/20 column, GE Healthcare), washed with lysis buffer and eluted with lysis buffer plus 1% maltose. The eluted protein was diluted with 25 mM sodium phosphate pH 7.0 and 5 mM β-mercaptoethanol to a final NaCl concentration of ∼50 mM (approximately 20-fold dilution by volume) and immediately loaded onto a strong cation-exchange column (8 ml Source 15S packed in a Tricorn 10/100 column, GE Heathcare) in buffer SA (25 mM sodium phosphate pH 7.0, 50 mM NaCl and 5 mM β-mercaptoethanol) and eluted with buffer SB (buffer SA with 1.0 M NaCl). The MBP fusion tag was removed prior to final purification by incubation with recombinant tobacco etch virus (rTEV) protease for 3–6 h at 277 K, which resulted in the presence of three additional residues (Gly-His-Met) at the N-terminus of the VP35 IID construct. Cleaved protein samples were further purified by 2× size-exclusion chromatography (Superdex 75 HR 10/300 GL, GE Healthcare) equilibrated with 20 mM Tris–HCl pH 7.0, 50 mM NaCl and 5 mM β-mecaptoethanol. The purity of the samples was assessed at each step by Coomassie staining of SDS–PAGE gels.
2.3. Dynamic light scattering
Protein samples were centrifuged for at least 10 min at 14 000g prior to dynamic light-scattering (DLS) experiments. DLS studies were performed on a DynaPro801 DLS instrument (Protein Solutions Inc.). DLS data were collected and analyzed using DYNAMICS v.6.3.01 software. All data were collected at 298 K and at least 20 scans were gathered for analysis.
2.4. Crystallization
Initial conditions for crystallization were identified using a commercial screen (Hampton Research) and optimized using solutions generated in-house. Native and selenomethionine-labeled (SeMet) crystals were grown at 298 K using the hanging-drop vapor-diffusion method with 17 mg ml−1 protein solution in the size-exclusion chromatographic buffer, which was diluted in a 1:1 ratio with the well solution. Crystals from the optimized solutions were soaked for approximately 60 s in a reservoir solution containing well solution plus glycerol to a final concentration of 25%(w/v) and vitrified in a nitrogen stream.
2.5. Data collection and processing
Diffraction data for native and SeMet proteins were collected from single crystals at the Advanced Light Source (beamline 4.2.2) at 100 K on a CCD detector. 360 frames were recorded at a crystal-to-detector distance of 95 mm using an oscillation range of 0.5°. For SeMet crystals, complete anomalous sets were obtained at wavelengths corresponding to the peak absorbance (0.9792 Å), the inflection point (0.9795 Å) and a remote wavelength (0.9643 Å) from the absorption edge of selenium. All diffraction data were indexed, integrated, scaled and merged using d*TREK (Pflugrath, 1999 ▶). Intensities were converted to structure factors using the CCP4 program TRUNCATE (Collaborative Computational Project, Number 4, 1994 ▶; French & Wilson, 1978 ▶).
3. Results and discussion
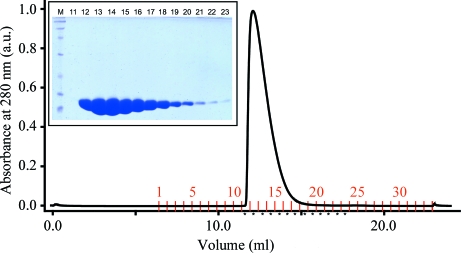
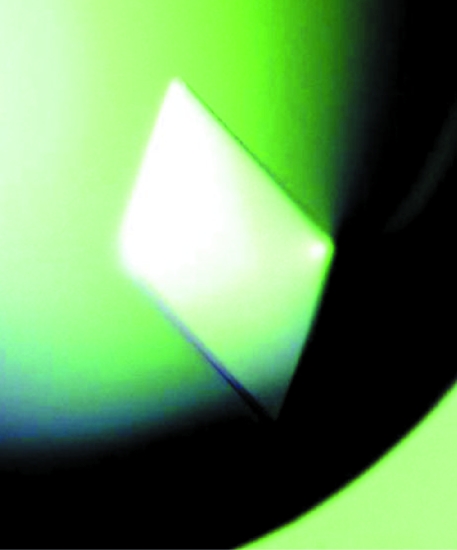
VP35 IID protein was purified using multiple affinity, ion-exchange and gel-filtration chromatographic steps, which produced a homogeneous protein sample as visualized by a Coomassie-stained SDS–PAGE assay (Fig. 1 ▶). We obtained final yields of ∼8 and 4 mg l−1 for the native and SeMet proteins, respectively. The sample homogeneity of purified VP35 IID was further assessed by DLS experiments, which showed near 100% monodispersity with a hydrodynamic radius of 19.5 Å and a calculated molecular weight of 16 kDa. Crystals grew within 2–4 d to dimensions of 40 × 100 × 200 µm in the best well solution condition, which contained 200 mM sodium citrate pH 5.8 and 11%(w/v) PEG 4000. The Matthews coefficient was determined to be 2.17 Å3 Da−1 (Matthews, 1968 ▶), with a solvent content of 43% and two molecules in the asymmetric unit cell. Both SeMet and native proteins produced crystal plates belonging to space group P212121 (Fig. 2 ▶). The native and SeMet crystals diffracted to 1.4 Å resolution at the ALS synchrotron. Data were collected to >96% completeness and the data-collection statistics are summarized in Table 1 ▶. A detailed description of the VP35 IID structure, including the determination of experimental phases, refinement and model building, is published elsewhere (Leung et al., 2009 ▶). Analysis of the VP35 IID structure will provide opportunities to identify the structural features that contribute to virulence and for targeted design of antiviral drugs against Ebola VP35.
Figure 1.
Gel-filtration chromatography and SDS–PAGE analysis. Chromatogram of the Superdex 75 10/300 GL gel-filtration column. Inset, SDS–PAGE analysis of the corresponding column fractions in the chromatogram.
Figure 2.
A representative native crystal of Ebola VP35 IID. The crystal dimensions are approximately 40 × 100 × 200 µm.
Table 1. Data collection.
Values in parentheses are for the highest resolution shell.
| SeMet | ||||
|---|---|---|---|---|
| Native | Peak | Inflection | Remote | |
| Space group | P212121 | P212121 | ||
| Unit-cell parameters (Å, °) | a = 51.49, b = 66.21, c = 72.13, α = β = γ = 90 | a = 51.50, b = 66.19, c = 72.74, α = β = γ = 90 | ||
| Wavelength (Å) | 0.9795 | 0.9792 | 0.9795 | 0.9643 |
| Resolution (Å) | 41.91–1.40 (1.45–1.40) | 42.03–1.40 (1.45–1.40) | 42.03–1.40 (1.45–1.40) | 42.03–1.40 (1.45–1.40) |
| Rmerge† (%) | 6.1 (44.1) | 8.5 (44.6) | 8.9 (46.9) | 9.6 (47.1) |
| Average I/σ(I) | 11.0 (2.4) | 9.7 (2.3) | 9.9 (2.1) | 9.2 (2.3) |
| Completeness (%) | 99.7 (98.8) | 96.7 (80.1) | 96.6 (79.9) | 97.5 (85.3) |
| Redundancy | 6.65 (4.91) | 6.43 (4.49) | 6.44 (4.51) | 6.52 (4.79) |
R
merge = 
 .
.
Acknowledgments
We thank Drs D. Klein, J. Rutter and D. Borek for support and discussions, Dr J. Hoy for assistance with initial X-ray data collection and Dr B. Fulton for initial NMR data collection. We also thank P. Ramanan, L. Helgeson, D. Peterson and M. Farahbakhsh for laboratory assistance and the Iowa State University Office of Biotechnology Facilities (DNA, Macromolecular X-ray Crystallography, Nuclear Magnetic Resonance and Protein Facilities). This work was supported in part by the Roy J. Carver Charitable Trust Research Grant 09-3271 (to GKA), a Roy J. Carver Charitable Trust Graduate Fellowship (to NDG) and National Institutes of Health Grant AI059536 (to CFB).
References
- Basler, C. F., Mikulasova, A., Martinez-Sobrido, L., Paragas, J., Muhlberger, E., Bray, M., Klenk, H. D., Palese, P. & Garcia-Sastre, A. (2003). J. Virol.77, 7945–7956. [DOI] [PMC free article] [PubMed]
- Basler, C. F., Wang, X., Muhlberger, E., Volchkov, V., Paragas, J., Klenk, H. D., Garcia-Sastre, A. & Palese, P. (2000). Proc. Natl Acad. Sci. USA, 97, 12289–12294. [DOI] [PMC free article] [PubMed]
- Cardenas, W. B., Loo, Y. M., Gale, M. Jr, Hartman, A. L., Kimberlin, C. R., Martinez-Sobrido, L., Saphire, E. O. & Basler, C. F. (2006). J. Virol.80, 5168–5178. [DOI] [PMC free article] [PubMed]
- Collaborative Computational Project, Number 4 (1994). Acta Cryst. D50, 760–763.
- Enterlein, S., Warfield, K. L., Swenson, D. L., Stein, D. A., Smith, J. L., Gamble, C. S., Kroeker, A. D., Iversen, P. L., Bavari, S. & Muhlberger, E. (2006). Antimicrob. Agents Chemother.50, 984–993. [DOI] [PMC free article] [PubMed]
- Feldmann, H., Klenk, H. D. & Sanchez, A. (1993). Arch. Virol. Suppl.7, 81–100. [DOI] [PubMed]
- Feng, Z., Cerveny, M., Yan, Z. & He, B. (2007). J. Virol.81, 182–192. [DOI] [PMC free article] [PubMed]
- French, S. & Wilson, K. (1978). Acta Cryst. A34, 517–525.
- Hartman, A. L., Bird, B. H., Towner, J. S., Antoniadou, Z. A., Zaki, S. R. & Nichol, S. T. (2008). J. Virol.82, 2699–2704. [DOI] [PMC free article] [PubMed]
- Hartman, A. L., Dover, J. E., Towner, J. S. & Nichol, S. T. (2006). J. Virol.80, 6430–6440. [DOI] [PMC free article] [PubMed]
- Hartman, A. L., Ling, L., Nichol, S. T. & Hibberd, M. L. (2008). J. Virol.82, 5348–5358. [DOI] [PMC free article] [PubMed]
- Hartman, A. L., Towner, J. S. & Nichol, S. T. (2004). Virology, 328, 177–184. [DOI] [PubMed]
- Leung, D. W., Ginder, N. D., Fulton, D. B., Nix, J., Basler, C. F., Honzatko, R. B. & Amarasinghe, G. K. (2009). Proc. Natl Acad. Sci. USA, 106, 411–416. [DOI] [PMC free article] [PubMed]
- Matthews, B. W. (1968). J. Mol. Biol.33, 491–497. [DOI] [PubMed]
- Moller, P., Pariente, N., Klenk, H. D. & Becker, S. (2005). J. Virol.79, 14876–14886. [DOI] [PMC free article] [PubMed]
- Pflugrath, J. W. (1999). Acta Cryst. D55, 1718–1725. [DOI] [PubMed]
- Reid, S. P., Cardenas, W. B. & Basler, C. F. (2005). Virology, 341, 179–189. [DOI] [PMC free article] [PubMed]