Abstract
A broad region on chromosome 12p13 has been intensely investigated for novel genetic variants associated with Alzheimer disease (AD). We examined this region with 23 microsatellite markers using 124 North European (NE) families and 209 Caribbean Hispanic families with late-onset AD (FAD). Significant evidence for linkage was present in a 5 cM interval near 20 cM in both the NE FAD (LOD=3.5) and the Caribbean Hispanic FAD (LOD=2.2) datasets. We further investigated these families and an independent NE case-control dataset using 14 single nucleotide polymorphisms (SNPs). The initial screening of the region at ~20 cM in the NE case-control dataset revealed significant association between AD and seven SNPs in several genes, with the strongest result for rs2532500 in TAPBPL (p=0.006). For rs3741916 in GAPDH, the C allele, rather than the G allele as was observed by Li and colleagues (2004), was the risk allele. When the two family datasets were examined, none of the SNPs were significant in NE families, but two SNPs were associated with AD in Caribbean Hispanics: rs740850 in NCAPD2 (p=0.0097) and rs1060620 in GAPDH (p=0.042). In a separate analysis combining the Caribbean Hispanic families and NE cases and controls, rs740850 was significant after correcting for multiple testing (empirical p=0.0048). Subsequent haplotype analyses revealed that two haplotype sets -- haplotype C-A at SNPs 6-7 within NCAPD2 in Caribbean Hispanics, and haplotypes containing C-A-T at SNPs 8-10 within GAPDH in Caribbean Hispanic family and NE case-control datasets -- were associated with AD. Taken together, these SNPs may be in linkage disequilibrium with a pathogenic variant(s) on or near NCAPD2 and GAPDH.
Keywords: Alzheimer disease, GAPDH, NCAPD2, linkage, association
INTRODUCTION
Alzheimer disease (AD) occurs as a late-onset disorder with increasing frequency after age 65 years. Genome-wide linkage analyses have identified several loci as potential sites containing late-onset AD susceptibility genes [1-3]. One of the most consistent results has been on chromosome 12p. Linkage was originally reported at ~45 cM (LOD=3.5) [3]. Previously, we confirmed this locus using 53 North European families multiply affected by AD with the strongest evidence for linkage near the markers D12S358 (26 cM) and D12S96 (68 cM) [4], and in 79 Caribbean Hispanic families where two-point linkage analysis using affected sib pairs yielded LOD scores of 3.15 at D12S1623 (16 cM) and 1.43 at D12S1042 (49 cM) [5]. Although several genes on chromosome 12 have been implicated as potential sites for AD-associated variants (http://www.alzgene.org), none have been confirmed.
It is not surprising that several independent studies have reported significant support for linkage in different but nearby regions of chromosome 12p, given the complex underlying biology of AD. It has been shown that the actual disease locus can reside as far as 20 cM away from the markers with the maximum LOD scores [6, 7]. For example, after fine mapping and genotyping additional family members [3], Scott and colleagues [8] found that support for linkage expanded to include ~40 cM over both arms of the chromosome between 26 cM (D12S358 LOD=2.5) and 67 cM (D12S390 LOD=2.0). Thus, potentially the AD-linked locus includes the entire 67 cM interval on chromosome 12p.
In this study, we fine mapped the chromosome 12 locus using linkage analysis with 23 microsatellite markers (14 cM - 83 cM) in two expanded groups of families multiply affected by late-onset AD (FAD) consisting of 124 North European and 209 Caribbean Hispanic pedigrees. In the regions with the highest support for linkage, we analyzed 14 single nucleotide polymorphisms (SNPs) that were chosen based on a report of significant association between sporadic AD and variations in or nearby the glyceraldehyde-3-phosphate dehydrogenase gene (GAPDH) [9].
MATERIALS AND METHODS
Participants
We used two FAD datasets: 124 North European families and 209 Caribbean Hispanic families. The North European FAD dataset included 685 family members, and the Caribbean Hispanic FAD dataset included 1,147 family members who were recruited in the Dominican Republic, Puerto Rico and in New York City. The clinical characteristics of the North European FAD [4] and Caribbean Hispanic FAD [10] datasets have been previously described and have a mean age at onset of 70 (SD=9) and 73 (SD=11) years, respectively. The third cohort consisted of 183 patients with AD (36% males) and 224 controls (41% males) of North European ancestry (Britain, France, Germany) drawn from the same populations as the North European FAD dataset [11]. The mean age-at-onset for cases was 76 years (SD=7), and the mean age at follow-up for unrelated controls was 73 years (SD=8). The characteristics of the three datasets are summarized in Table 1.
Table 1.
Demographic and clinical characteristics of the participants in the three independent datasets.
| Characteristic | Family Dataset | Case-Control Dataset | ||
|---|---|---|---|---|
| North European | Caribbean Hispanic | NE Cases | NE Controls | |
| Total number of AD families | 124 | 209 | N/A | N/A |
| Total Number of Subjects | 685 | 1,147 | 183 | 224 |
| Number Patients with AD | 321 | 589 | N/A | N/A |
| Number At-risk Relatives | 342 | 499 | N/A | N/A |
| Number Nuclear sibships (>2 siblings genotyped)* | 163 | 272 | N/A | N/A |
| Number Patients per family, mean (range) | 2.6 (1-6) | 2.85(1-12) | N/A | N/A |
| Number At-risk persons per family, mean (range) | 2.8 (0-18) | 2.41(0-15) | N/A | N/A |
| Mean age at onset (years) | 70 ± 9 | 73±11 | 76±7 | 73±8 |
| Autopsy Confirmation (one per family) | 58 (50%) | 5 (2.2%) | 0 (0%) | 0 (0%) |
The diagnosis of AD was based on direct examination, guided by the National Institute of Neurological Disorders and Stroke-Alzheimer's Disease and Related Disorders Association (NINCDS-ADRDA) diagnosis criteria [12]. Informed consent was obtained from all individuals participating in the study.
Genotyping
Genomic DNA was isolated from blood samples using a QIAGEN kit. Prior to performing linkage analysis, we checked for Mendelian inconsistencies in marker data using the PEDCHECK program [13]. In the North European and Caribbean Hispanic FAD datasets, we genotyped 23 microsatellite markers mapped on chromosome 12 at an average inter-marker distance of ~3.1 cM (14 cM - 83 cM) according to the Marshfield Map set 13 (http://research.marshfieldclinic.org/genetics) (Table 2). The PCR conditions for genotyping of the microsatellite makers were obtained from the Genome Database (www.gdb.org). For the SNP-based study, we focused on the 167 kb interval at 20 cM of chromosome 12p13, which was prioritized based on the strongest support for linkage in our FAD datasets and the report of significant association between sporadic AD and variations nearby GAPDH [9]. From a public database (www.ncbi.nlm.nih.gov/SNP/), we identified 14 SNPs in the intragenic sequence of seven genes with minor allele frequencies >8%. This selection shown in Table 3 included five SNPs that were significant in an earlier study [9]. SNP genotyping was performed using the GenomeLab SNPstream System (Beckman Coulter Inc., Fullerton, CA) [14]. Primer sequences were designed using software provided at www.autoprimer.com (Beckman Coulter Inc., Fullerton, CA) and are available upon request. APOE genotyping was performed as previously described [15]. A subset of 100 samples was genotyped twice for every SNP with a concordance rate of 99%.
Table 2.
Results for the two-point linkage analysis under autosomal dominant (A) and recessive (B) models (affecteds-only model) for the North European (NE) and Caribbean Hispanic (CH) samples. Markers with LOD>1 are in bold. (C) The position in cM is based on the Kosambi map.
| Order | Marker Name | cM(C) | North Europeans | Caribbean Hispanics | ||
|---|---|---|---|---|---|---|
| 2-pt-LOD Dom(A) | 2-pt-LOD Rec(B) | 2-pt-LOD Dom(A) | 2-pt-LOD Rec(B) | |||
| 1 | D12S356 | 14.23 | 0.23 | 0.71 | 0.57 | 0.71 |
| 2 | D12S374 | 14.53 | 0.58 | 0.37 | 0.15 | 0.00 |
| 3 | D12S1623 | 15.69 | 0.13 | 0.10 | 2.21 | 2.07 |
| 4 | D12S397 | 17.72 | 0.53 | 0.63 | 0.45 | 0.01 |
| 5 | D12S1695 | 19.68 | 3.46 | 2.92 | 0.00 | 0.00 |
| 6 | D12S77 | 20.27 | 0.40 | 0.61 | 1.83 | 1.53 |
| 7 | D12S89 | 23.41 | 0.00 | 0.01 | 0.89 | 0.67 |
| 8 | D12S391 | 26.23 | 0.05 | 0.24 | 0.02 | 0.00 |
| 9 | D12S358 | 26.33 | 0.04 | 0.14 | 0.44 | 0.11 |
| 10 | D12S269 | 30.60 | 0.14 | 0.12 | 0.58 | 0.13 |
| 11 | D12S373 | 36.06 | 0.00 | 0.00 | 0.00 | 0.00 |
| 12 | D12S1042 | 48.70 | 0.08 | 0.47 | 0.95 | 1.09 |
| 13 | D12S1090 | 56.38 | 0.00 | 0.00 | 0.00 | 0.00 |
| 14 | D12S1701 | 62.54 | 0.63 | 1.32 | 0.00 | 0.00 |
| 15 | D12S390 | 67.63 | 0.16 | 0.06 | 0.00 | 0.00 |
| 16 | D12S96 | 68.16 | 1.70 | 1.58 | 0.00 | 0.00 |
| 17 | D12S398 | 68.46 | 0.28 | 0.38 | 0.00 | 0.03 |
| 18 | D12S1632 | 71.61 | 1.91 | 1.60 | 0.00 | 0.00 |
| 19 | D12S90 | 72.61 | 0.00 | 0.00 | 0.00 | 0.00 |
| 20 | D12S83 | 75.17 | 0.56 | 0.59 | 0.00 | 0.00 |
| 21 | D12S375 | 80.52 | 0.04 | 0.00 | 0.00 | 0.03 |
| 22 | D12S1722 | 82.12 | 0.03 | 0.07 | 0.00 | 0.00 |
| 23 | D12S92 | 83.19 | 0.91 | 0.67 | 0.04 | 0.18 |
Table 3.
Single Nucleotide Polymorphisms (SNPs) used in this study and the association analysis (significant results are in bold). The results for the family-based association analysis of the North European (NE) and Caribbean Hispanic (CH) FAD families, and for the two-point analysis of the NE case-control dataset. (A) Minor Allele Frequency. (B) SNPs genotyped by Li et al study (2004). (C) SNPs genotyped by Lin et al. study (2006). (D) rs3741916 is now known as rs1136666. (E) Not Informative. SNP information was obtained from the NCBI site (www.ncbi.nlm.nih.gov/SNP, NCBI build 35; www.genome.ucsc.edu, Assembly May 2004).
| Order | Gene Name | SNP Function | RS # | Chrom. Position | Distance (kb) | NE Case Control Dataset | NE FAD | CH FAD | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| p-value |
|||||||||||||
| Genotype | Allele | MAFa Cases | MAF Controls | Associated Allele | p-value FBAT | p-value FBAT | Associated Allele | ||||||
| 1 | TAPBPL | V146M | 2532501 | 6,433,014 | 0.047 | 0.121 | 0.40 | 0.34 | A | 0.3570 | 0.1377 | A | |
| 2 | TAPBPL | A165T | 2532500 | 6,433,071 | 0.06 | 0.006 | 0.043 | 0.43 | 0.34 | T | 0.3670 | 0.3293 | T |
| 3 | PKP2P1 | 5'UTR | 2008134B,C | 6,458,706 | 25.64 | 0.975 | 0.881 | 0.29 | 0.30 | 0.9300 | 0.4169 | ||
| 4 | NCAPD2 | intronic | 2072373B | 6,502,149 | 43.44 | 0.109 | 0.289 | 0.12 | 0.14 | 0.4560 | 0.6144 | ||
| 5 | NCAPD2 | intronic | 7311174B,C | 6,505,828 | 3.68 | 0.561 | 0.784 | 0.29 | 0.28 | 0.3850 | 0.4629 | ||
| 6 | NCAPD2 | R1100R | 2072374B,C | 6,508,106 | 2.28 | 0.686 | 0.780 | 0.29 | 0.28 | 0.3960 | 0.4298 | ||
| 7 | NCAPD2 | N1161N | 740850 | 6,508,377 | 0.27 | 0.039 | 0.046 | 0.40 | 0.33 | A | 0.2950 | 0.0097 | A |
| 8 | GAPDH | 5'UTR | 3741916B,C,D | 6,514,252 | 5.88 | 0.054 | 0.027 | 0.14 | 0.21 | C | 0.2710 | 0.1982 | C |
| 9 | GAPDH | intron | 3741918 C | 6,514,517 | 0.27 | 0.032 | 0.030 | 0.14 | 0.17 | A | 0.1030 | 0.6071 | A |
| 10 | GAPDH | intron | 1060620 C | 6,514,983 | 0.47 | 0.432 | 0.795 | 0.17 | 0.18 | T | 0.5050 | 0.0418 | T |
| 11 | GAPDH | intron | 1060619 C | 6,515,042 | 0.06 | 0.180 | 0.254 | 0.19 | 0.19 | 0.1030 | 0.9802 | ||
| 12 | HOM-TES-103 | intron | 740852 | 6,520,587 | 5.55 | 0.180 | 0.970 | 0.08 | 0.07 | 0.1950 | 0.6076 | ||
| 13 | CHD4 | D139E | 1639122 | 6,581,408 | 60.82 | 0.065 | 0.018 | 0.24 | 0.33 | A | 0.6870 | 0.2248 | A |
| 14 | GPR92 | A153A | 3741924 | 6,600,217 | 18.81 | 0.025 | 0.004 | 0.04 | 0.07 | T | N/IE | N/IE | |
Statistical Analyses
Linkage analysis
We conducted two-point linkage analyses, considering both dominant and recessive modes of inheritance under an affecteds-only model [16, 17]. For all linkage analyses, we used microsatellite markers only, and assumed a susceptibility allele frequency of 0.001, and penetrance values of 0.001 for gene carriers and 0.0 for non-carriers. Although these affecteds-only parameters are artificial, they lead to statistical tests with properties that are comparable to `model-free' analyses when studying common diseases [18]. The analysis was implemented using the MLINK program from the FASTLINK package [19, 20].
We then performed a multipoint affected sibpair linkage analysis using GENEHUNTER (version 2.1) to increase the information content at a given chromosomal location. For this analysis, we combined the Hispanic and North European FAD datasets. To take into account differences in allele frequency between the ethnic groups, we treated one marker as two tightly linked markers (i.e., θ=0.0001 between members of each marker pair), and assigned one to each ethnic group. We then computed ethnic-specific allele frequencies for each marker. We used the weighted `all pairs' option, and set the increment function to scan at 1.0 cM. The sibpair analysis calculated the probability of sharing zero, one, or two alleles (z0, z1, or z2) identical by descent (IBD) between sib pairs, because susceptibility will have probabilities (z0, z1, and z2) that differ from the expected Mendelian proportions [21, 22]. The LOD score was computed under the assumption of dominance, but restricting the parameters to Holmans's Triangle Inequality: 2z0≤z1≤½ and z0+z1+z2=1 [23, 24].
Association analysis of the case-control data
Prior to the allelic and genotypic association analyses, we assessed the SNP genotype data for deviation from Hardy-Weinberg equilibrium in the controls from the case-control dataset and in the founders from the FAD datasets using the PEDSTAT program [25]. For all association analyses, only the SNP markers were used. We compared the allelic and genotypic association in the case-control dataset using the conventional χ2 test or Fisher's exact test when the expected frequency of one or more cells was fewer than five [26]. In addition, we adjusted for covariates APOE ε4, sex, and age-at-onset in patients, and age at the last examination in controls using a multivariate logistic regression analysis. For this analysis, we dichotomized the SNP genotype as either having at least one or no copy of the minor allele. Similarly, an APOE ε4 carrier was defined as an individual having one or more APOE ε4 alleles. The association was considered significant if the nominal p-value was below 0.05 in two independent datasets. Haplotype associations were assessed using HAPLO.STATS which computes a haplotype specific empirical p-value for each haplotype and a permutation-based global p-value for the haplotype set [27].
Association analysis of the family data
Association in the FAD data sets was evaluated using FBAT version 1.7.2 [28]. Because our previous studies showed significant linkage in this region [4, 5], we computed the empirical variance function in the FBAT program to test the null hypothesis of no association in the presence of linkage. An additive genetic model was assumed throughout the study. Haplotype analyses using a sliding window approach in which overlapping sets of two to three contiguous SNPs were conducted in the Northern European case-control dataset and Caribbean Hispanic FAD dataset because a number of SNPs in these two datasets showed nominally significant association in the two-point analysis. We computed empirical p-values for haplotype-specific p-values, and permutation based global p-values to adjust for multiple testing for a specific haplotype set. To minimize the risk of false positive findings from haplotype analysis, we performed a multi-marker analysis using SNPs that were significant in haplotype analysis. This analysis allows a test of the null hypothesis without the required assumption of no recombination between the SNPs [29]. We then conducted an APOE ε4-positive conditional analysis because earlier linkage analyses of the Caribbean Hispanic families [5] and Caucasian families [30] revealed possible influence of APOE on this region. For this purpose, we considered an individual with AD and at least one APOE ε4 allele affected. An individual was considered unknown otherwise. Conversely, we conducted an APOE ε4-negative conditional linkage analysis, in which an individual with AD was considered affected in the absence of an APOE ε4 allele.
Analysis of combined family and case-control data
We combined the Caribbean Hispanic families and Caucasian cases-controls which those two datasets had nominally significant associations for rs2532500 and rs740850. For this purpose, we performed the DFAM procedure of the PLINK package [31]. This approach combines family data with unrelated cases and controls. The family data were analyzed using the transmission disequilibrium test using sibships as in sibTDT [32], and the case-control data were analyzed using a clustered-analysis of the Cochran-Mantel-Haesnzel test, which assesses allelic association conditional on cluster based on affection status. We examined the SNPs after combining all three datasets for the sake of completeness.
Assessment of linkage disequilibrium (LD)
LD structure was examined with the program HAPLOVIEW version 3.32 [33] (Figure 2). Haplotype blocks were defined using a confidence-interval algorithm [34]. The default settings were used in these analyses, which create 95% confidence bounds on D' to define SNP pairs in strong LD. Haplotypes and their frequencies were estimated using an accelerated expectation-maximization (EM) algorithm similar to the partition/ligation method implemented in HAPLOVIEW [35].
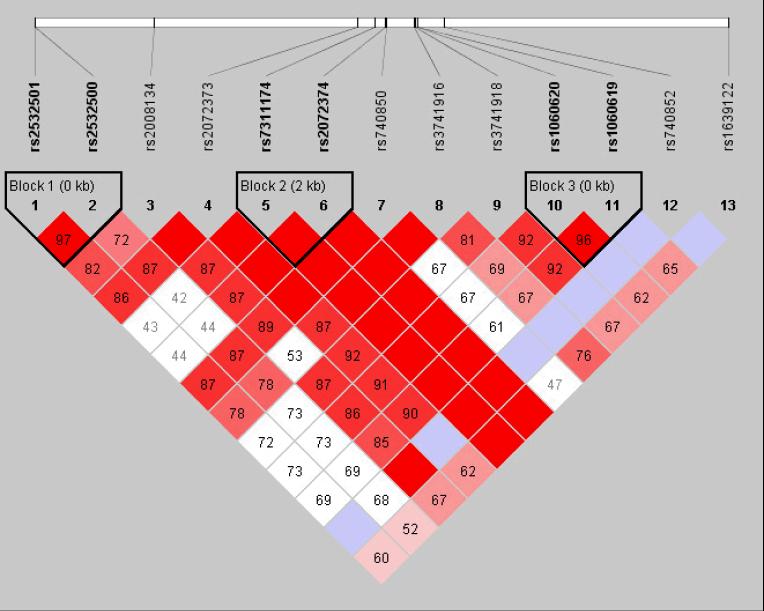
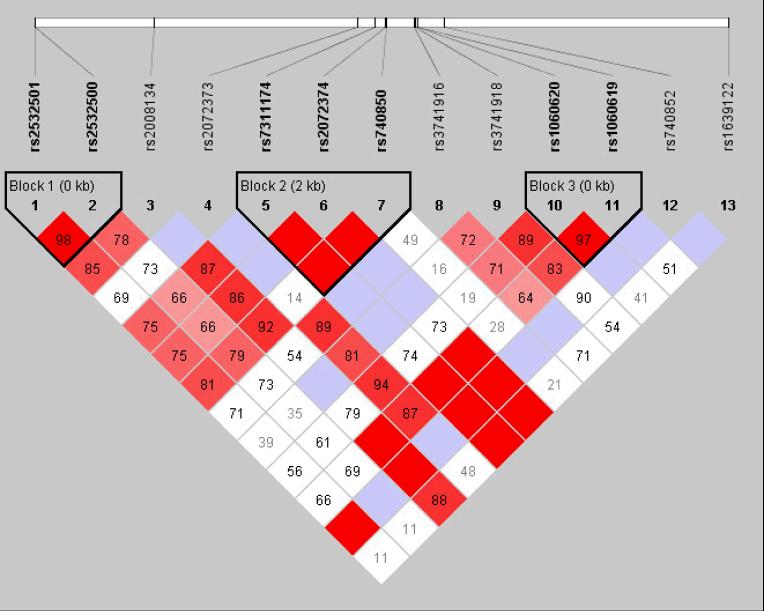
Figure 2.
The linkage disequilibrium (LD) pattern in the North European samples (A) and Caribbean Hispanic samples (B). LD was estimated across several single nucleotide polymorphisms in this chromosome region using the HAPLOVIEW software. The five-color scheme (white to red) represents the increasing strength of LD. Boxes with a D' of 1 are shaded in bright red. Cells with D' <1 are shades of pink or red. Blue represents D' = 1, but with a low confidence estimate for D'.
RESULTS
Linkage analysis
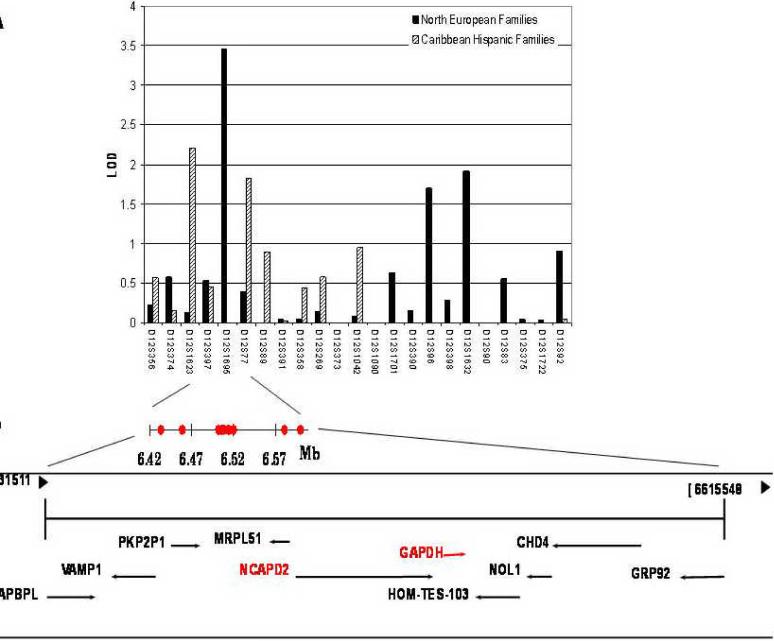
The analysis of the 23 microsatellite markers from the pericentromeric region of chromosome 12 generated a peak overall two-point LOD score of 3.46 at 19.7 cM in the North European FAD dataset (Table 2, Figure 1). In the Caribbean Hispanic FAD dataset, we observed two markers flanking the locus observed in the North European dataset, with LOD scores 2.21 at 15.7 cM and 1.83 at 20.3 cM. The multipoint linkage analysis of the combined datasets yielded a modest positive LOD score of 2.24 at 20.2 cM confirming the prior linkage support for this 5 cM region between 15 and 20 cM (Supplementary Figure 1). Thus, the two datasets independently support linkage to the chromosome 12p13 region. In addition, the North European FAD dataset also gave weakly positive scores for three markers in a four-Mb interval near 71 cM (LOD≤1.91, Table 2).
Figure 1.
(A) Results of the two-point linkage analysis (affecteds-only model) in the North European and Caribbean Hispanic families using 23 microsatellite markers on chromosome 12. LOD scores for autosomal dominant mode of inheritance are presented. (B) Genomic context and the positions of the selected single nucleotide polymorphisms for the prioritized chromosome 12p13 region.
Association analysis
For the 14 SNPs within the 6.4-6.6 Mb region (167 kb interval at 20 cM), we observed seven SNPs with nominal p≤0.05 in five different neighboring genes in at least one of the three datasets (Table 3): the TAP binding protein-like (TAPBPL); non-SMC condensin I complex, subunit D2 (NCAPD2, also known as CNAP1); GAPDH; cadherin 4, type 1, R-cadherin (CDH4); and G protein-coupled receptor 92 (GPR92). The strongest association with AD was observed for the T allele of a non-synonymous SNP, rs2532500, in the TAPBPL gene (genotype association p=0.006) in the North European case-control dataset. In the logistic regression analysis (Supplementary Table 1), three of the seven SNPs remained significant in the univariate model. However, these SNPs were no longer significant in a multivariate model that adjusted for age, sex, and APOE.
The same set of 14 SNPs were examined in the two FAD datasets using a single-point, family-based association analysis [28]. None of the SNPs were associated with FAD in the North European families. However, in the Caribbean Hispanic FAD dataset, there were two significant SNPs, one in the NCAPD2 (SNP 7: rs740850, p=0.0097), the other in GAPDH (SNP 10: rs1060620, p=0.0418). Both associations remained significant in the APOE conditional analysis in APOE ε4-positives (rs740850, p=0.015; and rs1060620, p=0.049).
We then performed association analysis after combining the two datasets that had positive allelic association, namely Caribbean Hispanic families and Caucasian cases and controls (Table 4). We found rs2532500 and rs740850 to be significantly associated with AD. For rs2532500, the T allele was significantly associated with AD after correcting for multiple testing (empirical p=0.0259); while for rs740850, the A allele was significantly associated with AD (empirical p=0.0048). However, when all three datasets were used for sake of completeness, none of the SNPs were found to be significant. Only rs3741916 (empirical p=0.0622), adjacent to rs740850 (empirical p=0.1115), approached experimental-wise significance.
Table 4.
Results of single point association analysis combining family and case-control data. For estimation of empirical p-value, we estimated empirical p-value based on 10,000 replicates.
| SNP # | SNP | Obs | Exp | Chi-sq | P | Pointwise Empirical P |
Studywise Empirical P |
|---|---|---|---|---|---|---|---|
| 1 | rs2532501 | 171 | 155.2 | 6.739 | 0.009434 | 0.0109 | 0.0649 |
| 2 | rs2532500 | 177 | 159.2 | 8.54 | 0.00347 | 0.0064 | 0.0259 |
| 3 | rs2008134 | 103 | 106.2 | 0.3203 | 0.5714 | 0.6123 | 0.9954 |
| 4 | rs2072373 | 57 | 64.78 | 2.711 | 0.09968 | 0.1093 | 0.4803 |
| 5 | rs7311174 | 99 | 98.95 | 8.68E-05 | 0.9926 | 0.9800 | 1.0000 |
| 6 | rs2072374 | 98 | 98.35 | 0.003902 | 0.9502 | 0.9620 | 1.0000 |
| 7 | rs740850 | 187 | 165.3 | 11.84 | 0.00058 | 0.0004 | 0.0048 |
| 8 | rs3741916 | 76 | 87.93 | 5.293 | 0.02141 | 0.0177 | 0.1361 |
| 9 | rs3741918 | 62 | 70.32 | 3.028 | 0.08183 | 0.0758 | 0.4076 |
| 10 | rs1060620 | 92 | 95.93 | 0.5488 | 0.4588 | 0.4650 | 0.9810 |
| 11 | rs1060619 | 89 | 92.26 | 0.3965 | 0.5289 | 0.5396 | 0.9937 |
| 12 | rs740852 | 30 | 28.44 | 0.2185 | 0.6402 | 0.6549 | 0.9988 |
Haplotype analysis was performed in the North European case-control and Caribbean Hispanic FAD datasets to follow up the significant findings in the single-point analysis (Table 5). Haplotype analysis was not performed in the North European FAD dataset because none of the SNPs showed significant association in the single-point analysis. In the North European case-control dataset, the haplotypes containing the A allele at rs740850 (SNP 7) were positively associated with AD, whereas the haplotypes containing the G allele at rs740850 (SNP 7) were protective. Although haplotypes containing the C allele at rs2072374 (SNP 6) or the G allele at rs3741916 (SNP 8) were significantly associated with AD, the direction of the haplotype association (i.e., whether a haplotype was deleterious or protective against AD) was inconsistent. In the Caribbean Hispanic FAD dataset, a similar pattern of haplotype association was observed. Haplotypes including rs740850 (SNP 7) were significant. Specifically, haplotype C-A at SNP 6-7 and haplotype C-A-C at SNP 6-7-8 were positively associated with AD (Z=1.96-2.12, both haplotype-specific empirical p and global p<0.05). On the other hand, haplotypes T-C-G at SNP 5-6-7 (rs7311174, rs2072374 and rs740850) showed a consistent protective effect on AD in the two datasets (North European case-control set, haplotype specific p-value=0.032; global p-value=0.080; Caribbean Hispanic set, haplotype specific p-value=0.005; global p-value=0.0074). The T-C-G haplotype frequencies for the North European case-control dataset and Caribbean Hispanic dataset were comparable (0.359 and 0.408, respectively).
Table 5. Haplotype analysis results: Family and case-control studies.
Results of the haplotype analysis in the North European case-control dataset and Caribbean Hispanic familial dataset with p-value<0.05. Haplotypes in bold were positively associated with AD. aGlobal p-values are based on permutation. In the Caribbean Hispanic dataset, SNP 14 was not informative.
| 1 rs2532501 | 2 rs2532500 | 3 rs2008134 | 4 rs2072373 | 5 rs7311174 | 6 rs2072374 | 7 rs740850 | 8 rs3741916 | 9 rs3741918 | 10 rs1060620 | 11 rs1060619 | 12 rs740852 | 13 rs1639122 | 14 rs3741924 | Haplotype frequency | Z-Score | Haplo-specific empirical p | Global pa |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| North European Case-control Dataset | |||||||||||||||||
| C | T | 0.365 | -1.89 | 0.04500 | 0.08600 | ||||||||||||
| T | C | G | 0.359 | -2.24 | 0.03200 | 0.08000 | |||||||||||
| C | G | 0.362 | -2.33 | 0.02600 | 0.08900 | ||||||||||||
| G | G | 0.175 | -2.54 | 0.01257 | 0.01547 | ||||||||||||
| C | G | G | 0.174 | -2.25 | 0.02100 | 0.17800 | |||||||||||
| G | A | 0.049 | -2.50 | 0.01300 | 0.02500 | ||||||||||||
| C | A | 0.794 | 2.63 | 0.01000 | 0.02500 | ||||||||||||
| G | G | A | 0.049 | -2.47 | 0.01412 | 0.00753 | |||||||||||
| A | G | 0.006 | 1.73 | 0.03578 | 0.01547 | ||||||||||||
| A | G | T | 0.006 | 1.87 | 0.01365 | 0.00753 | |||||||||||
| G | A | T | 0.050 | -2.55 | 0.01300 | 0.03700 | |||||||||||
| C | A | T | 0.762 | 2.00 | 0.04600 | 0.03700 | |||||||||||
| G | C | 0.291 | -2.25 | 0.02800 | 0.07700 | ||||||||||||
| G | A | 0.634 | 1.97 | 0.05000 | 0.07700 | ||||||||||||
| C | C | 0.013 | -2.43 | 0.00386 | 0.00617 | ||||||||||||
| A | T | 0.707 | 2.48 | 0.01620 | 0.00617 | ||||||||||||
| G | C | C | 0.013 | -2.43 | 0.00500 | 0.02200 | |||||||||||
| G | A | T | 0.632 | 2.12 | 0.03400 | 0.02200 | |||||||||||
| Caribbean Hispanic Family Dataset | |||||||||||||||||
| C | T | 0.397 | -2.46 | 0.01395 | 0.05340 | ||||||||||||
| C | C | T | 0.396 | -2.29 | 0.02216 | 0.05880 | |||||||||||
| C | T | C | 0.249 | -2.16 | 0.03063 | 0.16380 | |||||||||||
| C | A | 0.409 | 1.96 | 0.04953 | 0.01320 | ||||||||||||
| C | G | 0.400 | -2.72 | 0.00650 | 0.01320 | ||||||||||||
| T | C | A | 0.419 | 1.96 | 0.04985 | 0.00740 | |||||||||||
| T | C | G | 0.408 | -2.80 | 0.00504 | 0.00740 | |||||||||||
| A | C | 0.377 | 2.06 | 0.03985 | 0.01100 | ||||||||||||
| G | G | 0.157 | -2.37 | 0.01787 | 0.01100 | ||||||||||||
| C | A | C | 0.377 | 2.12 | 0.03406 | 0.02860 | |||||||||||
| A | C | A | 0.355 | 2.19 | 0.02845 | 0.02060 | |||||||||||
| A | T | 0.728 | 2.08 | 0.03719 | 0.09100 | ||||||||||||
| T | C | 0.146 | -2.06 | 0.03948 | 0.09100 | ||||||||||||
| C | A | T | 0.698 | 2.37 | 0.01656 | 0.09340 | |||||||||||
| G | T | C | 0.110 | -2.33 | 0.01968 | 0.09340 | |||||||||||
| T | A | 0.731 | 2.16 | 0.03094 | 0.02180 | ||||||||||||
| C | A | 0.022 | -1.98 | 0.04781 | 0.02180 | ||||||||||||
| T | C | A | 0.015 | -1.97 | 0.04898 | 0.17501 | |||||||||||
| C | A | G | 0.022 | -1.96 | 0.04722 | 0.06060 | |||||||||||
| G | C | 0.468 | 2.07 | 0.03841 | 0.18920 | ||||||||||||
Haplotypes with nominal p≤0.05 are shown; SNP 14 uninformative
Global p-values are based on permutation.
In the GAPDH gene, haplotypes that included the T allele at rs1060620 (SNP 10) were significantly associated with AD in both the North European case-control and Caribbean Hispanic family datasets. Haplotype C-A-T at SNP 8-9-10 was significantly associated with AD in both datasets (haplotype specific empirical p=0.046, 0.017, respectively), and haplotype T-A at SNP 10-11 (haplotype specific empirical p-value=0.03) was significantly associated with AD in the Caribbean Hispanic family dataset.
Lastly, the multi-marker analysis of Caribbean Hispanic families supported the associations observed in the above haplotype analysis for SNPs 7-8 (p=0.030), SNPs 9-10 (p=0.048), and SNPs 6-7-8 (p=0.015).
DISCUSSION
We conducted multi-stage fine mapping of the chromosome 12 locus between 12p13.2 and 12q24.2, spanning 70 cM, using two ethnically diverse FAD datasets and one case-control AD dataset. We examined these two datasets together, because we hypothesized that there may present a common susceptibility gene(s) for LOAD in this region, since multiple datasets from more than one ethnic background yielded strong support for linkage and some of those studies reported significant allelic association. The present linkage analyses of the NE Caucasian dataset as well as the Caribbean Hispanic FAD dataset continue to provide strong evidence for linkage at 15-20 cM on 12p. The subsequent association study of 14 SNPs spanning a 167 kb region under the linkage peak (Figure 1) revealed nominally significant results for seven SNPs in a case-control study. However, only two of these SNPs located in two adjacent genes (NCAPD2 and GAPDH) demonstrated nominally significant association with AD in one of two FAD datasets (Table 3). Those two SNPs (rs2532500 and rs740850) were significantly associated with AD, when we combined the Caribbean Hispanic family data with the NE Caucasian case-control data. Moreover, two haplotype sets spanning SNPs 6-10 were significant; particularly, haplotype C-A at SNPs 8-9 located in NCAPD2 were significantly associated with AD in both Caribbean Hispanic families and in North European case-control dataset.
Our findings localize to the same region that was identified by earlier reports, which detected a significant association between AD and six SNPs in or nearby GAPDH [9, 36]. Li and colleagues [9] observed somewhat different allelic associations with AD in three case-control series and one case-control series derived from a linkage study. This series-to-series heterogeneity of disease risk suggests that the observed AD associated SNPs are in fact in linkage disequilibrium with a disease-causing allele at a nearby site. All five SNPs that were significant in the study by Li and colleagues [9] were included in our study; however, only rs3741916 (SNP 8) generated a significant result. In our North European case-control dataset, we found the C allele of rs3741916 to be marginally associated with AD (p=0.027). Further, in the Caribbean Hispanic FAD and NE Caucasian case-control datasets, the haplotypes that include the C allele at rs3741916 (e.g., haplotype C-A-T at SNPs 8-10) were associated with AD (Table 5). This finding is consistent with the results of a recent report by Lin and colleagues [36.], which conducted a study of North American FAD and an AD case-control datasets using eight SNPs in the PKP2P1, NCAPD2, and GAPDH genes. Of those, seven of these SNPs were included in the current investigation (Table 3). Lin and colleagues [36] found no associations in the family-based study. However, they did observe the CC genotype at rs3741916 (p=0.021) to be marginally associated with sporadic AD, and the G allele at rs3741916 be protective (p=0.014). In contrast, Li and colleagues [9] observed the G allele at rs3741916 to be over-represented in AD cases when compared with the frequency the G allele in controls.
It has been suggested that the rs3741916 polymorphism may affect the transcription of GAPDH since it is located at -8 bp of the start codon [9]. The contradictory findings across studies at rs3741916 may be due to allelic heterogeneity, and argues against the functional significance of this SNP. Another possible explanation for the inconsistencies in allelic association was suggested by Lin and colleagues [37], in which effects from multiple loci and a varying degree of correlations among these variants can lead to changing direction of association in different samples. In our current datasets (Table 5), however, two haplotype sets were consistently associated with AD: (1) haplotype C-A at SNPs 6-7 was over-represented in AD in Caribbean Hispanics; and (2) haplotypes containing C-A-T at SNPs 8-10 were over-represented in both datasets. Although our findings from two datasets are consistent to some extent, further examination is necessary to probe the region surrounding SNPs 6-10.
A recent report by De Ferrari and colleagues [38] proposed the low density lipoprotein receptor-related protein 6 (LRP6), located at ~26 cM, as a susceptibility gene for late onset AD, and hypothesized that LRP6 alters function of Wnt signaling components in AD. In our case-control dataset, we examined two SNPs identified by the authors (rs1012672 in exon 18 and rs2302685 in exon 14), but neither were significantly associated with AD (data not shown).
The current results support the hypothesis that the region surrounding NCAPD2 and GAPDH at chromosome 12p13 may harbor an AD susceptibility gene(s), and the two potential risk haplotypes are haplotype C-A at SNP 6-7 within NCAPD2 and haplotype C-A-T at SNPs 8-10 within GAPDH. However, the LD for the GAPDH region extends for approximately 130 Kb (Figure 2), and has a dense genomic context, containing at least nine genes (Figure 1). Given significant results with several SNPs from the current study as well as that of two published reports [9, 36], future studies need to investigate a more dense and broad SNP coverage of the interval on 12p detected by current linkage studies.
Supplementary Material
ACKNOWLEDGEMENTS
This work was supported by grants from the National Institute of Health and the National Institute on Aging: R37-AG15473, P01-AG07232 (RM), R01-AG09029, P30-AG13846 (LF), P20RR11126 (IJ), the Alzheimer Association, The Blanchett Hooker Rockefeller Foundation and The Charles S. Robertson Gift from the Banbury Fund (RM). The laboratory under the direction of Dr. St George-Hyslop received additional support from the Alzheimer Society of Canada, Japan-Canada and Canadian Institutes of Health Research Joint Health Research Program (ER), the Canadian Institutes of Health Research, Alzheimer Society of Ontario, Howard Hughes Medical Institute, Canada Foundation for Innovation (PH), Fonds de la Recherche en Santé (YM).
REFERENCES
- 1.Blacker D, Bertram L, Saunders AJ, Moscarillo TJ, Albert MS, Wiener H, Perry RT, Collins JS, Harrell LE, Go RC, Mahoney A, Beaty T, Fallin MD, Avramopoulos D, Chase GA, Folstein MF, McInnis MG, Bassett SS, Doheny KJ, Pugh EW, Tanzi RE. Results of a high-resolution genome screen of 437 Alzheimer's Disease families. Hum Mol Genet. 2003;12(1):23–32. doi: 10.1093/hmg/ddg007. [DOI] [PubMed] [Google Scholar]
- 2.Kehoe P, Wavrant-De Vrieze F, Crook R, Wu WS, Holmans P, Fenton I, Spurlock G, Norton N, Williams H, Williams N, Lovestone S, Perez-Tur J, Hutton M, Chartier-Harlin MC, Shears S, Roehl K, Booth J, Van Voorst W, Ramic D, Williams J, Goate A, Hardy J, Owen MJ. A full genome scan for late onset Alzheimer's disease. Hum Mol Genet. 1999;8(2):237–45. doi: 10.1093/hmg/8.2.237. [DOI] [PubMed] [Google Scholar]
- 3.Pericak-Vance MA, Bass MP, Yamaoka LH, Gaskell PC, Scott WK, Terwedow HA, Menold MM, Conneally PM, Small GW, Vance JM, Saunders AM, Roses AD, Haines JL. Complete genomic screen in late-onset familial Alzheimer disease. Evidence for a new locus on chromosome 12. JAMA. 1997;278(15):1237–41. [PubMed] [Google Scholar]
- 4.Rogaeva E, Premkumar S, Song Y, Sorbi S, Brindle N, Paterson A, Duara R, Levesque G, Yu G, Nishimura M, Ikeda M, O'Toole C, Kawarai T, Jorge R, Vilarino D, Bruni AC, Farrer LA, St George-Hyslop PH. Evidence for an Alzheimer disease susceptibility locus on chromosome 12 and for further locus heterogeneity. JAMA. 1998;280(7):614–8. doi: 10.1001/jama.280.7.614. [DOI] [PubMed] [Google Scholar]
- 5.Mayeux R, Lee JH, Romas SN, Mayo D, Santana V, Williamson J, Ciappa A, Rondon HZ, Estevez P, Lantigua R, Medrano M, Torres M, Stern Y, Tycko B, Knowles JA. Chromosome-12 mapping of late-onset Alzheimer disease among Caribbean Hispanics. Am J Hum Genet. 2002;70(1):237–43. doi: 10.1086/324773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Roberts SB, MacLean CJ, Neale MC, Eaves LJ, Kendler KS. Replication of linkage studies of complex traits: an examination of variation in location estimates. Am J Hum Genet. 1999;65(3):876–84. doi: 10.1086/302528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sillanpaa MJ, Auranen K. Replication in genetic studies of complex traits. Ann Hum Genet. 2004;68(Pt 6):646–57. doi: 10.1046/j.1529-8817.2004.00122.x. [DOI] [PubMed] [Google Scholar]
- 8.Scott WK, Grubber JM, Conneally PM, Small GW, Hulette CM, Rosenberg CK, Saunders AM, Roses AD, Haines JL, Pericak-Vance MA. Fine mapping of the chromosome 12 late-onset Alzheimer disease locus: potential genetic and phenotypic heterogeneity. Am J Hum Genet. 2000;66(3):922–32. doi: 10.1086/302828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li Y, Nowotny P, Holmans P, Smemo S, Kauwe JS, Hinrichs AL, Tacey K, Doil L, van Luchene R, Garcia V, Rowland C, Schrodi S, Leong D, Gogic G, Chan J, Cravchik A, Ross D, Lau K, Kwok S, Chang SY, Catanese J, Sninsky J, White TJ, Hardy J, Powell J, Lovestone S, Morris JC, Thal L, Owen M, Williams J, Goate A, Grupe A. Association of late-onset Alzheimer's disease with genetic variation in multiple members of the GAPD gene family. Proc Natl Acad Sci U S A. 2004;101(44):15688–93. doi: 10.1073/pnas.0403535101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lee JH, Mayeux R, Mayo D, Mo J, Santana V, Williamson J, Flaquer A, Ciappa A, Rondon H, Estevez P, Lantigua R, Kawarai T, Toulina A, Medrano M, Torres M, Stern Y, Tycko B, Rogaeva E, St George-Hyslop P, Knowles JA. Fine mapping of 10q and 18q for familial Alzheimer's disease in Caribbean Hispanics. Mol Psychiatry. 2004;9(11):1042–51. doi: 10.1038/sj.mp.4001538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rogaeva EA, Premkumar S, Grubber J, Serneels L, Scott WK, Kawarai T, Song Y, Hill DL, Abou-Donia SM, Martin ER, Vance JJ, Yu G, Orlacchio A, Pei Y, Nishimura M, Supala A, Roberge B, Saunders AM, Roses AD, Schmechel D, Crane-Gatherum A, Sorbi S, Bruni A, Small GW, Conneally PM, Haines JL, Van Leuven F, St George-Hyslop PH, Farrer LA, Pericak-Vance MA. An alpha-2-macroglobulin insertion-deletion polymorphism in Alzheimer disease. Nat Genet. 1999;22(1):19–22. doi: 10.1038/8729. [DOI] [PubMed] [Google Scholar]
- 12.McKhann G, Drachman D, Folstein M, Katzman R, Price D, Stadlan EM. Clinical diagnosis of Alzheimer's disease: report of the NINCDS-ADRDA Work Group under the auspices of Department of Health and Human Services Task Force on Alzheimer's Disease. Neurology. 1984;34(7):939–44. doi: 10.1212/wnl.34.7.939. [DOI] [PubMed] [Google Scholar]
- 13.O'Connell JR, Weeks DE. PedCheck: a program for identification of genotype incompatibilities in linkage analysis. Am J Hum Genet. 1998;63(1):259–66. doi: 10.1086/301904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bell PA, Chaturvedi S, Gelfand CA, Huang CY, Kochersperger M, Kopla R, Modica F, Pohl M, Varde S, Zhao R, Zhao X, Boyce-Jacino MT, Yassen A. SNPstream UHT: ultra-high throughput SNP genotyping for pharmacogenomics and drug discovery. Biotechniques. 2002;Suppl:70–2. 74, 76–7. [PubMed] [Google Scholar]
- 15.Hixson JE, Vernier DT. Restriction isotyping of human apolipoprotein E by gene amplification and cleavage with HhaI. J Lipid Res. 1990;31(3):545–8. [PubMed] [Google Scholar]
- 16.Terwilliger JD, Ott J. Handbook of Human Genetic Linkage. The Johns Hopkins University Press; Baltimore: 1994. p. 307. [Google Scholar]
- 17.Goring HH, Terwilliger JD. Linkage analysis in the presence of errors I: complex-valued recombination fractions and complex phenotypes. Am J Hum Genet. 2000;66(3):1095–106. doi: 10.1086/302797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Goring HH, Terwilliger JD. Linkage analysis in the presence of errors IV: joint pseudomarker analysis of linkage and/or linkage disequilibrium on a mixture of pedigrees and singletons when the mode of inheritance cannot be accurately specified. Am J Hum Genet. 2000;66(4):1310–27. doi: 10.1086/302845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cottingham RW, Jr., Idury RM, Schaffer AA. Faster sequential genetic linkage computations. Am J Hum Genet. 1993;53(1):252–63. [PMC free article] [PubMed] [Google Scholar]
- 20.Lathrop GM, Lalouel JM, Julier C, Ott J. Strategies for multilocus linkage analysis in humans. Proc Natl Acad Sci U S A. 1984;81(11):3443–6. doi: 10.1073/pnas.81.11.3443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Risch N. Linkage strategies for genetically complex traits. III. The effect of marker polymorphism on analysis of affected relative pairs. Am J Hum Genet. 1990;46(2):242–53. [PMC free article] [PubMed] [Google Scholar]
- 22.Risch N. Linkage strategies for genetically complex traits. II. The power of affected relative pairs. Am J Hum Genet. 1990;46(2):229–41. [PMC free article] [PubMed] [Google Scholar]
- 23.Faraway JJ. Improved sib-pair linkage test for disease susceptibility loci. Genet Epidemiol. 1993;10(4):225–33. doi: 10.1002/gepi.1370100403. [DOI] [PubMed] [Google Scholar]
- 24.Holmans P. Asymptotic properties of affected-sib-pair linkage analysis. Am J Hum Genet. 1993;52(2):362–74. [PMC free article] [PubMed] [Google Scholar]
- 25.Wigginton JE, Abecasis GR. PEDSTATS: descriptive statistics, graphics and quality assessment for gene mapping data. Bioinformatics. 2005;21(16):3445–7. doi: 10.1093/bioinformatics/bti529. [DOI] [PubMed] [Google Scholar]
- 26.SPSS . SPSS 15.0 for Windows. SPSS; Chilcago, IL: 2007. [Google Scholar]
- 27.Schaid DJ, Rowland CM, Tines DE, Jacobson RM, Poland GA. Score tests for association between traits and haplotypes when linkage phase is ambiguous. Am J Hum Genet. 2002;70(2):425–34. doi: 10.1086/338688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Horvath S, Xu X, Lake SL, Silverman EK, Weiss ST, Laird NM. Family-based tests for associating haplotypes with general phenotype data: application to asthma genetics. Genet Epidemiol. 2004;26(1):61–9. doi: 10.1002/gepi.10295. [DOI] [PubMed] [Google Scholar]
- 29.Rakovski CS, Xu X, Lazarus R, Blacker D, Laird NM. A new multimarker test for family-based association studies. Genet Epidemiol. 2007;31(1):9–17. doi: 10.1002/gepi.20186. [DOI] [PubMed] [Google Scholar]
- 30.Scott WK, Grubber JM, Conneally PM, Small GW, Hulette CM, Rosenberg CK, Saunders AM, Roses AD, Haines JL, Pericak-Vance MA. Fine mapping of the chromosome 12 late-onsetAlzheimer disease locus: potential genetic and phenotypic heterogeneity. Am J Hum Genet. 2000;66(3):922–32. doi: 10.1086/302828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, Maller J, Sklar P, de Bakker PI, Daly MJ, Sham PC. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81(3):559–75. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Spielman RS, Ewens WJ. A sibship test for linkage in the presence of association: the sib transmission/disequilibrium test. Am J Hum Genet. 1998;62(2):450–8. doi: 10.1086/301714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21(2):263–5. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- 34.Gabriel SB, Schaffner SF, Nguyen H, Moore JM, Roy J, Blumenstiel B, Higgins J, DeFelice M, Lochner A, Faggart M, Liu-Cordero SN, Rotimi C, Adeyemo A, Cooper R, Ward R, Lander ES, Daly MJ, Altshuler D. The structure of haplotype blocks in the human genome. Science. 2002;296(5576):2225–9. doi: 10.1126/science.1069424. [DOI] [PubMed] [Google Scholar]
- 35.Qin ZS, Niu T, Liu JS. Partition-ligation-expectation-maximization algorithm for haplotype inference with single-nucleotide polymorphisms. Am J Hum Genet. 2002;71(5):1242–7. doi: 10.1086/344207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lin PI, Martin ER, Bronson PG, Browning-Large C, Small GW, Schmechel DE, Welsh-Bohmer KA, Haines JL, Gilbert JR, Pericak-Vance MA. Exploring the association of glyceraldehyde-3-phosphate dehydrogenase gene and Alzheimer disease. Neurology. 2006;67(1):64–8. doi: 10.1212/01.wnl.0000223438.90113.4e. [DOI] [PubMed] [Google Scholar]
- 37.Lin PI, Vance JM, Pericak-Vance MA, Martin ER. No gene is an island: the flip-flop phenomenon. Am J Hum Genet. 2007;80(3):531–8. doi: 10.1086/512133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.De Ferrari GV, Papassotiropoulos A, Biechele T, Wavrant De-Vrieze F, Avila ME, Major MB, Myers A, Saez K, Henriquez JP, Zhao A, Wollmer MA, Nitsch RM, Hock C, Morris CM, Hardy J, Moon RT. Common genetic variation within the low-density lipoprotein receptor-related protein 6 and late-onset Alzheimer's disease. Proc Natl Acad Sci U S A. 2007;104(22):9434–9. doi: 10.1073/pnas.0603523104. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.