Abstract
The nature of the nucleosomal barrier which regulates access to the underlying DNA during many cellular processes is not fully understood. Here we present a detailed map of histone-DNA interactions along the DNA sequence to near basepair accuracy by mechanically unzipping single molecules of DNA, each containing a single nucleosome. This interaction map revealed a distinct ~5 bp periodicity that was enveloped by three broad regions of strong interactions, with the strongest at the dyad and the other two at ~ ±40 bp from the dyad. Unzipping up to the dyad allowed recovery of a canonical nucleosome upon relaxation of the DNA, but unzipping beyond the dyad resulted in removal of histone octamer from its initial DNA sequence. These findings have significant implications for how RNA polymerase and other DNA-based enzymes may gain access to DNA associated with a nucleosome.
The nucleosome is the fundamental repeating unit of eukaryotic chromatin, consisting of ~147 bp of DNA wrapped ~1.7 times around a histone octamer1. Nucleosomes must be stable and yet dynamic structures, both maintaining eukaryotic DNA in a condensed state and also permitting regulated access to genetic information contained therein. During many important cellular processes, DNA-binding proteins must access specific genomic regions that are occluded by nucleosomes. In particular, in vitro studies show that RNA polymerase slows down, pauses, or stalls upon encountering a nucleosome2–7. The resistance that RNA polymerase encounters when transcribing a chromatin template should be largely dictated by both the strengths and locations of histone-DNA interactions in the nucleosome. Therefore a detailed map of these interactions would lay an important foundation for understanding the structural details of eukaryotic transcription and how gene expression may be regulated by histone modifications, DNA sequence, and nucleosome remodeling.
Analysis of the nucleosome crystal structure indicates that histone-DNA interactions are not uniform along the DNA1,8; however, experimental determination of this interaction map has proven to be challenging and is still largely controversial. Although it is well established that the overall stability of a nucleosome depends on its constituent DNA sequence and histone modifications9–11, the way in which specific interactions in a nucleosome lead to this stability is less well understood. The mechanical nature of this problem makes it ideally suited for investigation by single molecule manipulation approaches12–19. Previously, we have stretched single DNA molecules of chromatin and obtained data on the relative locations of strong histone-DNA interactions14,17. These data indicate the presence of three regions of strong interactions, consistent with those suggested by counting the number of apparent histone-DNA contacts seen in the nucleosome crystal structure20. However, subsequent single molecule stretching experiments challenged this interpretation and suggested that force signatures from stretching experiments can be attributed to the rotation of the spool geometry of the nucleosome rather than regions of strong histone-DNA interactions21. These studies favor a model in which histone-DNA interactions are uniform along the DNA22,23. The controversy arose because stretching experiments cannot readily separate contributions from geometry and interaction strengths nor can they quantitatively assay interaction strengths near the dyad.
Recently, we have developed a method to sequentially determine the absolute locations of histone-DNA interactions by mechanically unzipping a DNA molecule containing a nucleosome assembled with histones purified from HeLa cells16. However, the precision of that method was insufficient to map out all of the densely packed histone-DNA interactions in a nucleosome. In the current work, using an improved unzipping method, we have mapped the locations of the interactions to near bp accuracy along the DNA and quantitatively assayed the strengths of these interactions. The histone-DNA interaction map, together with mechanical invasion experiments, provide a simple explanation of the RNA polymerase pausing pattern within a nucleosome and makes testable predictions on the fate of histones during transcription.
RESULTS
Precision mapping of interactions to near bp
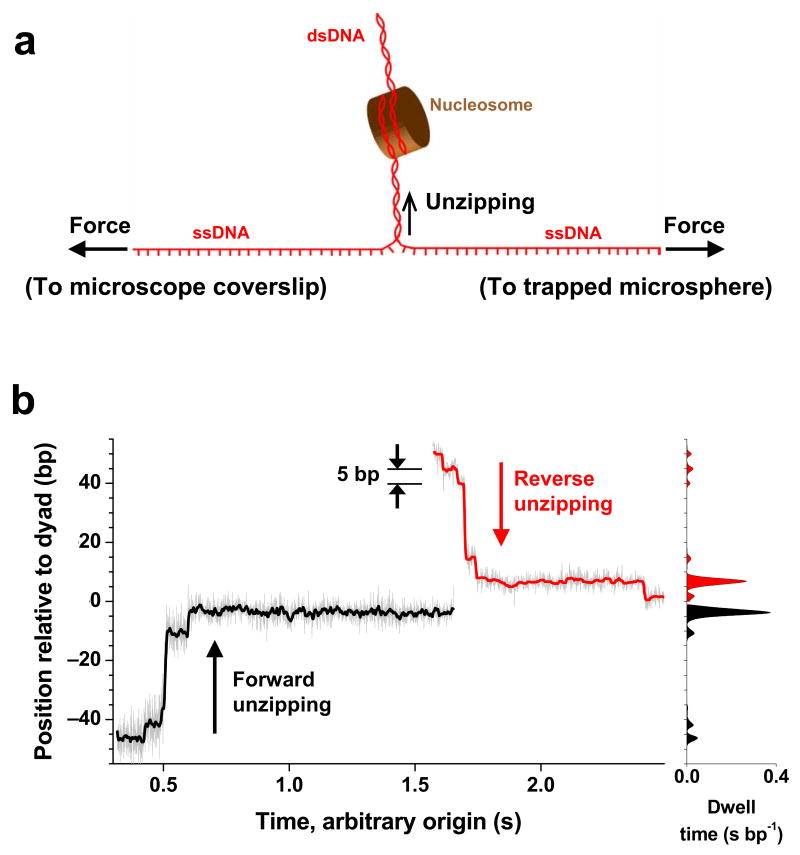
The experimental configuration is sketched in Figure 1a (also Methods and Supplementary Fig. 1). A DNA molecule containing a single nucleosome uniquely positioned at a 601 nucleosome positioning sequence24 was attached to the surface of a microscope coverslip via one of its strands and to a microsphere held in an optical trap via the other strand16. As the coverslip was moved away from the trapped microsphere, dsDNA was sequentially converted to ssDNA upon base pair separation. As the unzipping fork progressed through the nucleosome, it encountered resistance from histone-DNA interactions at well-defined locations and, because these interactions require dsDNA, they were sequentially disrupted. The magnitude of resistance should strongly correlate with histone-DNA affinity, and thus a histone-DNA interaction map was generated along the DNA. We improved the alignment method and showed that this technique achieved a resolution of better than 1 bp (Methods and Supplementary Fig. 2c). Its accuracy and precision of determining the absolute sequence position of an interaction were both ~1.5 bp (Methods and Supplementary Fig. 2b)
Figure 1.
Nucleosome disruptions under a constant unzipping force. (a) Experimental configuration. A DNA molecule was mechanically unzipped through a nucleosome uniquely positioned at a 601 sequence. (b) Representative traces for unzipping under a constant applied force (~ 28 pN). Two traces are shown: one from forward unzipping (black) and one from reverse unzipping (red). Both traces were low pass filtered from the raw traces (grey) to 60 Hz. The unzipping fork paused at specific locations, which are evident from both the traces (left) and their corresponding dwell time histograms (right).
Mapping strengths of histone-DNA interactions in a nucleosome
To quantitatively assay the strengths of the histone-DNA interactions, we unzipped through individual nucleosomal DNA molecules with a constant unzipping force of ~28 pN (Methods). Under a force clamp25, the dwell times at different sequence positions measure the strengths of interactions at those positions, provided that disruption of each interaction follows a similar energy landscape. Thus this method allows direct mapping of the strengths of interactions. Figure 1b shows example traces for unzipping DNA through a nucleosome under a constant force (Supplementary Fig. 3 for additional traces). DNA molecules were unzipped from both directions along the DNA (referred to as “forward” and “reverse”) (Methods and Supplementary Fig. 3). In both cases, the unzipping fork did not move through the nucleosomal DNA at a constant rate but instead dwelled at specific locations within the nucleosome, indicating the presence of strong interactions. In particular, these traces revealed that the fork dwelled with discrete steps spaced by ~5 bp and the longest dwell times tended to occur near the dyad.
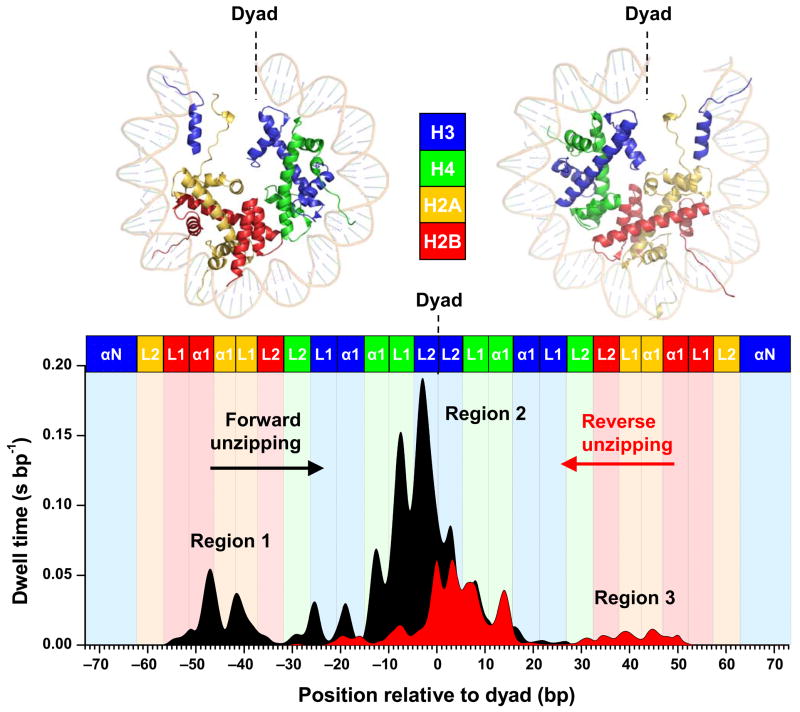
An interaction map was generated by averaging dwell time histogram measurements from many traces from both forward and reverse unzipping, as shown in Figure 2. Several features are evident from these plots. (1) There are three broad regions of strong interactions: one located at the dyad and two ~ ±40 bp from the dyad. (2) An ~5 bp periodicity occurred within each region of interaction. (3) The interactions near the entry and exit DNA are particularly weak. The unzipping fork did not dwell at a 20 bp region of both entry and exit DNA, indicating that the histones are only loosely bound to the DNA. (4) For unzipping in both the forward and reverse directions, the first two regions of interactions encountered were always detected, but not the last region. This indicates that once the dyad region of interactions was disrupted, the nucleosome became unstable and histones dissociated from the 601 sequence. (5) The total dwell time in the nucleosome was longer in the forward direction compared with that in the reverse direction, indicating nucleosomes were more difficult to disrupt when unzipped in the forward direction, likely reflecting the non-palindromic nature of the 601 sequence.
Figure 2.
Histone-DNA interaction map within a nucleosome core particle. Top Panel: Crystal structure of the nucleosome core particle1, where dots indicate regions where interactions between DNA and one of the core histones are likely to occur. The two halves of the nucleosome are shown separately for clarity. Bottom Panel: A histone-DNA interaction map constructed from the averaged dwell time histograms of the unzipping fork at constant force (~ 28 pN). Individual traces were low-pass filtered to 60 Hz and their dwell time histograms were binned to 1 bp. A total of 27 traces from the forward template and 30 traces from the reverse template were used for the construction. Each peak corresponds to an individual histone-DNA interaction and the heights are indicative of their relative strengths. Three regions of strong interactions are indicated: one located at the dyad (region 2) and two located off-dyad (regions 1 and 3). Colored boxes indicate predictions from the crystal structure where individual histone binding motifs are expected to interact with DNA. The H3 N-terminal alpha helices (αN) and the histone loops (L1, L2) and alpha helices (α1) that compose the L1L2 and α1α1 DNA-binding sites observed in the crystal structure1 are also indicated.
Highlighting histone-DNA interactions near entry/exit DNA
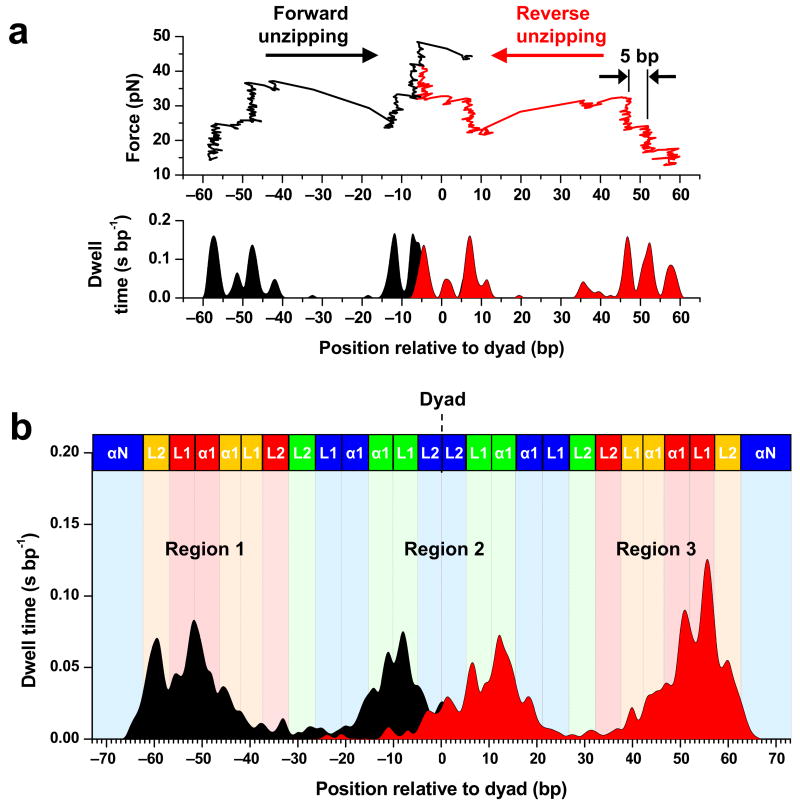
Because the entry and exit DNA regulate the initial invasion of a nucleosome by a motor protein, experiments were carried out starting from a lower unzipping force to specifically detect interactions at those locations and then the force was ramped up to allow complete unzipping through the nucleosomal DNA. We unzipped through nucleosomal DNA molecules under a constant loading rate (8 pN s−1), which highlighted the edge of the region first encountered16 (Methods). Figure 3a shows example traces of nucleosomes unzipped from both forward and reverse directions. Figure 3b shows the averaged dwell time histograms measured during both forward and reverse unzipping (Supplementary Fig. 4 for additional traces). Aside from the aforementioned bias in the dwell time histogram, many features are consistent with data from unzipping under a constant force. The interactions near the entry and exit DNA were more evident, still showing a clear ~5 bp periodicity. This indicates that DNA segments at least up to 60 bp from the dyad have substantial interactions with the histone core.
Figure 3.
Nucleosome disruptions under a constant loading rate. (a) Representative traces for unzipping under a constant loading rate (8 pN s−1). Two traces are shown: one from forward unzipping (black) and one from reverse unzipping (red). For clarity, the naked DNA signature before and after each nucleosome disruption event is not shown. The unzipping fork again paused at specific locations, which are evident from both the traces (top) and their corresponding dwell time histograms (bottom). (b) The average dwell time histograms of the unzipping fork under a constant loading rate. Individual traces such as those shown above were low-pass filtered to 60 Hz and their dwell time histograms were binned to 1 bp. A total of 36 traces from each direction were used for the construction. Other notations are the same as those of Figure 2.
Features shared by nucleosomes on arbitrary DNA sequences
To determine whether the conclusions above are also valid for nucleosomes of arbitrary DNA sequence or just for the 601 sequence, we assembled nucleosomes onto a DNA segment that does not contain any known positioning elements (Methods). The assembly condition was controlled to achieve a relatively low saturation level so that each DNA molecule had at most one nucleosome. When such nucleosomal DNA molecules were unzipped with a loading rate clamp using the same conditions as those of Figure 3, nucleosomes were found at various locations on the template (Supplementary Fig. 5), likely due to the lack of any known nucleosome positioning element on this DNA sequence. Each unzipping trace contains two major regions of strong interaction, with the second region presumably located near the dyad. These nucleosomes possessed essentially identical characteristics to those of the 601 sequence, except that their peak forces within each region were typically smaller by a few pN, reflecting weaker interactions of histone with non-positioning DNA sequences. The key features remained essentially identical: the three regions of strong interactions with the strongest at the dyad, the 5 bp periodicity, and the loss of nucleosome stability upon dyad disruption. These results indicate that the conclusions of this work are not restricted to nucleosomes on the 601 sequence but are general to nucleosomes on any sequence.
Mechanical invasion of a nucleosome
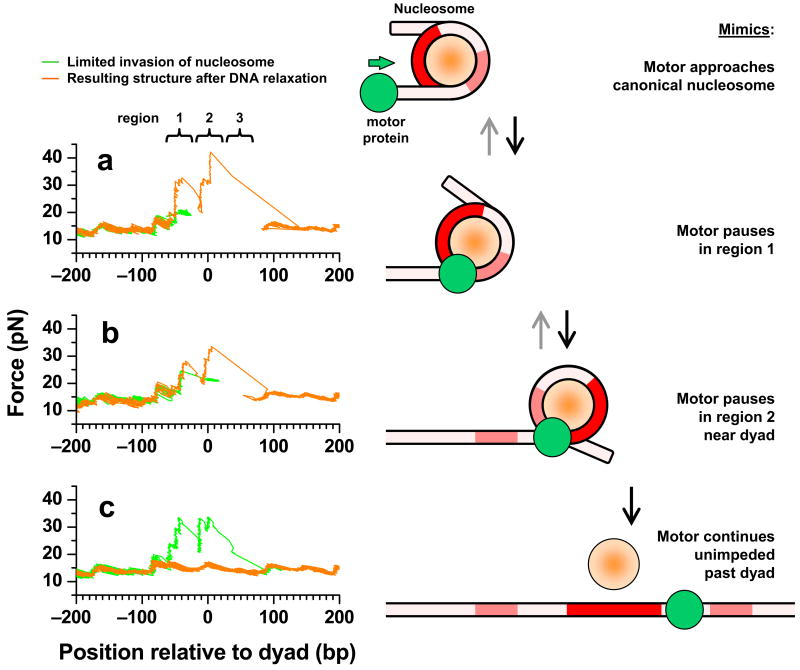
In order to mimic invasion by a motor protein as it progresses into a nucleosome, we carried out 3 sets of mechanical invasion experiments (Fig. 4). In the first set, unzipping was allowed to proceed into and then held within the first region of strong interactions, before the DNA was relaxed to allow rezipping (Fig. 4a). The state of the nucleosome was subsequently examined by unzipping through the entire 601 sequence. The majority of traces examined in this way (75%) showed a canonical nucleosome structure at the 601 sequence. The remaining 25% showed altered structures, likely due to incomplete re-annealing of the DNA in the presence of histones (Supplementary Fig. 6). In the second set, unzipping was allowed to proceed into and then held within the dyad region of interactions, before the DNA was relaxed to allow rezipping (Fig. 4b). The majority of the resulting structures (70%) again resembled a canonical nucleosome at the 601 sequence. In the third set, unzipping was allowed to proceed past the dyad region of interactions, before DNA was relaxed to allow rezipping (Fig. 4c). Subsequently, all traces showed force signatures indistinguishable from those of naked 601 sequence, indicating complete removal of the histone octamer from the 601 sequence. These results indicate that motor enzymes may be capable of accessing nearly half of the underlying DNA without resulting in histone dissociation.
Figure 4.
Mechanical unzipping (left) to mimic motor enzyme progression into a nucleosome (right). (a) DNA was unzipped with a loading rate clamp (8 pN s−1) until the unzipping force reached ~ 20 pN, which typically occurred within the 1st region of interactions (green curve). The unzipping force was then held at this force for 10 s, resulting in a horizontal force line due to the hopping of the unzipping fork among different positions within the first region. These steps mimic a motor invasion into the 1st region of interactions and subsequent pausing within the region (right). The tension in the DNA was then relaxed for ~ 3 s and the state of the nucleosome was determined by unzipping a second time (orange curve). (b) Similar to b, except that the unzipping force was held at ~ 21 pN immediately after the unzipping fork entered the dyad region of interactions. These steps mimic motor invasion into the dyad region of interactions before pausing (right). (c) Similar to b, except that DNA was unzipped past the dyad region of interactions. This mimics motor invasion past the dyad (right).
DISCUSSION
Histone-DNA interaction map of a nucleosome
This study presents a high resolution quantitative map of histone-DNA interactions in a nucleosome. It not only provides a direct measure of locations of interactions to near bp resolution, but also quantitatively assays the strengths of these interactions. The overall features of the interaction map are not specific to the 601 sequence but are shared by DNA of arbitrary sequence (Supplementary Fig. 5).
The histone-DNA interaction map reveals the existence of three regions of strong interactions. This is the most direct evidence that the histone-DNA interactions within a nucleosome are not uniform: the strongest region of interactions is located at the dyad and another two regions of strong interactions are ~ ±40 bp from the dyad. The locations of all three regions are strongly correlated with those estimated from the crystal structure of the nucleosome8,20. The central region is clearly the strongest and this observation explains why nucleosome stability has been shown to be most sensitive to DNA sequence near the dyad26. The locations of the off-dyad regions are also consistent with findings from our previous nucleosome stretching measurements14,17. This also indicates that in the single molecule stretching experiments, nucleosome spool geometry may not contribute significantly to force signatures or contribute in such a way that coincides with the effects due to the two regions of off-dyad interactions.A 5 bp periodicity in the interaction map was observed whereas prior to this work a 10 bp periodicity would have been expected. The crystal structure of the nucleosome shows that specific DNA-histone contacts are made each time the DNA minor groove faces the histone octamer surface, leading to binding sites spaced at ~10 bp1. Closer inspection shows that interactions from the two strands of the dsDNA completely stagger with each other and alternate between the two strands along the sequence at every 5 bp. However, in crystal structure analyses histone interaction with each minor groove of the DNA has been treated as a single binding site1,20,27. This is reasonable since disruption of a histone interaction with one of the DNA strands at a minor groove may result in a concurrent disruption of a histone interaction with the other strand. Prior to our experiments, we had anticipated observation of a 10 bp periodicity. The fact that we have actually observed a 5 bp periodicity indicates that the histone interactions with two strands of DNA at its minor groove are rather decoupled, and can thus be disrupted sequentially instead of simultaneously.
The interactions near exit and entrance DNA were found to be particularly weak, although they maintain the 5 bp periodicity. These weak interactions are expected to permit spontaneous peeling of DNA ends from the octamer surface as observed by equilibrium accessibility assays28,29.
Implication for transcription
Although RNA polymerases are known to be powerful molecular motors30,31, the presence of a nucleosome still presents a major obstacle2–7. The mechanical unzipping experiments described here resemble the action of RNA polymerase which opens up a transcription bubble and unzips the downstream DNA while advancing into a nucleosome (right panel of Fig. 4). The histone-DNA interaction map (Fig. 2) has significant implications for how RNA polymerases may gain access to DNA associated with a nucleosome. RNA polymerase is expected to initially proceed rather smoothly, but pause when it encounters the off-dyad interactions. Disruption of these interactions permits it to proceed toward the dyad. The polymerase will then pause most strongly within the dyad region of interactions. Once it overcomes the dyad interactions, it will proceed through the rest of the nucleosomal DNA with minimal resistance. The interaction map also predicts that the 601-positioned nucleosome acts as a polar barrier: transcription in the forward direction is less efficient than in the reverse direction. Likely, asymmetries of this sort exist in eukaryotic genomes, and may have functional importance for normal gene expression where positioned nucleosomes reside at key positions transited by Pol II32. Interestingly, many of these predictions have been verified by biochemical studies of Pol II or Pol III transcription through nucleosomes2–7. While the interaction map also suggests that transcription pausing may exhibit a finer ~5 bp periodic pattern, an ~10 bp periodicity has been observed5,6,33,34. Although this periodicity has been attributed to nucleosome restriction of RNAP rotation coupled with DNA loop formation, this work offers a simpler explanation. The ~10 bp periodicity in transcription pausing may be due to RNA polymerase cooperatively disrupting a pair of interactions located at each minor groove of DNA.
Although the pausing pattern of RNA polymerase is dictated by both the mechanical barriers encountered as well as its own motor properties, similarities between the dwell time in the histone-DNA interaction map (Fig. 2) and the polymerase pausing pattern within a nucleosome suggest that the barriers encountered by the polymerase are a major determinant of its pausing behavior. Thus, this explanation of the pausing pattern within a nucleosome provides a simpler explanation than existing models3,5,33. The consistency of the histone-DNA interaction map with biochemical assays of RNA polymerase pausing pattern is an indication that this map may also be used to predict how other motor enzymes pass through nucleosomes.
The results from nucleosome invasion experiments yield testable predictions regarding the fate of nucleosomes during transcription. If RNA polymerase backtracks before the dyad, histones will not dissociate from the DNA but will tend to reform a canonical nucleosome at the same location (Supplementary Discussion), perhaps encouraging further backtracking of the polymerase. Once the RNA polymerase passes the dyad, histones will most likely be removed from their original locations.
METHODS
Nucleosomal DNA templates
We prepared nucleosomal DNA templates using methods similar to those previously described16. Briefly, each DNA construct consisted of two separate segments (Supplementary Fig. 1a). An ~1.1 kbp anchoring segment was prepared by PCR from plasmid pRL57435 using a digoxigenin-labeled primer and then digested with BstXI (NEB) to produce a ligatable overhang. Each unzipping segment was prepared by PCR using a biotin-labeled primer and then digested with BstXI and dephosphorylated using CIP (NEB) to introduce a nick into the final DNA template. Nucleosomes were assembled from purified HeLa histones onto the unzipping fragment by a well established salt dialysis method36. The two segments were joined by ligation immediately prior to use. This produced the complete template that was labeled with a single dig tag on one end and a biotin tag located 7 bp after the nick in one DNA strand.
The forward 601 unzipping segment is ~ 0.8 kbp and was prepared by PCR from plasmid 60124 as described previously16. The reverse template is nearly identical to the forward template, except that the reverse unzipping segment was flipped so that the unzipping fork would approach the nucleosome from the opposite direction. To achieve this, the reverse segment was produced via different primers, such that the ligatable overhang produced through BstXI digestion and nick introduced via CIP were located on the end opposite that of the forward segment. The unzipping segment that does not contain any known nucleosome positioning element is ~ 0.8 kbp and was prepared by PCR from plasmid pBR322 (NEB).
Hairpin DNA templates
We prepared three different hairpin templates from the forward template (without nucleosomes) by truncating the unzipping segment at precise locations via restriction enzymes and ligating the same hairpin onto the end in each case. The lengths of the unzipping templates are indicated in Supplementary Fig. 2b.
Method of unzipping under constant force
For experiments involving unzipping through a nucleosome under a constant force, the unzipping started with a loading rate clamp (8 pN s−1) until the desired force of ~28 pN was reached within a nucleosome. The unzipping force was then held constant via feedback control of the coverslip position25. This force is much higher than the sequence-dependent unzipping force of the naked 601 sequence (13–16 pN), minimizing the dwell time contribution due solely to DNA base pairing interactions, but is small enough to allow sufficient dwell time at each DNA sequence position for detection. Upon reaching the end of the 601 sequence, the unzipping was continued under a loading rate clamp (8 pN s−1). Unzipping before and after the 601 segment under a constant loading rate generated distinct unzipping signatures that could be used for data alignment (see below).
Method of unzipping under constant loading rate
An optical trapping setup was used to unzip a single DNA molecule by moving the microscope coverslip horizontally away from the optical trap (Supplementary Fig. 1b). As barriers to fork progression were encountered, a computer-controlled feedback loop increased the applied load linearly with time (8 pN s−1) as necessary to overcome those barriers. Whenever the unzipping fork stopped, e.g., at an interaction, the unzipping force was ramped up linearly with time until the interaction was disrupted37. When two interactions occurred in close vicinity, upon the disruption of the first interaction the force was unable to relax back to the baseline before being ramped up again for the second interaction, subjecting this subsequent interaction to a higher initial force. Therefore, for each region of interactions, the dwell time histogram highlighted the edge of the region first encountered. Another feature of this method was the display of the distinctive force signature for a nucleosome, allowing for ease of identification of the nucleosome structure16 (compare traces in Supplementary Fig. 3 with Supplementary Fig. 4).
Data collection and alignment
Data were low pass filtered to 5 kHz, digitized at ~12 kHz and later filtered to 60 Hz. Previously, to improve the positional precision and accuracy, the experimental curves were aligned to the theoretical curve by cross-correlation of a region immediately preceding the nucleosome disruption16. In the current work, the precision and accuracy were further improved by an additional cross-correlation of a region immediately following the nucleosome disruption. To account for minor instrumental drift, trapping bead size variations, and DNA linker variations, the alignment allowed for a small additive shift (< 5 bp) and multiplicative linear stretch (< 2%) using algorithms similar to those previously described38.
Supplementary Material
Supplementary Information is linked to the online version of the paper at http://www.nature.com/nsmb.
Acknowledgments
We thank members of the Wang lab and Dr. B. Brower-Toland for critical reading of the manuscript, J. Jin for helpful advice with biochemical preparations, and Dr. D.S. Johnson for helpful discussions on instrumentation. We wish to acknowledge support from NIH grants (GM059849 to M.D.W; GM25232 to J.T.L.), the Keck Foundation (to M.D.W), the Cornell Nanobiotechnology Center (to M.D.W. and J.T.L.), and the Molecular Biophysics Training Grant Traineeship (to M.A.H.).
References
- 1.Luger K, Mader AW, Richmond RK, Sargent DF, Richmond TJ. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature. 1997;389:251–60. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- 2.Bondarenko VA, et al. Nucleosomes can form a polar barrier to transcript elongation by RNA polymerase II. Mol Cell. 2006;24:469–79. doi: 10.1016/j.molcel.2006.09.009. [DOI] [PubMed] [Google Scholar]
- 3.Kireeva ML, et al. Nature of the nucleosomal barrier to RNA polymerase II. Mol Cell. 2005;18:97–108. doi: 10.1016/j.molcel.2005.02.027. [DOI] [PubMed] [Google Scholar]
- 4.Kireeva ML, et al. Nucleosome remodeling induced by RNA polymerase II: loss of the H2A/H2B dimer during transcription. Mol Cell. 2002;9:541–52. doi: 10.1016/s1097-2765(02)00472-0. [DOI] [PubMed] [Google Scholar]
- 5.Studitsky VM, Kassavetis GA, Geiduschek EP, Felsenfeld G. Mechanism of transcription through the nucleosome by eukaryotic RNA polymerase. Science. 1997;278:1960–3. doi: 10.1126/science.278.5345.1960. [DOI] [PubMed] [Google Scholar]
- 6.Studitsky VM, Walter W, Kireeva M, Kashlev M, Felsenfeld G. Chromatin remodeling by RNA polymerases. Trends Biochem Sci. 2004;29:127–35. doi: 10.1016/j.tibs.2004.01.003. [DOI] [PubMed] [Google Scholar]
- 7.Walter W, Kireeva ML, Studitsky VM, Kashlev M. Bacterial polymerase and yeast polymerase II use similar mechanisms for transcription through nucleosomes. J Biol Chem. 2003;278:36148–56. doi: 10.1074/jbc.M305647200. [DOI] [PubMed] [Google Scholar]
- 8.Davey CA, Sargent DF, Luger K, Maeder AW, Richmond TJ. Solvent mediated interactions in the structure of the nucleosome core particle at 1.9 a resolution. J Mol Biol. 2002;319:1097–113. doi: 10.1016/S0022-2836(02)00386-8. [DOI] [PubMed] [Google Scholar]
- 9.Cosgrove MS, Boeke JD, Wolberger C. Regulated nucleosome mobility and the histone code. Nat Struct Mol Biol. 2004;11:1037–43. doi: 10.1038/nsmb851. [DOI] [PubMed] [Google Scholar]
- 10.Kouzarides T. Chromatin modifications and their function. Cell. 2007;128:693–705. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- 11.Segal E, et al. A genomic code for nucleosome positioning. Nature. 2006;442:772–8. doi: 10.1038/nature04979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bancaud A, et al. Nucleosome chiral transition under positive torsional stress in single chromatin fibers. Mol Cell. 2007;27:135–47. doi: 10.1016/j.molcel.2007.05.037. [DOI] [PubMed] [Google Scholar]
- 13.Bennink ML, et al. Unfolding individual nucleosomes by stretching single chromatin fibers with optical tweezers. Nat Struct Biol. 2001;8:606–10. doi: 10.1038/89646. [DOI] [PubMed] [Google Scholar]
- 14.Brower-Toland BD, et al. Mechanical disruption of individual nucleosomes reveals a reversible multistage release of DNA. Proc Natl Acad Sci U S A. 2002;99:1960–5. doi: 10.1073/pnas.022638399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cui Y, Bustamante C. Pulling a single chromatin fiber reveals the forces that maintain its higher-order structure. Proc Natl Acad Sci U S A. 2000;97:127–32. doi: 10.1073/pnas.97.1.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shundrovsky A, Smith CL, Lis JT, Peterson CL, Wang MD. Probing SWI/SNF remodeling of the nucleosome by unzipping single DNA molecules. Nat Struct Mol Biol. 2006;13:549–54. doi: 10.1038/nsmb1102. [DOI] [PubMed] [Google Scholar]
- 17.Brower-Toland B, et al. Specific contributions of histone tails and their acetylation to the mechanical stability of nucleosomes. J Mol Biol. 2005;346:135–46. doi: 10.1016/j.jmb.2004.11.056. [DOI] [PubMed] [Google Scholar]
- 18.Gemmen GJ, et al. Forced unraveling of nucleosomes assembled on heterogeneous DNA using core histones, NAP-1, and ACF. J Mol Biol. 2005;351:89–99. doi: 10.1016/j.jmb.2005.05.058. [DOI] [PubMed] [Google Scholar]
- 19.Pope LH, et al. Single chromatin fiber stretching reveals physically distinct populations of disassembly events. Biophys J. 2005;88:3572–83. doi: 10.1529/biophysj.104.053074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Luger K, Richmond TJ. DNA binding within the nucleosome core. Curr Opin Struct Biol. 1998;8:33–40. doi: 10.1016/s0959-440x(98)80007-9. [DOI] [PubMed] [Google Scholar]
- 21.Mihardja S, Spakowitz AJ, Zhang Y, Bustamante C. Effect of force on mononucleosomal dynamics. Proc Natl Acad Sci U S A. 2006;103:15871–6. doi: 10.1073/pnas.0607526103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kulic IM, Schiessel H. DNA spools under tension. Phys Rev Lett. 2004;92:228101. doi: 10.1103/PhysRevLett.92.228101. [DOI] [PubMed] [Google Scholar]
- 23.Sakaue T, Lowen H. Unwrapping of DNA-protein complexes under external stretching. Phys Rev E Stat Nonlin Soft Matter Phys. 2004;70:021801. doi: 10.1103/PhysRevE.70.021801. [DOI] [PubMed] [Google Scholar]
- 24.Lowary PT, Widom J. New DNA sequence rules for high affinity binding to histone octamer and sequence-directed nucleosome positioning. J Mol Biol. 1998;276:19–42. doi: 10.1006/jmbi.1997.1494. [DOI] [PubMed] [Google Scholar]
- 25.Johnson DS, Bai L, Smith BY, Patel SS, Wang MD. Single-molecule studies reveal dynamics of DNA unwinding by the ring-shaped T7 helicase. Cell. 2007;129:1299–309. doi: 10.1016/j.cell.2007.04.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Thastrom A, Bingham LM, Widom J. Nucleosomal locations of dominant DNA sequence motifs for histone-DNA interactions and nucleosome positioning. J Mol Biol. 2004;338:695–709. doi: 10.1016/j.jmb.2004.03.032. [DOI] [PubMed] [Google Scholar]
- 27.Muthurajan UM, et al. Crystal structures of histone Sin mutant nucleosomes reveal altered protein-DNA interactions. EMBO J. 2004;23:260–71. doi: 10.1038/sj.emboj.7600046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li G, Levitus M, Bustamante C, Widom J. Rapid spontaneous accessibility of nucleosomal DNA. Nat Struct Mol Biol. 2005;12:46–53. doi: 10.1038/nsmb869. [DOI] [PubMed] [Google Scholar]
- 29.Li G, Widom J. Nucleosomes facilitate their own invasion. Nat Struct Mol Biol. 2004;11:763–9. doi: 10.1038/nsmb801. [DOI] [PubMed] [Google Scholar]
- 30.Wang MD, et al. Force and velocity measured for single molecules of RNA polymerase. Science. 1998;282:902–7. doi: 10.1126/science.282.5390.902. [DOI] [PubMed] [Google Scholar]
- 31.Galburt EA, et al. Backtracking determines the force sensitivity of RNAP II in a factor-dependent manner. Nature. 2007;446:820–3. doi: 10.1038/nature05701. [DOI] [PubMed] [Google Scholar]
- 32.Albert I, et al. Translational and rotational settings of H2A.Z nucleosomes across the Saccharomyces cerevisiae genome. Nature. 2007;446:572–6. doi: 10.1038/nature05632. [DOI] [PubMed] [Google Scholar]
- 33.Studitsky VM, Clark DJ, Felsenfeld G. Overcoming a nucleosomal barrier to transcription. Cell. 1995;83:19–27. doi: 10.1016/0092-8674(95)90230-9. [DOI] [PubMed] [Google Scholar]
- 34.Bednar J, Studitsky VM, Grigoryev SA, Felsenfeld G, Woodcock CL. The nature of the nucleosomal barrier to transcription: direct observation of paused intermediates by electron cryomicroscopy. Mol Cell. 1999;4:377–86. doi: 10.1016/s1097-2765(00)80339-1. [DOI] [PubMed] [Google Scholar]
- 35.Schafer DA, Gelles J, Sheetz MP, Landick R. Transcription by single molecules of RNA polymerase observed by light microscopy. Nature. 1991;352:444–8. doi: 10.1038/352444a0. [DOI] [PubMed] [Google Scholar]
- 36.Lee KM, Narlikar G. Assembly of nucleosomal templates by salt dialysis. Curr Protoc Mol Biol. 2001 doi: 10.1002/0471142727.mb2106s54. Chapter 21, Unit 21 6. [DOI] [PubMed] [Google Scholar]
- 37.Koch SJ, Shundrovsky A, Jantzen BC, Wang MD. Probing protein-DNA interactions by unzipping a single DNA double helix. Biophys J. 2002;83:1098–105. doi: 10.1016/S0006-3495(02)75233-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Deufel C, Wang MD. Detection of forces and displacements along the axial direction in an optical trap. Biophys J. 2006;90:657–67. doi: 10.1529/biophysj.105.065458. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Information is linked to the online version of the paper at http://www.nature.com/nsmb.