Abstract
Aim
Type 1 diabetes (T1D) is a complex trait for which variation in the classical human leucocyte antigen (HLA) loci within the Major Histocompatibility Complex (MHC) significantly influences disease risk. To date, HLA class II DR-DQ genes confer the strongest known genetic effect in T1D. HLA loci may also influence T1D through additional inherited or non-inherited effects. Evidence for the role of increased maternal–offspring HLA compatibility, and both parent-of-origin (POO) and non-inherited maternal HLA (NIMA) effects in autoimmune disease has been previously established. The current study tested hypotheses that classical HLA loci influence T1D through these mechanisms, in addition to genetic transmission of particular risk alleles.
Methods
The Type 1 Diabetes Genetics Consortium (T1DGC) cohort was of European descent and consisted of 2271 affected sib-pair families (total n = 11 023 individuals). Class I genes HLA-A, Cw and B, and class II genes HLA-DRB1, DQA1, DQB1, DPA1 and DPB1 were studied. The pedigree disequilibrium test was used to examine transmission of HLA alleles to individuals with T1D. Conditional logistic regression was used to model compatibility relationships between mother–offspring and father–offspring for all HLA loci. POO and NIMA effects were investigated by comparing frequencies of maternal and paternal transmitted and non-transmitted HLA alleles for each locus. Analyses were also stratified by gender of T1D-affected offspring.
Results
Strong associations were observed for all classical HLA loci except for DPA1, as expected. Compatibility differences between mother–offspring and father–offspring were not observed for any HLA loci. Furthermore, POO and NIMA HLA effects influencing T1D were not present.
Conclusions
Maternal–offspring HLA compatibility, POO and NIMA effects for eight classical HLA loci were investigated. Results suggest that these HLA-related effects are unlikely to play a major role in the development of T1D.
Keywords: HLA, maternal–offspring HLA compatibility, non-inherited maternal, parent-of-origin, T1D
Introduction
Type 1 diabetes (T1D) (MIM 222100) is an autoimmune disease characterized by chronic T cell–mediated destruction of pancreatic insulin-producing β-cells [1]. While age-at-onset peaks in late childhood, adults also develop this disorder, and incidence rates for females and males are similar [2]. Incidence in the USA is estimated to be ~15 in 100 000 children per year; however, it varies widely around the world and has been increasing over the past decade [2]. Although the aetiology of T1D remains unknown, evidence for genetic susceptibility is well established [3,4]. Concordance for T1D in monozygotic twins is 70% compared with just 13% in dizygotic twins; the relative risk for sibs (λs) is approximately 15 in the US Caucasian population [5].
The human leucocyte antigen (HLA) class II genes HLA-DRB1, DQA1 and DQB1 in the Major Histocompatibility Complex (MHC) region (6p21) are directly involved; the MHC region accounts for 40–50% of the genetic susceptibility in individuals of Northern European Caucasian descent [6]. The majority of Caucasian individuals with T1D carry the HLA-DR3 (DRB1*0301-DQA1*0501-DQB1*0201) or DR4 (DRB1*04-DQA1*0301-DQB1*0302) class II haplotype, and approximately 30–50%of individuals are DR3/DR4 heterozygotes [7]. DR3/DR4 heterozygosity confers the highest diabetes risk [8]. Different class II HLA associations with T1D are present in non-Caucasian populations [9]. Class I HLA-B has also been associated with T1D risk, specifically the B*39 and B*18 alleles [10,11]. Interestingly, the class II HLA-DR2 (DRB1*1501-DQB1*0602) haplotype is protective in all populations studied to date [12]. Additional non-MHC genetic risk factors for T1D include PTPN22 (1p13), CTLA4 (2q33) and IDDM2 (11p15) [13–15]. Environmental factors have also been strongly implicated in both pathogenesis and outcome of T1D [16].
HLA loci may also influence T1D through additional inherited or non-inherited effects. Differences in maternal and paternal transmission rates, or parent-of-origin (POO) effects, have been observed in T1D. One potential mechanism is ‘genomic imprinting’, an epigenetic modification of the genome that results in unequal transcription of parental alleles and subsequent allele expression, depending on whether alleles were transmitted maternally or paternally.
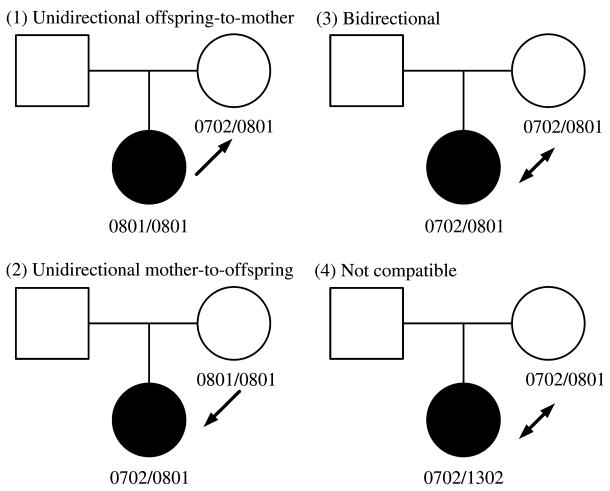
HLA compatibility between a mother and her offspring may also contribute to susceptibility to autoimmunity, possibly because HLA similarity between the mother and foetus may promote the persistence of foetal cells in the host or perhaps through specific exposure to non-inherited maternal HLA (NIMA) risk or protective alleles. Risk for an autoimmune disease would be potentially increased in either mother or offspring. Maternal–offspring HLA compatibility that increases disease risk in the mother could explain: (i) increased prevalence of autoimmune diseases in women following their child-bearing years and (ii) clinical similarities between scleroderma (systemic sclerosis (SSc)] and graft-vs.-host disease [17]. With regard to T1D, maternal–offspring HLA compatibility could affect risk in the offspring. Maternal–offspring cell trafficking is common and bidirectional; maternal nucleated cell and plasma DNA transfers into foetal circulation in 24 and 30% of offspring, respectively [18]. Maternal–offspring effects can present as excess HLA compatibility between the mother and affected offspring or excess maternal homozygosity. Possible maternal–offspring HLA compatibility relationships are illustrated in figure 1.
Fig. 1.
Illustration of possible maternal–offspring human leucocyte antigen (HLA) compatibility relationships using the HLA-B locus as an example.
Finally, non-host exposure during foetal development and potential long-term persistence of maternal cells in offspring may play a role in T1D pathogenesis [19–22]. The developing immune system of the foetus is exposed to NIMA in utero [21,23,24]. Decreased B-cell responses and cytotoxic T-cell activity to HLA class I NIMA have been reported [25–29]. NIMA can have a lifelong influence on the immune system and may tolerize or pre-dispose to autoimmune reactions [30–32].
The current study tested hypotheses that classical HLA loci influence T1D through these additional biological mechanisms in addition to genetic transmission of particular risk alleles.
Methods
Subjects
TheT1DGCcohort (release 2007.02.MHC) was of European descent and consisted of 2271 affected sibling-pair families with 11 023 individuals. This research resource has been previously described [33]. Briefly, the families were derived from multiple cohorts: Asia-Pacific (177), British Diabetes Association (BDA) (422), Danish (147), European (475), Human Biological Data Interchange (HBDI) (421), Joslin (117), North American (321), Sardinian (77) and UK (114). The mean number of individuals per family was 5 and ranged from 3 to 26. The mean number of generations per family was two and ranged from one to four.
A subset of 1780 affected sib pair (ASP) families had at least two-digit classical HLA genotypes available. Analyses were conducted on trio families consisting of two parents and one affected offspring; one affected offspring per family was chosen randomly from all affected offspring with HLA-DRB1 genotypes. We created two additional trio family samples for gender-stratified analyses. For the male sample, we randomly selected one male affected offspring per family. There were 1376 families with at least one male affected offspring. For the female sample, we randomly selected one female affected offspring per family. There were 1291 families with at least one female affected offspring. Parental genotypes were available to determine transmission for all affected offspring. A total of 6227 individuals were analysed, as presented in table 1.
Table 1.
Type 1 diabetes families with two-digit genotyping available for the classical human leucocyte antigen (HLA) loci used in analyses of parent-of-origin and non-inherited maternal HLA effects
| Sample | Families | Families with affected offspring of one gender only | Families with affected offspring of both genders | Affected offspring in analysis | Individuals in analysis |
|---|---|---|---|---|---|
| Overall | 1780 | 893 | 887 | 1780 | 5340 |
| Male | 1376 | 489 | 887 | 1376 | 4128 |
| Female | 1291 | 404 | 887 | 1291 | 3873 |
For the HLA compatibility analyses, we limited the samples described in the previous paragraph to 1213ASP families (5804 individuals) with complete four-digit classical HLA genotypes available. There were 954 families and 876 families in the male and female samples, respectively. A total of 4256 individuals were utilized for the analyses of compatibility patterns, as shown in table 2.
Table 2.
Subset of type 1 diabetes families with four-digit genotyping available for the classical human leucocyte antigen (HLA) loci used in the HLA compatibility analyses
| Sample | Families | Families with affected offspring of one gender only | Families with affected offspring of both genders | Affected offspring in analysis | Individuals in analysis |
|---|---|---|---|---|---|
| Overall | 1213 | 596 | 617 | 1213 | 3639 |
| Male | 954 | 337 | 617 | 954 | 2862 |
| Female | 876 | 259 | 617 | 876 | 2628 |
The families were collected for both linkage and association studies; the sample size was designed to be sufficiently large to detect associations as well as secondary gene effects in a region such as HLA. The data were generated as part of a high-density screen of the MHC designed for association and haplotype analysis and to detect genes in the HLA region additional to the well-documented HLADR-DQ effect. The nuclear family study design is advantageous because it precludes potential confounding from ethnic mismatching between patients and randomly ascertained controls because of population stratification, migration or admixture. This design also reduces the potential of misclassification error from genotyping because we can check the data for pedigree inconsistencies.
Classical HLA Genotyping
The T1DGC protocol has been previously described [34].
Statistical Analyses
We used PEDCHECK (v1.1) to identify pedigree inconsistencies in our overall sample of 1780 trio families [35]. For any pedigrees with an inconsistency, we “zeroed out” genotypes for the entire family at that specific locus only, assuming a genotyping error. MEGA2 (v3.0 R12) was used to manipulate data [36]. The pedigree disequilibrium test (PDT 6.0, build 5) was used to perform to examine frequencies of transmitted vs. non-transmitted alleles for each HLA locus [37]. The PDT is a powerful analytical method that uses genetic data from related nuclear families and discordant sibships within extended pedigrees.
We defined maternal–offspring compatibility categorically: (i) unidirectional child-to-mother compatible, (ii) unidirectional mother-to-offspring compatible, (iii) bidirectional and (4) incompatible. We modelled maternal–offspring compatibility in R (v2.6) using conditional logistic regression and for controls the compatibility of the affected child to the father was used [38]. The analysis was restricted to trios from the previously described overall, male and female samples that had complete four-digit genotyping information available.
We pair matched on family in a matched case–control analysis using conditional logistic regression; parent’s gender was the outcome [39].
In the above formula, n refers to the number of families in the matched analysis, unidirectional p. is unidirectional parent-to-child compatibility and unidirectional c. is unidirectional child-to-parent compatibility. These analyses were repeated with HLA compatibility as a binary exposure (any compatibility vs. none).
For the POO analyses, frequencies of maternal vs. paternal transmitted alleles were compared using a chi-squared contingency table test for heterogeneity in AFBAC (v1.13) [40]. The same test was used in the NIMA analyses to compare frequencies of maternal vs. paternal non-transmitted alleles. Both global and allele-specific analyses (when appropriate) were performed. Based on a Bonferroni correction for the total number of tests performed in this study (n = 24), a criterion of p < 0.002 was set for statistical significance.
Results
There were no pedigree inconsistencies for HLA-DQA1, DPA1 or DPB1. There were one, six and five families with pedigree inconsistencies at the HLA-DRB1, DQB1 and Cw loci, respectively. None of the families had more than one pedigree inconsistency, indicating genotyping error rather than non-paternity. As expected, the majority of classical HLA loci were strongly associated with T1D in both the overall and gender-stratified analyses. The only exception was the class II gene HLA-DPA1, which did not show evidence for association. Table 3 displays global p values. The primary disease genes are DR-DQ; we have not investigated whether the associations at the other loci are because of linkage disequilibrium with DR-DQ. Table S1 presents observed and expected transmitted and non-transmitted allele frequencies with the test statistic and odds ratio (OR).
Table 3.
Results from pedigree disequilibrium test analyses of transmission of HLA alleles to individuals with type 1 diabetes
| Global p values |
|||
|---|---|---|---|
| HLA locus | Overall* (n = 1780) |
Male† (n = 1376) |
Female‡ (n = 1291) |
| A | 7.2 × 10−9 | 2.3 ×10−8 | 1.4 × 10−5 |
| Cw | 7.2 × 10−11 | 8.7 ×10−11 | 4.8 × 10−11 |
| B | 8.2 × 10−11 | 8.9 ×10−11 | 9.1 × 10−11 |
| DRB1 | 8.4 × 10−11 | 9.2 ×10−11 | 8.7 × 10−11 |
| DQA1 | 7.6 × 10−11 | 8 ×10−11 | 7.4 × 10−11 |
| DQB1 | 6.6 × 10−11 | 8.4 ×10−11 | 9.5 × 10−11 |
| DPA1 | 0.03 | 0.14 | 0.36 |
| HLA-DPB1 | 4.9 × 10−11 | 1.3 ×10−8 | 6.6 × 10−11 |
HLA, human leucocyte antigen.
The overall sample (n = 1213) compares transmitted vs. non-transmitted HLA alleles using one affected offspring per family.
The male sample (n = 954) compares transmitted vs. non-transmitted HLA alleles using one affected offspring per family.
The female sample (n = 876) compares transmitted vs. non-transmitted HLA alleles using one affected offspring per family.
Unidirectional offspring-to-mother compatibility at the class I genes HLA-A, Cw and B and the class II genes HLA-DRB1, DQA1, DQB1, DPA1 and DPB1 was not associated with T1D. The corresponding OR, 95% confidence intervals and p values are listed in table 4. Unidirectional mother-to-offspring compatibility, bidirectional compatibility and any compatibility did not demonstrate association with T1D (data not shown). Table S2 provides frequencies of maternal–offspring and paternal–offspring HLA compatibility relationships.
Table 4.
Results from analyses of maternal vs. paternal unidirectional offspring-to-parent HLA compatibility in type 1 diabetes families
| HLA locus | Sample | OR (95% CI) | Global p value |
|---|---|---|---|
| A | Overall* | 0.74 (0.42–1.30) | 0.31 |
| Male† | 0.78 (0.41–1.46) | 0.44 | |
| Female‡ | 1.20 (0.61–2.34) | 0.59 | |
| Cw | Overall | 0.87 (0.42–1.82) | 0.71 |
| Male | 0.60 (0.25–1.43) | 0.25 | |
| Female | 0.76 (0.30–1.90) | 0.57 | |
| B | Overall | 0.41 (0.14–1.22) | 0.11 |
| Male | 0.18 (0.04–0.84) | 0.03 | |
| Female | 0.38 (0.11–1.26) | 0.11 | |
| DRB | Overall | 0.77 (0.37–1.58) | 0.48 |
| Male | 0.64 (0.29–1.42) | 0.28 | |
| Female | 0.60 (0.26–1.39) | 0.22 | |
| DQA | Overall | 0.99 (0.61–1.61) | 0.96 |
| Male | 0.85 (0.50–1.46) | 0.56 | |
| Female | 0.83 (0.47–1.47) | 0.53 | |
| DQB | Overall | 0.78 (0.46–1.33) | 0.35 |
| Male | 0.70 (0.38–1.28) | 0.25 | |
| Female | 0.63 (0.33–1.19) | 0.15 | |
| DPA | Overall | 0.97 (0.50–1.90) | 0.93 |
| Male | 0.66 (0.28–1.57) | 0.36 | |
| Female | 2.30 (0.97–5.45) | 0.05 | |
| DPB | Overall | 1.04 (0.68–1.58) | 0.86 |
| Male | 0.89 (0.55–1.43) | 0.63 | |
| Female | 0.83 (0.51–1.30) | 0.46 |
HLA, human leucocyte antigen.
The overall sample (n = 1213) compares maternal vs. paternal HLA compatibility using one affected offspring per family.
The male sample (n = 954) compares maternal vs. paternal HLA compatibility using one affected male offspring per family.
The female sample (n = 876) compares maternal vs. paternal HLA compatibility using one affected female offspr1ing per family.
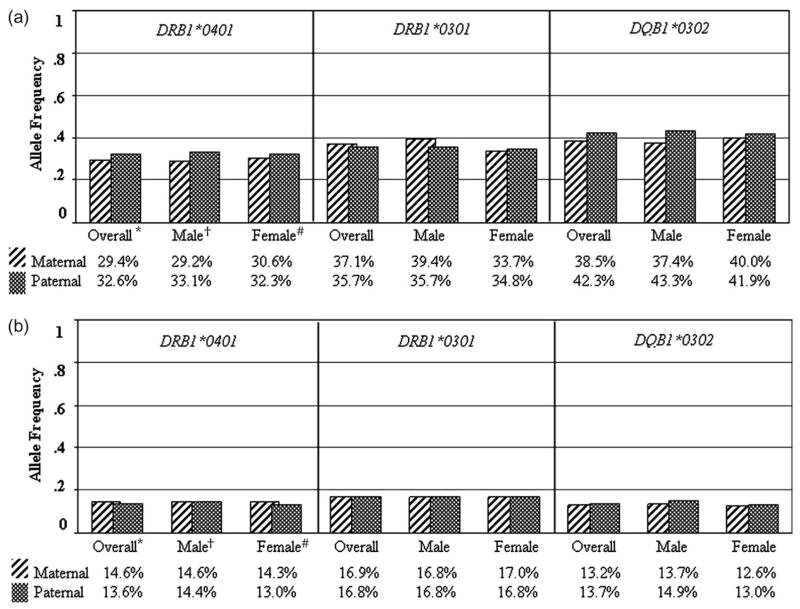
In addition, POO and NIMA effects were also examined for each classical HLA locus; evidence for involvement in T1D was not present (table 5) even when specific T1D risk alleles were examined separately, including DRB1*0401, DRB1*0301 and DQB1*0302. Table S3 shows observed and expected allele-specific transmitted maternal and paternal allele frequencies with ORs. Table S4 shows observed and expected allele-specific non-transmitted maternal and paternal allele frequencies with ORs. Comparison of paternal and maternal transmitted and paternal and maternal non-transmitted T1D risk HLA alleles in the overall data set and for male and female cases analysed separately revealed nearly identical frequencies (figure 2).
Table 5.
Results from analyses of parent-of-origin (POO) HLA effects and non-inherited maternal HLA (NIMA) effects (non-transmitted maternal vs. paternal alleles) in type 1 diabetes families
| Maternal vs. paternal |
|||||||
|---|---|---|---|---|---|---|---|
| Transmitted |
Non-transmitted |
||||||
| HLA locus | Sample | χ2 | d.f. | p | χ2 | d.f. | p |
| A | Overall* | 7.9 | 18 | 0.98 | 18.3 | 18 | 0.44 |
| Male† | 12.4 | 17 | 0.77 | 17.8 | 17 | 0.40 | |
| Female‡ | 15.1 | 18 | 0.66 | 17.0 | 18 | 0.53 | |
| Cw | Overall | 23.4 | 21 | 0.32 | 22.1 | 21 | 0.39 |
| Male | 41.2 | 21 | 0.01 | 20.1 | 21 | 0.51 | |
| Female | 7.1 | 20 | 1.00 | 15.7 | 20 | 0.73 | |
| B | Overall | 30.8 | 36 | 0.71 | 42.1 | 36 | 0.23 |
| Male | 46.6 | 36 | 0.11 | 41.9 | 36 | 0.23 | |
| Female | 29.8 | 33 | 0.63 | 36.2 | 33 | 0.32 | |
| DRB | Overall | 39.6 | 32 | 0.17 | 36.4 | 32 | 0.27 |
| Male | 33.9 | 31 | 0.33 | 19.6 | 31 | 0.94 | |
| Female | 29.3 | 32 | 0.60 | 45.9 | 32 | 0.05 | |
| DQA | Overall | 14.7 | 8 | 0.07 | 5.8 | 8 | 0.66 |
| Male | 13.2 | 7 | 0.07 | 2.2 | 7 | 0.95 | |
| Female | 5.9 | 8 | 0.66 | 9.8 | 8 | 0.28 | |
| DQB | Overall | 20.3 | 11 | 0.04 | 6.9 | 11 | 0.81 |
| Male | 18.1 | 11 | 0.08 | 2.7 | 11 | 0.99 | |
| Female | 14.8 | 11 | 0.19 | 11.6 | 11 | 0.39 | |
| DPA | Overall | 14.9 | 7 | 0.04 | 6.2 | 7 | 0.52 |
| Male | 7.5 | 7 | 0.38 | 7.1 | 7 | 0.42 | |
| Female | 12.1 | 6 | 0.06 | 4.7 | 6 | 0.58 | |
| DPB | Overall | 26.3 | 31 | 0.71 | 24.7 | 31 | 0.78 |
| Male | 15.4 | 27 | 0.96 | 18.5 | 27 | 0.89 | |
| Female | 25.3 | 31 | 0.75 | 23.1 | 31 | 0.85 | |
HLA, human leucocyte antigen.
The overall sample (n = 1780) compares maternal vs. paternal transmitted or non-transmitted alleles using one affected offspring per family.
The male sample (n = 1376) compares maternal vs. paternal transmitted or non-transmitted alleles using one affected male offspring per family.
The female sample (n = 1291) compares maternal vs. paternal transmitted or non-transmitted alleles using one affected female offspring per family.
Fig. 2.
(A) Transmitted paternal and maternal allele frequencies for type 1 diabetes (T1D) susceptibility alleles from analyses of parent-of-origin (POO) effects in T1D families; (B) non-transmitted paternal and maternal allele frequencies for T1D susceptibility alleles from analyses of non-inherited maternal HLA (NIMA) effects in T1D families. *The overall sample (n = 1780) compares maternal vs. paternal non-transmitted alleles using one affected child per family. †The male sample (n = 1376) compares maternal vs. paternal non-transmitted alleles using one affected male offspring per family. ‡The female sample (n = 1291) compares maternal vs. paternal non-transmitted alleles using one affected female offspring per family. HLA, human leucocyte antigen.
Discussion
To date, the MHC confers the strongest known genetic effect in T1D; associations are well established for class I and class II loci, particularly for the class II HLA-DRB1* 0301- and *04-associated haplotypes. HLA loci may also influence T1D through additional inherited or non-inherited effects. Evidence for increased maternal–offspring HLA class II compatibility has been reported for systemic lupus erythematosus (SLE) and SSc. Compared with controls, male SLE patients were more likely to have HLA class II genotypes identical to their mothers [41]. In addition, compared with controls, SSc patients exhibited increased HLA class II compatibility with their offspring or with their offspring or mother [42,43]. Taken together, these results suggest that HLA class II loci may be involved in the aetiology of both SLE and SSc through undefined mechanism(s) that are dependent on maternal–offspring compatibility. Several biological hypotheses have been proposed where increased compatibility could result in a small number of non-host cells that could ultimately (i) cause dysregulation among host cells, (ii) lead to presentation of non-host peptides by host cells to other host cells, (iii) inactivate T lymphocytes upon interaction or (iv) undergo differentiation and become targets of a later immune response [20,44–46].
The current study is the first to examine maternal–offspring HLA compatibility in T1D. Our results indicate that maternal–offspring compatibility at the MHC class I genes HLA-A, Cw and B and class II genes HLA-DRB1, DQA1, DQB1, DPA1 and DPB1 does not influence T1D. Our study used paternal–offspring HLA compatibility as controls in an analysis matched on family. Future studies would ideally test for differences in patterns of compatibility using an independent controls sample of mother–offspring pairs without T1D.
POO effects, potentially operating through the phenomenon of imprinting, have been observed previously in T1D and multiple sclerosis (MS) with respect to the inheritance of HLA class II alleles but results have been inconsistent. Excess paternal inheritance of the DR3 risk haplotype has been reported in female Sardinian MS patients [47]. More recently, excess maternal inheritance of HLA-DRB1*15 was observed in a larger study of 1515 MS families (p = 0.005) [48]. An early study of 107 T1D families reported increased paternal transmission of DR4 to affected and unaffected offspring (72.1%) compared with maternal transmission (55.6%) [49]. A study of 28 Japanese T1D families reported that the DQA1*0301-DQB1*0302 haplotype exhibited preferential maternal transmission and strong transmission disequilibrium with T1D positive for antibodies to glutamic acid decarboxylase [50]. Others have reported no POO effects in HLA class II alleles [51–53]; these studies examined 61, 108 and 282 T1D families, respectively. Many of these studies suffered from relatively small sample sizes and did not account for multiple testing when reporting statistical significance. In contrast, this study is the largest (table 2) to date to examine POO effects in the classical HLA loci and account for multiple statistical tests. Results do not support a role for HLA-associated POO effects in T1D, even for T1D risk alleles DRB1*0401, DQB1*0302 and DRB1*0301, and indeed are in agreement with others [51–53].
Interestingly, POO effects have also been examined in T1D for non-MHC genetic risk factors. There were no POO effects observed for PTPN22 in a study of 341 T1D families [54]. A study of the CTLA4 exon 1 polymorphism (49 A/G) in 70 T1D families showed increased maternal allele transmission of the G allele to T1D-affected offspring: 71% vs. the random 50% observed in unaffected offspring (p < 0.03). This distortion was stronger in T1D offspring with maternal inheritance of HLA-DRB1*03 (80%, p < 0.01) or variable number tandem repeats at the IDDM2 locus (80%, p < 0.02) [55]. A paternal origin effect has been observed for IDDM8 (6q27) and a maternal origin effect has been observed in 404 parent–offspring T1D trios for the IGF2R locus (6q26) [3,56]. Further research is needed to confirm these findings.
There is evidence that HLA alleles may also act as environmental risk factors. This current study tested the hypothesis that cells and antigens of the mother may modulate the antigen-specific reactivity of the foetal immune system. Exposure to NIMA via several different mechanisms may therefore shape the immune repertoire of the offspring and either predispose to or protect against future immune reactions. A tolerogenic effect may explain the longer survival of renal transplants from sibling donors expressing NIMA vs. non-inherited paternal antigens (NIPA) [57]. In the pre-cyclosporine era, breastfeeding exposure was associated with improved graft survival in recipients of maternal kidney transplants [58,59]. The role of breast milk in this observation was confirmed using a highly immunogenic heart allograft mouse model in which both in utero exposure and milk feeding were required for the NIMA effect [60].
In addition to maternal–offspring cell trafficking and oral exposure through breast milk, another potential mechanism for NIMA is maternal microchimerism, when a small population of cells or DNA in an individual is derived from their mother. Maternal cells have been detected in offspring several decades following birth [61]. Compared with healthy women, female SSc patients have increased frequencies of maternal cells in their peripheral blood cells [62].
An NIMA effect on risk for rheumatoid arthritis (RA) has been explored in several studies. An early study reported association between NIMA and RA for HLA-DR4 alleles [63]. Negative findings were later reported by a study of familial RA: frequencies of HLA-DRB1*04, *0401/*0404, and shared epitope (SE)-positive NIMA compared with NIPA were not increased in RA patients lacking these susceptibility alleles [64]. A later study reported an excess of DRB1*04 and SE NIMA (p = 0.05) compared with NIPA; a combined analysis with previous studies showed that mothers were more likely to carry a non-inherited DRB1*04 and SE alleles [65]. Recently, in the largest study of NIMA to date, the first evidence for a protective NIMA effect was reported: a mother carrying the protective amino acid sequence DERAA (HLA-DRB1*0103, *0402, *1103, *1301 and *1304) at the SE may transfer protection against RA to her DERAA-negative offspring [66].
To date, only one study of NIMA effects in T1Dhas been reported. T1D patients who did not carry any high-risk HLA alleles presented HLA DR3-DQ2 and DR4-DQ8 risk haplotypes more frequently as NIMA compared with NIPA [67]. The results from the current study, however, do not support a NIMA effect in T1D. A global test for NIMA effects, and a specific examination of the known T1D risk alleles DRB1*0401, DQB1*0302 and DRB1*0301 revealed negative results.
In conclusion, the largest study of T1D families, to date, does not support a major role for the classical HLA loci in disease susceptibility mediated through maternal–offspring compatibility, POO or NIMA effects.
Supplementary Material
Table S1 Allele-specific association results for classical human leucocyte antigen loci in type 1 diabetes families: observed transmitted and non-transmitted allele frequencies, expected allele frequency, test statistic, odds ratio and p value.
Table S2 Frequencies of human leucocyte antigen (HLA) maternal–offspring and paternal–offspring HLA compatibility relationships in type 1 diabetes families.
Table S3 Allele-specific results from parent-of-origin analyses for classical human leucocyte antigen loci in type 1 diabetes families: observed transmitted maternal vs. paternal allele frequencies, expected allele frequency, test statistic, odds ratio and p value.
Table S4 Allele-specific results from non-inherited maternal human leucocyte antigen (HLA) analyses for classical HLA loci in type 1 diabetes families: observed non-transmitted maternal vs. paternal allele frequencies, expected allele frequency, test statistic, odds ratio and p value.
Acknowledgments
The project described was supported by grant number F31AI075609 from the National Institute of Allergy and Infectious Diseases. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institute of Allergy and Infectious Diseases or the National Institutes of Health. This research used resources provided by the Type 1 Diabetes Genetics Consortium, a collaborative clinical study sponsored by the National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK), National Institute of Allergy and Infectious Diseases (NIAID), National Human Genome Research Institute (NHGRI), National Institute of Child Health and Human Development (NICHD), and Juvenile Diabetes Research Foundation International (JDRF) and supported by U01 DK062418.
Footnotes
Conflict of interest: The authors declare that they have no conflicts of interest in publishing this article.
Supporting Information: Additional Supporting Information may be found in the online version of this article.
References
- 1.Eisenbarth GS. Type I diabetes mellitus. A chronic autoimmune disease. N Engl J Med. 1986;314:1360–1368. doi: 10.1056/NEJM198605223142106. [DOI] [PubMed] [Google Scholar]
- 2.Incidence and trends of childhood Type 1 diabetes worldwide 1990–1999. Diabet Med. 2006;23:857–866. doi: 10.1111/j.1464-5491.2006.01925.x. [DOI] [PubMed] [Google Scholar]
- 3.McCann JA, Xu YQ, Frechette R, Guazzarotti L, Polychronakos C. The insulin-like growth factor-II receptor gene is associated with type 1 diabetes: evidence of a maternal effect. J Clin Endocrinol Metab. 2004;89:5700–5706. doi: 10.1210/jc.2004-0553. [DOI] [PubMed] [Google Scholar]
- 4.Lorenzen T, Pociot F, Hougaard P, Nerup J. Long-term risk of IDDM in first-degree relatives of patients with IDDM. Diabetologia. 1994;37:321–327. doi: 10.1007/BF00398061. [DOI] [PubMed] [Google Scholar]
- 5.Jahromi MM, Eisenbarth GS. Genetic determinants of type 1 diabetes across populations. Ann N Y Acad Sci. 2006;1079:289–299. doi: 10.1196/annals.1375.044. [DOI] [PubMed] [Google Scholar]
- 6.Noble JA, Valdes AM, Cook M, Klitz W, Thomson G, Erlich HA. The role of HLA class II genes in insulin-dependent diabetes mellitus: molecular analysis of 180 Caucasian, multiplex families. Am J Hum Genet. 1996;59:1134–1148. [PMC free article] [PubMed] [Google Scholar]
- 7.Rotter JI, Anderson CE, Rubin R, Congleton JE, Terasaki PI, Rimoin DL. HLA genotypic study of insulin-dependent diabetes – the excess of DR3/DR4 heterozygotes allows rejection of the recessive hypothesis. Diabetes. 1983;32:169–174. doi: 10.2337/diab.32.2.169. [DOI] [PubMed] [Google Scholar]
- 8.Thomson G, Robinson WP, Kuhner MK, et al. Genetic-heterogeneity, modes of inheritance, and risk estimates for a joint study of Caucasians with insulin-dependent diabetes-mellitus. Am J Hum Genet. 1988;43:799–816. [PMC free article] [PubMed] [Google Scholar]
- 9.Karvonen M, Tuomilehto J, Libman I, Laporte R. A review of the recent epidemiologic data on the worldwide incidence of type-1 (insulin-dependent) diabetes-mellitus. Diabetologia. 1993;36:883–892. doi: 10.1007/BF02374468. [DOI] [PubMed] [Google Scholar]
- 10.Valdes AM, Erlich HA, Noble JA. Human leukocyte antigen class I B and C loci contribute to type 1 diabetes (T1D) susceptibility and age at T1D onset. Hum Immunol. 2005;66:301–313. doi: 10.1016/j.humimm.2004.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nejentsev S, Howson JMM, Walker NM, et al. Localization of type 1 diabetes susceptibility to the MHC class I genes HLA-B and HLA-A. Nature. 2007;450:887–892. doi: 10.1038/nature06406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pugliese A. Genetic protection from insulin-dependent diabetes mellitus. Diabetes Nutr Metab. 1997;10:169–179. [Google Scholar]
- 13.Bottini N, Musumeci L, Alonso A, et al. A functional variant of lymphoid tyrosine phosphatase is associated with type I diabetes. Nat Genet. 2004;36:337–338. doi: 10.1038/ng1323. [DOI] [PubMed] [Google Scholar]
- 14.Nistico L, Buzzetti R, Pritchard LE, et al. The CTLA-4 gene region of chromosome 2q33 is linked to, and associated with, type 1 diabetes. Belgian Diabetes Registry Hum Mol Genet. 1996;5:1075–1080. doi: 10.1093/hmg/5.7.1075. [DOI] [PubMed] [Google Scholar]
- 15.Bennett ST, Lucassen AM, Gough SC, et al. Susceptibility to human type 1 diabetes at IDDM2 is determined by tandem repeat variation at the insulin gene minisatellite locus. Nat Genet. 1995;9:284–292. doi: 10.1038/ng0395-284. [DOI] [PubMed] [Google Scholar]
- 16.Adeghate E, Schattner P, Dunn E. An update on the etiology and epidemiology of diabetes mellitus. Ann N Y Acad Sci. 2006;1084:1–29. doi: 10.1196/annals.1372.029. [DOI] [PubMed] [Google Scholar]
- 17.Bianchi DW. Fetomaternal cell trafficking: a new cause of disease? Am J Med Genet. 2000;91:22–28. doi: 10.1002/(sici)1096-8628(20000306)91:1<22::aid-ajmg4>3.0.co;2-3. [DOI] [PubMed] [Google Scholar]
- 18.Lo YMD, Lau TK, Chan LYS, Leung TN, Chang AMZ. Quantitative analysis of the bidirectional fetomaternal transfer of nucleated cells and plasma DNA. Clin Chem. 2000;46:1301–1309. [PubMed] [Google Scholar]
- 19.Lambert NC, Stevens AM, Tylee TS, Erickson TD, Furst DE, Nelson JL. From the simple detection of microchimerism in patients with autoimmune diseases to its implication in pathogenesis. Ann N Y Acad Sci. 2001;945:164–171. doi: 10.1111/j.1749-6632.2001.tb03881.x. [DOI] [PubMed] [Google Scholar]
- 20.Nelson JL. Microchimerism and human autoimmune diseases. Lupus. 2002;11:651–654. doi: 10.1191/0961203302lu271oa. [DOI] [PubMed] [Google Scholar]
- 21.Nelson JL. Microchimerism and HLA relationships of pregnancy: implications for autoimmune diseases. Curr Rheumatol Rep. 2001;3:222–229. doi: 10.1007/s11926-001-0022-5. [DOI] [PubMed] [Google Scholar]
- 22.Holzgreve W, Hahn S, Zhong XY, et al. Genetic communication between fetus and mother: short- and long-term consequences. Am J Obstet Gynecol. 2007;196:372–381. doi: 10.1016/j.ajog.2006.12.013. [DOI] [PubMed] [Google Scholar]
- 23.Bianchi DDW, Zickwolf GGK, Weil GGJ, Sylvester SS, DeMaria MMA. Male fetal progenitor cells persist in maternal blood for as long as 27 years postpartum. Proc Natl Acad Sci U S A. 1996;93:705–708. doi: 10.1073/pnas.93.2.705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hall JM, Lingenfelter P, Adams SL, Lasser D, Hansen JA, Bean MA. Detection of maternal cells in human umbilical cord blood using fluorescence in situ hybridization. Blood. 1995;86:2829–2832. [PubMed] [Google Scholar]
- 25.Claas FH, van Leeuwen A, van Rood JJ. Hyperimmunized patients do not need to wait for an HLA identical donor. Tissue Antigens. 1989;34:23–29. doi: 10.1111/j.1399-0039.1989.tb01713.x. [DOI] [PubMed] [Google Scholar]
- 26.Claas FH, Gijbels Y, van der Velden-de Munck J, van Rood JJ. Induction of B cell unresponsiveness to non-inherited maternal HLA antigens during fetal life. Science. 1988;241:1815–1817. doi: 10.1126/science.3051377. [DOI] [PubMed] [Google Scholar]
- 27.Phelan D, Hadley G, Duffy B, Mohanam S, Mohanakumar T. Antiidiotypic antibodies to HLA class I alloantibodies in normal individuals: a mechanism of tolerance to non-inherited maternal HLA antigens. Hum Immunol. 1991;31:1–6. doi: 10.1016/0198-8859(91)90041-7. [DOI] [PubMed] [Google Scholar]
- 28.Zhang L, van Rood JJ, Claas FH. The T-cell repertoire is not dictated by self antigens alone. Res Immunol. 1991;142:441–445. doi: 10.1016/0923-2494(91)90044-j. [DOI] [PubMed] [Google Scholar]
- 29.Roelen DL, van Bree FP, van Beelen E, van Rood JJ, Claas FH. No evidence of an influence of the non-inherited maternal HLA antigens on the alloreactive T cell repertoire in healthy individuals. Transplantation. 1995;59:1728–1733. doi: 10.1097/00007890-199506270-00015. [DOI] [PubMed] [Google Scholar]
- 30.van Rood JJ, Claas F. Noninherited maternal HLA antigens: a proposal to elucidate their role in the immune response. Hum Immunol. 2000;61:1390–1394. doi: 10.1016/s0198-8859(00)00211-1. [DOI] [PubMed] [Google Scholar]
- 31.van Rood JJ, Claas F. Both self and non-inherited maternal HLA antigens influence the immune response. Immunol Today. 2000;21:269–273. doi: 10.1016/s0167-5699(00)01628-5. [DOI] [PubMed] [Google Scholar]
- 32.Stevens AM. Microchimeric cells in systemic lupus erythematosus: targets or innocent bystanders? Lupus. 2006;15:820–826. doi: 10.1177/0961203306070068. [DOI] [PubMed] [Google Scholar]
- 33.Rich SS, Concannon P, Erlich H, et al. The Type 1 Diabetes Genetics Consortium. Ann N Y Acad Sci. 2006;1079:1–8. doi: 10.1196/annals.1375.001. [DOI] [PubMed] [Google Scholar]
- 34.Erlich H, Valdes AM, Noble J, et al. HLA DR-DQ haplotypes and genotypes and type 1 diabetes risk: analysis of the type 1 diabetes genetics consortium families. Diabetes. 2008;57:1084–1092. doi: 10.2337/db07-1331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.O’Connell JR, Weeks DE. PedCheck: a program for identification of genotype incompatibilities in linkage analysis. Am J Hum Genet. 1998;63:259–266. doi: 10.1086/301904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mukhopadhyay N, Almasy L, Schroeder M, Mulvihill WP, Weeks DE. Mega2: data-handling for facilitating genetic linkage and association analyses. Bio-informatics. 2005;21:2556–2557. doi: 10.1093/bioinformatics/bti364. [DOI] [PubMed] [Google Scholar]
- 37.Martin ER, Monks SA, Warren LL, Kaplan NL. A test for linkage and association in general pedigrees: the pedigree disequilibrium test. Am J Hum Genet. 2000;67:146–154. doi: 10.1086/302957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.R Development Core Team. R: A Language and Environment for Statistical Computing. Vienna: R Foundation for Statistical Computing; 2007. [Google Scholar]
- 39.Kleinbaum D, Klein M. Logistic Regression: A Self-learning Text. 2. New York: Springer; 2002. p. 520. [Google Scholar]
- 40.Thomson G. Mapping disease genes: family-based association studies. Am J Hum Genet. 1995;57:487–498. [PMC free article] [PubMed] [Google Scholar]
- 41.Stevens AM, Tsao BP, Hahn BH, et al. Maternal HLA class II compatibility in men with systemic lupus erythematosus. Arthritis Rheum. 2005;52:2768–2773. doi: 10.1002/art.21256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Artlett CM, Welsh KI, Black CM, Jimenez SA. Fetal-maternal HLA compatibility confers susceptibility to systemic sclerosis. Immunogenetics. 1997;47:17–22. doi: 10.1007/s002510050321. [DOI] [PubMed] [Google Scholar]
- 43.Nelson JL, Furst DE, Maloney S, et al. Microchimerism and HLA-compatible relationships of pregnancy in scleroderma. Lancet. 1998;351:559–562. doi: 10.1016/S0140-6736(97)08357-8. [DOI] [PubMed] [Google Scholar]
- 44.Srivatsa B, Srivatsa S, Johnson KL, Samura O, Lee SL, Bianchi DW. Microchimerism of presumed fetal origin in thyroid specimens from women: a case-control study. Lancet. 2001;358:2034–2038. doi: 10.1016/S0140-6736(01)07099-4. [DOI] [PubMed] [Google Scholar]
- 45.Toda I, Kuwana M, Tsubota K, Kawakami Y. Lack of evidence for an increased microchimerism in the circulation of patients with Sjogren’s syndrome. Ann Rheum Dis. 2001;60:248–253. doi: 10.1136/ard.60.3.248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tanaka A, Lindor K, Gish R, et al. Fetal microchimerism alone does not contribute to the induction of primary biliary cirrhosis. Hepatology. 1999;30:833–838. doi: 10.1002/hep.510300410. [DOI] [PubMed] [Google Scholar]
- 47.Marrosu MG, Sardu C, Cocco E, et al. Bias in parental transmission of the HLA-DR3 allele in Sardinian multiple sclerosis. Neurology. 2004;63:1084–1086. doi: 10.1212/01.wnl.0000138493.04890.7c. [DOI] [PubMed] [Google Scholar]
- 48.Ramagopalan SV, Herrera BM, Bell JT, et al. Parental transmission of HLA-DRB1*15 in multiple sclerosis. Hum Genet. 2008;122:661–663. doi: 10.1007/s00439-007-0442-z. [DOI] [PubMed] [Google Scholar]
- 49.Vadheim CM, Rotter JI, Maclaren NK, Riley WJ, Anderson CE. Preferential transmission of diabetic alleles within the HLA gene complex. N Engl J Med. 1986;315:1314–1318. doi: 10.1056/NEJM198611203152103. [DOI] [PubMed] [Google Scholar]
- 50.Sasaki T, Nemoto M, Yamasaki K, Tajima N. Preferential transmission of maternal allele with DQA1*0301-DQB1*0302 haplotype to affected offspring in families with type 1 diabetes. J Hum Genet. 1999;44:318–322. doi: 10.1007/s100380050168. [DOI] [PubMed] [Google Scholar]
- 51.Martin-Villa JM, Vicario JL, Martinez-Laso J, et al. Lack of preferential transmission of diabetic HLA alleles by healthy parents to offspring in Spanish diabetic families. J Clin Endocrinol Metab. 1990;70:346–348. doi: 10.1210/jcem-70-2-346. [DOI] [PubMed] [Google Scholar]
- 52.Bain SC, Rowe BR, Barnett AH, Todd JA. Parental origin of diabetes-associated HLA types in sibling pairs with type I diabetes. Diabetes. 1994;43:1462–1468. doi: 10.2337/diab.43.12.1462. [DOI] [PubMed] [Google Scholar]
- 53.Undlien DE, Akselsen HE, Joner G, et al. No difference in the parental origin of susceptibility HLA class II haplotypes among Norwegian patients with insulin-dependent diabetes mellitus. Am J Hum Genet. 1995;57:1511–1514. [PMC free article] [PubMed] [Google Scholar]
- 54.Ladner MB, Bottini N, Valdes AM, Noble JA. Association of the single nucleotide polymorphism C1858T of the PTPN22 gene with type 1 diabetes. Hum Immunol. 2005;66:60–64. doi: 10.1016/j.humimm.2004.09.016. [DOI] [PubMed] [Google Scholar]
- 55.Fajardy I, Vambergue A, Stuckens C, Weill J, Danze PM, Fontaine P. CTLA-4 49 A/G dimorphism and type 1 diabetes susceptibility: a French case-control study and segregation analysis. Evidence of a maternal effect. Eur J Immunogenet. 2002;29:251–257. doi: 10.1046/j.1365-2370.2002.00309.x. [DOI] [PubMed] [Google Scholar]
- 56.Paterson AD, Petronis A. Sex of affected sibpairs and genetic linkage to type 1 diabetes. Am J Med Genet. 1999;84:15–19. doi: 10.1002/(sici)1096-8628(19990507)84:1<15::aid-ajmg4>3.0.co;2-o. [DOI] [PubMed] [Google Scholar]
- 57.Burlingham WJ, Grailer AP, Heisey DM, et al. The effect of tolerance to noninherited maternal HLA antigens on the survival of renal transplants from sibling donors. N Engl J Med. 1998;339:1657–1664. doi: 10.1056/NEJM199812033392302. [DOI] [PubMed] [Google Scholar]
- 58.Campbell DA, Jr, Lorber MI, Sweeton JC, Turcotte JG, Niederhuber JE, Beer AE. Breast feeding and maternal-donor renal allografts. Possibly the original donor-specific transfusion. Transplantation. 1984;37:340–344. doi: 10.1097/00007890-198404000-00004. [DOI] [PubMed] [Google Scholar]
- 59.Kois WE, Campbell DA, Lorber MI, Sweeton JC, Dafoe DC. Influence of breast-feeding on subsequent reactivity to a related renal-allograft. J Surg Res. 1984;37:89–93. doi: 10.1016/0022-4804(84)90166-5. [DOI] [PubMed] [Google Scholar]
- 60.Andrassy J, Kusaka S, Jankowska-Gan E, et al. Tolerance to noninherited maternal MHC antigens in mice. J Immunol. 2003;171:5554–5561. doi: 10.4049/jimmunol.171.10.5554. [DOI] [PubMed] [Google Scholar]
- 61.Adams KM, Nelson JL. Microchimerism: an investigative frontier in autoimmunity and transplantation. JAMA. 2004;291:1127–1131. doi: 10.1001/jama.291.9.1127. [DOI] [PubMed] [Google Scholar]
- 62.Lambert NC, Erickson TD, Yan Z, et al. Quantification of maternal microchimerism by HLA-specific real-time polymerase chain reaction: studies of healthy women and women with scleroderma. Arthritis Rheum. 2004;50:906–914. doi: 10.1002/art.20200. [DOI] [PubMed] [Google Scholar]
- 63.van der Horst-Bruinsma IE, Hazes JM, Schreuder GM, et al. Influence of non-inherited maternal HLA-DR antigens on susceptibility to rheumatoid arthritis. Ann Rheum Dis. 1998;57:672–675. doi: 10.1136/ard.57.11.672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Barrera P, Balsa A, Alves H, et al. Noninherited maternal antigens do not increase the susceptibility for familial rheumatoid arthritis. European Consortium on Rheumatoid Arthritis Families (ECRAF) J Rheumatol. 2001;28:968–974. [PubMed] [Google Scholar]
- 65.Harney S, Newton J, Milicic A, Brown MA, Wordsworth BP. Non-inherited maternal HLA alleles are associated with rheumatoid arthritis. Rheumatology (Oxford) 2003;42:171–174. doi: 10.1093/rheumatology/keg059. [DOI] [PubMed] [Google Scholar]
- 66.Feitsma AL, Worthington J, van der Helm-van Mil AHM, et al. Protective effect of noninherited maternal HLA-DR antigens on rheumatoid arthritis development. Proc Natl Acad Sci U S A. 2007;104:19966–19970. doi: 10.1073/pnas.0710260104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Pani MA, Van Autreve J, Van der Auwera BJ, Gorus FK, Badenhoop K. Non-transmitted maternal HLA DQ2 or DQ8 alleles and risk of Type I diabetes in offspring: the importance of foetal or post partum exposure to diabetogenic molecules. Diabetologia. 2002;45:1340–1343. doi: 10.1007/s00125-002-0900-z. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1 Allele-specific association results for classical human leucocyte antigen loci in type 1 diabetes families: observed transmitted and non-transmitted allele frequencies, expected allele frequency, test statistic, odds ratio and p value.
Table S2 Frequencies of human leucocyte antigen (HLA) maternal–offspring and paternal–offspring HLA compatibility relationships in type 1 diabetes families.
Table S3 Allele-specific results from parent-of-origin analyses for classical human leucocyte antigen loci in type 1 diabetes families: observed transmitted maternal vs. paternal allele frequencies, expected allele frequency, test statistic, odds ratio and p value.
Table S4 Allele-specific results from non-inherited maternal human leucocyte antigen (HLA) analyses for classical HLA loci in type 1 diabetes families: observed non-transmitted maternal vs. paternal allele frequencies, expected allele frequency, test statistic, odds ratio and p value.