Abstract
The POU-IV or “Brn-3” class of POU-domain transcription factors is represented in Drosophila by I-POU and twin-of-I-POU, alternative splice products of the I-POU gene. I-POU has been previously reported to inhibit DNA binding by the POU-III class factor drifter/Cf1a via the formation of heterodimeric complexes. Here we report that expression of the I-POU/tI-POU message is maximal late in the embryonic phase of Drosophila development, and I-POU is the preferred splice variant. Although I-POU lacks two basic amino acid residues in the POU-homeodomain found in tI-POU and Brn-3.0, these three POU-IV class proteins exhibit very similar DNA-binding specificity. In contrast to previously published reports, the results presented here show no effect of I-POU on DNA binding by drifter, and no evidence for I-POU/drifter dimerization. These results suggest that the I-POU/tI-POU gene products function by transcriptional mechanisms similar to those of the homologous POU-IV class factors expressed in other species, not by a unique inhibitory mechanism.
The POU-domain proteins are a large family of transcriptional regulators, many of which exhibit specific expression in differentiating tissues. Based on the degree of sequence similarity, six subclasses of POU factors have been identified. In mammals, the POU-III and POU-IV subclasses appear to have special significance for development of the nervous system. The POU-III class includes the mammalian genes Brn-1, Brn-2, SCIP/Tst-1/Oct-6, and Brn-4. Brn-2 is required for normal development of the hypothalamus in mice (1, 2) and mutations at the Brn-4 locus in humans (3) have been linked to congenital deafness. The mammalian POU-IV class genes include Brn-3.0, Brn-3.1, and Brn-3.2 (also designated Brn-3a, Brn-3c, and Brn-3b, respectively), and expression of these factors is almost entirely restricted to terminally differentiating neurons of several types (4–7). Recent targeted mutations in mice have shown that Brn-3.1 is necessary for correct development of the auditory system (8), and that Brn-3.2 is required for differentiation of retinal ganglion cells (8, 9).
Consistent with the duplication and divergence of structures noted for many gene families, the POU-III and POU-IV classes are each represented by a single known Drosophila gene (10–12). The Drosophila POU-III gene, originally designated cf1a, and more recently as drifter (dfr) and ventral veinless, was originally identified by interaction with regulatory elements in the Drosophila dopamine decarboxylase gene. dfr/Cf1a has been shown to play a role in differentiation of the trachea, midline glia, and wing veins in Drosophila development (13–15).
The Drosophila representative of the POU-IV class was originally identified based on its homology to Brn-3.0 (12). The product of this gene, known as “I-POU” for inhibitory POU, was initially reported to differ from Brn-3.0 by the lack of two usually conserved basic amino acids located at the amino terminus of the POU-homeodomain. I-POU was reported to have no intrinsic DNA binding capacity, but instead to form a high-affinity one-to-one complex with dfr in the absence of DNA, which completely inhibited the DNA binding and transactivation ability of dfr. In a subsequent report (16), the I-POU genomic locus was shown to contain a possible alternate splice acceptor that could restore the “missing” amino acids, producing “twin of I-POU” (tI-POU). The addition of the basic amino acids eliminated binding to dfr, but restored the DNA binding and transactivation properties of I-POU (17).
Advances in understanding the conformation of the POU-domain on DNA (18–22), and ongoing work in our laboratory on the DNA binding specificity of the POU domain proteins, have led us to reconsider the properties of the I-POU/tI-POU proteins. Specifically, we have reexamined the developmental expression of the I-POU/tI-POU mRNA, the DNA-binding properties of the two proteins, and the ability of I-POU to dimerize with and inhibit DNA binding by dfr. Here we show that I-POU and tI-POU are alternate splice variants of the Drosophila POU-IV class gene, both of which bind DNA with high affinity and with site specificity similar to their mammalian counterpart Brn-3.0. In contrast to the previously reported results, in these experiments I-POU did not inhibit DNA binding by dfr, and no interaction between I-POU and dfr could be detected by any of several methods, including those used in the original studies.
MATERIALS AND METHODS
RNase protections were performed as described in ref. 6, with hybridizations carried out at 45°C. Electrophoretic mobility-shift assays (EMSAs) were carried out in 6% polyacrylamide gels as described previously (6), in a 20 μl assay mixture containing 20 mM Tris (pH 8.0), 100 mM KCl, 5 mM MgCl2, 2 mM EDTA, 100 μg/ml poly(dI-dC), 100 μg/ml BSA, 10% glycerol, 1 mM DTT, and the amount of POU protein, 32P-radiolabeled oligonucleotide, and competitor oligonucleotide stated in the figures. For random oligonucleotide EMSAs, the random template oligonucleotide CCAGGCTCGAGGTCTCGN16GCACGCTCGAGGAGTCC was annealed to the primer GGACTCCTCGAGCGTGC and filled by Klenow fragment DNA polymerase, with 32P added to the primer by polynucleotide kinase prior to annealing. Specific oligonucleotides used in EMSAs included the octamer/heptamer (o/h) and octamer/mutant heptamer (oct) sites from the IgG promoter (23), and a series of specific POU-IV class binding sites that included an optimal Brn-3.0 site (Site 1), and several variants of this sequence. The determination of Site 1 by random oligonucleotide selection will be described elsewhere. Site 1 strongly resembles a regulatory element from the rat CRH gene promoter region previously shown to bind to Brn-3.0 and I-POU. The sequence of the site 1 oligonucleotide was GATCTCTCCTGCATAATTAATTACGCCCGGATC, and in each of the variant oligonucleotides derived from this site, the underscored sequence was replaced by the sequences shown in Fig. 2B.
Figure 2.
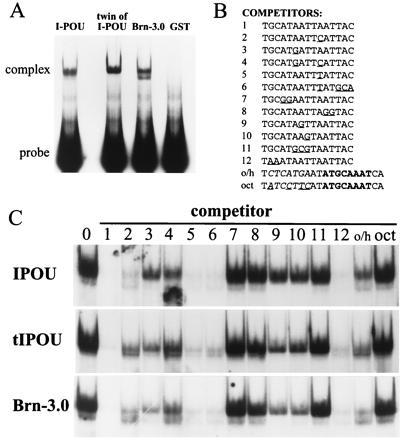
Interaction of POU-IV class proteins with DNA. (A) Binding of I-POU, tI-POU, and Brn-3.0 POU-domain GST-fusion proteins to an oligonucleotide containing an internal 16 base pair random sequence. Each EMSA shown used a similar amount of active POU-protein, as determined by binding to a specific oligonucleotide (Site 1, below). (B) Specific sites tested for binding to POU-IV class proteins. Site 1 is an optimal Brn-3.0 recognition element determined by random oligonucleotide selection, and Sites 2–12 are systematic mutations of this sequence. Octamer (oct) and octamer/heptamer (o/h) elements are derived from Oct-2 recognition sequences in the IgG heavy chain promoter (23). (C) Competition EMSAs of I-POU, tI-POU, and Brn-3.0 binding to specific recognition sites. Each assay contains 1.25 nM of 32P-labeled Site 1, 50 nM of the stated cold competitor oligo, and an equal amounts of the appropriate bacterially expressed POU-protein. Loss of signal indicates effective competition and thus high affinity binding by the competitor. For example, Site 1 competitor abolishes binding to the radiolabeled Site 1, but the oct site does not compete effectively. An artifact appears below the EMSA band in I-POU, lane 4.
For immunoprecipitation, 1–3 μl of each translation product or 4 μl of cotranslation product was incubated 30 min at room temperature in an incubation mix containing 250 mM NaCl, 5 mM EDTA, 10 mM Tris (pH 7.5), 0.25% Nonidet P-40, and 0.01% SDS, as described by Treacy et al. (12, 17). Following the initial incubation, 0.2 μg mouse monoclonal anti-Flag antibody was added and incubated 30 min at 4°C, followed by 5 μl sheep anti-mouse paramagnetic bead suspension (Dynal, Great Neck, NY). After a 30-min incubation at 4°C with mixing, the beads were separated magnetically and washed twice in the binding buffer, and the bound proteins were eluted at 90°C in SDS/PAGE sample buffer.
Crosslinking reactions were performed in 20 mM Hepes (pH 7.8)/20% glycerol as described previously (12, 17). In vitro-translated proteins (1–4 μl) were incubated together for 30 min in a 30 μl reaction volume, followed by a 20-min incubation in the presence of the stated concentration of bis-maleimidohexane (BMH), and subsequent quenching with 560 mM 2-mercaptoethanol. Immunoprecipitation or crosslinking products were electrophoresed in SDS/12% PAGE gels, and fluorographed 24–48 hr with Entensify (NEN).
RESULTS
The previously reported sequence of the Drosophila genomic region that includes the amino terminal region of the I-POU homeodomain (17) indicates the presence of potential alternate splice acceptors (Fig. 1A), which could produce mRNA encoding I-POU and tI-POU proteins that differ by two amino acids. However, data on the relative usage of these splice acceptors with probes for each of the two forms have not been reported. To assay the expression of the possible alternatively spliced I-POU/tI-POU mRNA species by RNase protection, three antisense riboprobes were designed (Fig. 1B). One probe, complementary to the POU-specific domain, hybridizes with both splice forms. In addition, I-POU- and tI-POU-specific homeodomain probes were designed to span the alternate splice acceptor, and thus give full-length protection with only one splice variant.
Figure 1.
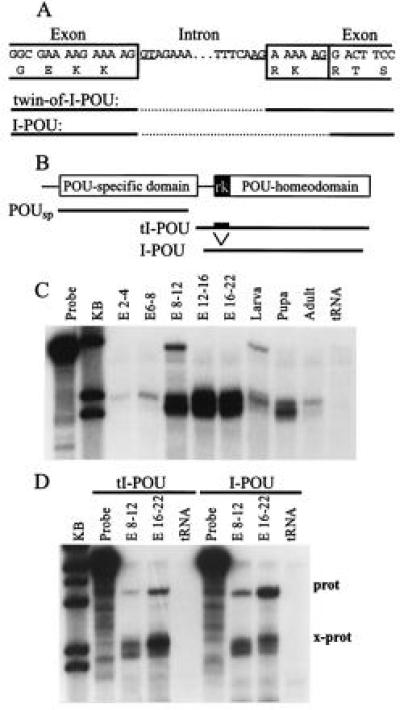
Developmental expression of I-POU/tI-POU. (A) Diagram of the intron-exon boundaries at the amino terminus of the I-POU homeodomain. Alternative splicing includes (tI-POU) or excludes (I-POU) a six-base sequence encoding Arg-Lys. (B) RNase protection assay probes used for I-POU/tI-POU detection. The POU-specific domain probe includes sequences common to both splice forms, whereas the POU-homeodomain probes yield full-length protected fragments only with I-POU or tI-POU. (C) Expression of I-POU/tI-POU in Drosophila development assayed with the POU-specific probe. Embryonic ages (E) are stated in hours. The appearance of a closely spaced doublet varied according to the conditions of RNase digestion and does not appear to represent alternate forms of the message. (D) Relative expression of tI-POU and I-POU, using splice form-specific POU-homeodomain probes. In the 8–12 h and the 16–22 h embryos, both forms could be detected, but I-POU mRNA was the predominant species, as demonstrated by the more abundant full-length I-POU protection product (prot) and the more abundant tI-POU cross-protection product (x-prot).
In developing Drosophila, overall expression of I-POU + tI-POU transcripts is minimal in the early embryo, but increases strongly at mid-embryogenesis, peaking in late embryonic development (Fig. 1C). I-POU + tI-POU expression declines with respect to the total RNA pool in later developmental stages, but this may in part represent growth and development of non-expressing tissues. These results are in general agreement with the expression pattern previously reported (17). Protection assays with probes specific for the alternative splice forms (Fig. 1D) demonstrated that I-POU is the preferred splice variant in the embryo, but that tI-POU expression is also detected.
DNA binding of I-POU and tI-POU proteins were initially assessed in EMSAs using an oligonucleotide probe containing an internal 16 base pair random sequence (Fig. 2A). I-POU, tI-POU and the murine POU-IV class protein Brn-3.0 showed similar binding to the random oligonucleotide, in contrast to the previously published reports that I-POU did not bind a random oligonucleotide in random oligonucleotide blots (12) or random oligonucleotide EMSAs (12, 17).
Competition EMSAs with a variety of oligonucleotides (Fig. 2B) were then used to compare the specific DNA binding properties of the I-POU and tI-POU POU-domains in detail, and examine the evolutionary conservation of DNA recognition in this gene family by comparison to the murine Brn-3.0 POU-domain. In each competition assay, an optimal Brn-3.0 recognition site (site 1, Fig. 2B) was radiolabeled, and inhibition of binding by a 40-fold molar excess of the competitor oligonucleotide was assayed. Overall, the DNA recognition profiles of I-POU, tI-POU, and Brn-3.0 were extremely similar, with only minor relative differences in affinity for some competitors. However, these results with a limited number of sites do not exclude small differences in DNA-binding specificity or overall affinity between these proteins, and do not address possible differences in other functional properties.
I-POU has been previously reported to form a 1:1 complex with dfr, which completely inhibits dfr binding to DNA, and this inhibitory activity was deletion mapped to the I-POU homeodomain (17). Because this inhibitory complex was reported to form in the absence of DNA, inhibition of dfr DNA binding by I-POU should be independent of the dfr recognition site used. We examined the effect of the I-POU and tI-POU domains on DNA binding by dfr in EMSAs. The IgG oct/hep recognition element (23) was used as a model for dfr DNA binding in these assays because this site is somewhat selective for dfr in the presence of I-POU/tI-POU, thus allowing the dfr band to be visualized in the presence of excess I-POU/tI-POU protein. To create a difference in the electrophoretic mobility between the POU proteins used, bacterially expressed I-POU and tI-POU were removed from the glutathione S-transferase (GST) moiety by thrombin proteolysis, and the GST–dfr fusion protein was used uncleaved. As shown in Fig. 3, I-POU present in up to 40-fold molar excess over dfr had no effect whatsoever on dfr binding to its recognition site, and tI-POU gave similar results in this binding inhibition assay.
Figure 3.
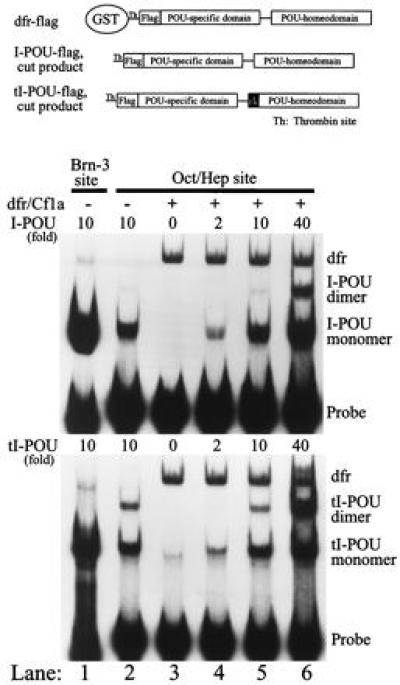
I-POU and tI-POU do not inhibit DNA binding by dfr. Bacterially expressed dfr and I-POU/tI-POU proteins of distinct electrophoretic mobilities were generated by thrombin proteolysis of I-POU/tI-POU GST fusion proteins at the site shown. In each experiment, increasing stoichiometric equivalents of I-POU (Upper) or tI-POU (Lower) were tested for the ability to block dfr binding to an octamer/heptamer site (lanes 4–6). The dfr complex was unaffected, even at a 40-fold molar excess of I-POU. Controls: lanes 1 and 2 show I-POU/tI-POU high affinity binding to an optimal POU-IV class recognition element (Site 1, Fig. 2B) and moderate affinity binding to the oct/hep site. Lane 3: dfr binding to the oct/hep site in the absence of I-POU/tI-POU. The dfr (0.4 ng) and I-POU/tI-POU (0.8–16 ng) proteins were preincubated together 1 h before the addition of oligonucleotide to allow interaction of dfr and I-POU/tI-POU in the absence of DNA. Two molecules of I-POU/tI-POU were observed to bind to the oct/hep site at very high protein concentrations (lanes 5 and 6). This dimerization was not cooperative, and results from the presence of a second weak (heptamer) binding element in the oct/hep oligonucleotide.
Immunoprecipitation of in vitro-translated proteins was then used to further assess potential interaction between I-POU and dfr. A “Flag” sequence was included in the I-POU in vitro transcription/translation construct to allow immunoprecipitation by anti-Flag antisera. The inclusion of the Flag sequence and a longer carboxyl-terminal domain in the I-POU expression construct were sufficient to allow the I-POU and dfr translation products to be distinguished by apparent molecular weight on denaturing polyacrylamide gels. When in vitro-translated I-POU-Flag and dfr were incubated together and immunoprecipitated under the conditions used in the prior studies (12, 17), quantitative immunoprecipitation of I-POU-Flag was observed, without any detectable coprecipitation of dfr (Fig. 4B). Because heterodimerization of some transcription factors of the helix–loop–helix family is enhanced by cotranslation of the interacting proteins, immunoprecipitation of I-POU–flag was also performed in the presence of cotranslated dfr, again without detectable precipitation of the latter protein. As a positive control for the Flag immunoprecipitation method, the basic helix–loop–helix/zip protein Mad, which has been shown to form heterodimers in the absence of DNA with the related protein Max (24), was effectively coprecipitated with Max–flag protein (Fig. 4C).
Figure 4.
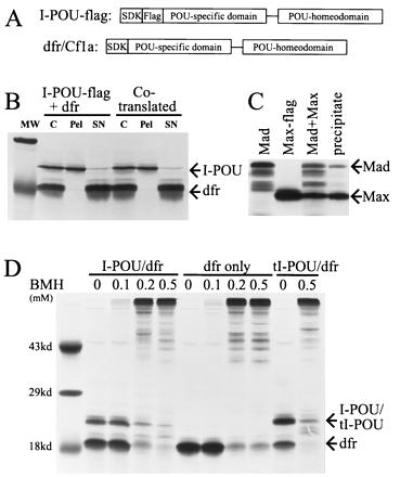
Immunoprecipitation and crosslinking of I-POU and dfr. (A) I-POU/tI-POU and dfr proteins were expressed by in vitro translation in the presence of [35S]methionine in a reticulocyte lysate translation system acceding to manufacturer’s instructions (Promega). An amino-terminal Kozak consensus sequence containing an initial methionine residue was added to facilitate translation. The I-POU/tI-POU expression construct included a “Flag” peptide sequence (DYKDDDDK) to allow immunoprecipitation by an epitope-specific monoclonal antibody (IBI/Kodak). (B) Immunoprecipitation of I-POU-flag in the presence of dfr. In the first set, separate translation reactions were mixed and incubated 1 h before precipitation. In the second set, I-POU-Flag and dfr were cotranslated. Proteins were then immunoprecipitated using Flag-specific antibodies, and separated in denaturing polyacrylamide gels (Methods). MW, molecular weight standard (29 and 19 kDa shown); C, control; pel, immunoprecipitate; SN, supernatant. (C) Immunoprecipitation of Max-Flag in the presence of Mad (positive control). Max-Flag and Mad expression constructs were generated as shown for I-POU-Flag and dfr, and were in vitro transcribed and cotranslated [parent Mad and Max plasmids were a gift of R. Eisenman (24)]. In vitro translation of Mad consistently produced several truncated translation products which did not coprecipitate with Max. (D) Chemical crosslinking of I-POU/tI-POU and dfr with BMH. In vitro translation products for I-POU+dfr (set 1), dfr alone (set 2), and tI-POU+dfr (set 3) were exposed to the stated concentrations of BMH as described, and the products were separated by SDS/PAGE. No specific heterodimeric complexes were observed under any of the conditions tested.
The principal method previously used to demonstrate I-POU/dfr dimerization was chemical crosslinking of in vitro-translated proteins in reticulocyte lysate by the chemical crosslinker BMH, which is reactive with free sulfhydryl groups. However, when we attempted to crosslink I-POU and dfr with 1 mM BMH under the exact conditions described previously, extensive nonspecific crosslinking resulted, presumably to various reticulocyte lysate proteins, and the expected dimeric complex was not observed. Careful titration of the amount of BMH in the reaction did not reveal any concentration that resulted in specific crosslinking (Fig. 4D). Lysates containing I-POU plus dfr, dfr alone, and tI-POU plus dfr gave identical results in the crosslinking assay. BMH crosslinking of lysates containing Mad + Max proteins also gave only nonspecific crosslinking to lysate proteins, and we conclude that BMH may not be a specific enough reagent to give specific crosslinking products in crude reticulocyte lysates.
Further crosslinking experiments were performed under a variety of different reaction conditions (not shown), such as with the low concentration of glutaraldehyde previously shown to specifically crosslink protein dimers of the basic helix–loop–helix family (25). No conditions were found that yielded specific I-POU/dfr heterodimers. Taken together with the immunoprecipitation data, these results indicate that it is highly unlikely that I-POU and dfr form stable dimers in solution.
DISCUSSION
The present study examines in detail the DNA and protein binding properties of I-POU and tI-POU, which represent the POU-IV class of transcription factors in Drosophila. Although mutations in Drosophila have led directly or indirectly to the identification of most mammalian transcription factors, I-POU is unusual in that it was identified based on structural homology to previously discovered genes in mammals and Caenorhabditis elegans, and I-POU mutations have not yet been identified. However, based on the function of the POU-IV class factors in nematodes and vertebrates and on the expression of I-POU in specific developing neurons (12), it appears likely that I-POU will play a role in neural development.
Initial data suggested that I-POU functioned by the formation of an inhibitory complex with another POU factor, dfr/Cf1a, but this is not supported by the present results. Instead, consistent with the extremely similar primary structures of the POU-IV factors, I-POU exhibits DNA-binding properties that resemble closely tI-POU and Brn-3.0.
Recent advances in understanding the structure of the homeodomain are helpful in understanding the very similar DNA-binding properties of the I-POU, tI-POU and Brn-3.0 POU domains observed in this study. Fig. 5 summarizes protein-DNA contacts for the antennapedia (26), engrailed (18), and Oct-1 (20) homeodomains. The principal contacts are highly conserved in the POU-IV class proteins, and suggest that the interaction of the POU-IV homeodomain with DNA will be very similar to that of the previously solved structures. The deletion of the Arg-Lys (RK) residues at homeodomain positions 3 and 4 in I-POU relative to tI-POU results in the occurrence of Gly-Glu residues in positions 1 and 2, and substitution of Lys for Arg in position 3. The amino acid residues at homeodomain positions 1 and 2 do not contact DNA in the known homeodomain structures and vary widely in the known sequences (22). Thus, it is not surprising that a substitution at this position does not eliminate DNA binding. The substitution of Lys for Arg at position 3 in I-POU would also not be expected to alter DNA binding, as either residue may occur at this position in the various POU proteins.
Figure 5.
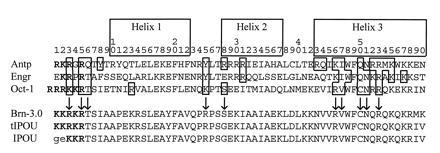
Conserved elements of homeodomain structure. Alignment of known homeodomain structures illustrate the homeodomain protein-DNA contacts, and suggest that conserved residues will mediate DNA binding by the POU-IV class proteins. Boxed residues indicate protein–DNA contacts, including those with the major and minor grooves and phosphate backbone. The likely conserved DNA contacts for the POU-IV class proteins are indicated by arrows. Boldface type indicates basic amino acids among residues 1–5 of the homeodomain, and the Arg-Lys residues altered by alternative splicing of I-POU/tI-POU are underscored.
Because critical DNA contacts do not differ between I-POU and tI-POU, the principal effect of the Arg-Lys deletion in I-POU may be effectively to shorten the variable “linker” region that connects the amino terminus of the POU-homeodomain to the POU-specific domain (27). Use of the 3′ splice acceptor in I-POU effectively shortens the linker from 20 to 18 amino acids. However, the Brn-3.0 linker consists of only 17 amino acids, suggesting that the loss of two residues from this region does not impose a critical constraint on DNA binding. In summary, the Arg-Lys residues “missing” from I-POU relative to tI-POU are not in a position highly conserved among homeodomain proteins, are not a part of a crucial helical structure, are not necessary for a conserved DNA contact, and would not be expected to constrain the spacing between the POU-specific and POU-homeodomains by critically shortening the linker region. Thus, the similar DNA-binding characteristics of the POU-IV class proteins are consistent with the current understanding of POU-domain structure-function relationships.
The POU-IV class proteins are of intrinsic interest because they exhibit remarkable conservation of structure across diverse phyla including nematodes, insects, and vertebrates. In C. elegans and mice, the critical role of these genes in development of specific sets of neurons has been established (8, 9, 28, 29). It is likely that I-POU/tI-POU will also play a role in neurodevelopment, but in the context of the present results, the hypothesis that I-POU functions by the unusual mechanism of a specific inhibitory interaction with dfr/Cf1a should be discarded. Instead, it appears likely that I-POU will regulate gene expression by mechanisms similar to those used by its homologues in other species.
Acknowledgments
We thank Jerry M. Rhee for technical assistance, Dr. Gordon Gill for helpful discussions, and Drs. J. Posakony and J. Kadonaga for advice on Drosophila culture. This work was supported in part by the Pfizer New Faculty Award, Department of Veterans Affairs MERIT funding, and the Scottish Rite Schizophrenia Research Program. E.E.T. is a Young Investigator of the National Association for Research on Schizophrenia and Depression.
Footnotes
Abbreviations: I-POU, inhibitory POU; tI-POU, twin of I-POU; EMSA, electrophoretic mobility-shift assay; GST, glutathione S-transferase; BMH, bis-maleimidohexane.
References
- 1.Schonemann M D, Ryan A K, McEvilly R J, O’Connell S M, Arias C A, Kalla K A, Li P, Sawchenko P E, Rosenfeld M G. Genes Dev. 1995;9:3122–3135. doi: 10.1101/gad.9.24.3122. [DOI] [PubMed] [Google Scholar]
- 2.Nakai S, Kawano H, Yudate T, Nishi M, Kuno J, Nagata A, Jishage K, Hamada H, Fujii H, Kawamura K, Shiba K, Noda T. Genes Dev. 1995;9:3109–3121. doi: 10.1101/gad.9.24.3109. [DOI] [PubMed] [Google Scholar]
- 3.de Kok Y, van der Maarel S, Bitner-Glindzicz M, Huber I, Monaco A, Malcol S, Pemprey M, Ropers H, Cremers F. Science. 1995;267:685–688. doi: 10.1126/science.7839145. [DOI] [PubMed] [Google Scholar]
- 4.Gerrero M R, McEvilly R, Turner E, Lin C, O’Connell S, Jenne K, Hobbs M V, Rosenfeld M G. Proc Natl Acad Sci USA. 1993;90:10841–10845. doi: 10.1073/pnas.90.22.10841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Xiang M, Zhou L, Peng Y-W, Eddy R, Shows T B, Nathans J. Neuron. 1993;11:689–701. doi: 10.1016/0896-6273(93)90079-7. [DOI] [PubMed] [Google Scholar]
- 6.Turner E E, Jenne K, Rosenfeld M G. Neuron. 1994;12:205–218. doi: 10.1016/0896-6273(94)90164-3. [DOI] [PubMed] [Google Scholar]
- 7.Fedtsova N, Turner E E. Mech Devel. 1995;53:291–304. doi: 10.1016/0925-4773(95)00435-1. [DOI] [PubMed] [Google Scholar]
- 8.Erkman L, McEvilly R J, Luo L, Ryan A K, Hooshmand F, O’Connell S M, Keithley E M, Rapaport D H, Ryan A F, Rosenfeld M G. Nature (London) 1996;381:603–606. doi: 10.1038/381603a0. [DOI] [PubMed] [Google Scholar]
- 9.Gan L, Xiang M, Zhou L, Wagner D S, Klein W H, Nathans J. Proc Natl Acad Sci USA. 1996;93:3920–3925. doi: 10.1073/pnas.93.9.3920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Johnson W A, Hirsh J. Nature (London) 1990;343:467–470. doi: 10.1038/343467a0. [DOI] [PubMed] [Google Scholar]
- 11.Billin A N, Cockerill K A, Poole S J. Mech Dev. 1991;34:75–84. doi: 10.1016/0925-4773(91)90045-8. [DOI] [PubMed] [Google Scholar]
- 12.Treacy M N, He X, Rosenfeld M G. Nature (London) 1991;350:577–584. doi: 10.1038/350577a0. [DOI] [PubMed] [Google Scholar]
- 13.Anderson M G, Perkins G L, Chittick P, Shrigley R J, Johnson W A. Genes Dev. 1995;9:123–137. doi: 10.1101/gad.9.1.123. [DOI] [PubMed] [Google Scholar]
- 14.de Celis J F, Llimargas M, Casanova J. Development (Cambridge, UK) 1995;121:3405–3416. doi: 10.1242/dev.121.10.3405. [DOI] [PubMed] [Google Scholar]
- 15.Billin A N, Poole S J. Roux Arch Dev Biol. 1995;204:502–508. doi: 10.1007/BF00360858. [DOI] [PubMed] [Google Scholar]
- 16.Herr W. Nature (London) 1991;350:554–555. doi: 10.1038/350554a0. [DOI] [PubMed] [Google Scholar]
- 17.Treacy M N, Neilson L I, Turner E E, He X, Rosenfeld M G. Cell. 1992;68:491–505. doi: 10.1016/0092-8674(92)90186-g. [DOI] [PubMed] [Google Scholar]
- 18.Kissinger C R, Liu B, Martin-Blanco E, Kornberg T B, Pabo C O. Cell. 1990;63:579–590. doi: 10.1016/0092-8674(90)90453-l. [DOI] [PubMed] [Google Scholar]
- 19.Wolberger C, Vershon A K, Liu B, Johnson A D, Pabo C O. Cell. 1991;67:517–528. doi: 10.1016/0092-8674(91)90526-5. [DOI] [PubMed] [Google Scholar]
- 20.Klemm J D, Rould M A, Aurora R, Herr W, Pabo C O. Cell. 1994;77:21–32. doi: 10.1016/0092-8674(94)90231-3. [DOI] [PubMed] [Google Scholar]
- 21.Gehring W J, Qian Y Q, Billeter M, Furukubo-Tokunaga K, Schier A F, Resendez-Perez D, Affolter M, Otting G, Wuthrich K. Cell. 1994;78:211–223. doi: 10.1016/0092-8674(94)90292-5. [DOI] [PubMed] [Google Scholar]
- 22.Gehring W J, Affolter M, Burglin T. Annu Rev Biochem. 1994;63:487–426. doi: 10.1146/annurev.bi.63.070194.002415. [DOI] [PubMed] [Google Scholar]
- 23.Poellinger L, Roeder R G. Mol Cell Biol. 1989;9:747–756. doi: 10.1128/mcb.9.2.747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ayer D E, Kretzner L, Eisenman R N. Cell. 1993;72:211–222. doi: 10.1016/0092-8674(93)90661-9. [DOI] [PubMed] [Google Scholar]
- 25.Klein E S, Simmons D M, Swanson L W, Rosenfeld M G. Genes Dev. 1993;7:55–71. doi: 10.1101/gad.7.1.55. [DOI] [PubMed] [Google Scholar]
- 26.Qian Y Q, Billeter M, Otting G, Muller M, Gehring W J, Wuthrich K. Cell. 1989;59:573–580. doi: 10.1016/0092-8674(89)90040-8. [DOI] [PubMed] [Google Scholar]
- 27.Herr W, Cleary M A. Genes Dev. 1995;9:1679–1693. doi: 10.1101/gad.9.14.1679. [DOI] [PubMed] [Google Scholar]
- 28.Finney M, Ruvkun G, Horvitz H R. Cell. 1988;55:757–769. doi: 10.1016/0092-8674(88)90132-8. [DOI] [PubMed] [Google Scholar]
- 29.Finney M, Ruvkun G. Cell. 1990;63:895–905. doi: 10.1016/0092-8674(90)90493-x. [DOI] [PubMed] [Google Scholar]


