Summary
The Male-specific lethal (MSL) complex up-regulates the single male X chromosome to achieve dosage compensation in Drosophila. We have proposed that MSL recognition of specific entry sites on the X is followed by local targeting of active genes marked by H3K36 trimethylation. Here we analyze the role of the MSL3 chromodomain in the second targeting step. Using ChIP-chip analysis, we find that MSL3 chromodomain mutants retain binding to chromatin entry sites, but show a clear disruption in the full pattern of MSL targeting in vivo, consistent with a loss of spreading. Furthermore, when compared to wild-type, chromodomain mutants lack preferential affinity for nucleosomes containing H3K36me3 in vitro. Our results support a model in which activating complexes, like their silencing counterparts, use the nucleosomal binding specificity of their respective chromodomains to spread from initiation sites to flanking chromatin.
Keywords: MSL3 chromodomain, X chromosome dosage compensation
Introduction
Dosage compensation is an essential chromatin-mediated process in Drosophila melanogaster, required in males to upregulate the single X chromosome by two-fold to match the gene expression from two Xs in females. At least five proteins and two non-coding roX RNAs are members of the MSL (male-specific lethal) complex that acetylates histone H4 at lysine 16. It has been known for some time that the MSL complex specifically targets the polytene X chromosome at hundreds of sites in a reproducible, banded pattern, but the rules that govern specific targeting are only recently becoming evident with the application of high resolution approaches such as ChIP-chip.
Drosophila MSL3 (male-specific lethal 3) is a conserved protein containing an N-terminal chromodomain1,2. In the absence of MSL3, partial MSL complexes can only target a subset of sites on X, termed high affinity or chromatin entry sites (CES), originally mapped at the cytological level to approximately 35–70 locations3–6. Initial targeting of chromatin entry sites is postulated to provide the X chromosome-specificity of dosage compensation. In a two-step model, this is followed by spreading in cis to active genes, independently of sequence. Full targeting correlates with the 3’ biased H3K36me3 general mark on active genes, and is diminished in set2 mutants which lack the enzyme responsible for this mark, providing evidence for a sequence-independent mechanism7–9. Alternatively, secondary sites may be recognized through DNA sequences of lesser affinities acting cooperatively6,10–14 but such sequences have yet to be defined or mutated to demonstrate function.
The concept of sequence-independent spreading is further supported by the identification of ectopic binding sites seen when a roX2 transgene is inserted on an autosome8. In the absence of a roX gene in cis, autosomes would never normally be targeted by the MSL complex. However, when MSL complex is ectopically localized on autosomes, flanking secondary sites display the same characteristics of typical targets seen on the X, correlating with 3’ enrichment of H3K36me3 on active genes. Formally, it is still possible that there is a sequence component to 3’ recognition, but if so, it is not specific to X chromosomal genes.
Spreading is a concept resting on a foundation of observations from the study of gene silencing and heterochromatin. The idea originated from the discovery and analysis of position-effect variegation in Drosophila15,16. The molecular mechanism of spreading is most clearly defined by the study of silencing of mating type genes in S. pombe, in which Swi6 and Clr4 are chromodomain proteins that play prominent roles17,18. Likewise, the founding chromodomain proteins, HP1 and Polycomb, both function in repressive complexes in Drosophila that are implicated in creating silent domains19,20. We previously demonstrated that formation of the complete MSL pattern on the X was dependent on MSL3 and H3K36me3, providing a circumstantial case for the involvement of the MSL3 chromodomain in spreading of the dosage compensation complex3,8. Yet, the chromodomain of MSL3 has been mutated and deemed dispensable for MSL targeting at the cytological level21. Here, by high resolution ChIP-chip, we demonstrate that MSL3 chromodomain mutants in Drosophila actually fail to bind the majority of genes on the X. Our results support a model in which initial sequence-specific targeting of chromatin entry sites is followed by spreading in cis, mediated predominantly by the MSL3 chromodomain.
Results
Developmental delay of MSL3 chromodomain mutant males
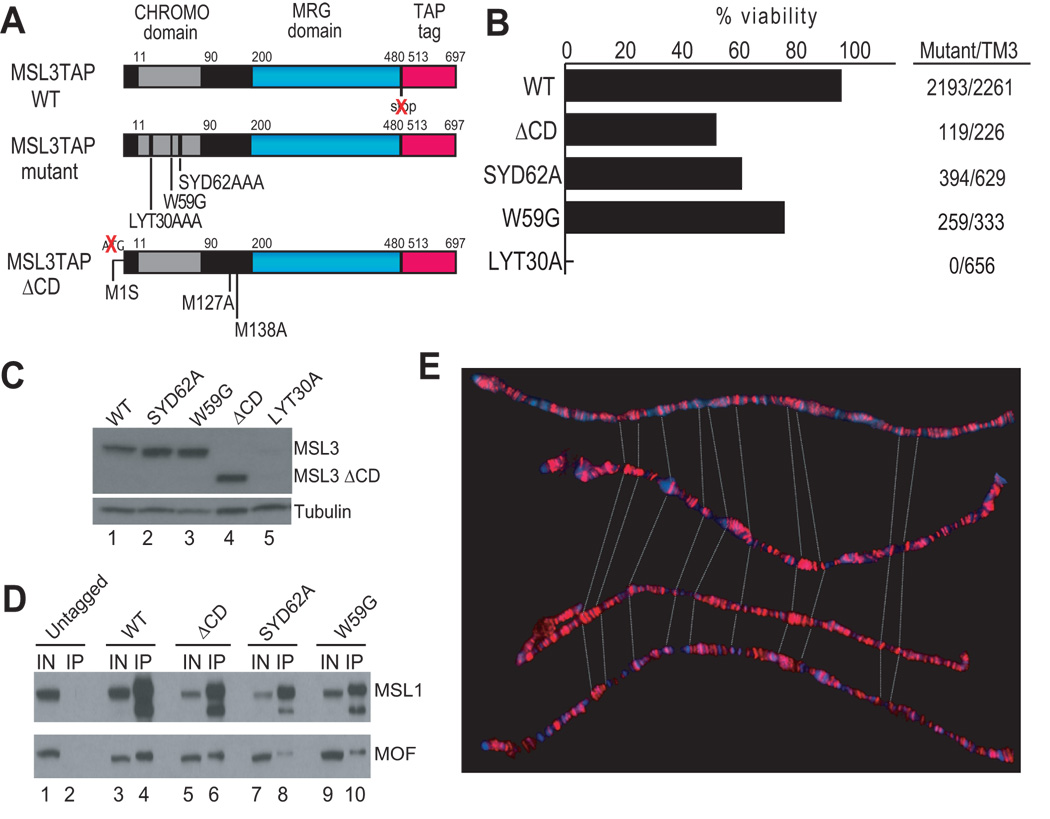
In order to study the contribution of the MSL3 chromodomain to targeting of the dosage compensation complex, we created transgenic lines expressing a deletion of the domain, as well as a set of targeted point mutants in potential methyl-lysine binding aromatic residues identified by alignment with other chromodomains22,23. The mutants were constructed in the context of a wild-type transgene fused to the TAP (Tandem Affinity Purification) epitope tag (Fig. 1a). The chromodomain deletion was created by mutating the start codon for full-length MSL3, ablating its expression and leading to overproduction of a naturally-occuring short form initiating at Met127, downstream of the chromodomain. The peptide composition of the short form was confirmed by TAP-tag affinity purification followed by mass spectrometry (Supplementary Fig. 1). The site-directed mutants designed to disrupt chromodomain function altered the following residues: 1) Y31 along with its neighboring residues L30 and T32 into alanines to obtain the LYT30A mutant; 2) Y63 along with S62 and D64 into alanines resulting in the SYD62A mutant; and 3) W59 into a glycine to create the W59G mutant (Fig. 1a).
Figure 1. Characterization of MSL3 chromodomain mutants.
(a) Schematic of the structure of the MSL3TAP transgenic protein, with mutations indicated below. To force expression of only the short isoform, the start codon for MSL3 was changed to a serine (M1S). (b) Complementation analysis. Rescue frequencies were calculated by comparing the number of msl31 homozygous mutant males expressing one copy of the transgene to their brothers with the TM3, Sb, msl3+ balancer. Number of progeny scored for each genotype is noted on the right. Chromodomain mutant males also exhibited a 2-day developmental delay with respect to their msl3+ brothers. (c) Protein expression was assayed on Western blots, using anti-PAP antibody against the TAP epitope. All but the LYT30A construct were stably expressed. (d) Chromodomain mutants assemble into MSL complexes. S2 cells expressing msl3tap constructs were used to immunoprecipitate MSL3TAP. Immunoprecipitations (IP) were then analyzed by Western blot for MSL1 and MOF. Even lanes: Input sample; odd lanes: IP sample. The lower band in the MSL1 IP may be a degradation product. (e) Chromodomain mutants are indistinguishable from WT in their binding patterns on the male X chromosome at the resolution of polytene staining (red: MSL3TAP). Stainings were done in the absence of endogenous msl3. From top to bottom: WT; SYD62A; ΔCD; W59G.
We inserted all mutant transgenes, as well as the WT construct, into the same genomic location on chromosome 2L (55C-D) using the ΦC31 mediated attB/attP site-specific recombination system to avoid position effects on transgene expression24. To assess the function of the mutants, we scored the ability of these transgenes to rescue msl31 mutant males (Fig. 1b). The WT construct could rescue msl31 mutant males efficiently (97%). It was previously shown that an msl3 mutant partially deleted for the chromodomain was sub-viable, with only 7% of mutant males reaching adulthood21. We observed higher viability for our ΔCD construct; roughly half of mutant males (53%) survived into adulthood, with the rest dying as late pupae. Although viable, all ΔCD mutant males were developmentally delayed by 2 days with respect to their heterozygous brothers carrying the msl3+ TM3 balancer chromosome, and displayed phenotypes such as held-out wings. Adult males were unhealthy and infertile. We observed similar results with two of the chromodomain missense mutants, SYD62A and W59G. Their mutant rescue was 62% and 77% respectively, and mutant males were developmentally delayed by 2 days. In contrast, the LYT30A mutant failed to complement msl31.
The lack of a functional chromodomain in MSL3 might lead to lower levels of stable MSL complex, and if this were the case, it would be difficult to assign a specific function to the conserved motif. Therefore, we performed Western analysis of males expressing the transgenes in the absence of endogenous msl3 to determine the levels of the MSL3-TAP mutant proteins. The expression of the LYT30A mutant was assessed in an msl3+ background, due to the inviability of these males. Consistent with its lack of rescue, the LYT30A mutant protein appeared to be highly unstable, and thus was deemed uninformative (Fig. 1c). In contrast, we found that the W59G, SYD62A, and ΔCD mutant proteins were expressed at levels comparable to WT MSL3. To assay for complex assembly, we affinity purified the W59G, SYD62A, and ΔCD mutant proteins from cell lysates using the TAP epitope, and in each case we could detect co-immunoprecipitation of MSL1 and MOF by Western analysis (Fig. 1d). Although this was a qualitative rather than quantitative assessment, the result is in agreement with previous studies, in which an intact MRG domain preserved functional interactions with the other components of the MSL complex, even in the absence of the chromodomain25.
ChIP-chip analysis of chromodomain mutant binding
Polytene chromosome binding patterns of the ΔCD, SYD62A and W59G mutant proteins on the X chromosome were indistinguishable from WT (Fig. 1e). This was previously the basis for the conclusion that MSL targeting was normal in the absence of the MSL3 chromodomain21. However, we reasoned that differences in binding might only be seen at the level of chromatin immunoprecipitation (ChIP) as observed for MSL binding in a set2 mutant8. Immunostaining of polytene chromosomes is non-linear and its resolution cannot distinguish between association with single sites and binding to multiple, tightly-clustered genes. Therefore, we performed ChIP-chip analysis using a mixed-population of msl31 mutant male and msl3+ female embryos expressing the WT, ΔCD or SYD62A constructs (see Methods for crosses). Since females lack MSL2 and hence do not have functional MSL complexes26, they do not contribute positively to the ChIP signal. We performed ChIP using the TAP epitope to immunoprecipitate chromatin fragments bound by the WT, ΔCD and SYD62A proteins, and characterized the resulting DNA on our previously described custom Nimblegen tiling arrays, which cover the entire euchromatic X chromosome and the left arm of chromosome 2 as a negative control7 (388,000 × 50mer probes tiled at 100bp resolution).
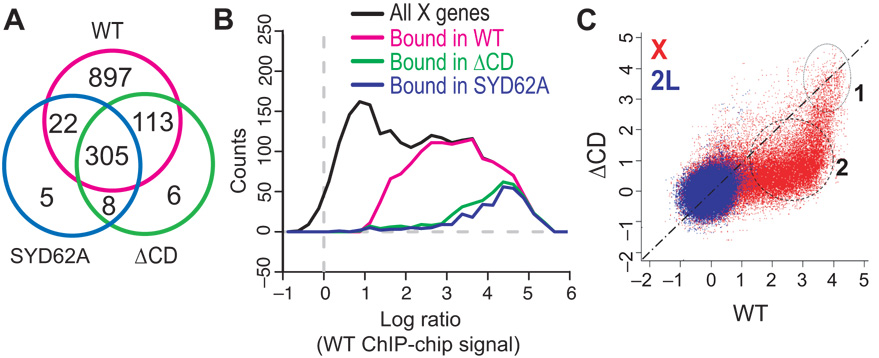
Each experiment was performed in duplicate per genotype and the signal intensities from both profiles were used to calculate a mean signal intensity value for each probe on the array. A computational algorithm was applied to locate statistically significant clusters of binding, assessing the minimum level of enrichment and the size of cluster (see Methods). The total number of bound clusters was calculated for each genotype and the number of bound genes within the clusters was determined. Using these criteria, WT MSL3-TAP protein bound to 1337 genes within 694 clusters on the X and 9 clusters were bound on chromosome arm 2L. Chromodomain mutants retained a strong bias for the X, but bound to only a subset of the WT targets. 340 genes were mapped within 338 clusters in the SYD62A mutant (with 13 clusters on 2L) and 432 genes were mapped within 444 clusters in the ΔCD mutant (with 15 clusters on 2L). Therefore, at least two-thirds of WT target genes were scored as unbound in the SYD62A and ΔCD mutants (Fig. 2a). The clusters in chromodomain mutants were also smaller and contained fewer genes on average than WT (~1.0 gene per cluster for SYD62A or ΔCD; ~1.9 genes per cluster for WT).
Figure 2. High resolution ChIP-chip mapping of MSL3 mutant binding sites.
(a) The number of genes within bound clusters on the X chromosome was compared among experiments in a Venn diagram. 98% (418/432) of ΔCD bound genes and 96% (327/340) of SYD62A bound genes are also WT sites; 92% (313/340) of SYD62A bound genes belong to the ΔCD set. (b) Distributions of WT log-ratios for genes from different categories. Among all X-linked genes (black), those identified as bound in WT (magenta) have high log-ratios. Importantly, most of the genes bound in ΔCD (green) or SYD62A (blue) mutants have the highest signals in WT. The color scheme corresponds to that in Fig. 2a; for this plot, we defined the signal for a gene as the maximum log-ratio over the gene. (c) Scatter-plot of ChIP-chip signals between WT and ΔCD at probe level. If the probe is equally bound in both WT and ΔCD experiments, the representative point should fall near the diagonal. The presence of two separate clusters of red probes on the X suggests that there are two types of bound sites in WT: chromodomain-independent (cluster 1) and chromodomain-dependent sites (cluster 2).
What characterizes the chromodomain-independent genes on the X? One possibility is that those are the genes that exhibit the highest occupancy or signal for the WT complex. When we plotted histograms of MSL3 occupancy (i.e. signal intensities from the WT ChIP-chip experiment) over each gene on the X, we found that the genes bound by the chromodomain mutants do fall within the region of high signal (Fig. 2b). To exclude the possibility that the smaller amount of binding observed in the MSL3 mutants was a consequence of experimental or statistical procedure, we compared the signal intensity of each probe in the WT (x-axis) to its intensity in the ΔCD experiment (y-axis) (Fig. 2c). If the signal on one array is simply a muted version of the signal on the other array, the probe values should be scattered along a single line. Instead we see that there are two separate probe clusters. This clearly shows that only a specific subclass of probes remains bound by the ΔCD mutant rather than a general decrease across all probes.
MSL3 chromodomain mutants retain entry site binding
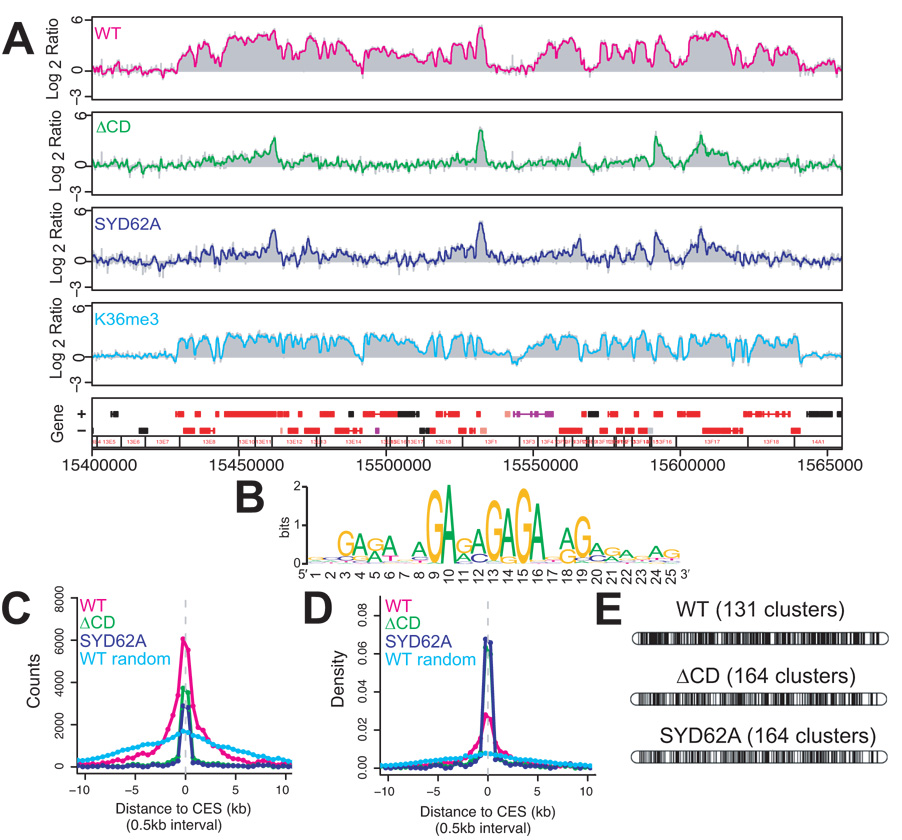
Interestingly, when individual ChIP-chip profiles of chromodomain mutant binding sites are compared to WT, they show graphically what might be expected for initial recognition sites of the MSL complex: discrete peaks of binding in place of coverage of whole genes and neighboring genes (Fig. 3a). Noting the pattern, we wondered whether the discrete peaks might reflect binding to chromatin entry sites, with a loss of spreading to cover whole genes and flanking targets. Chromatin entry sites (CES) were originally defined as the 35–70 sites seen on polytene chromosomes of msl3 null mutant larvae. However, we recently proposed that this was an underestimate based on ChIP-chip experiments of msl31 null mutant embryos, in which we identified a set of at least 150 candidate entry sites containing a GA- or TC–rich MSL recognition element (MRE) required for MSL binding27. Therefore, we examined whether the ~400 binding sites detected in our chromodomain mutants included the 150 newly defined chromatin entry sites. We found that 98% of the CES were contained within the SYD62A and ΔCD sets of binding clusters. Furthermore, when we searched for ~8–25bp motifs using the MEME algorithm28, the top sequence identified using 500 bp peaks from either SYD62A or ΔCD clusters was a GA-rich motif that aligns extremely well with the MRE27 (Fig. 3b). However, not all ΔCD sites have MREs. When we surveyed 500 bp segments centered on the peaks within SYD62A or ΔCD clusters, ~190 sites in both cases contain an MRE at a p value of 10−5. Using a 1 Kb window, these numbers were ~240; further increasing the window size had minimal effect. This indicates that a substantial number of binding sites in chromodomain mutants (up to 200) do not encompass an MRE. These sites generally displayed a lower signal in our mutant ChIP-chip analysis (data not shown). Therefore, we wondered whether these additional binding sites might be the result of a limited amount of spreading retained in the chromodomain mutants.
Figure 3. The MSL3 chromodomain is important for spreading.
(a) 200kb of sample binding data from WT and mutants. The chromodomain mutant proteins bind in specific peaks, but fail to cover all WT sites. The H3K36 methylation profile from male tissue culture cells (S2) over the same region is shown for comparison. The H3K36me3 profile is very similar to the profile for WT MSL binding, extending beyond the chromatin entry sites. Boxes below the profiles represent annotated genes; red: transcribed, black: non-transcribed. (b) GA-rich motif identified by MEME from the SYD62A binding peaks. (c) The distribution of distances to the nearest chromatin entry site (CES) for the probes bound by WT (magenta), the ΔCD mutant (green) and the SYD62A mutant (navy), or the average of 10 random circular permutations of WT (light blue). The number of probes bound by the mutants (y axis) falls more sharply than WT as one moves farther from CES (X axis). (d) Alternative representation of the data in (c). The y-axis represents the % of bound probes (i.e. density) rather than absolute count. The binding of chromodomain mutants is clustered at or near the chromatin entry sites as shown by increased density of binding. (e) Representation of ChIP-chip data of MSL3TAP binding on polytene chromosomes shows that patterns are not distinguishable at 30kb resolution. Each black bar represents a cluster bound by MSL3TAP after merging neighboring clusters with gaps smaller than 30kb.
To investigate this, we probed the spatial relationship between the 150 CES defined by Alekseyenko et al.27 and the additional sites bound by SYD62A and ΔCD mutants. We computed the distribution of the distances of bound probes to their nearest CES in WT and each chromodomain mutant (Fig. 3c,d). This analysis indicated that spreading in WT may generally occur within 5–10 kb on either side of a CES, roughly corresponding to the size of an interband or a thin band on polytene chromosomes29. In contrast, binding in the SYD62A and ΔCD mutants clustered almost exclusively around the 150 CES. Although the two mutants differed in the absolute number of binding sites scored as positive in the ChIP-chip experiments, the distribution of those sites shows that the mutants have quite similar profiles that differ substantially from the wild-type profile. In the mutants, the number of bound probes fell sharply past 1 kb on either side of the proposed chromatin entry sites (Fig. 3c,d), suggesting an appreciable reduction in spreading.
While this analysis showed a clear difference between mutants and wild-type in the extent of MSL binding at a distance from CES, differences between WT and chromodomain mutant patterns disappear when plotted at 30 kb resolution, since the majority of MSL binding sites are clustered within a few kilobases of an entry site (Fig. 3e). This explains the previous inability to detect differences in immunostaining at the level of polytene chromosomes21. We conclude that chromodomain mutants recognize a defined set of chromatin entry sites through the MRE motif, with a limited amount of additional binding that may represent very local (<2 kb) spreading in cis.
MSL3 chromodomain mutants are defective in spreading
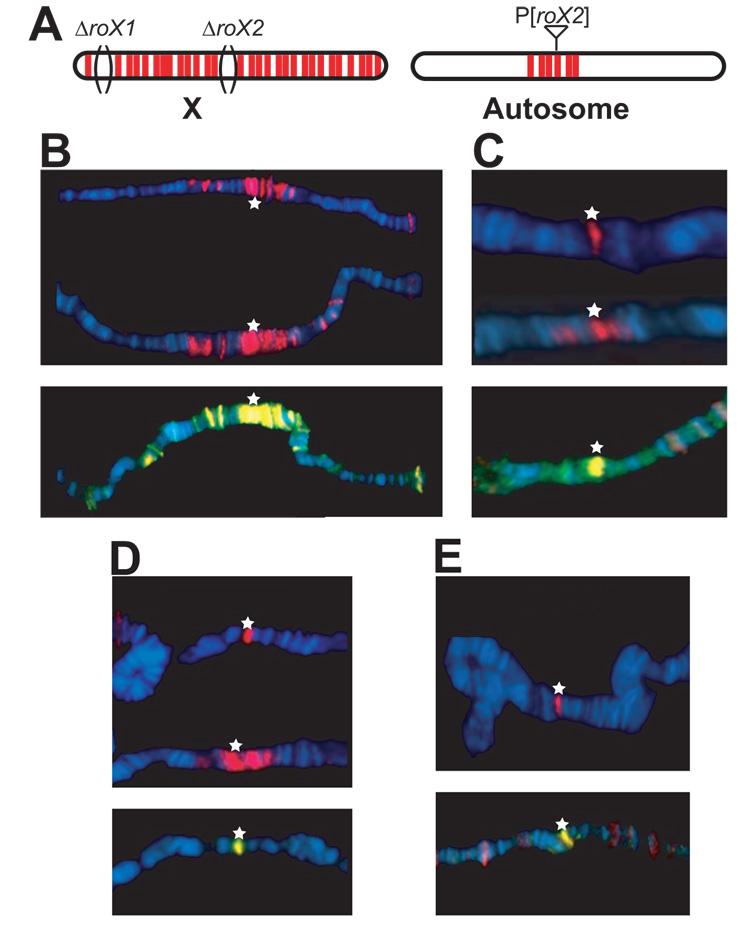
To further probe the ability of the SYD62A and ΔCD proteins to direct spreading, we asked whether expression of the mutant proteins might visibly interfere with the extensive wild-type spreading seen from an autosomal roX transgene insertion on polytene chromosomes. We used a transgenic line carrying a GMroX2 transgene at 26D8, which shows extensive spreading in the absence of roX1 and roX2 genes on the X (ref. 8, Fig. 4a). We analyzed the behavior of the mutant proteins and their effects on wild-type complexes, in a dominant assay with one functional copy of the endogenous wild-type msl3+ gene present. When stained for the TAP epitope, transgenic WT MSL3 showed consistent and extensive spreading around the roX2 insertion in close to 100% of nuclei, as expected (Fig. 4b). However, the chromodomain mutants failed to do so to varying extents. In msl31 heterozygotes carrying a ΔCD transgene, about half of all nuclei showed spreading of both MSL3TAP and MSL2, albeit not as extensive as the WT control (Fig. 4c). The remaining nuclei did not show any spreading for either MSL3TAP or MSL2 although both proteins were detected at the transgene insertion site. In the W59G point mutant, most nuclei showed spreading for MSL3TAP and MSL2 (Fig. 4d). The most pronounced defect was in the SYD62A mutant, where almost all nuclei lacked spreading for both MSL3TAP and MSL2 even though wild-type MSL3 protein was present (Fig. 4e). Therefore, chromodomain mutant proteins can interfere with ectopic spreading of endogenous MSL complexes, even in the presence of wild-type MSL3. This argues against a model in which simply lowering the level of complex results in the loss of binding seen in ΔCD mutants. Rather, the lack of an intact chromodomain is specifically implicated in diminished spreading from chromatin entry sites.
Figure 4. MSL3 chromodomain mutants disrupt spreading in vivo.
Chromosomes from roX1 roX2 deficient 3rd instar larvae with the GMroX2-26D8 transgene on chromosome 2L were stained for either the TAP epitope (red) alone or for both anti-MSL2 (red) and anti-MSL3 (green) whose overlap is shown by yellow. Anti-MSL3 staining shows both TAP-tagged and untagged MSL3 proteins. The transgene insertion is indicated by a star and the chromosomes are aligned by the location of the transgene for comparison. (a) When a transgene harboring roX2 is put on an autosome in the absence of both roX genes on the X, the MSL complex spreads extensively around the transgene insertion. (b) In the presence of the WT transgene (red), 100% of nuclei show extensive spreading. Three representative chromosomes are shown. (c) ΔCD (red) shows mosaic spreading. Three representative nuclei are shown. One shows spreading, albeit limited, whereas the other two show binding to the transgene but no spreading. Endogenous complexes (yellow) are also affected. (d) W59G (red) also shows mosaic spreading, but a majority of the nuclei show limited spreading. (e) SYD62A generally fails to spread and interferes with endogenous spreading.
Defective H3K36me3 binding by MSL3 chromodomain mutants
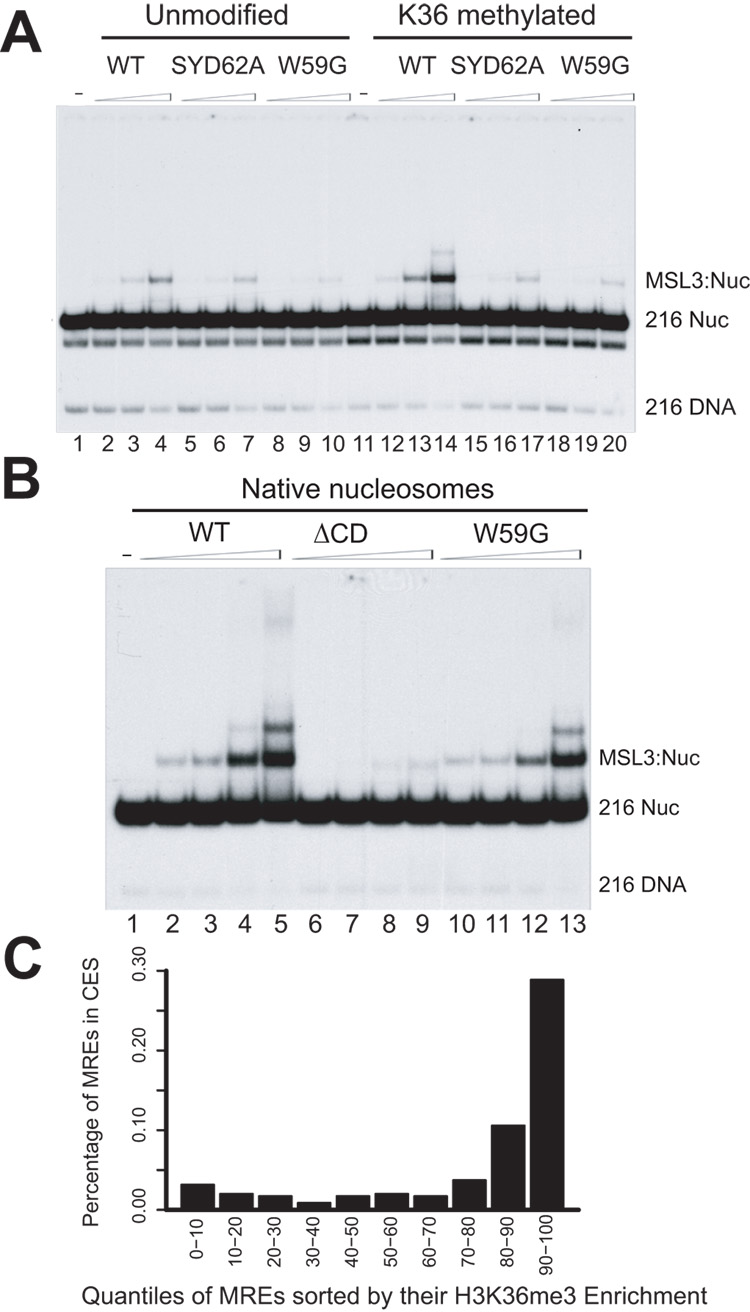
How does the MSL3 chromodomain mediate local spreading? One possibility is that it binds to a specific histone mark. MSL3 homologues in yeast (Eaf3) and humans (hMRG15) have been implicated in H3K36me3 interaction via their chromodomains22,30–32. Recombinant wild-type MSL3 binds nucleosomes, with a preference for methylated H3K36 in vitro8, thus, the MSL3 chromodomain is clearly a strong candidate to mediate this interaction. To specifically address this possibility, we purified TAP-tagged recombinant WT, ΔCD, SYD62A and W59G proteins using the baculovirus expression system and tested them for binding to nucleosomes using in vitro gel shift assays.
When tested for binding to methylated H3K36, WT protein shows increased binding to modified nucleosomes vs. unmodified ones (Fig. 5a, comparing lane 14 to lane 4), consistent with previous data that MSL3 has a higher affinity for nucleosomes with H3K36 methylation8. The SYD62A and the W59G mutants showed no difference in binding to methylated vs. unmodified nucleosomes, suggesting that general affinity for nucleosomes was retained, but specificity for H3K36 methylation in these mutants is lost (Fig. 5a). The ΔCD recombinant protein lacked even general binding to nucleosomes (Fig. 5b), as reported previously21. These data demonstrate that recombinant MSL3 recognizes nucleosomes via its chromodomain, with increased affinity for octamers containing the methylated H3K36 mark.
Figure 5. Chromodomain mutants show loss of preference for H3K36me3 containing nucleosomes.
A range of concentrations of recombinant MSL3 proteins was tested for binding to nucleosomes in gel shift assays. (a) Binding to recombinant nucleosomes methylated at H3K36. SYD62A and W59G point mutants can still bind unmodified nucleosomes albeit to a lesser extent than the WT (compare lanes 4, 7 and 10). WT has higher affinity for modified nucleosomes (compare lanes 4 and 14). The point mutants show no difference in binding to modified vs. unmodified nucleosomes (compare lanes 7 and 17 for SD62A; 10 and 20 for W59G). (b) WT MSL3 can bind native nucleosomes efficiently in gel shift assays. W59G has decreased binding and ΔCD has no detectable binding. (c) Chromatin entry sites are preferentially found in H3K36me3-enriched regions. Sequences matching the consensus for MSL recognition elements (MREs) were ordered by their enrichment for H3K36me3 and grouped into 10 bins (x-axis). The likelihood of an MRE being in a functional chromatin entry site (CES) (y-axis) increases as the level of H3K36me3 enrichment increases. H3K36me3 enrichment was computed by averaging the signal of the 3 probes nearest to each MRE.
In the simplest model, absence of the MSL3 chromodomain or lack of the H3K36me3 mark would result in similar MSL binding profiles, with retention of intial binding at entry sites, but a defect in subsequent spreading. The inviability of set2 mutants precluded collection of sufficient material for comparative ChIP-chip analysis8. However, the individual target genes previously characterized as requiring the Set2 H3K36 methyltransferase for optimal MSL binding8 were also chromodomain-dependent in our current study (binding to 7 of 8 genes was strongly reduced, Supplementary Fig. 2). Furthermore, apparent spreading in wild type clearly correlated with the H3K36me3 mark surrounding chromatin entry sites (see Fig. 3a for an example).
The hypothesis that H3K36me3 functions in spreading led us to wonder whether chromatin entry sites might be more likely to be located in regions of the genome enriched for this mark. We found that this is indeed the case (ref. 27, Fig. 5c). When X chromosome locations with potential MSL recognition elements (MREs) are characterized with respect to the average density of the H3K36me3 mark, it is clear that MREs in regions with higher H3K36me3 signal are the most likely to fall within functional chromatin entry sites (Fig. 5c). Together, these data further support a model in which the MSL3 chromodomain mediates MSL spreading through association with the H3K36me3 mark.
Discussion
In this study, we provide evidence that initial targeting and spreading aspects of the MSL complex are separable steps required to achieve the final MSL binding pattern on the X, the latter of which is mediated by the chromodomain of the MSL3 component. By making stable but functionally defective MSL3 mutants, we were able to uncouple these two activities and study them in isolation.
By comparing ChIP-chip data obtained from WT and MSL3 chromodomain mutants, we identified 2 classes of target genes on the X; a chromodomain-independent group (~25% of all targets) and a larger chromodomain-dependent group (~75% of all targets). The chromodomain-independent group encompasses a larger set of chromatin entry sites than originally postulated from analyses of polytene chromosomes in msl3 null mutants. The difference could be related to a reduction in overall MSL complex levels when MSL3 protein is completely absent. Thus, it is possible that there is an entry site within the majority of active gene clusters (Fig. 3e). If spreading of MSL complex is generally a short-range phenomenon (Fig. 3c), this could also explain why the complex seems to fail to spread at the cytological level into autosomal fragments inserted on the X and vice versa10,11. Having hundreds rather than a more limited set of initiation sites would make Drosophila dosage compensation targeting more similar to the C.elegans model than previously suspected33,34.
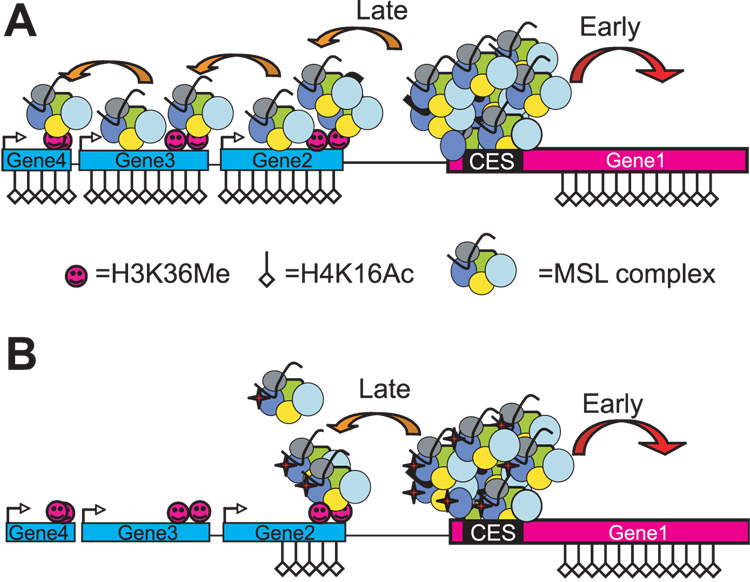
The data presented here are summarized in a two-step model for targeting of the MSL complex to the X chromosome (Fig. 6). In this model, there are 2 classes of MSL target genes on the X, sequence-dependent chromatin entry sites and chromodomain-dependent active genes. Initially, MSL complex recognizes specific MREs within entry sites on the X chromosome, leading to local acetylation of nearby targets27,35. The local acetylation at these sites is sufficient for marginal survival and this step is independent of the MSL3 chromodomain. In a second step, the MSL3 chromodomain directs the complex to more distant genes by recognizing H3K36 methylated nucleosomes (Fig. 6a), probably in conjunction with general DNA and nucleosomal binding21,25. Whether MSL complex actually traverses intergenic regions or releases and reattaches to nearby active genes is not known. Furthermore, chromodomain defects may lead to secondary effects on spreading through other MSL subunits, although our nucleosomal binding analysis using recombinant MSL3 protein strongly suggests a direct effect. Regardless, in the absence of the MSL3 chromodomain, the spreading step is defective (Fig. 6b). How far the complex will travel without the chromodomain may depend on the local MSL concentration at the nearest chromatin entry site.
Figure 6. A model for MSL3 chromodomain-dependent spreading.
(a) An initial group of sequence-dependent targets recruit the complex and lead to upregulation of transcription via local acetylation, which is sufficient for marginal survival. A second class of genes is targeted by spreading mediated by the MSL3 chromodomain, partly via the recognition of H3K36Me mark at 3’ ends of active genes. (b) In the chromodomain mutants, targeting by spreading is defective. The complex is not stabilized at 3’ ends of targets and diffuses away.
This work, together with a large body of gene-silencing literature, suggests an important parallel between principles for organizing chromatin into active and silent domains. Groups studying the mechanism of heterochromatin initiation and propagation have long embraced the spreading concept, while it has been less evident that similar mechanisms function to organize active chromatin. In the field of Drosophila dosage compensation, the spreading idea continues to be debated. Here we show that the chromodomain can be an important facilitator for establishment of an upregulated, active chromatin state. Our results support a genomic organization model in which chromodomains are key componenets for establishment of both active and silent chromatin domains.
Methods
Fly genetics and transgenesis
We carried out mutagenesis using a derivative of Invitrogen’s GATEWAY™ entry vector pENTR™4 containing a 6 kb BamHI msl3-tap fragment (pENTR4-MSL3TAP), and the Stratagene XL mutagenesis kit. Primers used for mutagenesis are available upon request. The pGreeni-RfA-attB destination vector was constructed using an EGFP gene driven in the eye by the Pax6 promoter36, the Gateway® RfA recombination cassette containing a ccdB and a chloramphenicol resistance gene, and the 293bp phiC31 attB sequence. The RfA cassette was exchanged with MSL3TAP derivatives from pENTR4-MSL3TAP by LR recombination (Invitrogen).
We created transgenic flies using phiC31 integrase-mediated attB/attP recombination. We performed injections as described previously24. 200 ng µl−1 of each pGreeni plasmid containing attB and msl3-tap was mixed with 0.5 µg µl−1 phiC31 integrase mRNA, and injected into fly embryos carrying the attP1 docking site on chromosome 2R. mRNA for the integrase gene was synthesized using the mMessage mMachine kit (Ambion) after linearization of 1µg of pET11phiC31. We identified transgenics by eye-specific GFP expression. Transgenics were crossed to msl31 mutants to establish stocks. For complementation, we crossed homozygous msl31 mutant virgins (y w; msl31/msl31) to transgenic males heterozygous for msl31 (y w; attP1 y+{p[gfp+ MSL3TAP-pGreeni]}; msl31/TM3,Sb) and calculated the rescue rates by dividing the number of rescued males by the number of their TM3-balanced brothers. The genotype of rescued males are as follows: y w; attP1 y+{p[gfp+ MSL3TAP-WT-pGreeni]}/+ ; msl31/msl31 with MSL3TAP-WT replaced with ΔCD, SYD62A, W59G and LYT30A, respectively.
We performed polytene chromosome immunostaining as previously described27,37. Primary antibodies were: affinity purified anti-MSL2 and anti-MSL3 at 1:500 dilution and anti-PAP antibody (Sigma) at 1:250 dilution. Secondary antibodies were: anti-mouse AlexaFluor594 (1:500); anti-goat AlexaFluor488 (Invitrogen). Hoechst dye (Invitrogen) was used to stain DNA (1:10,000). To test general cross-reactivity of the Protein A moiety of the TAP tag to antibodies, we performed immunostaining with various alternative primary and secondary antibodies, but only the anti-PAP antibody (Sigma) gave positive results.
Characterization of MSL3TAP complexes in Schneider cells
We performed transfections of 10 µg of pGreeni-MSL3TAP plasmids using the Calcium Phosphate Transfection Kit (Invitrogen). We co-transfected 1µg of plasmid encoding the hygromycin resistance gene (pCoHygro) to select for stable transformant growth in Schneider Drosophila Medium (Gibco) containing 300 µg µl−1 hygromycin (Invitrogen). We performed TAP purification from whole cell extracts as described previously27,36,37. 5 mg of total protein were used for each pulldown. We performed Western blots using the following primary antibodies: rabbit anti-MSL1 (1:2000); rabbit anti-MOF (1:5000); goat anti-MSL3 (1:3000); mouse anti-tubulin (1:10000; Sigma); rabbit anti-PAP (1:2000; Sigma). HRP-conjugated secondary antibodies and the Dura kit were used for visualization (Pierce).
Chromatin preparation and ChIP-chip analysis
We performed preparation of chromatin from embryos as described previously7. For a single experiment, we collected 0.5 g of 12- to 17 hour mixed sex embryos from WT MSL3-TAP, ΔCD and SYD62A from the following crosses: we crossed homozygous virgin mothers carrying the MSL3TAP transgene on the second chromosome in a homozygous msl31 mutant background (y w; attP1 y+{p[gfp+ MSL3TAP-pGreeni]}; msl31) to msl31 mutant males carrying a wild-type msl3+ cosmid on their X chromosome (y w p[cos8-1-msl3+]; +; msl31). Male progeny inherit the Y chromosome from their fathers rather than the msl3+ cosmid and thus are dependent on their respective MSL3TAP transgenes for function. Females inherit the msl3+ cosmid, but lack functional MSL complexes because of female-specific translational repression of MSL2. We performed chromatin immunoprecipitation using the TAP-tag as described previously37. The resulting amplified input and IP’ed DNA were labeled and hybridized to custom microarrays by NimbleGen Systems, Inc. as described previously7.
We performed two replicates for each genotype. For each two-channel Nimblegen array (ChIP vs. input), we corrected for dye-specific bias from the average intensity vs. log-ratio plot using a combination of rotation and curve-fitting39. We computed the noise level (σ*) of each array based on the median absolute deviation of the differences between neighboring probes. A signal above 2σ* was considered to be statistically significant (σ* is normalized so that it becomes standard deviation when data are Gaussian). To identify binding clusters, we first took the mean of the two replicates after rescaling based on their noise levels, and applied running median smoothing with window size of 7 to the mean log-ratios along their chromosomal locations. Based on the DNA fragment size and the enrichment of the X chromosome compared to 2L, clusters consisting of at least 8 probes above the enrichment threshold were counted as significant.
Assembly of nucleosomes and gel shift assays
We produced MSL3TAP proteins in baculovirus-infected SF9 cells as previously described, using the ‘Bac-toBac’ expression system (Invitrogen)8. Mono-nucleosomes carrying specific modification were prepared as described40. 216 bp DNA templates were PCR amplified from the pGEM-601R plasmid using one 5’ biontinylated primer and one regular primer. Purified DNA probes were reconstituted into nucleosomes with recombinant Xenopus core histones41. Resulting mono- nucleosomes were then immobilized onto streptavidin-coated magnetic Dynal 280 beads and subjected to histone methylation reactions using human recombinant human Set2 (HYPB) in the presence or absence of 5 µM cold SAM as described previously42. Modified nucleosomes were finally released from magnetic beads by overnight incubation with EcoRV. Gel purification of nucleosomes was performed subsequent to release from beads. EMSA experiments were performed at 30°C in 15 µl of the EMSA buffer (10 mM HEPES pH 7.8, 50 mM KCl, 4 mM MgCl2, 5 mM DTT, 0.25 mg ml−1 BSA, 5% (v/v) Glycerol and 0.1 mM PMSF). The total reactions were directly loaded onto a 3.5– 5% native polyacrylamide gel (37.5:1) in 0.3 × TBE. Electrophoresis was carried out at 4°C for 4–5 hours.
Supplementary Material
Acknowledgements
Special thanks to A.A. Alekseyenko (Harvard-Partners Center for Genetics and Genomics) for important advice, discussions, and protocols and to members of the Kuroda lab for critical comments on the manuscript, H. Oh (currently at Rutgers University), for a key fly stock, J. Racine and A. Sarovschii for technical assistance and the Taplin Biological Mass Spectrometry Facility (Harvard Medical School) for peptide identification. This work was supported by the National Institutes of Health (GM45744 to M.I.K and GM67825 to P.J.P). B.L. and J.L.W. were supported by GM47867 from the NIH, and by funding from the Stowers Institute for Medical Research. ChIP-chip data have been deposited in the NCBI GEO public repository, accession number GSE11817.
References
- 1.Koonin EV, Zhou S, Lucchesi JC. The chromo superfamily: new members, duplication of the chromo domain and possible role in delivering transcription regulators to chromatin. Nucleic Acids Res. 1995;23:4229–4233. doi: 10.1093/nar/23.21.4229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Marin I, Baker BS. Origin and evolution of the regulatory gene male-specific lethal-3. Mol. Biol. Evol. 2000;17:1240–1250. doi: 10.1093/oxfordjournals.molbev.a026407. [DOI] [PubMed] [Google Scholar]
- 3.Palmer MJ, Richman R, Richter L, Kuroda MI. Sex-specific regulation of the male-specific lethal-1 dosage compensation gene in Drosophila. Genes Dev. 1994;8:698–706. doi: 10.1101/gad.8.6.698. [DOI] [PubMed] [Google Scholar]
- 4.Lyman LM, Copps K, Rastelli L, Kelley RL, Kuroda MI. Drosophila male-specific lethal-2 protein: structure/function analysis and dependence on MSL-1 for chromosome association. Genetics. 1997;147:1743–1753. doi: 10.1093/genetics/147.4.1743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gu W, Szauter P, Lucchesi JC. Targeting of MOF, a putative histone acetyl transferase, to the X chromosome of Drosophila melanogaster. Dev. Genet. 1998;22:56–64. doi: 10.1002/(SICI)1520-6408(1998)22:1<56::AID-DVG6>3.0.CO;2-6. [DOI] [PubMed] [Google Scholar]
- 6.Demakova OV, et al. The MSL complex levels are critical for its correct targeting to the chromosomes in Drosophila melanogaster. Chromosoma. 2003;112:103–115. doi: 10.1007/s00412-003-0249-1. [DOI] [PubMed] [Google Scholar]
- 7.Alekseyenko AA, Larschan E, Lai WR, Park PJ, Kuroda MI. High-resolution ChIP-chip analysis reveals that the Drosophila MSL complex selectively identifies active genes on the male X chromosome. Genes Dev. 2006;20:848–857. doi: 10.1101/gad.1400206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Larschan E, et al. MSL complex is attracted to genes marked by H3K36 trimethylation using a sequence-independent mechanism. Mol Cell. 2007;28:121–133. doi: 10.1016/j.molcel.2007.08.011. [DOI] [PubMed] [Google Scholar]
- 9.Bell O, et al. Transcription-coupled methylation of histone H3 at lysine 36 regulates dosage compensation by enhancing recruitment of the MSL complex in Drosophila. Mol Cell Biol. 2008;28:3401–3409. doi: 10.1128/MCB.00006-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fagegaltier D, Baker BS. X Chromosome Sites Autonomously Recruit the Dosage Compensation Complex in Drosophila Males. PLoS Biol. 2004;2:1854–1861. doi: 10.1371/journal.pbio.0020341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Oh H, Bone JR, Kuroda MI. Multiple classes of MSL binding sites target dosage compensation to the X chromosome of Drosophila. Curr Biol. 2004;14:481–487. doi: 10.1016/j.cub.2004.03.004. [DOI] [PubMed] [Google Scholar]
- 12.Dahlsveen IK, Gilfillan GD, Shelest VI, Lamm R, Becker PB. Targeting determinants of dosage compensation in Drosophila. PLoS Genet. 2006;2:e5. doi: 10.1371/journal.pgen.0020005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gilfillan GD, et al. Cumulative contributions of weak DNA determinants to targeting the Drosophila dosage compensation complex. Nucleic Acids Res. 2007;35:3561–3572. doi: 10.1093/nar/gkm282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kind J, Akhtar A. Cotranscriptional recruitment of the dosage compensation complex to X-linked target genes. Genes Dev. 2007;21:2030–2040. doi: 10.1101/gad.430807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Muller HJ, Altenburg E. The Frequency of Translocations Produced by X-Rays in Drosophila. Genetics. 1930;15:283–311. doi: 10.1093/genetics/15.4.283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schotta G, Ebert A, Dorn R, Reuter G. Position-effect variegation and the genetic dissection of chromatin regulation in Drosophila. Semin Cell Dev Biol. 2003;14:67–75. doi: 10.1016/s1084-9521(02)00138-6. [DOI] [PubMed] [Google Scholar]
- 17.Grewal SI, Jia S. Heterochromatin revisited. Nat Rev Genet. 2007;8:35–46. doi: 10.1038/nrg2008. [DOI] [PubMed] [Google Scholar]
- 18.Zhang K, Mosch K, Fischle W, Grewal SIS. Roles of the Clr4 methyltransferase complex in nucleation, spreading and maintenance of heterochromatin. Nat. Structural & Mol. Biol. 2008;15:381–388. doi: 10.1038/nsmb.1406. [DOI] [PubMed] [Google Scholar]
- 19.Elgin SC, Grewal SI. Heterochromatin: silence is golden. Curr Biol. 2003;13:R895–R898. doi: 10.1016/j.cub.2003.11.006. [DOI] [PubMed] [Google Scholar]
- 20.Schwartz YB, Pirrotta V. Polycomb silencing mechanisms and the management of genomic programmes. Nat Rev Genet. 2007;8:9–22. doi: 10.1038/nrg1981. [DOI] [PubMed] [Google Scholar]
- 21.Buscaino A, Legube G, Akhtar A. X-chromosome targeting and dosage compensation are mediated by distinct domains in MSL-3. EMBO Rep. 2006;7:531–538. doi: 10.1038/sj.embor.7400658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang P, et al. Structure of human MRG15 chromo domain and its binding to Lys36-methylated histone H3. Nucleic Acids Res. 2006;34:6621–6628. doi: 10.1093/nar/gkl989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Flanagan JF, et al. Molecular implications of evolutionary differences in CHD double chromodomains. J Mol Biol. 2007;369:334–342. doi: 10.1016/j.jmb.2007.03.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Groth AC, Fish M, Nusse R, Calos MP. Construction of transgenic Drosophila by using the site-specific integrase from phage phiC31. Genetics. 2004;166:1775–1782. doi: 10.1534/genetics.166.4.1775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Morales V, Regnard C, Izzo A, Vetter I, Becker PB. The MRG domain mediates the functional integration of MSL3 into the dosage compensation complex. Mol Cell Biol. 2005;25:5947–5954. doi: 10.1128/MCB.25.14.5947-5954.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lucchesi JC, Kelly WG, Panning B. Chromatin remodeling in dosage compensation. Annu Rev Genet. 2005;39:615–651. doi: 10.1146/annurev.genet.39.073003.094210. [DOI] [PubMed] [Google Scholar]
- 27.Alekseyenko AA, et al. A sequence motif within chromatin entry sites directs MSL establishment on the Drosophila X chromosome. Cell. 2008;134:599–609. doi: 10.1016/j.cell.2008.06.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bailey TL, Elkan C. Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Proc Int Conf Intell Syst Mol Biol. 1994;2:28–36. [PubMed] [Google Scholar]
- 29.Beermann W. Chromomeres and genes. Results Probl Cell Differ. 1972;4:1–33. doi: 10.1007/978-3-540-37164-9_1. [DOI] [PubMed] [Google Scholar]
- 30.Carrozza MJ, et al. Histone H3 methylation by Set2 directs deacetylation of coding regions by Rpd3S to suppress spurious intragenic transcription. Cell. 2005;123:581–592. doi: 10.1016/j.cell.2005.10.023. [DOI] [PubMed] [Google Scholar]
- 31.Joshi AA, Struhl K. Eaf3 chromodomain interaction with methylated H3-K36 links histone deacetylation to Pol II elongation. Mol Cell. 2005;20:971–978. doi: 10.1016/j.molcel.2005.11.021. [DOI] [PubMed] [Google Scholar]
- 32.Keogh MC, et al. Cotranscriptional set2 methylation of histone H3 lysine 36 recruits a repressive Rpd3 complex. Cell. 2005;123:593–605. doi: 10.1016/j.cell.2005.10.025. [DOI] [PubMed] [Google Scholar]
- 33.McDonel P, Jans J, Peterson BK, Meyer BJ. Clustered DNA motifs mark X chromosomes for repression by a dosage compensation complex. Nature. 2006;444:614–618. doi: 10.1038/nature05338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ercan S, Giresi PG, Whittle CM, Zhang X, Green RD, Lieb JD. X chromosome repression by localization of the C. elegans dosage compensation machinery to sites of transcription initiation. Nat Genet. 2007;39:403–408. doi: 10.1038/ng1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yokoyama R, Pannuti A, Ling H, Smith ER, Lucchesi JC. A plasmid model system shows that Drosophila dosage compensation depends on the global acetylation of histone H4 at lysine 16 and is not affected by depletion of common transcription elongation chromatin marks. Mol. Cell. Biol. 2007;27:7865–7870. doi: 10.1128/MCB.00397-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nagel AC, Maier D, Preiss A. Green fluorescent protein as a convenient and versatile marker for studies on functional genomics in Drosophila. Dev Genes Evol. 2002;212:93–98. doi: 10.1007/s00427-002-0210-y. [DOI] [PubMed] [Google Scholar]
- 37.Kelley RL, et al. Epigenetic spreading of the Drosophila dosage compensation complex from roX RNA genes into flanking chromatin. Cell. 1999;98:513–522. doi: 10.1016/s0092-8674(00)81979-0. [DOI] [PubMed] [Google Scholar]
- 38.Puig O, et al. The tandem affinity purification (TAP) method: a general procedure of protein complex purification. Methods. 2001;24:218–229. doi: 10.1006/meth.2001.1183. [DOI] [PubMed] [Google Scholar]
- 39.Peng S, Alekseyenko AA, Larschan E, Kuroda MI, Park PJ. Normalization and experimental design for ChIP-chip data. BMC Bioinformatics. 2007;8:219. doi: 10.1186/1471-2105-8-219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Li B, et al. Combined action of PHD and chromo domains directs the Rpd3S HDAC to transcribed chromatin. Science. 2007;316:1050–1054. doi: 10.1126/science.1139004. [DOI] [PubMed] [Google Scholar]
- 41.Li B, et al. Preferential occupancy of histone variant H2AZ at inactive promoters influences local histone modifications and chromatin remodeling. Proc Natl Acad Sci U S A. 2005;102:18385–18390. doi: 10.1073/pnas.0507975102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Li B, Howe L, Anderson S, Yates JR, Workman JL. The Set2 histone methyltransferase functions through the phosphorylated carboxyl-terminal domain of RNA polymerase II. J Biol Chem. 2003;278:8897–8903. doi: 10.1074/jbc.M212134200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.