Abstract
In this work three DNA-chitosan nanoparticle formulations (Np), differing in the molecular weight (MW; 150 kDa, 400 kDa, and 600 kDa) of the polysaccharide, were prepared and administered by two different administration routes: the hydrodynamics-based procedure and the intraduodenal injection. After the hydrodynamic injection, DNA-chitosan nanoparticles were predominantly accumulated in the liver, where the transgene was expressed during at least 105 days. No significant influence of MW was observed on the levels of luciferase expression. The curves of bioluminescence versus time obtained using the charge-coupled device (CCD) camera were described and divided in three phases: (i) the initial phase, (ii) the sustained release step and (iii) the decline phase (promotor inactivation, immunological and physiological processes). From these curves, which describe the transgene expression profile, the behavior of the different formulations as gene delivery systems was characterized. Therefore, the following parameters such as Cmax (maximum level of detected bioluminescence), AUC (area under the bioluminescence-time curve) and MET (mean time of the transgene expression) were calculated. This approach offers the possibility of studying and comparing transgene expression kinetics among a wide variety of gene delivery systems. Finally, the intraduodenal administration of naked DNA permitted the gene transfer in a dose dependent manner quantifiable with the CCD camera within 3 days. Nevertheless, the same administration procedure of the three formulations did not improve the levels of transgene expression obtained with naked DNA. This fact could be explained by the rapid physiological turn-over of enterocytes and by the ability of chitosan nanoparticles to control the DNA release.
Keywords: chitosan, nanoparticles, gene delivery, hydrodynamics-based procedure, bioluminescence, intestine
Introduction
Chitosan is a partially deacetylated polymer of N-acetyl glucosamine. It is usually prepared from chitin [(1→ 4)-2-acetamido-2-deoxy-beta-D-glucan], the second most universally abundant biopolymer after cellulose. The presence of amino groups on chitosan backbone confers a high positive charge density dependent on its deacetylation degree (Illum et al 1994), and makes it behave as a polycation at acidic and neutral pH (Schipper et al 1996). Many chitosans of different molecular weights and degrees of deacetylation are commercially available, and their low manufacturing costs render their use attractive. These cationic polymers can efficiently condense plasmid DNA permitting its protection during the intracellular transport (Dunlap et al 1997). Furthermore, through their biodegradation, polymers can control and modulate the release of the plasmid DNA and, thus, prolong and sustain the expression of the encoded protein.
DNA-chitosan nanoparticles were first described as a non-viral delivery system for plasmids in 1995 (Mumper et al 1995). Several promising studies have been reported describing the use of chitosans in animal models. Following topical application of DNA-chitosan nanoparticles, transgene expression was detected in skin with significant levels of antigen-specific IgG in blood (Cui et al 2001). After intratracheal administration, Koping-Hoggard and colleagues (2001) obtained a mid-airways distribution of the DNA-chitosan complexes, where transgene expression was observed in virtually every epithelial cells. On the other hand, the major success of this nanoparticulate system has been to open a new modality of administration to gene therapy that cannot be performed by another gene delivery system, viral included: the oral gene therapy. Benefiting from chitosan mucoadhesive properties (Janes et al 2001; Kato et al 2003), therapeutic approaches with DNA-chitosan based vaccines have been applied successfully, offering a new immunoprophylactic strategy for oral allergen-gene immunization (Roy et al 1999; Chew et al 2003).
However, levels of transgene expression remain critical for many therapeutic applications. A few years ago, a new way of administering naked DNA was developed: the hydrodynamics-based procedure (Liu et al 1999; Zhang et al 1999). This administration procedure induces very high levels and long term transgene expression in the liver. For this reason it has been widely used to evaluate a number of aspects including the therapeutic activity of determined genes (Liu and Knapp 2001), the analysis of regulatory functions of DNA sequences (Kramer et al 2003), and the design of RNAs interference (RNAi) used to silence the expression of targeted genes (Song et al 2003).
Despite the lack of information regarding the mechanisms of the hydrodynamics-based procedure, taking advantage of the quantification of a long term transgene expression allows the estimation of the kinetic parameters leading to a better understanding of the behavior of the gene delivery systems in vivo. The calculation of parameters such as area under the bioluminescence-time curve (AUC) results in valuable information for comparative studies (Koping-Hoggard et al 2004).
In the present report, following the physicochemical characterization of the different DNA-chitosan formulations (150, 400, and 600 kDa), the effect of chitosan molecular weight (MW) on the levels of transgene expression was first analyzed after hydrodynamic injection. Then the kinetic parameters of the different transgene expressions were calculated. In a second step, we visualized and quantified the ability of the three formulations to transduce the enterocyte monolayer over time after intraduodenal administration in mice.
Materials and methods
Materials
Chitosans were purchased from Fluka (Sigma-Aldrich, St. Louis, MO, USA) in MWs of 150, 400, and 600 kDa and 85% deacetylation degree. The plasmid pEGFP-N1 (4.7 kbp) encoding the green fluorescent protein (GFP) was obtained from Clontech Laboratories Inc (Palo Alto, CA, USA), and the plasmid pX2-Luc (6.15 kbp) encoding the luciferase under the control of CMV promotor was kindly provided by Dr Novo (University of Navarra, Pamplona, Spain). Plasmids were purified with the QIAfilter plasmid Giga kit that was purchased from Qiagen (Mississauga, ON, Canada).
D-luciferin was obtained from Xenogen (Alameda, CA, USA), DNAse I from Invitrogen (Barcelona, Spain), PicoGreen assay from Molecular Probes (Leiden, The Netherlands), and the chitosanase from Roche Farma (Madrid, Spain). OCT compound was purchased from Tissue-Tek (Zoeterwoude, The Netherlands). The chemical reagents, such as acetic acid glacial (99.5%), ethanol, and NaOH were purchased from PANREAC (Madrid, Spain). All other chemicals used were of reagent grade and obtained from Merck (Darmstadt, Germany).
Preparation of DNA-chitosan nanoparticles
DNA-chitosan nanoparticles were performed as previously described by Mao and colleagues (2001). The three different chitosans were dissolved in 1% acetic acid solution under gentle heating to form a 0.5% (w/v) stock solution of each polysaccharide. For the preparation of nanoparticles, stock solutions were diluted to 0.02% chitosan (w/v) with 5 mM of sodium acetate and the pH adjusted to 5.5 with NaOH 2M. After, a DNA solution (100 μg/ml) in 25 mM sodium sulfate was prepared. Both solutions were heated at 55 °C, mixed together and vortexed during 20 seconds. This mixture containing the nanoparticles was used without any further modification.
Physicochemical characterization of the nanoparticles
Particle size, zeta potential, and morphology
The size and zeta potential of the nanoparticles were measured by photon correlation spectroscopy and laser Doppler anemometry respectively, using a Zetamaster analyser system (Malvern Instruments, Southborough, MA, USA). Size measurements were performed at 25 °C at a 90° scattering angle, and each measurement was recorded during 90s. The mean hydrodynamic diameter was generated by cumulative analysis. The zeta potential measurements were performed with an aqueous dip cell in automatic mode.
Moreover, the morphology and size of the DNA-chitosan nanoparticles were examined by scanning electron microscopy (SEM) in a Zeiss DSM-940A digital scanning electron microscope (Oberkochen, Germany).
DNA loading efficacy
Following synthesis, 200 μl of a dispersion of DNA-chitosan nanoparticles were centrifuged at 25,000 rpm during 30 min in a 3K30 centrifuge (rotor 12154-H, Sigma). Unbound DNA fraction in the supernatant (100 μl of the upper phase) was determined with the PicoGreen assay according to the manufacturer’s procedure. The fluorescence was measured in a Tecan GENios microplate reader (Barcelona, Spain) at excitation and emission wavelengths of 485 and 535 nm, respectively. The DNA loading efficiency was calculated as the ratio between the bound DNA and the total DNA amount used to prepare nanoparticles.
In vivo transgene expression monitorization and in situ localization
Hydrodynamic injection of DNA-chitosan nanoparticles
These studies were performed in compliance with the regulations of the responsible committee of the University of Navarra in line with the European legislation on animal experiments (86/609/EU).
Female Balb/c mice 6–8 weeks old (Harland, Barcelona, Spain), were injected via the tail vein in a volume equivalent to 10% of the body weight in 5–8s (containing 20 μg of DNA) according to the previously published procedure (Liu et al 1999). Saline was used as a carrier solution for injection.
Intraduodenal administration of DNA-chitosan nanoparticles
An aqueous suspension containing 50 μg DNA loaded in chitosan nanoparticles was administered in the upper small intestine of mice that had been previously fasted overnight. In brief, this procedure involved inhalatory anesthetizing the animal, opening the abdomen and locating the region of the intestine tract for direct administration into the intestinal lumen.
Inhalatory anesthesia was performed by placing the mice in an induction chamber to which 4% isoflurane (Forane, Abbott Laboratories, Madrid, Spain) in a continuous oxygen flow of 3 l/min was directed (Isoflurane Vaporizer, Medical supplier and services international limited, Barcelona, Spain). Two to three minutes later, the inspired isoflurane concentration was reduced to 2%–2.5%.
Transgene expression monitorization and localization
All mice were imaged using a cooled CCD camera (Xenogen IVIS, Alameda, CA, USA). The mice were previously anesthetized (ketamine-xylazine, 10:1) and immediately after, 100 μl of D-luciferin (30 mg/ml) was injected ip, The animals were placed in a light-light chamber, and a gray scale reference image was obtained under low-level illumination. Photons emitted from cells expressing the luciferase protein in the mice were collected for a period of time (1 sec to 5 min). Bioluminescence levels were integrated in region of interest (ROI) drawn by hand around bioluminescence zones corresponding to the liver or the gastrointestinal tract. Background bioluminescence was subtracted according to values obtained in ROI drawn on untreated control animals. The gray scale photographic images and bioluminescence color images were superimposed and analyzed using the LIVINGIMAGE v.2.11 software overlay (Xenogen) and IGOR image analysis software (V. 4.02 A, WaveMetrics, Lake Oswego, OR, USA).
In order to localize the cells expressing the transgene (GFP) in treated mice, sections of the proximal and median regions of the small intestine were collected and placed in OCT 24 h post administration. The frozen sections of the different regions of the small intestine collected were viewed and pictured with a fluorescence microscope ECLIPSE E800 (Nikon, Madrid, Spain).
Determination of the transgene expression kinetic parameters
After monitorization and quantification of transgene expression with the CCD camera, transgene expression was plotted versus time. From these curves, the following parameters describing the levels of transgene expression were calculated: Emax, t1/2 Ke, Ke, MET (mean time of the transgene expression [relative duration]), and AUC. Cmax was defined as the maximum level of detected bioluminescence during the transgene expressions, whereas Ke is a constant related to the rate at which the DNA is expressed in the organism; t1/2 Ke was considered as the transgene expression halftime in the body. Both t1/2 Ke, and Ke were calculated using data for the elimination phase. AUC was evaluated by the trapezoidal rule up to which denoted the last sampling point. In addition, MET has been calculated as follows:
| (1) |
in which AUMC is the area under the first moment of the curve. Both parameters were approximated by the trapezoidal rule and extrapolated to infinity. In addition, AUC was accepted if at least 80% of its value was incorporated by AUC (0–tz).
All the parameters were calculated using the WINNONLIN 1.5 software (Pharsight Corp, Mountain View, CA, USA).
Statistical method
The Mann–Whitney U-test was performed on the transgene expression parameters to determine statistical significance. P < 0.05 was considered to be significant, and P < 0.01 very significant.
Results and discussion
Characterisation of nanoparticles
Table 1 summarizes the main physicochemical characteristics of the resulting DNA-chitosan nanoparticles prepared with a ratio N/P of 3.3 (Mao et al 2001). In all cases, the size of the resulting particles was close to 200 nm, although the use of chitosan with higher molecular weight (MW) slightly increases the size of the resulting carriers. The particles were positively charged and no remarkable influence of chitosans MW was observed.
Table 1.
Physicochemical characteristics of the DNA-chitosan nanoparticles. (pLuc: plasmid encoding for luciferase, pEGFP: plasmid encoding for GFP). Experimental conditions: ratio N/P of 3.3. Data express the mean ± standard deviation (n = 3)
| Batch | Size (nm) | Zeta potential (mV) | DNA loading (%) |
|---|---|---|---|
| pLuc-Np 150 | 194 ± 14 | 14.3 ± 0.8 | 96 ± 4 |
| pLuc-Np 400 | 210 ± 18 | 13.7 ± 2.7 | 96 ± 3 |
| pLuc-Np 600 | 235 ± 12 | 12.9 ± 3.6 | 97 ± 3 |
| pEGFP-Np 150 | 182 ± 13 | 8.9 ± 5.1 | 96 ± 3 |
| pEGFP-Np 400 | 181 ± 8 | 10.5 ± 1.5 | 96 ± 4 |
| pEGFP-Np 600 | 227 ± 16 | 16.1 ± 4.2 | 97 ± 2 |
The quantification of DNA loaded in nanoparticles was carried out by PicoGreen assay. As shown in Table 1, the preparative process appeared to be highly efficient to induce the DNA condensation with the polysaccharide. Thus, more than 95% of the DNA was found to be associated with the three molecules of chitosan used.
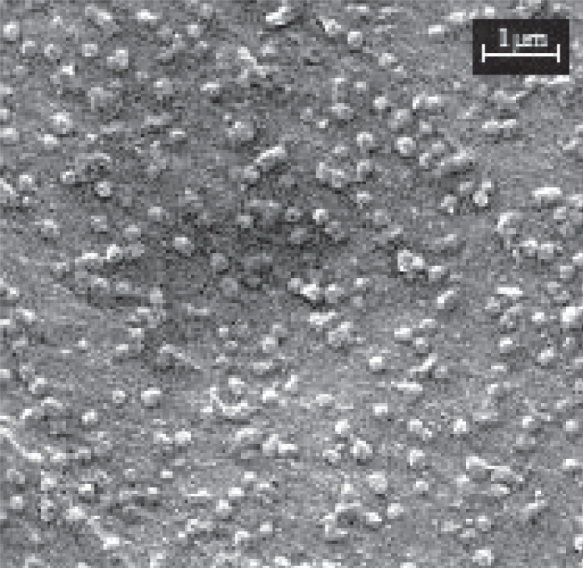
SEM analysis revealed that DNA-loaded chitosan nanoparticles with a size of about 200 nm presented a fairly spherical shape with some irregularities (Figure 1). These results were similar to those obtained by photon correlation spectroscopy and showed a homogeneous size distribution.
Figure 1.
Scanning electron microscopy (SEM) of DNA-chitosan nanoparticles (Np400).
In vitro release studies
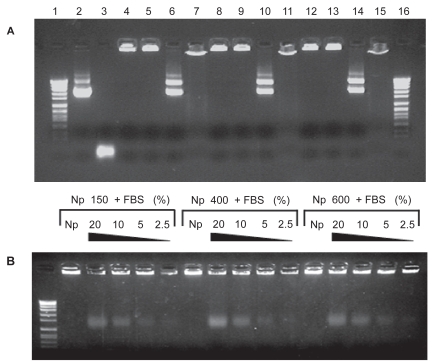
The next objective of our study was to evaluate the integrity of DNA after the preparative process, its release from the chitosan nanoparticles as well as the protection offered by these carriers to the loaded plasmid. Therefore, gel electrophoresis experiments were performed (Figure 2). Loaded DNA-chitosan nanoparticles displayed a pattern of bands co-localized with the loading well, meaning that encapsulated DNA was unable to migrate into the gel due to the strong interactions with the different chitosans (Figure 2A; Lanes 4, 8, and 12). This ionic interaction was independent from the molecular weight of the polysaccharide. The protection of DNA was verified with DNAse I as model enzyme. Naked DNA showed a complete degradation (Figure 2A; lane 3), whereas encapsulated DNA was preserved (at the concentration used) into the loading wells without any longer migration (Figure 2A; lanes 5, 9, and 13). Furthermore, when nanoparticles were digested with chitosanase, the released DNA displayed the same electrophoretic migration pattern as the naked DNA (Figure 2A; lanes 6, 10, 14, and 2). This result confirmed that the preparative process of nanoparticles preserved the integrity of DNA. This result agrees well with previous observations reported by Mao and colleagues (2001). To reinforce the fact that the release of DNA can be achieved by charge interaction disruption, nanoparticles were incubated with either NaOH or FBS. In the first case, the released DNA showed a retarded electrophoretic mobility into the gel, indicating that interaction disruption in our experimental conditions were only partial and chitosan molecules remained bound to DNA (Figure 2A; lanes 7, 11, and 15). In the second case, the incubation between nanoparticles and FBS for 2 h led to a partial release of DNA with an enhanced electrophoretic mobility (Figure 2B). These results point out the strong interaction between DNA and chitosan by means of ionic interactions and confirmed the data described by Liu and colleagues (2005).
Figure 2.
Electrophoresis of chitosan-DNA nanoparticles to determine effective encapsulation, DNAse I protection (digestion with DNAse I), plasmid integrity following release (digestion with chitosanase), and release after charge interaction disruption with NaOH or FBS. (A) Lanes 1 and 16: Molecular weight marker VII; Lane 2: pX2-Luc plasmid DNA; Lane 3: + DNAse I; Lane 4-8-12: nanoparticles composed of 150, 400 or 600 kDa chitosan respectively; Lane 5-9-13: + DNAse I; Lane 6-10-14: + chitosanase; Lane 7-11-15: + NaOH. (B) Nanoparticles composed of 150, 400, or 600 kDa chitosan incubated 2 h (37 °C) with FBS at untreated 20%, 10%, 5%, and 2.5%, respectively.
Hydrodynamic administration of DNA-chitosan nanoparticles
The hydrodynamics-based procedure allows high levels of transgene expression in the liver, and is therefore an interesting tool for investigating the behavior of the gene delivery in vivo. We took advantage of this administration procedure to follow a long term gene expression. The analysis of the transgene expression profiles obtained with the DNA-chitosan nanoparticles were used to determine the kinetic parameters of DNA expression.
Molecular imaging with the charge-coupled device (CCD) camera, using the luciferase as reporter gene permits the bioluminescence detection, spatial localization and its quantification over time in a sensitive and accurate manner (Bloquel et al 2006).
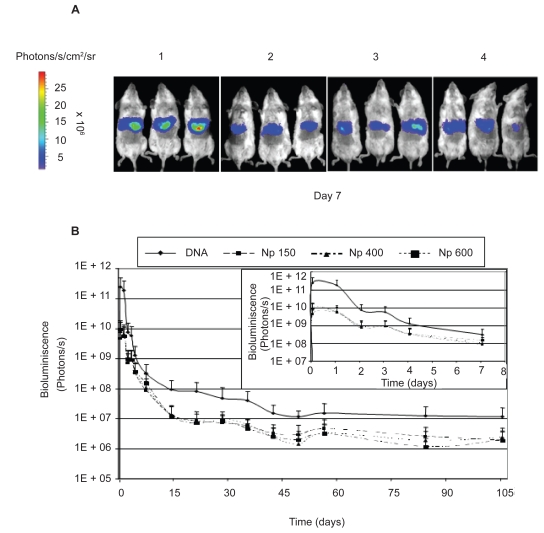
Figure 3A shows the levels of bioluminescence obtained in groups of animals treated with the 3 different formulations and the naked DNA (7 days post administration). These levels of luciferase expression at each time point were used to plot the curves of bioluminescence versus time (Figure 3B). With these curves, it is possible to differentiate the three following phases: the initial phase, the sustained release step and the decline phase.
Figure 3.
(A) Bioluminescent imaging of hepatic luciferase expression in living mice using a cooled CCD camera. The figure shows the mice 7 days after the hydrodynamic injection. (A1) Animals treated with naked DNA, (A2) Np150, (A3) Np400 and (A4) Np600. (B) Evolution of the transgene expression (in bioluminescence units) versus time. In the upper right hand of the figure, time course of the transgene expression is represented from day 0 to day 7.
Notes: Data express the mean ± standard deviation (n = 3).
In the initial phase or the transgene expression 24 h post administration, the plasmid expression was found to be much higher in the case of naked DNA than in DNA-chitosan nanoparticles. This observation is in agreement with Rosmanith and colleagues (2002) who reported important decreases (and even inhibition) of plasmid expressions when loaded in liposomes. In our case, no statistical differences in the levels of transgene expression were observed for the three types of nanoparticle formulations tested.
Mechanistically, the entrance of DNA into the hepatocytes, as shown by Zhang and colleagues (2004) using TEM, might be explained by the mechanism of hydroporation caused by the hydrodynamics-based procedure. The entrance effectiveness of the nanoparticulate systems could be discriminated by the dimension of the holes generated in the hepatocytes’ membrane. This hypothesis was confirmed by Kobayashi and colleagues (2004), which showed that particles with a size of 50 nm were delivered intracellularly, while nanoparticles with a size of 200 nm and 500 nm hardly entered into the hepatocytes and stayed pasted to the plasmatic membrane.
The second phase would be characterized by a sustained and prolonged expression of the transgene due to the release of encapsulated DNA. In fact, at day 7, marked differences of transgene expression still appeared between naked DNA treated animals and the 3 groups administered with the nanoparticle formulations. However, the differences in the intensities of bioluminescence between naked DNA and nanoparticle-treated animals were reduced to 3-fold while they were at 50-fold on day 1 (see Figure 3B). This fact can only be explained by a sustained release of DNA from nanoparticles located in liver cells. Meanwhile the hepatocytes can proceed to the extracellular particles endocytosis, this added expression is only transient (days 3–9), and could find an explanation by the phagocytosis of a great number of the injected nanoparticles by the Kupffer cells, implying their activation and that of the competent immune cells (Sebestyen et al 2006), which led to the disappearance of part of the bioluminescence. This phenomenon has already been described for lipoplexes after intravenous injection (Litzinger et al 1996; Nishikawa et al 2005).
The last phase would be characterized by the progressive silencing of the promotor CMV (Gerolami et al 2000; Aliño et al 2003; Nash et al 2004) and, probably to some extent, also due to the immune response generated by the presence of CpG activating motives (Hodges et al 2004) and the directed response against the foreign reporter protein (Chu et al 2005). All of these processes would start just after the initiation of the gene transfer. However, the maintenance of plasmid expressing liver cells is a fact that allows the detection of the bioluminescence during more than 105 days after the hydrodynamic injection. This long term expression was not affected by the MW of chitosan used.
In order to characterize the kinetic profile of transgene expression, we proposed the following methodology. In preamble, we assumed that the detected bioluminescence can be related to the effective amount of luciferase expressed (Boquel et al 2006). Therefore, the curves describing the transgene expressions of the different formulations administered by hydrodynamics-based procedure could be analysed to estimate some parameters that are able to quantify the intensity, extent and duration of the gene transfer phenomenon occurring in the liver. These parameters are: Emax, t1/2 Ke, Ke, AUC, and MET. In principle, Cmax can be defined as the maximum level of gene expression, while AUC, the amount of transgene expressed, would describe the intensity of the transduction phenomenon. Finally, MET would be able to estimate the relative duration of the transgene expression. For the different formulations tested, Table 2 summarizes all of these parameters.
Table 2.
Kinetic parameters of transgene expression obtained for the different formulations tested. Data express the mean ± standard deviation (n = 3)
| pLuc | Cmax | Ke (days−1) | t1/2 Ke (days) | AUC (Photons/s)xdays | MET (days) |
|---|---|---|---|---|---|
| Naked DNA | 2.7 × 1011 ± 1.4 × 1011 | 0.024 ± 0.002 | 35.61 ± 18.45 | 3.8 × 1011 ± 1.2 × 1011 | 2.30 ± 1.17 |
| Np150 | 4.6 × 109 ± 2.7 × 109 | 0.020 ± 0.016 | 29.00 ± 3.51 | 1.1 × 1010 ± 4.0 × 109 | 3.43 ± 0.40 |
| Np400 | 7.7 × 109 ± 3.0 × 109 | 0.015 ± 0.007 | 45.49 ± 12.43 | 1.3 × 1010 ± 3.7 × 109 | 3.60 ± 0.65 |
| Np600 | 9.8 × 109 ± 3.6 × 109 | 0.019 ± 0.004 | 36.58 ± 12.55 | 1.2 × 1010 ± 3.6 × 109 | 3.79 ± 2.08 |
Concerning Cmax, it was clear that the administration of naked DNA permitted to obtain a transgene expression which was quantified to be between 20- and 50-times higher than for DNA – chitosan formulations. On the other hand, by increasing the MW of chitosan to prepare nanoparticles, a slight increase on the Cmax was observed. Similarly, AUC which quantified the global transgene expression phenomenon was found to be about 30 times higher for naked DNA than for DNA – chitosan formulations. On the contrary, MET (or the relative duration of the transgene expression) was found to be higher for chitosan nanoparticles than for naked DNA. In fact, the use DNA – chitosan nanoparticles enabled us to prolong the expression of the transgene between 26.12 h and 35.76 h more than by using naked DNA.
In summary, despite initial high differences of transgene expression (Cmax) correlated to the mechanism of entrance into the hepatocytes (DNA versus formulations), a further compensation of global transgene expression (AUC) seemed to be due to the sustained controlled release (MET), indistinctly of the MW used.
Intraduodenal administration of DNA-chitosan nanoparticles
Taking advantage of the mucoadhesive properties of chitosans (Janes et al 2001; Kato et al 2003) several investigators have used the β-Galactosidase to observe the introduction of a foreign reporter gene in the intestinal tract of mice (Roy et al 1999; Chen et al 2004). However, the β-Galactosidase is largely expressed in the intestine due to its bacterial origin and shouldn’t be therefore considered as an appropriate reporter gene for the gastrointestinal tract. In the present study, we have chosen two different foreign transgenes: the green fluorescent protein (GFP) and the luciferase (Luc) to contrast the results previously reported. The DNA-chitosan nanoparticles were administered directly in the upper part of the duodenum, avoiding any acidic denaturation after the stomach passage and allowing therefore an estimation of the full potential of our gene delivery systems in the intestine.
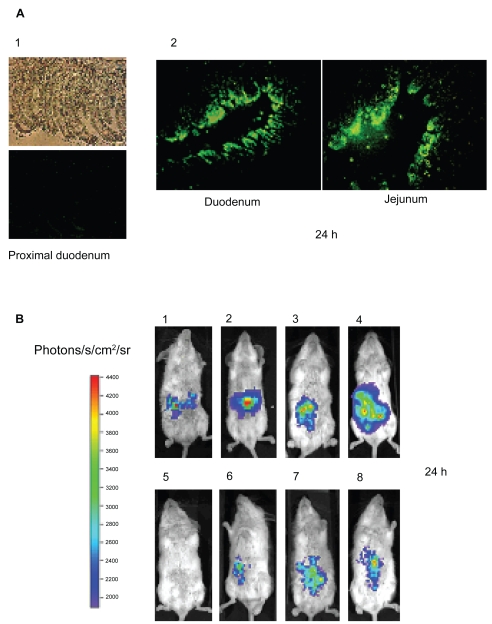
Figure 4A shows the GFP expression in two distant sections of the intestine (duodenum and ileum) 24 hours post administration of DNA loaded in Np400. These photographs show that our nanoparticles were able to transduce the enterocyte monolayer along the intestinal tract, and with high intensity in some sections. These data confirmed the great expectative projected by several studies previously published (Roy et al 1999; Cheng et al 2004; Guliyeva et al 2006; MacLaughlin et al 1998).
Figure 4.
(A) Intestinal sections expressing GFP, 24 h post intraduodenal administration. (A1) 1 ml of saline as control, (A2) Np400 corresponding to a dose of 50 μg plasmid (pEGFP) in 1 ml of formulation. (B) Bioluminescent imaging of intestinal luciferase expression in living mice using a cooled CCD camera. (B1) 25 μg naked DNA in a volume of 500 μl saline; (B2) 50 μg naked DNA in a volume of 500 μl saline; (B3) 50 μg naked DNA in a volume of 1 ml saline; (B4) 100 μg naked DNA in a volume of 1 ml saline. (B5) Untreated animal. (B6) Amount of Np150 corresponding to 50 μg of DNA in 1 ml saline (B7) Np400 corresponding to 50 μg of DNA in 1 ml saline, and (B8) Np600 corresponding to 50 μg of DNA in 1 ml saline.
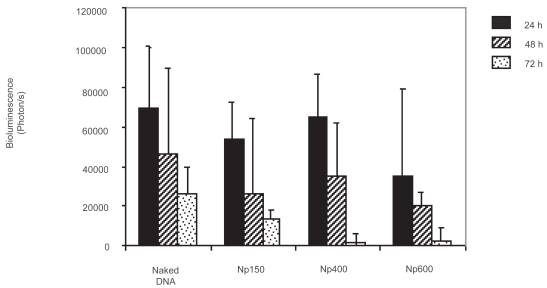
Bioluminescence monitorized by the CCD camera gave some evidence regarding the intensity of the transgene expression phenomenon over time (localization and quantification). Interestingly, after direct injection of naked DNA in the intestinal lumen, transgene expression could be detected and quantified in the intestine in a dose dependent manner (Figure 4B). This expression reached its maximum point at 24 h and quickly decreased to become hardly detectable after 72 h. These results correlate with the enterocytes’ lifetime determined by Ferraris and colleagues (1992). In the same way, the three different formulations were administered without any further modification after the coacervation process. Luciferase expression could be also followed within 3 days (Figure 5).
Figure 5.
Evolution of the luciferase expression in the intestine over the time.
Notes: Data express the mean ± standard deviation (n = 4).
Unfortunately, in comparison with naked DNA, similar results were obtained without any improvement in the intestinal tract distribution and global bioluminescence quantification. Moreover, no statistical difference was noted between the 3 formulations. This phenomenon can be related with the very short lifetime of the mice enterocytes. In fact, the enterocytes would not be a good cell type target for such retarded sustained delivery systems as observed in hydrodynamics-based procedure.
Controversially, Guliyeva and colleagues (2006) were able to see β-Galactosidase expression in the gastrointestinal tract at the fifth day post oral administration using microparticles made of chitosan. Nevertheless, it could be explained by the fact that their microparticulate system could be able to reach and transduce the enterocytes precursors located in the intestinal crypts, that it could maybe also cross the epithelial barrier disrupting tight junctions (Smith et al 2004), or get to the Peyer patches via the M cells (Van Der Lubben et al 2001).
Conclusions
In conclusion, chitosan nanoparticles can be prepared by coacervation with a high reproducibility. No remarkable influence of MW and plasmid size was observed on the formation and physicochemical properties of these carriers.
After the hydrodynamic injection, DNA-chitosan nanoparticles were predominantly accumulated in the liver, where the transgene is expressed for at least 105 days. The curves of luciferase expression observed with the CCD camera can be divided in 3 phases. Moreover, no effect of the chitosan MW was observed in the kinetic profiles of transgene expression. On the other hand, the proposed methodology can permit, in an easy way, the calculation of kinetic parameters that characterize the transgene expressions and allow further comparisons between wide varieties of gene delivery systems.
Finally, the intraduodenal administration of naked DNA allowed the gene transfer in a dose dependent manner quantifiable within 3 days with the CCD camera. The same administration of the three formulations induced similar patterns of transgene expression without any improvement in the duration and intestinal distribution compared to naked DNA. This fact could be explained by the enterocytes’ rapid physiological turn-over and by significantly high MET that can be related with the ability of chitosan nanoparticles to sustain and prolong the release of DNA.
Acknowledgments
This research was supported by “Programa Nacional de Proyectos de Investigación Aplicada” (ORDEN PRE/621/2008). Miguel N Centelles was supported by a fellowship grant from the ‘Asociación de Amigos de la Universidad de Navarra’. We would like to thank Prof Rafael Jordana, who has performed scanning electron microscopy of DNA-chitosan nanoparticles.
References
- Alino SF, Crespo A, Dasi F. Long-term therapeutic levels of human alpha-1 antitrypsin in plasma after hydrodynamic injection of nonviral DNA. Gene Ther. 2003;10:1672–9. doi: 10.1038/sj.gt.3302065. [DOI] [PubMed] [Google Scholar]
- Bloquel C, Trollet C, Pradines E, et al. Optical imaging of luminescence for in vivo quantification of gene electrotransfer in mouse muscle and knee. BMC Biotechnol. 2006;8:6–16. doi: 10.1186/1472-6750-6-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Yang WL, Li G, et al. Transfection of mEpo gene to intestinal epithelium in vivo mediated by oral delivery of chitosan-DNA nanoparticles. World J Gastroenterol. 2004;10:112–16. doi: 10.3748/wjg.v10.i1.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chew JL, Wolfowicz CB, Mao HQ, et al. Chitosan nanoparticles containing plasmid DNA encoding house dust mite allergen, Der p 1 for oral vaccination in mice. Vaccine. 2003;21:2720–9. doi: 10.1016/s0264-410x(03)00228-7. [DOI] [PubMed] [Google Scholar]
- Chu Q, Joseph M, Przybylska M, et al. Transient siRNA-mediated attenuation of liver expression from an alpha-galactosidase A plasmid reduces subsequent humoral immune responses to the transgene product in mice. Mol Ther. 2005;12:264–73. doi: 10.1016/j.ymthe.2005.04.007. [DOI] [PubMed] [Google Scholar]
- Cui Z, Mumper RJ. Chitosan-based nanoparticles for topical genetic immunization. J Control Release. 2001;75:409–19. doi: 10.1016/s0168-3659(01)00407-2. [DOI] [PubMed] [Google Scholar]
- Dhawan S, Singla AK, Sinha VR. Evaluation of mucoadhesive properties of chitosan microspheres prepared by different methods. AAPS PharmSciTech. 2004;5(4):e67. doi: 10.1208/pt050467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunlap DD, Maggi A, Soria MR, et al. Nanoscopic structure of DNA condensed for gene delivery. Nucleic Acids Res. 1997;25:3095–101. doi: 10.1093/nar/25.15.3095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferraris RP, Villenas SA, Diamond J. Regulation of brush-border enzyme activities and enterocyte migration rates in mouse small intestine. Am J Physiol. 1992;262:1047–59. doi: 10.1152/ajpgi.1992.262.6.G1047. [DOI] [PubMed] [Google Scholar]
- Gerolami R, Uch R, Jordier F, et al. Gene transfer to hepatocellular carcinoma: transduction efficacy and transgene expression kinetics by using retroviral and lentiviral vectors. Cancer Gene Ther. 2000;7:1286–92. doi: 10.1038/sj.cgt.7700225. [DOI] [PubMed] [Google Scholar]
- Guliyeva U, Oner F, Ozsoy S, et al. Chitosan microparticles containing plasmid DNA as potential oral gene delivery system. Eur J Pharm Biopharm. 2006;62:17–25. doi: 10.1016/j.ejpb.2005.08.006. [DOI] [PubMed] [Google Scholar]
- Hodges BL, Taylor KM, Joseph MF, et al. Long-term transgene expression from plasmid DNA gene therapy vectors is negatively affected by CpG dinucleotides. Mol Ther. 2004;10:69–78. doi: 10.1016/j.ymthe.2004.04.018. [DOI] [PubMed] [Google Scholar]
- Illum L, Farraj NF, Davis SS. Chitosan as a novel nasal delivery system for peptide drugs. Pharm Res. 1994;11:1186–9. doi: 10.1023/a:1018901302450. [DOI] [PubMed] [Google Scholar]
- Janes KA, Calvo P, Alonso MJ. Polysaccharide colloidal particles as delivery systems for macromolecules. Adv Drug Deliv Rev. 2001;47:83–97. doi: 10.1016/s0169-409x(00)00123-x. [DOI] [PubMed] [Google Scholar]
- Kato Y, Onishi H, Machida Y. Application of chitin and chitosan derivatives in the pharmaceutical field. Curr Pharm Biotechnol. 2003;4:303–9. doi: 10.2174/1389201033489748. [DOI] [PubMed] [Google Scholar]
- Kobayashi N, Hirata K, Chen S, et al. Hepatic delivery of particulates in the submicron range by a hydrodynamics-based procedure: implications for particulate gene delivery systems. J Gene Med. 2004;6:455–63. doi: 10.1002/jgm.531. [DOI] [PubMed] [Google Scholar]
- Koping-Hoggard M, Tubulekas I, Guan H, et al. Chitosan as a nonviral gene delivery system. Structure-property relationships and characteristics compared with polyethylenimine in vitro and after lung administration in vivo. Gene Ther. 2001;8:1108–21. doi: 10.1038/sj.gt.3301492. [DOI] [PubMed] [Google Scholar]
- Koping-Hoggard M, Varum KM, Issa M, et al. Improved chitosan-mediated gene delivery based on easily dissociated chitosan polyplexes of highly defined chitosan oligomers. Gene Ther. 2004;11:1441–52. doi: 10.1038/sj.gt.3302312. [DOI] [PubMed] [Google Scholar]
- Kramer MG, Barajas M, Razquin N, et al. In vitro and in vivo comparative study of chimeric liver-specific promoters. Mol Ther. 2003;7:375–85. doi: 10.1016/s1525-0016(02)00060-6. [DOI] [PubMed] [Google Scholar]
- Litzinger DC, Brown JM, Wala I, et al. Fate of cationic liposomes and their complex with oligonucleotide in vivo. Biochim Biophys Acta. 1996;1281:139–49. doi: 10.1016/0005-2736(95)00268-5. [DOI] [PubMed] [Google Scholar]
- Liu D, Knapp JE. Hydrodynamics-based gene delivery. Curr Opin Mol Ther. 2001;3:192–97. [PubMed] [Google Scholar]
- Liu F, Song Y, Liu D. Hydrodynamics-based transfection in animals by systemic administration of plasmid DNA. Gene Ther. 1999;6:1258–66. doi: 10.1038/sj.gt.3300947. [DOI] [PubMed] [Google Scholar]
- Liu W, Sun S, Cao Z, et al. An investigation on the physicochemical properties of chitosan/DNA polyelectrolyte complexes. Biomaterials. 2005;26:2705–11. doi: 10.1016/j.biomaterials.2004.07.038. [DOI] [PubMed] [Google Scholar]
- MacLaughlin FC, Mumper RJ, Wang J, et al. Chitosan and depolymerized chitosan oligomers as condensing carriers for in vivo plasmid delivery. J Control Release. 1998;56:259–72. doi: 10.1016/s0168-3659(98)00097-2. [DOI] [PubMed] [Google Scholar]
- Mumper RJ, Wang JJ, Claspell JM, et al. Novel polymeric condensing carriers for gene delivery. Proc Int Symp Controlled Release Bioact Mater. 1995;22:178–79. [Google Scholar]
- Nash KL, Jamil B, Maguire J, et al. Hepatocyte-specific gene expression from integrated lentiviral vectors. J Gene Med. 2004;6:974–83. doi: 10.1002/jgm.591. [DOI] [PubMed] [Google Scholar]
- Nishikawa M, Takakura Y, Hashida M. Pharmacokinetics of plasmid DNA-based non-viral gene medicine. Adv Genet. 2005;53:47–68. [PubMed] [Google Scholar]
- Rossmanith W, Chabicovsky M, Herkner K, et al. Cellular gene dose and kinetics of gene expression in mouse livers transfected by high-volume tail-vein injection of naked DNA. DNA Cell Biol. 2002;21:847–53. doi: 10.1089/104454902320908496. [DOI] [PubMed] [Google Scholar]
- Roy K, Mao HQ, Huang SK, et al. Oral gene delivery with chitosan-DNA nanoparticles generates immunologic protection in a murine model of peanut allergy. Nat Med. 1999;5:387–91. doi: 10.1038/7385. [DOI] [PubMed] [Google Scholar]
- Schipper NG, Varum KM, Artursson P. Chitosans as absorption enhancers for poorly absorbable drugs. 1: Influence of molecular weight and degree of acetylation on drug transport across human intestinal epithelial (Caco-2) cells. Pharm Res. 1996;13:1686–92. doi: 10.1023/a:1016444808000. [DOI] [PubMed] [Google Scholar]
- Sebestyen MG, Budker VG, Budker T, et al. Mechanism of plasmid delivery by hydrodynamic tail vein injection. I. Hepatocyte uptake of various molecules. J Gene Med. 2006;8:852–73. doi: 10.1002/jgm.921. [DOI] [PubMed] [Google Scholar]
- Smith J, Wood E, Dornish M. Effect of chitosan on epithelial cell tight junctions. Pharm Res. 2004;21:43–9. doi: 10.1023/b:pham.0000012150.60180.e3. [DOI] [PubMed] [Google Scholar]
- Song E, Lee SK, Wang J, et al. RNA interference targeting Fas protects mice from fulminant hepatitis. Nat Med. 2003;9:347–51. doi: 10.1038/nm828. [DOI] [PubMed] [Google Scholar]
- Van Der Lubben IM, Konings FA, Borchard G, et al. In vivo uptake of chitosan microparticles by murine Peyer’s patches: visualization studies using confocal laser scanning microscopy and immunohistochemistry. J Drug Target. 2001;9:39–47. doi: 10.3109/10611860108995631. [DOI] [PubMed] [Google Scholar]
- Yang PL, Althage A, Chung J, et al. Hydrodynamic injection of viral DNA: a mouse model of acute hepatitis B virus infection. Proc Natl Acad Sci U S A. 2002;99:13825–30. doi: 10.1073/pnas.202398599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang G, Budker V, Wolff JA. High levels of foreign gene expression in hepatocytes after tail vein injections of naked plasmid DNA. Hum Gene Ther. 1999;10:1735–7. doi: 10.1089/10430349950017734. [DOI] [PubMed] [Google Scholar]
- Zhang G, Gao X, Song YK, et al. Hydroporation as the mechanism of hydrodynamic delivery. Gene Ther. 2004;11:675–82. doi: 10.1038/sj.gt.3302210. [DOI] [PMC free article] [PubMed] [Google Scholar]