Abstract
Purine nucleoside analogues of medicinal importance, such as cladribine, require phosphorylation by deoxycytidine kinase (dCK) for pharmacological activity. Structural studies of ternary complexes of human dCK show that the enzyme conformation adjusts to the different hydrogen-bonding properties between dA and dG and to the presence of substituent at the 2-position present in dG and cladribine. Specifically, the carbonyl group in dG elicits a previously unseen conformational adjustment of the active site residues Arg104 and Asp133. In addition, dG and cladribine adopt the anti conformation, in contrast to the syn conformation observed with dA. Kinetic analysis reveals that cladribine is phosphorylated at the highest efficiency with UTP as donor. We attribute this to the ability of cladribine to combine advantageous properties from dA (favorable hydrogen-bonding pattern) and dG (propensity to bind to the enzyme in its anti conformation), suggesting that dA analogues with a substituent at the 2-position are likely to be better activated by human dCK.
Introduction
Deoxycytidine kinase (dCKa) plays a pivotal role in the supply of precursors for DNA synthesis, as well as being required for the activation of numerous medicinally relevant nucleoside analogue (NA) prodrugs. Physiologically, as the initial enzyme in the nucleotide salvage pathway, dCK is responsible for converting the pyrimidine dC and the purines dA and dG into their monophosphate forms. Subsequently, the products of the dCK-catalyzed reaction are converted to their triphosphorylated form, allowing their incorporation into DNA by polymerases. Moreover, dCK is required for the initial phosphorylation of NAs clinically used in both antiviral (e.g., ddC and 3TC against HIV) and anticancer (e.g., AraC and cladribine) therapy. In fact, dCK is most often the rate-limiting enzyme in the overall pathway that converts the administered nucleoside to its triphosphorylated metabolite. Therefore, understanding the structural parameters that allow for efficient phosphorylation of nucleosides by this enzyme is of prime importance for the further development of efficacious NA-based therapeutics.
In addition to being highly promiscuous at the nucleoside acceptor site, dCK can utilize several nucleoside triphosphates as phosphoryl group donors. Indeed, previous work has suggested that UTP can be utilized by dCK in a similar1 or even better efficiency2,3 than the more abundant ATP. Interestingly, in complementary work that focuses on the nucleoside dA bound to dCK in the presence of ADP or UDP,4 we observed that the identity of the nucleotide at the donor site can affect the conformation of dA at the acceptor site. Therefore, it is important to study each nucleoside in the presence of both ATP and UTP.
In this paper we present the crystal structures of human dCK in complex with its physiological substrate dG at the acceptor site and UDP at the phosphoryl donor site. We also present the ternary complex of dCK with the nucleoside analogue 2-chloro-2′-deoxyadenosine (cladribine, CLAD) and either UDP or ADP bound at the donor site of the enzyme. We observe that unlike the situation seen for the complex of dA + UDP, in which the enzyme adopted a more open conformation not compatible with catalysis, dG binds to the enzyme in its closed and active conformation. Furthermore, the dA-analogue cladribine acts similarly to dG, also adopting the closed conformation. Our work reveals the structural adjustments required by the dCK active site to accommodate purines with differing hydrogen-bonding properties (i.e., dA versus dG) or different steric properties (i.e., dA versus cladribine). In addition, the dependence between the nucleoside properties and the establishment of the closed and active conformation are revealed. This information can be incorporated into the design process of novel dCK-activated nucleoside analogues.
Results and Discussion
The promiscuity of dCK extends to both the nucleoside acceptor and the nucleotide donor binding sites.5 Table 1 presents steady state kinetic data for wild type human dCK measured for the three purines dA, dG, and cladribine as phosphoryl acceptors, with either ATP or UTP as phosphoryl donors. It is apparent from these data that communication between the donor site and acceptor site exists. Invariably, the Km of the nucleoside is dramatically decreased (~5- to 15-fold) in the presence of UTP versus its Km in the presence of ATP. However, this lowering of Km is concomitant with a decrease in kcat. Thus, the combined effect on efficiency (kcat/Km) of replacing ATP by UTP is ~3-fold increase for dA and no increase for dG. In contrast, in the case of cladribine, there is ~10-fold increase in phosphorylation efficiency with UTP over ATP (Table 1). Since the efficiency of activation of NAs such as cladribine can strongly influence their therapeutic effect, one of the goals of this work is to decipher the factors that endow this compound with favorable phosphorylation kinetics by human dCK.
Table 1.
Kinetic Analysis of Human Wild Type dCK Using ATP or UTP as Donors and dA, dG, or Cladribine as Acceptors
| substrates | kcat (s−1) | Km (μM) | kcat/Km (×10−3) |
|---|---|---|---|
| dA + ATP | 2.1 ± 0.1 | 115 ± 18 | 18.2 |
| dA + UTP | 0.37 ± 0.01 | 8.5 ± 1.4 | 43.5 |
| dG + ATP | 2.5 ± 0.1 | 181 ± 14 | 13.8 |
| dG + UTP | 0.53 ± 0.04 | 41 ± 7 | 12.9 |
| CLAD + ATP | 0.6 ± 0.04 | 78 ± 16 | 7.7 |
| CLAD + UTP | 0.37 ± 0.01 | 5.1 ± 0.9 | 72.6 |
In our recently reported three-dimensional structure of dCK in complex with its physiological substrate dA and ADP or UDP,4 we observed a previously undetected enzyme conformation when bound to UDP, whereas the expected conformation was observed with ADP. Moreover, the nature of the nucleotide affected the conformation of the nucleoside dA (syn with UDP, anti with ADP) and of the enzyme (open with UDP, closed with ADP). This prompted us to analyze the analogous complexes with additional purines, namely, dG and cladribine. Note that our studies employ the diphosphate forms of the nucleotide, since crystals were of lower quality when nonhydrolyzable analogues of UTP or ATP were used. Previous fluorescence studies indicated that UDP can elicit the same change in tryptophan fluorescence as UTP, suggesting that the structures obtained in the presence of diphosphates can be used to approximate the structures in the presence of triphosphates.4
Adaptation of Ternary dCK + dG + UDP Complex to the Closed Enzyme Conformation
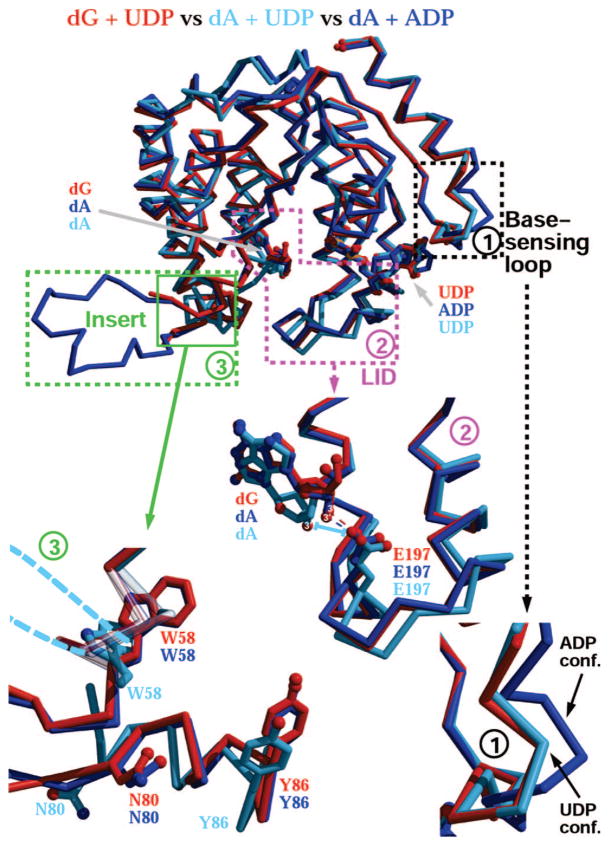
The residues in dCK that are sensitive to the identity of the nucleotide belong to three main regions, designated as (1) base-sensing loop, (2) LID area, and (3) insert region (Figure 1). The base-sensing loop is an area extending from residues 240 to 254 that directly interacts with the base moiety of the nucleotide and adopts a different main chain conformation in the presence of ADP from that with UDP. Comparison of the dG + UDP structure to that of dA + UDP reveals the same conformation for that region (Figure 1, base-sensing loop zoom 1). For comparison, the alternative conformation of this region adopted in the dA + ADP structure is also shown (we were not successful in obtaining diffraction quality crystals for the complex of dG + ADP).
Figure 1.
Both the identity of the nucleoside at the phosphoryl acceptor site and that of the nucleotide at the donor site determine the conformation of dCK, as manifested in three regions: the base-sensing loop (1), the LID (2), and the insert (3). Top panel is a superposition of three dCK ternary complex structures: dG + UDP (red), dA + UDP (cyan), and dA + ADP (blue). For each enzyme region a zoomed panel is also shown: panel 1 illustrates how the base-sensing loop changes conformation upon binding of UDP versus ADP; panel 2 depicts the opening of the LID region occurring in the complex of dA + UDP (cyan). In that structure, the stabilizing hydrogen bond between Glu197 and the 3′-hydroxyl group of the nucleoside occurring when dCK is in its closed state (dG + UDP and dA + ADP structures) is lost (cyan bar). Finally, panel 3 shows the insert region. Despite the fact that the insert in the dG + UDP structure was not completely traceable, the flanking residues suggest that the insert adopts the same conformation seen in the dA + ADP complex. For the dA + UDP structure, Trp58, Asn80, and Tyr86 move considerably, and we predict that the insert would undergo a conformation that is distinct from the one shown for the other two complexes (dashed cyan line). The rmsd values for the overlays are as follows: dG + UDP versus dA + UDP, 0.88 Å on 200 atoms; dG + UDP versus dA + ADP, 0.67 Å on 213 atoms. All structure figures were drawn with Molscript20,21 and Raster3D.22
Whereas in the presence of UDP, the base-sensing region, regardless if the structure was solved with the nucleoside dA or dG, adopts the same conformation, the LID region of the dG + UDP complex adopts a different conformation from that seen in the dA + UDP complex. In fact, the LID’s conformation of the dG + UDP complex is similar to that seen in the dA + ADP structure (Figure 1, LID zoom 2). In the LID conformation seen in the dA + ADP structure, the LID residue Glu197 makes a hydrogen bond to the 3′-hydroxyl group of dA (Figure 1, LID zoom 2). In contrast, the LID conformation adopted in the dA + UDP precludes such an interaction. Yet, despite dG being crystallized with UDP, the LID adopted the conformation as seen in the dA + ADP complex, a conformation that permits the interaction between Glu197 and the 3′-hydroxyl of dG.
An additional ramification for the presence of UDP in the case of the dA complex was the adoption of the syn conformation by the nucleoside. In contrast, despite the presence of UDP in the dG complex, the nucleoside is observed in the anti conformation. This is the conformation that was also seen for dA when ADP was present instead of UDP. In other words, despite the presence of UDP, which resulted in dA adopting the syn conformation, dG is found in the anti conformation, which is the conformation observed for dA with ADP. Importantly, the syn conformation is concomitant with a more open active site, thereby precluding a catalytically important interaction with Glu53 (the catalytic base). Thus, dG, in the presence of UDP, binds to the enzyme in its active conformation.
In the dA + UDP complex, as a consequence of both dA being in the syn conformation and the opening of the LID, the third region, the insert (extending from residue 54 to 90), also assumed a conformation different from that seen for dA + ADP. The particular LID conformation is transmitted to the insert region via an interaction between Asn80 of the insert and Glu196 of the LID. Residue Asn80 underwent a change in conformation that correlated with the LID conformation and so did Trp58 and Tyr86 (Figure 1, insert zoom 3). Despite the fact that in the dA + UDP complex the insert area was not completely traceable, it is apparent from the position of flanking residues that the insert would have assumed an entirely different conformation compared to the one described for all the ADP complexes that we had analyzed thus far. Since dG binds in the anti conformation, despite the presence of UDP, and the LID adopts the more closed conformation, the insert region of the dG + UDP complex is more similar to that seen in the dA + ADP complex structure (Figure 1, insert zoom 3).
Thus, the overall conformation of dCK in complex with dG + UDP, excluding the base-sensing loop, resembles the one that the enzyme assumed in all the previous structures that we have solved with ADP at the donor site and dC or dA at the acceptor binding site. In other words, the base-sensing loop shows the most stringent selection, adopting a UDP-specific or an ADP-specific conformation, regardless of the nature of the nucleoside. In contrast, while the LID and insert regions are affected by the nature of the nucleotide, the identity of the nucleoside is also a factor that determines their conformation. If the nucleoside is dA, UDP elicits an open LID conformation and a particular insert fold. If the nucleoside is dG (or a pyrimidine, for that matter), regardless of UDP’s presence, the LID adopts the closed conformation, as seen with ADP. As stated previously, only in the closed LID conformation can the important Glu197-nucleoside 3′-OH interaction take place (in addition to the Glu53-5′-OH interaction; see below). Thus, the dG + UDP complex revealed the closed and active conformation.
Active Site Adjustments Required To Bind dG Instead of dA
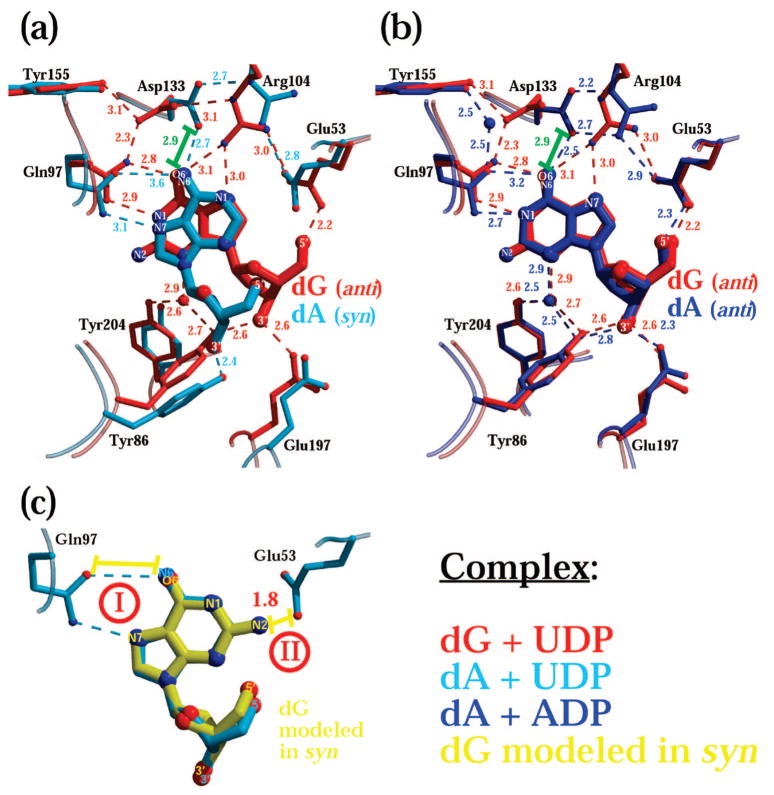
Detailed comparison of the residues involved in binding of dG and dA shows how the enzyme is adept at binding substrates that differ in their hydrogen-bonding pattern (Figure 2). Most noticeable is the adjustment of Asp133 in response to the group at position 6 of the nucleoside: an amino group in the case of adenine, a carbonyl group in the case of guanine (for schematic drawings of the nucleosides and atom numbering see Figure S1). The amino group in dA, either in syn or in anti, Figure 2a and Figure 2b, respectively, makes a favorable hydrogen bond to Asp133. In contrast, this arrangement would be detrimental in the case of dG because of the repulsion between the carbonyl group of the guanine base and the carboxyl group of Asp133 (distance as close as 2.9 Å, green bar in Figure 2a and Figure 2b). Consequently, Asp133, which in the dA complex makes two tight hydrogen bonds with Arg104, rotates by approximately 60° away from the arginine and establishes two alternative hydrogen-bonding interactions with Tyr155 and Gln97. To compensate for this movement of the side chain of Asp133, Arg104 in the dCK-dG complex approaches the guanine base, enabling its side chain to function as a hydrogen-bonding donor toward the carbonyl group in position 6 and the nitrogen in position 7. Notably, despite this adjustment of the side chain of Arg104, its interaction with the catalytic Glu53 is maintained.
Figure 2.
Binding by dCK to purines that differ in their hydrogen-bonding properties requires adjustment of active site residues. The nucleoside binding site of the dG + UDP complex is depicted in red, the dA + UDP complex in cyan, and the dA + ADP complex in blue. Color-coded dashes represent hydrogen bonds with the distance shown in angstroms. The green bar represents a putative repulsive interaction. (a) Overlay of the dG + UDP and the dA + UDP structures. In the presence of UDP, dG binds in the anti conformation while dA binds in syn. This difference in nucleoside conformation is accompanied by a change in enzyme conformation. Note the difference in Tyr86 belonging to the insert region and Glu197 and Tyr204 of the LID. (b) Overlay of the dG + UDP and the dA + ADP structures shows that the anti conformation of dG is similar to that of dA when the latter is paired with ADP. The identical nucleoside conformation results in a similar protein conformation involving residues (Tyr86, Glu197, and Tyr204) that are close/interact with the sugar moiety of the nucleoside. However, the residues that are close/interact with the base moiety (Gln97, Arg104, and Asp133) adjust to a hydrogen-bonding pattern that is different between dG and dA. Specifically, in dA, atoms N1 and N7 are hydrogen bond acceptors and the amino group in position 6 is a hydrogen bond donor. In contrast, in dG, the NH group in position 1 is a donor and the carbonyl group in position 6 an acceptor (see Figure S1 for atom numbering convention for dA and dG). When dA binds either in syn (panel a) or in anti (panel b), the amide group of the side chain of Gln97 maintains its orientation. This is possible because either N1 or N7 acts as acceptor to the glutamine amino group, and the NH2 group of dA retains its position regardless of the nucleoside conformation enabling an interaction with the glutamine carbonyl group. However, in the presence of dG, the amide group of Gln97 is forced to rotate by 180° in order to accommodate the opposite hydrogen-bonding properties of dG versus dA. This induces Asp133 to approach and interact with the rotated Gln97, with an additional interaction made with Tyr155. As a consequence, Arg104 moves closer to the guanine ring providing hydrogen bonds with atoms O6 and N7 of the purine. Panel c illuminates the structural reasons that prevent dG from binding in syn. Modeling of dG in the syn conformation (yellow) reveals that since both atoms O6 and N7 in dG are hydrogen bond acceptors, it is impossible to have a fully productive interaction between the guanine moiety in syn and the side chain of Gln97 (yellow bar I). Additionally, the amino group in position 2 of the guanine ring would be too close to Glu53 if dG would bind in syn (1.8 Å, yellow bar II).
Another important difference between the binding by dCK of dG versus dA is seen in the positioning of residue Gln97. Discrimination between the oxygen atom and the nitrogen atom, as present in the glutamine side chain, is not readily established by X-ray diffraction (a difference of only one electron). However, on the basis of hydrogen bond considerations, the amide group must flip from its assumed orientation with an adenine base in order to maintain hydrogen bond acceptor/donor interactions with the guanine base, where the acceptor is the carbonyl group in position 6 and the donor is the NH group in position 1.
These base-dependent (dG versus dA) interactions affect the conformations of Gln97, Arg104, and Asp133. In addition, the conformation of the base (syn versus anti) affects the positioning of residues in proximity to the sugar moiety of the nucleoside. Thus, residues Glu53, Tyr86, Glu197, and Tyr204 of the dG + UDP complex adopt a different position in comparison to the dA + UDP complex (Figure 2a). These differences are a consequence of the transition of the overall enzyme conformation from closed to open, as reflected in the movement of the LID and of the insert (see above). In contrast, since the dG in the dG + UDP structure and the dA in the dA + ADP structure both adopt the anti conformation, these active site residues are maintained in the same position (Figure 2b). In fact, in the complexes where the nucleosides dG and dA are in the anti conformation, both their sugar and base moieties overlay very well.
At this point an obvious question arises: why in the complex with UDP does dG bind in anti and not in syn like dA? Modeling of dG in the syn conformation based on the syn-dA structure (Figure 2c), shows that there are two main reasons that are not favorable toward the binding of dG in syn. First, the two productive interactions of anti-dG with Gln97 cannot be made with syn-dG. Gln97 provides one hydrogen bond donor site and one hydrogen bond acceptor site. In its syn conformation, dG presents two hydrogen bond accepting moieties. Thus, the carbonyl group of Gln97 cannot participate in stabilizing dG in its syn conformation.
A second reason disfavoring the syn-dG conformation has to do with the amino group at the 2-position. In the syn conformation, this amino group (N2) would be too close to Glu53 (1.8 Å to side chain of Glu53 if the Glu53 would be in the dA + UDP conformation).
Structure of dCK in Complex with Cladribine and Either UDP or ADP
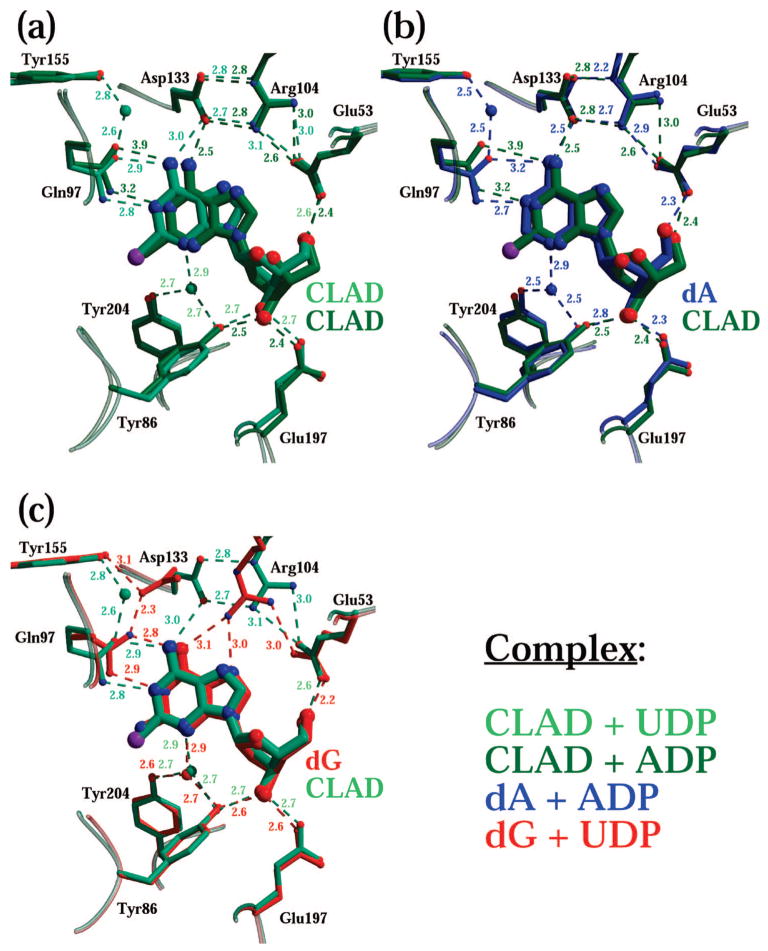
The dA-analogue cladribine, used to treat various leukemias, requires dCK for its activation.6 In this paper we present the ternary complex of dCK with cladribine (CLAD) and either UDP or ADP, solved at 1.8 and 2.5 Å resolution, respectively. Despite the fact that cladribine is an analogue of dA, the conformation we observe in our complexes mimics that seen for dG (and the rationalization for this conformation is similar to that made previously for dG). That is, cladribine binds in the anti conformation in the presence of ADP (as seen with dA) or UDP (in contrast to dA). An overlay of the two cladribine structures, and their comparison to the dG + UDP and dA + ADP structures, is presented in Figure 3. Recall that in the dA complex with UDP, the nucleoside dA was observed in the syn conformation and the enzyme is in an open/inactive state. We observe cladribine in its anti conformation in both the UDP and ADP ternary complexes (Figure 3a). The fact that the base moiety of cladribine when comparing the UDP and ADP complex structures does not precisely overlap can be attributed to a different chlorine occupancy (see Experimental Section). Positioning of residues in proximity to the nucleoside, such as Gln97, Asp133, Arg104, and Glu53, is basically the same in the cladribine structures compared to the dA/ADP structures (Figure 3b, dA shown in blue). This can be attributed to the amino group in position 6 of the base, present in both cladribine and dA.
Figure 3.
The dA-analogue cladribine mimics the dG state when binding to dCK. Shown is a comparison of the active sites of the CLAD + UDP complex (light-green), CLAD + ADP (dark-green), dA + ADP (blue), and dG + UDP (red). (a) Cladribine binds in the anti conformation in either the UDP or ADP ternary complex. Consequently, positioning of residues belonging to the insert and the LID regions is very similar, corresponding to the closed conformation of the enzyme. (b) Since cladribine is a dA analogue, the side chain of Gln97 does not need to rotate as in the case of dG (panel c). Cladribine binding in syn is made less favored because of a close interaction that would occur between the carboxyl group of Glu53 and the chlorine atom present in cladribine.
In terms of ligand-dependent protein conformation, the cladribine complexes mimic the closed conformation as seen with dG (Figure 3c and Figure S2). Specifically, the residues belonging to the insert (Trp58 and Tyr86; the former is not shown in the figure) and the LID (Glu197 and Tyr204) maintain the same position in the two cladribine complexes compared to both the dA + ADP and the dG + UDP structures (Figure 3b and Figure 3c). In fact, inspection of the superposed dG + UDP, CLAD + UDP, and CLAD + ADP structures (Figure S2) show that for all of them both the insert and the LID adopt the same conformation (i.e., the closed and active conformation). The sole conformation difference between these three complexes is found in the base-sensing loop, which adopts either the ADP-specific or UDP-specific conformation.
Conclusions
The work described in this paper extends and complements our previous studies of dCK4,7–10 and provided us with several unexpected results. The first surprising result is that dG binds in a different conformation than dA in the presence of UDP at the donor site. The structures with dA + UDP exposed an inactive and open dCK conformation, with the nucleoside in the syn conformation. In contrast, despite the presence of UDP, dG promotes the active and closed enzyme conformation, and the nucleoside adopts the anti conformation. Is the difference in the observed nucleoside conformation due to an intrinsic preference for a particular conformation (syn versus anti), or is it due to the dCK active site? Extensive experimental data showed that in solution purines undergo rapid transitions between the two states.11 In fact, dG has a preference for the syn conformation because of the ability of the guanine base to interact with the sugar moiety when in syn.11 Thus, in solution the enzyme dCK would encounter both conformations of the purines dA and dG. The conclusion is that the specific nucleoside conformation is dictated by the dCK active site.
Previous studies of UTP utilization by dCK indicated that (i) UTP binds prior to the nucleoside12 and (ii) its binding promotes an open/inactive dCK conformation.4 For a nucleoside such as dA, upon the formation of the ternary complex (dCK + UTP + dA), this open/inactive state is relatively stable, and this allowed us to crystallize dCK + UDP + dA in this state. In contrast, for a nucleoside such as dG, we propose that such a state only forms transiently, and it rapidly converts to the closed/active conformation. We identify the different hydrogen-bonding properties of dG and its amino group at the 2-position as likely factors behind this difference to dA.
The second surprising result presented here is that the dA-analogue cladribine actually behaves more like dG. The cladribine + UDP structure shows a closed/active conformation, with the NA in the anti conformation, analogous to the state seen in the dG + UDP structure. In this case, the overriding factor is the chlorine atom at the 2-position in cladribine. This chlorine, similarly to the amino group in dG, would clash with the side chain of Glu53 if adopting the syn conformation (Figure 2c).
Thus, our work demonstrates the relationship between the natures of the nucleotide at the donor site (UTP versus ATP), the nucleoside, and the conformation of dCK. This also serves to identify structural elements in nucleoside substrates (e.g., dA-like hydrogen-bonding pattern, dG-like substituent at the 2-position) that promote efficient phosphorylation by human dCK. Specifically, a bulky substituent such as a chlorine (as in cladribine) or amino (as in dG) promote the selection of nucleoside’s anti conformation by the dCK active site. Consistent with this is the fact that clofarabine is a good dCK substrate, and structural examination of its binding to dCK reveals the anti conformation.13 At the same time, a small substituent in this position, such as a fluorine atom as present in fludarabine, may not to be large enough to induce the anti conformation, and this could explain why this NA is not as efficiently phosphorylated by dCK.3
Experimental Section
Site-Directed Mutagenesis, Protein Expression, Purification, and Crystallization
As we previously reported,4 the C4S-dCK (C9S/C45S/C59S/C146S) and the C4S-E247A-dCK mutants were engineered using the QuickChange site-directed mutagenesis kit from Stratagene, using as template the wild type dCK gene cloned into the pET14b vector. The BL21(DE3) E. coli strain carrying the recombinant plasmid coding for His-tagged C4S-dCK was grown in 2YT media at 37 °C, induced with 0.1 mM IPTG, and harvested after 4 h. The cell pellet was sonicated, and the lysate was loaded onto a HisTrap HP column (Ni Sepharose high-performance resin from Amersham Biosciences). The washing buffer contained 20 mM imidazole, pH 7.5, while the elution buffer was made of 200 mM imidazole, 50 mM HEPES, pH 7.5, and 500 mM NaCl. EDTA (2 mM) was immediately added to the eluted protein to avoid degradation by traces of proteases. The protein (with His tag) was further purified on a S-200 gel filtration column (Amersham Biosciences) using as gel filtration buffer 20 mM HEPES, pH 7.5, 200 mM sodium citrate, and 2 mM EDTA. Complexes of dCK with dG + UDP, CLAD + UDP, and CLAD + ADP were then prepared (all nucleosides were purchased from Sigma) by mixing 5 mM nucleoside (dG or CLAD) with 5 mM nucleotide (ADP or UDP) and the enzyme (15–20 mg/mL in gel filtration buffer plus 5 mM MgCl2).
Crystals for the different complexes were obtained by vapor diffusion from hanging drops (1 μL of protein/nucleoside/nucleotide/MgCl2 mix plus 1 μL of reservoir) using a reservoir solution that contained 0.90–1.5 M trisodium citrate dihydrate and 100 mM HEPES, pH 7.5. The crystals containing UDP were grown at 12 °C (dG + UDP and CLAD + UDP), while the CLAD + ADP crystal was grown at room temperature (22 °C). The dG + UDP complex crystallized in space group P212121, CLAD + UDP in P21, and CLAD + ADP in C2221.
Crystallographic Data Collection and Processing
X-ray data were collected at the Advanced Photon Source, Argonne National Laboratories, using the SERCAT beamline BM-22 for the dG + UDP and the CLAD + UDP complexes and using beamline ID-22 for the CLAD + ADP complex. The data were collected at 100 K (the cryoprotectant solution for the crystals was mineral oil purchased from Sigma), and they were indexed, scaled, and merged using XDS and XSCALE14 (see Table 2 for data collection statistics).
Table 2.
Data Collection and Refinement Statistics
| dCK mutant | C4S | C4S | C4S+E247A |
| complex | dG + UDP | CLAD + UDP | CLAD + ADP |
| PDB codes | 2ZI7 | 2ZIA | 2ZI9 |
| cryst growth temp (°C) | 12 | 12 | 22 |
| X-ray source and detector | APS (SERCAT BM-22) MARCCD 225 | APS (SERCAT BM-22) MARCCD 225 | APS (SERCAT ID-22) MARCCD 300 |
| wavelength (Å) | 1.0 | 1.0 | 1.0 |
| temp (K) | 100 | 100 | 100 |
| resolna (Å) | 1.97 (1.97–2.0) | 1.80 (1.80–1.90) | 2.51 (2.51–2.66) |
| no. obsd reflns | 253226 | 318836 | 79207 |
| no. unique reflns | 36226 | 52169 | 20415 |
| completeness (%) | 96.0 (79.4) | 99.4 (96.9) | 98.4 (93.6) |
| Rsym (%) | 9.1 (45.8) | 11.4 (62.7) | 8.4 (64.0) |
| I/σ(I) | 13.5 (3.10) | 11.7 (3.3) | 11.5 (2.1) |
| space group | P212121 | P21 | C2221 |
| unit cell | |||
| a (Å) | 43.80 | 67.8 | 56.10 |
| b (Å) | 107.50 | 44.1 | 132.40 |
| c (Å) | 110.40 | 94.6 | 156.90 |
| β (deg) | 90.0 | 95.7 | 90.0 |
| refinement program | REFMAC5 | REFMAC5 | REFMAC5 |
| refinement statistics | |||
| Rcryst (%) | 20.6 | 20.2 | 22.8 |
| Rfree (%) | 25.9 | 25.0 | 33.2 |
| resoln range (Å) | 30-1.97 | 30-1.80 | 30-2.51 |
| molecules per au | 2 | 2 | 2 |
| no. of atoms | |||
| protein | 1732, 1793 | 1842, 1805 | 1981, 1861 |
| dG + CLAD | 19 × 2 | 19 × 2 | 19 × 2 |
| ADP + UDP | 25 × 2 | 25 × 2 | 27 × 2 |
| water | 84 | 297 | 162 |
| rms deviation | |||
| bond length (Å) | 0.016 | 0.015 | 0.012 |
| bond angles (deg) | 1.635 | 1.552 | 1.636 |
| av B-factors (Å2)/chain | |||
| protein | 31, 27 | 23, 23 | 53, 65 |
| dG/CLAD | 18, 16 | 18, 19 | 59, 84 |
| ADP/UDP | 21, 23 | 17, 17 | 48, 60 |
| waters | 30 | 30 | 52 |
| Ramachandran plot (%) | |||
| most favored regions | 92.1 | 91.2 | 79.3 |
| additionally allowed regions | 7.3 | 8.1 | 18.9 |
| generously allowed regions | 0.5 | 0.5 | 1.4 |
| disallowed regions | 0.1 | 0.2 | 0.5 |
Last shell in parentheses.
Structure Determination and Refinement
The structures of dG + UDP and CLAD + UDP were solved by molecular replacement using as initial model PDB code 1P60, while the structure with CLAD + ADP was solved applying rigid body refinement. For both scenarios, the initial model was PDB code 1P60.
In order to avoid bias, residues 55–82 of the insert region were omitted from each monomer of dCK dimer 1P60. The same was repeated for residues 238–254 of the base-sensing loop. Restrained refinement for the three structures was carried out using REF-MAC.15 The dG + UDP, CLAD + UDP, and CLAD + ADP structures were refined to 1.97, 1.80, and 2.51 Å, respectively. Both the 2Fobs − Fcalc and the Fobs − Fcalc electron density maps for the nucleosides dG and CLAD are shown in Figure S3 (CLAD of the CLAD + UDP complex is shown panel b, while the one of the CLAD + ADP complex is shown in panel c). The 2Fobs − Fcalc map is contoured at 2σ for the structures with UDP and at 1.5σ for the CLAD + ADP structure, while the Fobs − Fcalc map is contoured at −3σ. Note the strong peak at the chlorine atom present in the Fobs − Fcalc map contoured at −3σ (structures refined assigning full occupancy to the chlorine atom). A strong peak in a difference map contoured at negative σ levels indicates the presence of atom(s) in the model that is not supported by the data. In other words, the data are not consistent with the strong scattering expected at that position due to the chlorine atom. Lower than expected electron density at this position can be explained by radiation-induced damage occurring during data collection. This phenomenon is particularly well documented for halogens covalently bound to aromatic rings,16–18 such as are present in cladribine. Despite lacking atomic resolution that would be required to independently refine the B-factors and occupancy of atoms, we did attempt to approximate the partial occupancy of the chlorine. This was accomplished by evaluating the B-factor of the halogen as a function of its occupancy combined with inspection of the Fobs − Fcalc map contoured at −3σ. This procedure suggested a chlorine occupancy of ~0.2 for the CLAD + UDP structure. The lower resolution of the CLAD + ADP complex precludes such an analysis.
The structures were deposited in the PDB with accession codes 2ZI7 for the dG + UDP, 2ZIA for the CLAD + UDP, and 2ZI9 for the CLAD + ADP.
Steady-State Kinetic Assay
A spectroscopic enzyme-coupled assay was used to determine the activity of both wild type and mutant C4S-dCK as described previously19 in 50 mM Tris-HCl, pH 7.5, 100 mM KCl, and 5 mM MgCl2 at 37 °C with 0.35 μM dCK, 1 mM ATP-Mg, and 1 mM UTP-Mg. Concentrations of dG and CLAD varied between 50 and 600 μM when ATP was used as phosphate donor and between 10 and 200 μM with UTP. Kinetic data for the wild type protein are shown in Table 1, and kinetic data for the crystallization mutant C4S-dCK are shown in Table S1.
Supplementary Material
Kinetic analysis results of C4S dCK using ATP or UTP as donors and dA, dG, or cladribine as acceptors (Table S1), a schematic representation of the chemical structures of dA, dG, and cladribine (Figure S1), superposition of the three structures reported here (Figure S2), and electron density for the nucleosides (Figure S3). This material is available free of charge via the Internet at http://pubs.acs.org.
Acknowledgments
This work was supported by an NIH grant (E.S., S.H., and A.L.) and the Max-Planck-Society (M.K.). We thank the staff at the SER-CAT beamline for help in data collection.
Footnotes
Abbreviations: NAs, nucleoside analogues; dCK, deoxycytidine kinase; dGK, deoxyguanosine kinase; dC, deoxycytidine; dA, deoxyadenosine; dG, deoxyguanosine; CLAD, cladribine; C4S-dCK, C9S/C45S/C59S/C146S dCK; ATP, adenosine triphosphate; ADP, adenosine diphosphate; UTP, uridine triphosphate; UDP, uridine diphosphate; PDB, Protein Data Bank (http://www.rcsb.org).
References
- 1.Goody RS, Muller B, Restle T. Factors contributing to the inhibition of HIV reverse transcriptase by chain-terminating nucleotides in vitro and in vivo. FEBS Lett. 1991;291:1–5. doi: 10.1016/0014-5793(91)81089-q. [DOI] [PubMed] [Google Scholar]
- 2.White JC, Capizzi RL. A critical role for uridine nucleotides in the regulation of deoxycytidine kinase and the concentration dependence of 1-beta-D-arabinofuranosylcytosine phosphorylation in human leukemia cells. Cancer Res. 1991;51:2559–2565. [PubMed] [Google Scholar]
- 3.Shewach DS, Reynolds KK, Hertel L. Nucleotide specificity of human deoxycytidine kinase. Mol Pharmacol. 1992;42:518–524. [PubMed] [Google Scholar]
- 4.Sabini E, Hazra S, Ort S, Konrad M, Lavie A. Structural basis for substrate promiscuity of dCK. J Mol Biol. 2008;378:607–621. doi: 10.1016/j.jmb.2008.02.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Datta NS, Shewach DS, Mitchell BS, Fox IH. Kinetic properties and inhibition of human T lymphoblast deoxycytidine kinase. J Biol Chem. 1989;264:9359–9364. [PubMed] [Google Scholar]
- 6.Kawasaki H, Carrera CJ, Piro LD, Saven A, Kipps TJ, et al. Relationship of deoxycytidine kinase and cytoplasmic 5′-nucleotidase to the chemotherapeutic efficacy of 2-chlorodeoxyadenosine. Blood. 1993;81:597–601. [PubMed] [Google Scholar]
- 7.Sabini E, Ort S, Monnerjahn C, Konrad M, Lavie A. Structure of human dCK suggests strategies to improve anticancer and antiviral therapy. Nat Struct Biol. 2003;10:513–519. doi: 10.1038/nsb942. [DOI] [PubMed] [Google Scholar]
- 8.Godsey MH, Ort S, Sabini E, Konrad M, Lavie A. Structural basis for the preference of UTP over ATP in human deoxycytidine kinase: illuminating the role of main-chain reorganization. Biochemistry. 2006;45:452–461. doi: 10.1021/bi0518646. [DOI] [PubMed] [Google Scholar]
- 9.Sabini E, Hazra S, Konrad M, Burley SK, Lavie A. Structural basis for activation of the therapeutic L-nucleoside analogs 3TC and troxacitabine by human deoxycytidine kinase. Nucleic Acids Res. 2007;35:186–192. doi: 10.1093/nar/gkl1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sabini E, Hazra S, Konrad M, Lavie A. Nonenantioselectivity property of human deoxycytidine kinase explained by structures of the enzyme in complex with L- and D-nucleosides. J Med Chem. 2007;50:3004–3014. doi: 10.1021/jm0700215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Saenger W. Principles of Nucleic Acid Structure. Springer-Verlag; New York: 1984. [Google Scholar]
- 12.Hughes TL, Hahn TM, Reynolds KK, Shewach DS. Kinetic analysis of human deoxycytidine kinase with the true phosphate donor uridine triphosphate. Biochemistry. 1997;36:7540–7547. doi: 10.1021/bi970059r. [DOI] [PubMed] [Google Scholar]
- 13.Zhang Y, Secrist JA, Ealick SE. The structure of human deoxycytidine kinase in complex with clofarabine reveals key interactions for prodrug activation. Acta Crystallogr, Sect D: Biol Crystallogr. 2006;62:133–139. doi: 10.1107/S0907444905034293. [DOI] [PubMed] [Google Scholar]
- 14.Kabsch W. Automatic processing of rotation diffraction data from crystals of initially unknown symmetry and cell constants. J Appl Crystallogr. 1993;26:795–800. [Google Scholar]
- 15.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. 1997;D53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 16.Chai JJ, He C, Li M, Tang L, Luo M. Crystal structure of carboxypeptidase A complexed with an inactivator in two crystal forms. Protein Eng. 1998;11:841–845. doi: 10.1093/protein/11.10.841. [DOI] [PubMed] [Google Scholar]
- 17.Ennifar E, Carpentier P, Ferrer JL, Walter P, Dumas P. X-ray-induced debromination of nucleic acids at the Br K absorption edge and implications for MAD phasing. Acta Crystallogr, Sect D: Biol Crystallogr. 2002;58:1262–1268. doi: 10.1107/s0907444902009526. [DOI] [PubMed] [Google Scholar]
- 18.Olieric V, Ennifar E, Meents A, Fleurant M, Besnard C, et al. Using X-ray absorption spectra to monitor specific radiation damage to anomalously scattering atoms in macromolecular crystallography. Acta Crystallogr, Sect D: Biol Crystallogr. 2007;63:759–768. doi: 10.1107/S0907444907019580. [DOI] [PubMed] [Google Scholar]
- 19.Agarwal KC, Miech RP, Parks RE., Jr Guanylate kinases from human erythrocytes, hog brain, and rat liver. Methods Enzymol. 1978;51:483–490. doi: 10.1016/s0076-6879(78)51066-5. [DOI] [PubMed] [Google Scholar]
- 20.Kraulis PJ. MOLSCRIPT: a program to produce both detailed and schematic plots of protein structures. J Appl Crystallogr. 1991;24:946–950. [Google Scholar]
- 21.Esnouf R. An extensively modified version of molscript that includes greatly enhanced coloring capabilities. J Mol Graphics. 1997;15:133–138. doi: 10.1016/S1093-3263(97)00021-1. [DOI] [PubMed] [Google Scholar]
- 22.Merrit EA, Murphy MEP. Raster3D version 2.0, a program for photorealistic molecular graphics. Acta Crystallogr. 1994;D50:869–873. doi: 10.1107/S0907444994006396. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Kinetic analysis results of C4S dCK using ATP or UTP as donors and dA, dG, or cladribine as acceptors (Table S1), a schematic representation of the chemical structures of dA, dG, and cladribine (Figure S1), superposition of the three structures reported here (Figure S2), and electron density for the nucleosides (Figure S3). This material is available free of charge via the Internet at http://pubs.acs.org.