Abstract
Notch3 gene amplification has recently been identified in ovarian cancer but the Notch3 effectors that are involved in the development of ovarian cancer remain elusive. In this study, we have identified Pbx1, a proto-oncogene in hematopoietic malignancy, as a Notch3 target gene. Pbx1 expression is transcriptionally regulated by Notch3 activation, and Notch3/CSL protein complex directly binds to the Pbx1 promoter segment harboring the CSL binding sequence. The growth-inhibitory effect of gamma-secretase inhibitor could be partially reversed by ectopic Pbx1 expression. Furthermore, functional studies by Pbx1 shRNA knockdown demonstrate that Pbx1 is essential for cell proliferation and tumorigenicity. Taken together, the above findings indicate that Pbx1 is a direct Notch3-regulated gene that mediates the survival signal of Notch3 in ovarian cancer.
Introduction
Cancer development has been known to share many molecular frameworks with embryonic development, tissue renewal and differentiation (1). For example, in normal tissues the Notch signaling pathway is activated under strict temporal and spatial control during tissue regeneration and cell fate determination. In contrast, during tumor development Notch is constitutively activated and its sustained activation could be a result of genetic or epigenetic alterations. In human cancer, activation of the Notch receptor due to point mutations and gene translocations has been found in T-cell leukemia/lymphoma, non-small-cell lung cancer, and breast carcinoma (2–5). Based on a genome-wide analysis of DNA copy number changes using both digital karyotyping (6) and SNP array analysis (7), we have identified Notch3 gene amplification in high-grade ovarian serous carcinomas (8). Notch3 gene amplification correlated with gene overexpression and pathway activation in ovarian serous carcinoma tissues. Functional inactivation of Notch3 either by γ-secretase inhibitor or by Notch3-specific siRNA resulted in suppression of cell proliferation and induction of apoptosis, suggesting that Notch3 activation is an important survival signal in ovarian cancer cells.
The Notch signaling pathway is evolutionally conserved. The primary members of this signaling pathway include Notch ligands (Delta and Jagged), Notch receptors (Notch1–4), the nuclear transcription factors such as CSL (also known as RBP-J and CBF1) that bind to Notch intracellular fragment, and the target genes that are controlled by Notch3/CSL co-activator. Activation of Notch signaling is initiated by receptor-ligand interaction, which leads to proteolytic cleavages that liberate the Notch intracellular cytoplasmic domain (NICD) from the plasma membrane. NICD translocates to the nucleus and binds to transcription complexes that contain CSL. NICD-binding converts the CSL complex from a transcription repressor to a transcription activator, thereby initiating transcription of downstream effectors. The diverse biological activities of Notch signaling are thought to be mediated by the context-dependent expression of a heterogeneous group of downstream effectors.
Although several downstream genes in the Notch pathway have been reported (9–12), it is not known which of them are regulated by Notch3 in ovarian cancer cells. We have analyzed the known Notch target genes represented in the Serial Analysis of Gene Expression (SAGE) database in ovarian tissues. We found that the expression level of most of the known target genes, such as the Hes gene family, was extremely low and did not correlate with the expression level of Notch3 in ovarian cancer tissues. This suggested that the Notch3 targets in ovarian cancer were distinct from other known Notch targets. In order to identify candidate Notch3 targets we analyzed the genes that were downregulated following functional inactivation of Notch3 by treatment of ovarian cancer cells with γ-secretase inhibitor in this study. We selected one of the most promising candidate genes, Pbx1, for further characterization.
Materials and Methods
Affymetix GeneChip analysis
Cancer cell lines including OVCAR3, A2780 and MCF7 were cultured in the presence of 1 µM GSI (γ-secretase inhibitor I, EMD Chemicals, San Diego, CA) for 48 h. Control cells were cultured in the presence of DMSO under the same experimental condition. Total RNA was purified using an RNA purification kit (Qiagen, Valencia, CA) and RNA samples were hybridized onto the GeneChip arrays, HG-U133 Plus 2.0 (Affymetrix, Santa Clara, CA), that were spotted with over 47,000 human transcripts. The dCHIP software package1 was used for data analysis. By comparing global gene expression profiles between GSI-treated and mock-treated cells, we selected the differentially expressed genes with levels greater than 2 fold in all three tested cell lines. The differentially expressed genes were presented as individual boxes with pseudo-color to indicate gene expression levels.
Quantitative real-time PCR
Relative transcript expression levels were measured by quantitative real-time PCR using method previously described (13). The primer sequences were shown in supplementary Table 1. PCR reactions were performed in triplicates using an iCycler (BioRad, Hercules, CA). The amplified products were quantified by fluorescence intensity of SYBR Green I (Molecular Probes, Eugene, OR). Average fold changes were calculated by differences in threshold cycles (Ct) between pairs of samples to be compared. β-amyloid precursor gene was used for normalizing the cDNA concentration of each sample.
Western blot analysis
Protein lysates were prepared by resuspending cell pellets in Laemmli sample buffer containing 5 % β-mercaptoethanol. Protein lysates were separated by 4–12 % Tris-glycine gel electrophoresis, and transferred onto PVDF membrane using semi-dry apparatus (Bio-rad). The membrane was blocked with 5% non-fat dry milk in TBST(20 mM Tris-HCl, 0.5 M NaCl, 0.1% Tween20), incubated with primary antibody at room temperature for 3 hour, followed by washing with TBST. Subsequently the membrane was incubated with horse radish peroxidase-conjugated secondary antibody and detected with ECL solution (Thermo Scientific Cat No 34080). Antibodies used in this study include: anti-NICD3 (sc-5593, Santa Cruz); anti-N-terminal extra-cellular subunit of Notch3 (Abnova Cat No H00004854-M01); anti-Pbx1 (sc-28313, Santa Cruz); anti-Pbx1b (gift from Dr. Michael Cleary, Stanford University).
Chromatin immunoprecipitation (ChIP) assay
Chromatin immunoprecipitation (ChIP) was performed using the ChIP-IT enzymatic kit (Active motif, Carlsbad CA). HEK293 cells were infected with control pBabe-puro retrovirus and pBabe-CSL/V5 retrovirus for 24 h prior to fixation with 1% paraformaldehyde. The lysates of fixed cells were treated with an enzymatic shearing cocktail (Active Motif, Carlsbad, CA) and the soluble fraction was incubated with 4 µL of anti-V5 antibody or normal mouse IgG and precipitated with protein-G magnetic beads at 4°C for overnight. The crosslinks of the DNA eluate was reversed by heating at 65°C for 6 hours, and the samples were treated with proteinase K (10 µg/ml) at 37°C for 1 hr. Quantitative PCR was performed using SYBR Green I-based detection system as described above with the PCR primers that amplified different Pbx1 promoter regions (primer sequences shown in supplementary Table 1). The input DNA was defined as an aliquot of sheared chromatin prior to immunoprecipitation, and was used to normalize the amount of chromatin used in each experiment as previously described (14).
Electrophoretic mobility shift assays
Nuclear extracts were prepared using the NE-PER nuclear and cytoplasmic extraction reagents (Pierce, Rockford, IL). Electrophoretic mobility shift assay (EMSA) was performed using the LightShift chemiluminescent EMSA kit (Pierce Biotechnology, Rockford IL) according to manufacturer’s protocol. DNA fragments for two Pbx1 promoter regions (−254bp to −111bp and −354bp to −254bp) were PCR amplified using biotinylated PCR primers. The biotin-labeled DNA fragments were incubated with nuclear lysate from 293 cell transfected with SG5-flag-CSL and/or infected with NICD3 retrovirus. Nuclear protein (5 µg) was incubated with 20 fmol of biotin-labeled DNA probe for 20 min at room temperature. DNA probe/protein complexes were separated by electrophoresis, transferred to a nylon membrane, incubated with streptavidin-horse radish peroxidase, and visualized by chemiluminescence.
Immunohistochemistry
A rabbit polyclonal anti-NICD3 antibody (Santa Cruz Biotechnology, Santa Cruz, CA) and a mouse anti-Pbx1b primary antibody against the Pbx1b (kind gift from Dr. Michael Cleary, Stanford University, CA (15)) were used in immunohistochemistry. Tissue microarrays containing 81 high-grade ovarian serous carcinoma tissues per slide were de-paraffinized and antigen retrieved in a citrate buffer (pH 6.0) for 10 minutes. Immunostaining was performed using an EnVision+System peroxidase kit (DAKO, Carpinteria, CA). Immunointensity was independently scored by two investigators based on nuclear immunoreactivity. For discordant cases, a third investigator scored and the final intensity score was determined by the majority votes.
Production of Pbx1 and CSL retroviruses and rescue assay
The PCR products of the Pbx1a, Pbx1b, and CSL were cloned into an expression vector, pCDNA6 with a V5 tag. The coding sequence of Pbx1a-V5, Pbx1b-V5 and CSL-V5 was subcloned into the pBabe-puro retroviral vector and high titer retroviral stocks were generated using Phoenix ecotropic packaging cell line. Retroviral gene transfer was performed as previously described (16). For rescue assay, cells were infected with Pbx1a retrovirus and the empty pBabe-puro retrovirus. Cells were cultured in the presence of 0.7 µM, 1.0 µM, 1.5 µM, and 2.0 µM of GSI for 24 h. Mock-treated cells were cultured in the presence of DMSO vehicle control. Average cell counts and standard deviations were derived from 5 replicates. Student’s t-test was used to determine statistical significance.
Luciferase reporter assay
Immortalized ovarian surface epithelial cells (OSE10) were transfected with pGL3 plasmid (Promega, Madison, WI) or pGL3-Pbx1 which contains 3.0 kb Pbx1 promoter (gift from Dr. S. Higashiyama, Japan (17)). pRL-Renilla reporter plasmid (Promega) which serves to monitor the transfection efficiency was co-transfected with the pGL3 plasmid. Transfection was carried out by Lipofectamine 2000 (Invitrogen, Carlsbad, CA) and luciferase expression was determined by Dual-Glo luciferase reagent (Promega, Madison, WI). The firefly luciferase activity was normalized to luminescence from Renilla luciferase and the ratio of luminescence from the experimental reporter to luminescence from the control reporter was calculated.
Transfection and cell growth assay
The sequences of Notch3-specific siRNAs and transfection method have been previously described (8). Pbx1 shRNA plasmid that targeted Pbx1 coding sequence 5’-GCAAGCGACAGAAATCCTGAA-3’ was purchased from the Mission® shRNA collection at Sigma-Aldrich (St. Louis, MO). The shRNA sequence was subcloned into a pLKO.1-puro vector. For cell proliferation assays, cells were ded in 24-well culture plate, transfected with 1 µg Pbx1 shRNA plasmid using Lipofectamine 2000 (Invitrogen, Carlsbad, CA). One day after transfection, cells were plated into 96-well plates at a density of 3,000 cells/well and cell number was measured at different time point by the incorporation of SYBR Green I using a fluorescence microplate reader (Fluostar from BMG, Durham, NC). To determine if Pbx1 expression is essential for tumor cell survival in vivo, we transfected A2780 ovarian cancer cells with Pbx1 shRNA. Two millions of transfected cells were injected into the subcutaneous tissue of nude mice. Tumor volume was measured every other day for 18 days.
Results
Identification and validation of Notch3 regulated genes in cancer cells
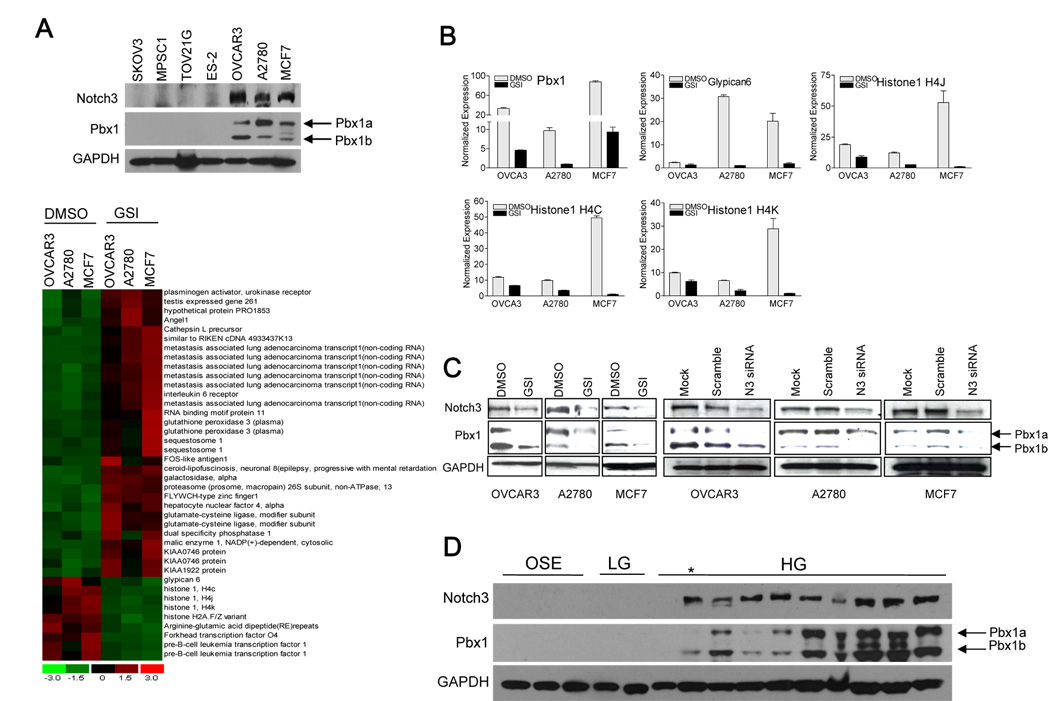
To elucidate the mechanisms by which Notch3 signaling promotes tumor growth and survival, we first sought to identify Notch3-regulated genes by comparing gene expression between cancer cells treated either with γ-secretase inhibitor (GSI) or with DMSO vehicle control. The release of Notch intracellular cytoplasmic domain (NICD), which is required for Notch signaling, is mediated by γ-secretase. GSI has been shown to potently inhibit γ-secretase, thereby inactivating Notch signaling (18, 19). Ovarian cancer cell lines including OVCAR3, A2780 and a breast cancer cell line, MCF7, overexpressing Notch3 were selected for Affymetrix GeneChip analysis (upper panel in Fig. 1A). Genes that demonstrated 2-fold differential expression from all three cell lines were identified using the dCHIP software (bottom panel in Fig 1A,). We specifically focused on genes that were suppressed by GSI because those genes were potentially upregulated by Notch3. Among those genes were Pbx1, glypican-6, histone 1 H4C, 4K and 4J, H2A member V and a gene containing the RERE repeats. Using quantitative real-time PCR, we were able to confirm 5 of them (Fig 1B). Pbx1 (Pre-B-cell leukemia transcription factor 1) was selected for further characterization for a number of reasons. First, by GeneChip analysis, Pbx1 showed the most dramatic and consistent downregulation by GSI in all 3 cell lines tested. Second, Pbx1 protein expression correlated with Notch3 expression in cell lines based on Western blot analysis (upper panel, Fig. 1A). Third, Pbx1 fusion protein, E2A-Pbx1, is known as a proto-oncogene in leukemia (20, 21). Lastly, the Pbx1 promoter contained a consensus sequence for CSL/NICD binding.
Fig. 1. Identification of Pbx1 as a putative Notch3 target gene.
A. Upper panel: Western blot was performed to assess Notch3 protein expression in a panel of cancer cell lines. OVCAR3, A2780, and MCF7, which express abundant Notch3 proteins, were selected for microarray analysis. GAPDH protein expression was used as loading control. Of note, the three cell lines with Notch3 overexpression also show Pbx1 overexpression. Lower panel: Affymetrix Genechip analysis was performed to identify genes differentially expressed between GSI-treated (1 µM) versus DMSO-treated cancer cells. dChip software was used to analyze the data. A cut-off ratio of 2 was used to select the GSI-sensitive genes. Columns represent experimental samples (cell lines), while rows represent genes. The expression level of each gene in an individual specimen is shown as a pseudo-color gradient based on the relative expression level normalized for each gene, where red indicates high and green indicates low expression. B. Quantitative real-time PCR was performed to confirm the candidate GSI-sensitive genes identified by Genechip analysis. The genes that can be confirmed by quantitative PCR include Pbx1, glypican-6, histone 4C, histone 4K, and histone 4J. C. Western blot analysis demonstrates that Pbx1 protein is downregulated by either GSI or Notch3 siRNA. In contrast, mock (no transfection) and scramble siRNA do not have significant effects on Pbx1 protein expression. Cells were incubated with GSI or Notch siRNA for 48 hours before assay. D. Western blot analysis of Pbx1 expression in a panel of ovarian tissues. OSE: ovarian surface epithelial cell line; LG: low-grade ovarian serous carcinoma; HG: high-grade ovarian serous carcinoma. * indicates samples with protein degradation. Pbx1a was found to be more susceptible to protein degradation.
Two alternatively spliced variants of Pbx1, Pbx1a and Pbx1b, have been previously reported and both shares same sequences except an additional exon in Pbx1a. Pbx1a encodes 430 amino acids while Pbx1b encodes 347 amino acids. Pbx1b shares the same amino acid sequences as Pbx1a from 1–333 amino acids but differs in its last 14 amino acids. Using Western blot and RT-PCR analyses, we found that both splicing variants were present in ovarian cancers, with Pbx1a appearing as the predominant form (Fig 1A and supplementary Fig 1).
The results of Genechip analysis (based on 2-fold differential expression) did not identify the Notch downstream target genes previously reported in other organ systems. However, when the selection criteria were relaxed, we found that several of the known Notch target genes were moderately down-regulated by GSI (supplementary Fig. 2A). Quantitative real-time PCR was performed on eight of the previously known target genes including HeyL, Hey2, TLE2, TLE4, and c-Myc in the same three cell lines selected for Genechip analysis. The real-time PCR results showed that a consistent down-regulation of gene expression upon GSI treatment could be observed in TLE2 and TLE4 (supplementary Fig. 2B). In addition, c-Myc and Cyclin A2 were significantly decreased by GSI in 2 of the 3 cell lines. These results suggested that these four genes may also have a role in mediating the function of Notch3 in some cancer cell lines.
Notch3-dependent Pbx1 expression
To determine whether Pbx1 expression is associated with Notch3 expression, we used the following approaches. First, we inhibited NICD3 expression using GSI and Notch3-specific siRNA in the Notch3 expressing cell lines, OVCAR3, A2780 and MCF7. As shown in Fig. 1C, both Notch3 and Pbx1 protein levels (both splicing isoforms) were significantly reduced by either GSI or Notch3 siRNA treatment. We found that GSI inhibited the expression of NICD3 in a time dependent fashion (supplementary Fig. 3A). Similar findings were previously reported (22–24).
To determine whether Pbx1 protein expression correlates with Notch3 protein expression in ovarian cancer tissues, Western blot was performed in a panel of protein lysates prepared from immortalized OSEs, low-grade serous carcinoma tumor tissues, and high-grade serous carcinoma tumor tissues. The result demonstrated that OSEs and low-grade tumors expressed low abundance of Pbx1 or Notch3. In comparison, most high-grade tumors expressed high levels of Notch3 and Pbx1 (Fig. 1D).
To determine if protein synthesis was required for suppression of Pbx1 transcription by GSI, cycloheximide was added to the cell culture system to block protein synthesis. Cells overexpressing Notch3 were treated with 1µM GSI for 6 hr to down-regulate Pbx1 expression. Cells were then washed and incubated with medium with (mock washout) or without GSI. As shown in supplementary Fig. 4, in the absence of GSI, a rapid upregulation of Pbx1 transcripts was detected even in the presence of cycloheximide. In contrast, continuous presence of GSI suppressed the Pbx1 transcription. These results indicated that Pbx1 transcription regulation by Notch3 did not require de novo protein synthesis.
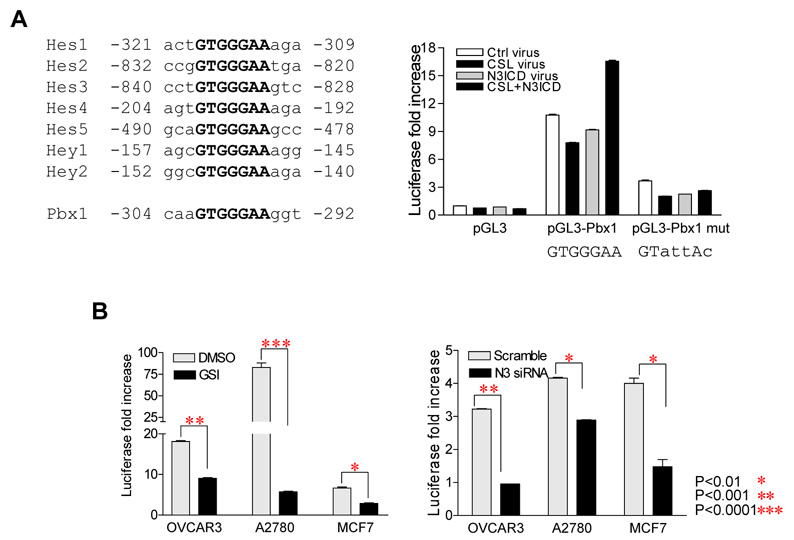
Next, we asked if Notch3 was directly involved in transcriptional regulation of Pbx1. Using a Pbx1 promoter-lucieferase reporter construct (17), we measured Pbx1 promoter activity in OSE cells derived from normal ovarian surface epithelium, This promoter construct, pGL3-Pbx1, includes a 3.0 kb Pbx1 promoter region containing one CSL consensus binding site located at −304 to −292 bp upstream of the translation initiation codon (Fig. 2A, left panel). pGL3-Pbx1 was transfected into OSE cells, which were previously engineered to express CSL, NICD3, or both. As shown in the right panel of Fig. 2A, luciferase activity was significantly higher in pGL3-Pbx1 transfected groups compared to pGL3 transfected groups. A low level of luciferase activity was detected in pGL3-Pbx1 transfected cells even in the absence of CSL or NICD3 virus transduction, likely due to endogenous CSL and NICD3 in OSE cells. Luciferase activity was further enhanced in cells co-transduced with CSL and NICD3. To test the specificity of the promoter activity, we created a promoter construct which carried mutated CSL binding site (GTAATAC). When assayed in OSE cells, luciferase activity driven by the pGL3-Pbx1 mutant reporter was significantly reduced under all experimental conditions indicating that a functional CSL site was required for promoter activity.
Fig. 2. Notch3 induces Pbx1 expression through promoter activation.
A. Left Panel: Examples of the conserved CSL binding site in Notch-regulated genes in human. Consensus CSL binding sequence is bolded. The consensus CSL binding site is located at −304 to −292 at Pbx1 promoter. Right Panel: Pbx1 promoter assay shows increased reporter activity in cells transfected with the wild-type Pbx1 promoter construct (pGL3-Pbx1) as compared to cells transfected with vector only control (pGL3). In pGL3-Pbx1 transfected groups, cells co-transduced with both CSL and NICD3 retroviruses demonstrate the highest promoter activity. Cells transduced with vector only virus and pGL3-Pbx1 also showed above background luciferase activity, likely due to the endogenous background expression of CSL and Notch in OSE. In contrast, cells transfected with the mutant Pbx1 promoter construct (pGL3-Pbx1 mut) containing the mutant CSL binding site demonstrate a significantly lower luciferase activity as compared to that of wild-type Pbx1 promoter.
B. The pGL3-Pbx1 promoter construct was transfected into 3 cancer cell lines overexpressing Notch3. Pbx1 promoter activity was high in each of these transfected lines, but was significantly reduced by either GSI or Notch3 siRNA.
Pbx1 promoter reporter construct was also employed to test promoter activity in cell lines (OVCAR3, A2780 and MCF7) that expressed both Notch3 and Pbx1. In these experiments, we determined if inactivating Notch3 using GSI or Notch3 siRNA would lead to inhibition of luciferase reporter activity. Indeed, in all Notch3-expressing cell lines examined, GSI and Notch3 siRNA treatment led to a significant suppression in Pbx1 promoter reporter activity (Fig. 2B).
Correlation of Pbx1b expression and NICD3 immunoreactivity in tumor samples
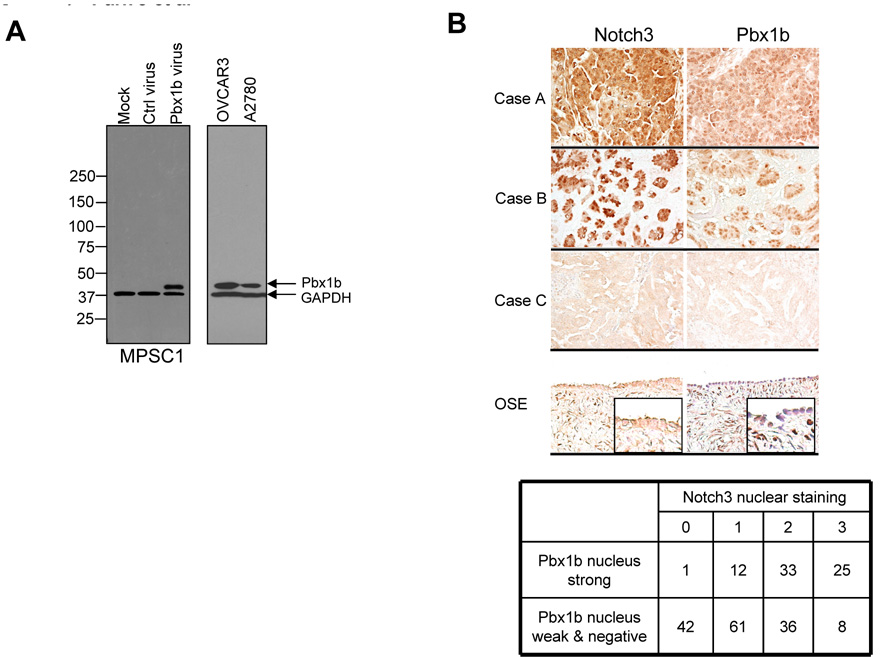
The above results demonstrated that Pbx1 expression was transcriptionally regulated by Notch3 pathway activation. To determine if this association also occurred in vivo, we performed immunohistochemistry to assess Notch3 and Pbx1 expression levels in a series of ovarian serous carcinoma tissues. After an extensive search for Pbx1 antibodies that worked in formalin-fixed, paraffin-embedded tissues, we only identified an antibody against the C-terminus of Pbx1b (gift from Dr. Michael Cleary, Stanford University) that recognized the antigen in paraffin sections. The specificity of this antibody was confirmed by demonstrating that a single protein band, corresponding to the molecular mass of Pbx1b protein, was detected in Pbx1b gene-transduced MPSC1 cells, but not in vector-transduced or in parental MPSC1 cells (Fig. 3A). Since Western blot and RT-PCR both demonstrated co-expression of Pbx1a and Pbx1b in ovarian tissues, Pbx1b immuoreactivity was used as the surrogate marker for Pbx1 expression. The specificity of the Notch3 antibody and its application in immunohistochemistry has been shown in previous reports (8, 16). Because nuclear localization of Notch is an indication of pathway activation and Pbx1b is a nuclear transcription factor, we focused on evaluating nuclear immunoreactivity of both proteins. Pbx1b nuclear immunoreactivity was always either intense or very weak/undetectable while Notch3 nuclear immunoreactivity showed a range of intensity (0, 1+, 2+ and 3+). Representative immunostaining results on tissue sections are shown in Fig. 3B. Using the Cochran-Armitage trend test, we found that there was a significant trend between Pbx1b and Notch3 nuclear co-overexpression in ovarian serous carcinoma tissues (p<0.0001, Fig 3B).
Fig. 3. Pbx1b is over-expressed in high-grade ovarian serous tumors and its nuclear expression level is positively correlated with Notch3 activation.
A. Western blot was performed to determine the specificity of the employed anti-Pbx1b antibody. A predominant protein band corresponding to Pbx1b (~ 38.4 kD) is detected in MPSC-1 transduced with Pbx1b retrovirus, but not in MPSC-1 transduced with control virus or in non-transduced MPSC-1 cells. A single band corresponding to Pbx1b protein can also be detected in ovarian cancer cell lines including OVCAR3 and A2780. GAPDH was used as the loading control.
B. Immunohistochemistry of Notch3 and Pbx1b in ovarian high-grade serous carcinoma tissues and in normal ovaries. Top: immunostaining of Notch3 and Pbx1b was performed on the same set of high-grade serous ovarian tumors. Three representative specimens with high Notch3 and Pbx1b expression (case A), medium Notch3 and Pbx1b expression (case B) and undetectable Notch3 and Pbx1b expression (case C) are shown. Normal ovarian surface epithelium (OSE) does not show nuclear Notch3 or Pbx1b immunoreactivity. Bottom: summary of immunohistochemistry result. Data demonstrate a significant trend of Notch3 and Pbx1b co-expression in nucleus (Cochran-Armitage trend test, p< 0.0001).
To further determine the Pbx1 expression pattern in OSE, low-grade and high-grade serous carcinoma tissues, we performed quantitative RT-PCR using PCR primers that amplified both Pbx1a and Pbx1b. The data demonstrated significantly higher Pbx1 mRNA levels in high-grade serous carcinoma tissues than in normal ovarian surface epithelial cells (OSE) or low-grade serous carcinomas (supplementary Fig. 5), a finding supporting the results using Western blot analysis shown in Fig. 1D.
Direct interaction between CSL/NICD3 protein complex and the Pbx1 promoter region
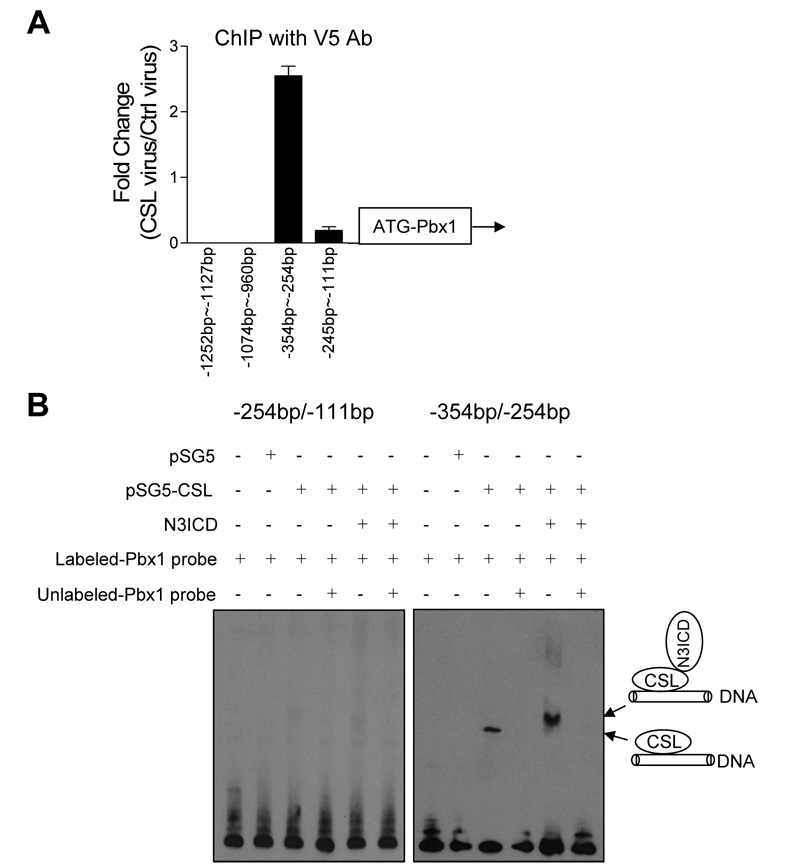
The above findings indicated transcriptional regulation of Pbx1 by Notch3 and were suggestive of direct interaction of Notch3/CSL complexes with the Pbx1 promoter. In order to determine if this is the case, we performed chromatin immunoprecipitation (ChIP) and electrophoretic mobility shift assay (EMSA). ChIP using an anti-V5 tag antibody in CSL-V5 transduced HEK293 cells compared to vector transduced HEK293 cells, demonstrated that among different Pbx1 promoter segments, a fragment spanning nucleotides −354 to −254 bp (−354/−254) containing the canonical consensus CSL-binding sequence bound most robustly to the CSL protein (Fig. 4A). Immunoprecipitation of the CSL-containing DNA segment appeared to be specific, as ChIP using control mouse IgG instead of anti-V5 antibody did not bring down this fragment (data not shown).
Fig. 4. Chromatin immunoprecipitation and electrophoretic mobility shift assay of Pbx1 promoter region.
A: Chromatin immunoprecipitation (ChIP) assay was performed to determine the CSL binding region at the Pbx1 promoter. Real time PCR was performed on fragmented chromatin precipitated by an anti-V5 antibody from 293T cells previously infected with CSL-V5 virus or control virus. PCR primers were designed to amplify four different regions at the Pbx promoter (−254/−111 bp, −354/−254 bp, −1074/−960 bp, and −1252/−1127 bp). Results were normalized to the total input of chromatin DNA and were presented as the fold change of CSL-V5 infected cells relative to control virus infected cells. Error bars represent standard deviations from triplicate experiments. B: Electrophoretic mobility shift assay (EMSA) was performed on two adjacent regions at the Pbx1 promoter, −254/−111 bp and −354/−254 bp. The −354/−254 bp region contains the CSL consensus binding sequence. Biotin-labeled DNA probe for each promoter region was incubated with nuclear lysates from HEK293 cells transfected with CSL and/or transduced with NICD3 virus. The specificity of CSL binding was determined in a competition reaction in which a 100-fold molar excess of unlabeled DNA probe was added to the binding reaction. The binding complex was resolved by electrophoresis on a 4–12% TBE gel. The position of biotin-labeled probe was detected with avidin-HRP followed by chemiluminescence.
EMSA was performed to determine if the NICD3/CSL protein complex directly bound to the Pbx1 promoter region containing the CSL binding sequence. For this purpose, a DNA probe spanning the −354/−254 region at the Pbx1 promoter containing the canonical CSL consensus site was generated. An adjacent DNA segment (−254/−111) was used as a control. As shown in Figure 4B, a band shift can be detected in the CSL-expressing group only when the −354/−254 probe was used in the assay. Binding of the CSL complex to the labeled probe could be competitively inhibited by 100-fold molar excess of unlabeled probes (Fig. 4B). Furthermore, when NICD3 was co-expressed with CSL in the same cells, the band shifted to a higher molecular weight, corresponding to the NICD3/CSL complex. This higher molecular weight band is more intense than the band formed in the absence of Notch3, suggesting that the NICD3/CSL forms a more stable complex with the Pbx1 promoter. A similar effect in the molecular weight shift has been reported in the Notch1/CSL complex (9).
Functional roles of Pbx1 in Notch3 signaling
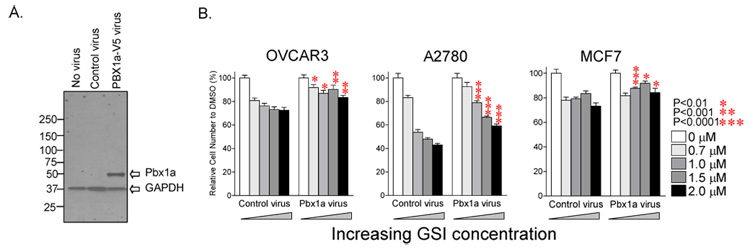
To determine the biological effects of Pbx1 in mediating Notch3 signaling, we examined if ectopic expression of Pbx1 (Fig. 5A) could reverse GSI-mediated growth inhibition in A2780, OVCAR3 and MCF7 cells. We found that as compared to control virus-expression, constitutive expression of ectopic Pbx1 could significantly protect the cells from the growth inhibitory effect of GSI at 1–2 µM concentration (Fig. 5B). However, ectopic expression of Pbx1 did not increase cell number in the same cell lines (data not shown), suggesting that Pbx1 alone is not sufficient to promote cell growth.
Fig. 5. Constitutive Pbx1a expression rescues cancer cells from GSI-induced growth inhibition.
A: Pbx1a cDNA tagged with a V5 epitope was cloned into the pBabe retroviral vector (controlled by a CMV promoter). Cells were transduced with the resulting Pbx1a retrovirus and analyzed by Western blot. Pbx1a protein was detected using an anti-V5 antibody. B: Notch3 expressing cancer cell lines (including OVCAR3, A2780, and MCF7) transduced with Pbx1a or control retrovirus were treated with various concentrations of GSI. The results demonstrate that ectopic Pbx1a expression reduces sensitivity to growth inhibition at 1–2 µM of GSI.
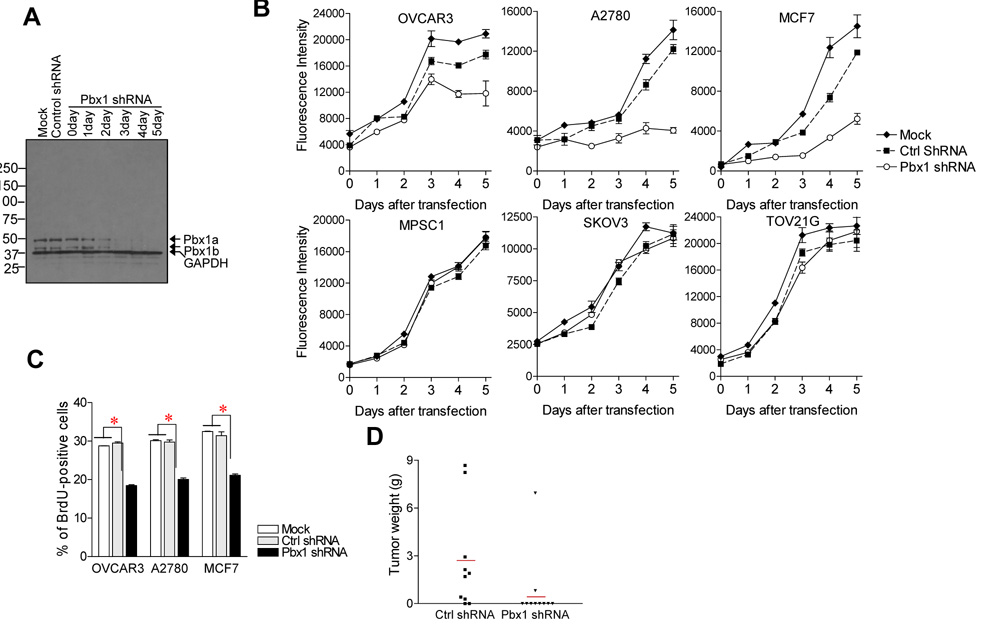
In order to assess if Pbx1 was essential for ovarian cancer cell growth, we examined the effect of Pbx1 shRNA on cell proliferation. Pbx1 shRNA was transfected into A2780, OVCAR3 and MCF7 cells, all of which expressed both Notch3 and Pbx1. The efficiency of Pbx1 shRNA in suppressing Pbx1 protein expression was demonstrated by Western blot analysis (Fig. 6A). Among all three cell lines, as compared to cells with mock transfection or control shRNA transfection, Pbx1 shRNA transfected cells showed a significant reduction in cell number (Fig. 6B). The decrease in cell number in Pbx1 shRNA treated cells was due to suppression of DNA synthesis as evidenced by reduced BrdU uptake (Fig. 6C). Increased apoptosis did not appear to be a factor because the percentage of apoptotic cells was similar between Pbx1 shRNA and control shRNA treated cells (data not shown). In ovarian cancer cells, including MPSC1, SKOV3, and TOV21G, that did not express detectable levels of Pbx1 or Notch3, Pbx1 shRNA did not have significant effects on cell growth (Fig. 6B).
Fig. 6. Pbx1 knockdown by shRNA inhibits cell proliferation.
A: Pbx1 shRNA was found to specifically suppress Pbx1 protein expression during the course of the experiment (2–5 days after transfection). Control shRNA or mock transfection did not have such effect. B: Pbx1 shRNA suppresses the growth of tumor cell lines including OVCAR3, A2780, and MCF7 that express both Notch3 and Pbx1 in vitro. In contrast, Pbx1 shRNA does not show significant growth-inhibitory effects on MPSC1, SKOV3 and TOV21G ovarian cancer cells that do not express detectable levels of Pbx1. C: BrdU uptake assay showed a reduction in DNA synthesis in cancer cells treated with Pbx1 shRNA as compared to control shRNA and mock-transfected groups. *: p< 0.001. D: Pbx1 gene knockdown reduces tumorigenecity of A2780 tumors in nude mice. Red bars: average of tumor weight.
In order to determine the role of Pbx1 on tumor development, we used shRNA to knock down Pbx1 expression in A2780 cells and measured their ability to form subcutaneous tumors in athymic nu/nu mice. We observed a difference in forming subcutaneous tumors between Pbx shRNA versus control shRNA-treated groups (Fig. 6D). Eight out of 10 mice in the control shRNA group grew tumors, while only 2 out of 10 mice in the Pbx1-shRNA-treated group grew tumor (p<0.05, Fisher’s exact test). The two tumors grew in the Pbx1 shRNA-transfected group were examined for Pbx1 expression by immunohistochemistry and quantitative RT-PCR and were found to harbor significant Pbx1 expression. This result suggests that tumor cell formation in these animals was likely the result of clonal selection in favor of Pbx1-expressing cells. However, there is no significant difference in tumor volume between Pbx1 shRNA versus control shRNA-treated group, possibly due to high variance of tumor size.
Discussion
Notch signaling involves in diverse cellular functions including cell fate determination, morphogenesis and oncogenesis which are mediated by unique Notch downstream effectors. Expression of specific Notch downstream genes is tissue and context dependent. Although a repertoire of Notch-controlled genes has been identified, the Notch effectors that are involved in the development of human cancer remain elusive. In this study, we report a novel Notch3-regulated gene, Pbx1, in ovarian cancer cells, and demonstrate that transcriptional regulation of Pbx1 is a direct consequence of Notch3 activation. Our functional studies provide evidence that Pbx1 mediates, at least in part, the effect of Notch activation in the development of ovarian cancer.
Several approaches were used in this study to demonstrate that Pbx1 was the direct downstream target gene of the Notch3 signaling pathway. First, Pbx1 promoter activity increased upon ectopic expression of CSL and NICD3 and decreased after treating cells with either GSI or Notch3-specific siRNA. Furthermore, mutating CSL consensus binding site at Pbx1 promoter leads to an abolishment of Pbx1 promoter activity. Therefore, our data demonstrated a significant association between Notch3 pathway activation and Pbx1 transcription activity. Second, chromatin immunoprecipitation and gel shift assays showed that there was a direct interaction between the NICD3/CSL complex and the Pbx1 promoter sequence containing the consensus CSL binding site. Third, constitutive expression of Pbx1 could rescue cells, at least partially, from the growth suppressive effects of GSI. Furthermore, our recent study using a co-culture system also demonstrated that Pbx1 promoter activity was stimulated upon Notch3 and Jagged-1 binding, and this activity was attenuated by Notch3 siRNA (25).
In this study, GSI treatment in Notch3-expressing cell lines was associated with a time-dependent decrease of NICD3 protein. This observation is similar to those previously reported (22–24). Furthermore, we also found that the expression of full-length Notch3 as well as N-terminal extracellular subunit of Notch3 were also down-regulated by GSI (supplementary Fig. 3B). This could be due to Notch3 transcription is regulated by NICD3 through a positive feedback loop. Accordingly, when NICD3 generation was inhibited by GSI, it also affected the transcription of Notch3, leading to further reduction of the Notch3 protein expression. Indeed, several studies have demonstrated the positive feedback regulation of Notch expression by its own signaling in a variety of organisms including C. elegans (26), Drosophila (27), and mouse (28). Although the above is our preferred explanation, it remains possible that GSI can directly affect transcription of certain genes in addition to its effect in inhibiting γ-secretase.
In this study Pbx1 shRNA was found to inhibit proliferation but did not have a significant effect on apoptosis. Our earlier study, however, demonstrated that inhibiting Notch3 signaling affected both proliferation and apoptosis (8). These results suggest that Pbx1 mediates part, but not all of the effects of the Notch3 pathway activation. Toward this end, we have identified other candidate Notch3-regulated genes in ovarian carcinoma through microarray analysis. Although confirmation is required, it is possible that some of these genes work in concert with Pbx1 to mediate the tumor survival of Notch3 activation.
Pbx1 is a member of the TALE (three-amino acid loop extension) class of homeodomain transcription factors, and can form multimeric transcriptional complexes with other homeodomain proteins such as Hox (29) and MyoD (30), and can participate in a variety of transcriptional regulatory processes. Deletion of Pbx1 resulted in organ hypoplasia and defects in B-cell differentiation indicating that Pbx1 expression is essential for organ development (31–35). Pbx1 was initially identified as a proto-oncogene in human leukemia cells harboring the translocation t(1;19)(q23;p13.3) that produced an oncogenic E2A-Pbx1 fusion protein (20, 36). The fusion of the potent transcriptional activation domain of E2A in chromosome 19 onto the DNA-binding domain of Pbx1 in chromosome 1 contributes to the acute lymphoblastic phenotype by modifying expression of genes normally responsive to Pbx1. In addition to leukemia, Pbx1 is highly expressed in melanoma cells and Pbx1 gene knockdown leads to growth suppression in melanoma cells (17). Consistent with this result, we demonstrated in this report that knockdown of Pbx1 significantly reduced cellular proliferation and tumorigenicity in cancer cells overexpressing both Pbx1 and Notch3, suggesting that Pbx1 expression provides a survival signal in cancer cells that are dependent on Notch3 pathway.
Identification of Pbx1 as a Notch3-regulated gene in cancer cells has the following biological and clinical implications. First, our results further support the view that Notch3 signaling is mediated by different downstream targets in different pathophysiologic settings. In ovarian cancer, Pbx1 appears to be one of the downstream targets that mediate Notch3 function. Second, Pbx1 may function as a molecular hub for different pathways including Notch and Hox. It has been shown that Pbx1 can directly interact with Hox (37) and other homeodomain proteins including Meis (38). The Pbx1-Meis complex functions as a cofactor with Hox to further enhance DNA binding activity (15). The critical role of Hox has been well-documented in a variety of human neoplastic diseases including leukemia (39, 40) and ovarian cancer (41–43). Our findings suggest that Notch3 activation potentially modulates the function of Hox protein through transcriptional upregulation of Pbx1. Further experiments are needed to more fully establish the putative link between Notch3 and Hox in the tumorigenesis of ovarian cancer. Third, the findings that Pbx1 is essential for ovarian cancer cell survival and proliferation suggest that Pbx1 might be a potential target for therapeutics in ovarian cancer in which Notch3 is activated. These tumors may have developed Notch3-Pbx1 pathway-dependent cellular machinery for growth and survival. If so, withdrawal or inhibition of Pbx1 expression may have a potent growth inhibitory effect. The cytotoxicity associated with GSI, especially on the gastrointestinal tract, is a major obstacle of its application to cancer patients (19, 44). Alternative strategies to specifically block Notch downstream targets such as Pbx1 are warrant to be developed.
Supplementary Material
Acknowledgements
This study is supported by Individual Investigator Grant from Ovarian Cancer Research Foundation (TLW), New Investigator Research Grant W81XWH from Department of Defense (TLW), career development award from NIH P50CA098252 (TLW), Institutional Research Grant from American Cancer Society (TLW), and NIH grants CA129080 & CA103937 (IMS). We thank Dr. Michael Cleary at Stanford University for the anti-Pbx1 antibody, Dr. S. Higashiyama at Ehime University in Japan for the pGL3-Pbx1 promoter construct and Dr. Diane Hayward at The Johns Hopkins University for the SG5-flag-CSL plasmid. We also thank Dr. Ching-Hung Lin and Dr. Kuan-Ting Kuo for help in statistical analyses.
Footnotes
References
- 1.Beachy PA, Karhadkar SS, Berman DM. Tissue repair and stem cell renewal in carcinogenesis. Nature. 2004;432:324–331. doi: 10.1038/nature03100. [DOI] [PubMed] [Google Scholar]
- 2.Weng AP, Ferrando AA, Lee W, et al. Activating mutations of NOTCH1 in human T cell acute lymphoblastic leukemia. Science. 2004;306:269–271. doi: 10.1126/science.1102160. [DOI] [PubMed] [Google Scholar]
- 3.Ellisen LW, Bird J, West DC, et al. TAN-1, the human homolog of the Drosophila notch gene, is broken by chromosomal translocations in T lymphoblastic neoplasms. Cell. 1991;66:649–661. doi: 10.1016/0092-8674(91)90111-b. [DOI] [PubMed] [Google Scholar]
- 4.Dang TP, Gazdar AF, Virmani AK, et al. Chromosome 19 translocation, overexpression of Notch3, and human lung cancer. J Natl Cancer Inst. 2000;92:1355–1357. doi: 10.1093/jnci/92.16.1355. [DOI] [PubMed] [Google Scholar]
- 5.Wood LD, Parsons DW, Jones S, et al. The genomic landscapes of human breast and colorectal cancers. Science. 2007;318:1108–1113. doi: 10.1126/science.1145720. [DOI] [PubMed] [Google Scholar]
- 6.Wang TL, Maierhofer C, Speicher MR, et al. Digital karyotyping. Proc Natl Acad Sci U S A. 2002;99:16156–16161. doi: 10.1073/pnas.202610899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Matsuzaki H, Dong S, Loi H, et al. Genotyping over 100,000 SNPs on a pair of oligonucleotide arrays. Nat Methods. 2004;1:109–111. doi: 10.1038/nmeth718. [DOI] [PubMed] [Google Scholar]
- 8.Park JT, Li M, Nakayama N, et al. Notch-3 gene amplification in ovarian cancer. Cancer Res. 2006;66:6312–6318. doi: 10.1158/0008-5472.CAN-05-3610. [DOI] [PubMed] [Google Scholar]
- 9.Weng AP, Millholland JM, Yashiro-Ohtani Y, et al. c-Myc is an important direct target of Notch1 in T-cell acute lymphoblastic leukemia/lymphoma. Genes Dev. 2006;20:2096–2109. doi: 10.1101/gad.1450406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Talora C, Cialfi S, Oliviero C, et al. Cross talk among Notch3, pre-TCR, and Tal1 in T-cell development and leukemogenesis. Blood. 2006;107:3313–3320. doi: 10.1182/blood-2005-07-2823. [DOI] [PubMed] [Google Scholar]
- 11.Jarriault S, Brou C, Logeat F, Schroeter EH, Kopan R, Israel A. Signalling downstream of activated mammalian Notch. Nature. 1995;377:355–358. doi: 10.1038/377355a0. [DOI] [PubMed] [Google Scholar]
- 12.Ronchini C, Capobianco AJ. Induction of cyclin D1 transcription and CDK2 activity by Notch(ic): implication for cell cycle disruption in transformation by Notch(ic) Mol Cell Biol. 2001;21:5925–5934. doi: 10.1128/MCB.21.17.5925-5934.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Buckhaults P, Zhang Z, Chen YC, et al. Identifying tumor origin using a gene expression-based classification map. Cancer Res. 2003;63:4144–4149. [PubMed] [Google Scholar]
- 14.Wysocka J, Swigut T, Xiao H, et al. A PHD finger of NURF couples histone H3 lysine 4 trimethylation with chromatin remodelling. Nature. 2006;442:86–90. doi: 10.1038/nature04815. [DOI] [PubMed] [Google Scholar]
- 15.Jacobs Y, Schnabel CA, Cleary ML. Trimeric association of Hox and TALE homeodomain proteins mediates Hoxb2 hindbrain enhancer activity. Mol Cell Biol. 1999;19:5134–5142. doi: 10.1128/mcb.19.7.5134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dang L, Fan X, Chaudhry A, Wang M, Gaiano N, Eberhart CG. Notch3 signaling initiates choroid plexus tumor formation. Oncogene. 2006;25:487–491. doi: 10.1038/sj.onc.1209074. [DOI] [PubMed] [Google Scholar]
- 17.Shiraishi K, Yamasaki K, Nanba D, et al. Pre-B-cell leukemia transcription factor 1 is a major target of promyelocytic leukemia zinc-finger-mediated melanoma cell growth suppression. Oncogene. 2007;26:339–348. doi: 10.1038/sj.onc.1209800. [DOI] [PubMed] [Google Scholar]
- 18.Geling A, Steiner H, Willem M, Bally-Cuif L, Haass C. A gamma-secretase inhibitor blocks Notch signaling in vivo and causes a severe neurogenic phenotype in zebrafish. EMBO Rep. 2002;3:688–694. doi: 10.1093/embo-reports/kvf124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shih IM, Wang TL. Notch signaling, gamma-secretase inhibitors, and cancer therapy. Cancer Res. 2007;67:1879–1882. doi: 10.1158/0008-5472.CAN-06-3958. [DOI] [PubMed] [Google Scholar]
- 20.Nourse J, Mellentin JD, Galili N, et al. Chromosomal translocation t(1;19) results in synthesis of a homeobox fusion mRNA that codes for a potential chimeric transcription factor. Cell. 1990;60:535–545. doi: 10.1016/0092-8674(90)90657-z. [DOI] [PubMed] [Google Scholar]
- 21.Kamps MP, Murre C, Sun XH, Baltimore D. A new homeobox gene contributes the DNA binding domain of the t(1;19) translocation protein in pre-B ALL. Cell. 1990;60:547–555. doi: 10.1016/0092-8674(90)90658-2. [DOI] [PubMed] [Google Scholar]
- 22.Minter LM, Turley DM, Das P, et al. Inhibitors of gamma-secretase block in vivo and in vitro T helper type 1 polarization by preventing Notch upregulation of Tbx21. Nat Immunol. 2005;6:680–688. [PubMed] [Google Scholar]
- 23.O'Neil J, Grim J, Strack P, et al. FBW7 mutations in leukemic cells mediate NOTCH pathway activation and resistance to gamma-secretase inhibitors. J Exp Med. 2007;204:1813–1824. doi: 10.1084/jem.20070876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lewis HD, Leveridge M, Strack PR, et al. Apoptosis in T cell acute lymphoblastic leukemia cells after cell cycle arrest induced by pharmacological inhibition of notch signaling. Chem Biol. 2007;14:209–219. doi: 10.1016/j.chembiol.2006.12.010. [DOI] [PubMed] [Google Scholar]
- 25.Choi JH, Park JT, Davidson B, Morin PJ, Shih IM, Wang TL. Jagged-1 and Notch3 juxtacrine loop regulates ovarian tumor growth and adhesion. Cancer Res. 2008;68 doi: 10.1158/0008-5472.CAN-08-0001. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wilkinson HA, Fitzgerald K, Greenwald I. Reciprocal changes in expression of the receptor lin-12 and its ligand lag-2 prior to commitment in a C. elegans cell fate decision. Cell. 1994;79:1187–1198. doi: 10.1016/0092-8674(94)90010-8. [DOI] [PubMed] [Google Scholar]
- 27.Carmena A, Buff E, Halfon MS, et al. Reciprocal regulatory interactions between the Notch and Ras signaling pathways in the Drosophila embryonic mesoderm. Dev Biol. 2002;244:226–242. doi: 10.1006/dbio.2002.0606. [DOI] [PubMed] [Google Scholar]
- 28.Robey E, Chang D, Itano A, et al. An activated form of Notch influences the choice between CD4 and CD8 T cell lineages. Cell. 1996;87:483–492. doi: 10.1016/s0092-8674(00)81368-9. [DOI] [PubMed] [Google Scholar]
- 29.Mann RS, Chan S-K. Extra specificity from extradenticle: the partnership between HOX and PBX/EXD homeodomain proteins. Trends in Genetics. 1996;12:258. doi: 10.1016/0168-9525(96)10026-3. [DOI] [PubMed] [Google Scholar]
- 30.Berkes CA, Bergstrom DA, Penn BH, Seaver KJ, Knoepfler PS, Tapscott SJ. Pbx Marks Genes for Activation by MyoD Indicating a Role for a Homeodomain Protein in Establishing Myogenic Potential. Molecular Cell. 2004;14:465. doi: 10.1016/s1097-2765(04)00260-6. [DOI] [PubMed] [Google Scholar]
- 31.Selleri L, Depew MJ, Jacobs Y, et al. Requirement for Pbx1 in skeletal patterning and programming chondrocyte proliferation and differentiation. Development. 2001;128:3543–3557. doi: 10.1242/dev.128.18.3543. [DOI] [PubMed] [Google Scholar]
- 32.DiMartino JF, Selleri L, Traver D, et al. The Hox cofactor and proto-oncogene Pbx1 is required for maintenance of definitive hematopoiesis in the fetal liver. Blood. 2001;98:618–626. doi: 10.1182/blood.v98.3.618. [DOI] [PubMed] [Google Scholar]
- 33.Kim SK, Selleri L, Lee JS, et al. Pbx1 inactivation disrupts pancreas development and in Ipf1-deficient mice promotes diabetes mellitus. Nat Genet. 2002;30:430. doi: 10.1038/ng860. [DOI] [PubMed] [Google Scholar]
- 34.Schnabel CA, Godin RE, Cleary ML. Pbx1 regulates nephrogenesis and ureteric branching in the developing kidney. Developmental Biology. 2003;254:262. doi: 10.1016/s0012-1606(02)00038-6. [DOI] [PubMed] [Google Scholar]
- 35.Sanyal M, Tung JW, Karsunky H, et al. B-cell development fails in the absence of the Pbx1 proto-oncogene. Blood. 2007;109:4191–4199. doi: 10.1182/blood-2006-10-054213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kamps MP, Look AT, Baltimore D. The human t(1;19) translocation in pre-B ALL produces multiple nuclear E2A-Pbx1 fusion proteins with differing transforming potentials. Genes Dev. 1991;5:358–368. doi: 10.1101/gad.5.3.358. [DOI] [PubMed] [Google Scholar]
- 37.LaRonde-LeBlanc NA, Wolberger C. Structure of HoxA9 and Pbx1 bound to DNA: Hox hexapeptide and DNA recognition anterior to posterior. Genes Dev. 2003;17:2060–2072. doi: 10.1101/gad.1103303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schnabel CA, Jacobs Y, Cleary ML. HoxA9-mediated immortalization of myeloid progenitors requires functional interactions with TALE cofactors Pbx and Meis. Oncogene. 2000;19:608–616. doi: 10.1038/sj.onc.1203371. [DOI] [PubMed] [Google Scholar]
- 39.Argiropoulos B, Yung E, Humphries RK. Unraveling the crucial roles of Meis1 in leukemogenesis and normal hematopoiesis. Genes Dev. 2007;21:2845–2849. doi: 10.1101/gad.1619407. [DOI] [PubMed] [Google Scholar]
- 40.Wong P, Iwasaki M, Somervaille TC, So CW, Cleary ML. Meis1 is an essential and rate-limiting regulator of MLL leukemia stem cell potential. Genes Dev. 2007;21:2762–2774. doi: 10.1101/gad.1602107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Naora H, Montz FJ, Chai CY, Roden RB. Aberrant expression of homeobox gene HOXA7 is associated with mullerian-like differentiation of epithelial ovarian tumors and the generation of a specific autologous antibody response. Proc Natl Acad Sci U S A. 2001;98:15209–15214. doi: 10.1073/pnas.011503998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cheng W, Liu J, Yoshida H, Rosen D, Naora H. Lineage infidelity of epithelial ovarian cancers is controlled by HOX genes that specify regional identity in the reproductive tract. Nat Med. 2005;11:531–537. doi: 10.1038/nm1230. [DOI] [PubMed] [Google Scholar]
- 43.Miao J, Wang Z, Provencher H, et al. HOXB13 promotes ovarian cancer progression. Proc Natl Acad Sci U S A. 2007;104:17093–17098. doi: 10.1073/pnas.0707938104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.van Es JH, van Gijn ME, Riccio O, et al. Notch/gamma-secretase inhibition turns proliferative cells in intestinal crypts and adenomas into goblet cells. Nature. 2005;435:959–963. doi: 10.1038/nature03659. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.