Abstract
Chondrocytes express RANKL, but their role in osteoclastogenesis is not clear. We report that hypertrophic chondrocytes induce osteoclast formation through RANKL production stimulated by BMP2 and Runx2/Smad1 and thus they may regulate resorption of calcified matrix by osteoclasts at growth plates.
Introduction
Bone morphogenetic protein (BMP) signaling and Runx2 regulate chondrogenesis during bone development and fracture repair and RANKL expression by osteoblast/stromal cells. Chondrocytes express RANKL, and this expression is stimulated by vitamin D3, but it is not known if chondrocytes directly support osteoclast formation or if BMPs or Runx2 is involved in this potential regulation of osteoclastogenesis.
Material and Methods
The chondrocyte cell line, ATDC5, primary mouse sternal chondrocytes, and chick sternal chondrocytes were used. Cells were treated with BMP2, and expression of RANKL and chondrocyte marker genes was determined by real-time RT-PCR and Western blot. Chondrocytes and spleen-derived osteoclast precursors ± BMP2 were co-cultured to examine the effect of chondrocyte-produced RANKL on osteoclast formation. A reporter assay was used to determine whether BMP2-induced RANKL production is through transcriptional regulation of the RANKL promoter and whether it is mediated by Runx2.
Results
BMP2 significantly increased expression of RANKL mRNA and protein in all three types of chondrocytes, particularly by Col X-expressing and upper sternal chondrocytes. Chondrocytes constitutively induced osteoclast formation. This effect was increased significantly by BMP2 and prevented by RANK:Fc. BMP2 significantly increased luciferase activity of the RANKL-luc reporter, and Smad1 increased this effect. Deletion or mutation of Runx2 binding sites within the RANKL promoter or overexpression of a dominant negative Runx2 abolished BMP2- and Smad1-mediated activation of RANKL promoter activity.
Conclusions
Hypertrophic chondrocytes may regulate osteoclastogenesis at growth plates to remove calcified matrix through BMP-induced RANKL expression.
Key words: osteoclasts, bone morphogenetic protein 2, chondrocytes, Runx2
INTRODUCTION
During embryonic development, the long bones of the skeleton are formed by endochondral ossification, a process in which bones are first formed as cartilage templates, which are replaced subsequently by bone. The process is initiated by aggregation of mesenchymal cells, which differentiate into proliferating chondrocytes to form the cartilaginous templates. In the centers of these elongating templates, the proliferating chondrocytes express predominantly collagen type II and differentiate into type X collagen-expressing hypertrophic chondrocytes. The cartilaginous matrix expressed by hypertrophic chondrocytes in the centers of these templates becomes mineralized and subsequently is invaded by blood vessels to form a medullary cavity. The cartilage is partly replaced by bone, which is laid down by osteoblasts and subsequently is resorbed by osteoclasts to maintain a cavity for bone marrow.(1)
Osteoclasts are multinucleated cells with the ability to resorb mineralized matrix and are formed by fusion of hematopoietic precursor cells in the monocyte/macrophage lineage.(2) Osteoclast formation requires the expression of RANKL by supporting cells, such as marrow stromal cells, osteoblasts, and T cells,(3) and of RANK, a member of the TNF receptor family, by osteoclast precursors. Osteoclast formation and activation are negatively regulated by osteoprotegerin (OPG), a protein that also belongs to the TNF receptor family. OPG is a soluble decoy receptor secreted by osteoblasts and stromal cells that prevents RANKL/RANK interaction by binding to RANKL.(4)
RANKL and OPG are also expressed by hypertrophic chondrocytes in the growth plate during embryonic development.(5) Recently, Masuyama et al.(6) showed, using wildtype (WT) and chondrocyte-specific vitamin D receptor knockout mice, that vitamin D3 stimulates chondrocytes to produce RANKL and thus promotes osteoclast formation. However, little is known about other factors that might influence chondrocyte RANKL expression and its potential role in hypertrophic chondrocyte regulation of osteoclastogenesis. This may be an important function of chondrocytes because, during endochondral bone formation, osteoclast precursors are recruited to the junction between hypertrophic chondrocytes and newly formed bone. There they remodel this bone to facilitate the formation of the marrow cavity and prevent the development of osteopetrosis. Interestingly, osteoclasts are not required for the removal of hypertrophic chondrocytes during vascular invasion or for the resorption of calcified cartilage matrix at the junction between the growth plate and newly formed bone because these take place in mice and humans who do not form osteoclasts.(7–9) These activities presumably are carried out by chondroclasts, which remain poorly defined cells that may be in the mononuclear/phagocyte lineage.(10,11)
Bone morphogenetic proteins (BMPs) and their Smad signaling molecules are important regulators of most aspects of endochondral bone formation during embryonic development, including the appearance and shape of mesenchymal condensations, skeletal patterning, induction of chondrocyte hypertrophy, and joint formation.(12) BMP2 induces expression of several markers of maturation in multiple models of chondrocyte differentiation.(13) It is expressed by osteoblast/stromal cells and directly enhances RANKL expression by these cells. In this way, it induces osteoclast formation and activity of osteoclasts, which express BMP receptors and Smads.(14,15) These reports suggest that BMP2 signaling may influence osteoclastogenesis through both direct and indirect mechanisms. However, it is not known if BMP2 affects chondrocyte expression of RANKL or OPG and thereby osteoclastogenesis.
In this study, we performed a series of in vitro experiments that define chondrocytes as another cell type that directly supports osteoclastogenesis in response to BMP2. BMP2 and its downstream signaling molecule, Smad1, activate the RANKL promoter predominantly in hypertrophic chondrocytes to induce osteoclast formation, an effect that is mediated through Runx2 binding sites in the promoter.
MATERIALS AND METHODS
Reagents
BMP2, human macrophage-colony stimulating factor (M-CSF), and RANK-Fc were purchased from R&D systems (Minneapolis, MN, USA); 1α25-dihydroxyvitamin D3 (vitamin D) was from Roche (Indianapolis, IN, USA); and rabbit anti- RANKL antibody was from Calbiochem (San Diego, CA, USA).
Chondrocyte cultures
Three sources of chondrocytes were used, including ATDC5 cells (cloned from a mouse teratocarcinoma) and primary sternal chondrocytes from mice and chickens.
ATDC5 cells were cultured in a 1:1 mixture of DMEM and Ham's F-12 medium (DMEM/F-12; Invitrogen, Carlsbad, CA, USA) containing 5% FBS (Invitrogen), 50 units/ml penicillin, 50 μg/ml streptomycin, 10 μg/ml human transferrin (Roche Molecular Biochemicals, Indianapolis, IN, USA), and 3 × 10−8 M sodium selenite (Sigma, St Louis, MO, USA) according to published protocols.(16) Primary mouse sternal chondrocytes were isolated from costal cartilage of newborn ICR mice through two enzymatic digestions (3 mg/ml of collagenase D; Sigma). Cells were plated at a density of 10,000 cells/cm2 and cultured for 5–6 days to reach confluence and were treated with BMP2 or PBS for various times in the presence of ascorbic acid (0.05 mg/ml) and β-glycerophosphate (10 mM) according to a published protocol.(16)
Primary chick chondrocytes were isolated from 15-day-old chick embryonic upper and lower sterna by digestion for 4 h at 37°C, with 5% CO2 in Hanks' balanced salt solution (HBSS) containing 0.05% collagenase D (Sigma) and 0.25% trypsin (Sigma) as described previously.(17) Chick chondrocytes were plated in 100-mm plates at a density of 3 × 106 cells/plate. After primary culture for 7 days, cells were harvested, and secondary cultures were placed in 6-well plates at a density of 5 × 105 cells/well for gene expression experiments.
Real-time RT-PCR
Cells were homogenized using 1 ml of TRIzol reagent (Invitrogen), and total RNA was extracted according to the manufacturer's protocol. cDNA was synthesized using 20 μl of reverse transcription reaction solution as we described previously.(18) Quantitative real-time RT-PCR amplifications were performed in an iCycler real-time PCR machine using iQ SYBR Green supermix (both from Bio-Rad Laboratories) according to the manufacturer's instructions. The quantity of mRNA of each gene was normalized using the CT (threshold cycle) value obtained for β-actin or GAPDH mRNA amplifications as we described previously.(18) The sequences for individual primers are shown in Table 1.
Table 1.
Sequences of primers used in the real-time PCR
| Genes | Sequences of primers | GenBank accession number | Target sites on genes | Product sizes (bp) |
| Mouse | ||||
| RANKL | F: 5’ CCAAGATCTCTAACATGACG 3’ | BC125603 | 635–774 | 140 |
| R: 5’ CACCATCAGCTGAAGATAGT 3’ | ||||
| OPG | F: 5’ CAGAGCGAAACACAGTTTG 3’ | U94331 | 697–497 | 201 |
| R: 5’ CACACAGGGTGACATCTATTC 3’ | ||||
| Col2 | F: 5’ GATGACATTATCTGTGAAG 3’ | NM_031163 | 345–494 | 150 |
| R: 5’ ATCTCTGATATCTCCAGG 3’ | ||||
| ColX | F: 5’ CTTTGTGTGCCTTTCAATCG 3’ | NM_009925 | 3540–3677 | 138 |
| R: 5’ GTGAGGTACAGCCTACCAGTT 3’ | ||||
| OC | F: 5’ CTTGGTGCACACCTAGCAGA 3’ | L24429 | 25–176 | 152 |
| R: 5’ ACCTTATTGCCCTCCTGCTT 3’ | ||||
| β-actin | F: 5’ ACCCAGATCATGTTTGAGAC 3’ | X03765 | 280–503 | 224 |
| R: 5’ GTCAGGATCTTCATGAGGTAGT 3’ | ||||
| Chicken | ||||
| RANKL | F: 5’ ACACGCCCTTTGAAAATCAG 3’ | XM425625 | 305–500 | 196 |
| R: 5’ GCAAAAGGTTGCTTCTCTGG 3’ | ||||
| Runx2 | F: 5’ ACTTTGACAATAACTGTCCT 3’ | AF445419 | 511–702 | 192 |
| R: 5’ GACCCCTACTCTCATACTGG 3’ | ||||
| Col2 | F: 5’ AGAAAGGAATCCAGCCCAAT 3’ | NM_204426 | 4384–4621 | 238 |
| R: 5’ ACACCTGCCAGATTGATTCC 3’ | ||||
| ColX | F: 5’ ACATGCATTTACAAATATCGTTAC 3’ | J04194 | 1340–1500 | 161 |
| R: 5’ AAAATAGTAGACGTTACCTTGACTC 3’ | ||||
| GAPDH | F: 5’ TATGATGATATCAAGAGGGTAGT 3’ | K01458 | 813–1011 | 199 |
| R: 5’ TGTATCCAAACTCATTGTCATAC 3’ | ||||
F, forward primer; R, reverse primer.
Western blot analysis
Cells were washed with cold PBS, and whole cell lysates were prepared by the addition of M-PER mammalian protein extraction reagent (Pierce, Rockford, IL, USA) containing a protease inhibitor mixture (Roche Applied Science). Twenty micrograms of protein was loaded per lane and separated on a 10% polyacrylamide gel, followed by transfer to a nitrocellulose membrane (Bio-Rad, Hercules, CA, USA) by electroblotting. Membranes were blocked for nonspecific binding in 3% nonfat dry milk and followed by incubation with an antibody at 4°C. After membranes were washed, the blots were probed with a horseradish peroxidase–conjugated secondary antibody (Bio-Rad) and visualized by an enhanced chemiluminescence system (Amersham Biosciences, Buckinghamshire, UK) according to the manufacturer's instructions. The intensity of bands in the film was determined by densitometric analysis using NIH Image software.
In vitro osteoclastogenesis assay
ATDC5 or primary mouse sternal chondrocytes (5000 cells/well) were plated in 48-well plates for 12 h and treated with or without BMP2 (100 ng/ml). Spleen cells were isolated from 2-mo-old C57BL/6 mice and cultured with M-CSF (10 ng/ml) for 3 days to generate osteoclast precursors as we described previously.(19) These osteoclast precursors (50,000 cells/well) were removed and placed on top of the BMP2-treated chondrocytes. Cells were co-cultured for an additional 10 days in the presence of M-CSF with or without BMP2. In some experiments, RANK-Fc (50 ng/ml) was added to block RANKL signaling. Cells were fixed with formalin and stained for TRACP activity. Multinucleated TRACP+ cells were counted as osteoclasts. For resorption pit formation assays, chondrocytes and splenocytes were co-cultured on slices of bovine cortical bone in 96-well plates. After 14 days of culture, bone slices were washed with PBS and stained with 0.1% toluidine blue. Pit numbers and area were quantified under a light microscope using an eyepiece reticule and standard stereometry methods.
Plasmid construction and luciferase assay
The following plasmid constructs were used in transient transfection assays. We used Smad1, Smad6, Smurf1, and dominant negative Runx2 expression vectors, as described previously.(18,20) The RANKL-luciferase reporter constructs and constructs with mutated Runx2 binding sites in RANKL promoter plasmids were obtained from Dr Charles O'Brien.(21) The nested deletion constructs of RANKL-luciferase plasmids were generated by PCR amplification of the cloned DNA fragments according to the sequence of the mouse rankl promoter (accession number BC125603). The following oligonucleotides were used to make deletion mutants: −649/−630 to +111, 5′-TTAGCTCTGCTAACAGCTC-3′; −227/−208 to +111, 5′-AAAGGATAGGGGCCAGCCT-3′; −144/−125 to +111, 5′-AAGGAAAGGAAGGAGGGCA-3′. All mutated constructs were verified by DNA sequence analysis. ATDC5 cells were co-transfected with 0.5 μg RANKL-luciferase reporter, 0.05 μg pRL-Renilla vector as an internal control, and 0.45 μg expression vector using FuGENE6 (Roche). Cells were treated with PBS or BMP2 (100 ng/ml) for 48 h. Luciferase activities were assayed using a Dual-Luciferase Reporter Assay System (Promega, Madison, WI, USA) according to the manufacturer's instructions.
Statistical evaluations
The data are presented as mean ± SE of three experiments, and all experiments were performed at least twice. Statistical analysis was performed using Mann-Whitney's U-test with Statview software; p < 0.05 was considered statistically significant.
RESULTS
BMP2 stimulates RANKL expression in chondrocytes
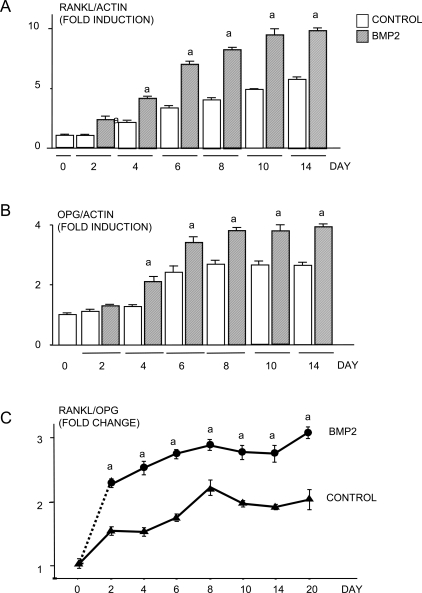
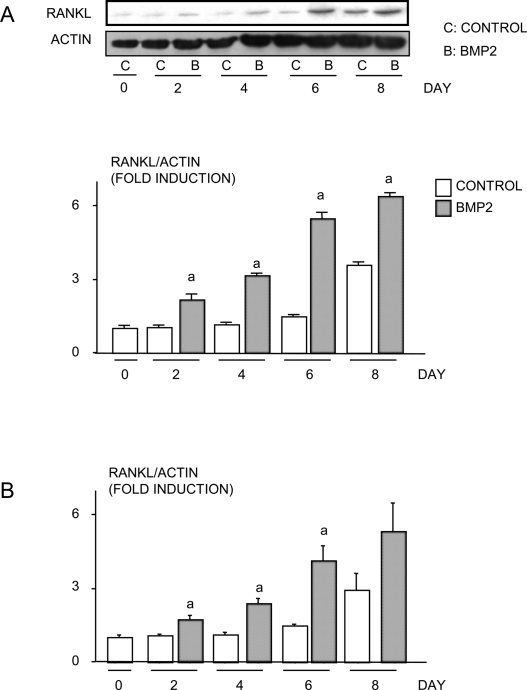
To determine whether BMP2 stimulates chondrocytes to produce RANKL, ATDC5 cells were treated with or without BMP2 (100 ng/ml) for various times. RANKL mRNA levels assessed using real-time RT-PCR increased progressively with time in control groups, and BMP2 treatment enhanced RANKL expression levels at each time-point (Fig. 1A). OPG mRNA expression levels also increased in control groups with time, and this effect was enhanced by BMP2 (Fig. 1B). Because BMP2 increased expression of both RANKL and OPG, we calculated the RANKL/OPG ratio in these cells, because this ratio is known to be a major determinant of osteoblast-regulated osteoclastogenesis,(22) and found that it was increased significantly. Furthermore, BMP2 treatment increased RANKL protein levels in both ATDC5 cells and primary mouse sternal chondrocytes (Figs. 2A and 2B).
FIG. 1.
BMP2 increases RANKL mRNA production in chondrocytes. ATDC5 cells were treated with BMP2 (100 ng/ml) for various times. The expression of (A) RANKL, (B) OPG, and β-actin mRNA was determined by real-time RT-PCR. The relative expression levels of RANKL and OPG were normalized by β-actin in the same samples. The fold induction of RANKL mRNA was calculated as follows: relative RANKL level in a given time/relative RANKL level in day 0. The RANKL/OPG ratio in each time-point was calculated by dividing relative RANKL level with the relative OPG level in the same sample (C). The cells treated with BMP2 at the various times indicated came from the same pool of cells as the control cultures. This is indicated by the broken line from 0 to 2 h. a p < 0.05 vs. control value at same time-point.
FIG. 2.
BMP2 increases RANKL protein expression in chondrocytes. (A) ATDC5 cells or (B) primary mouse sternal chondrocytes were treated with or without BMP2 (100 ng/ml) for various times. RANKL and β-actin protein levels were examined by Western blot analysis. A representative Western blot from ATDC5 cells is shown (top panels, A). Quantitation of protein bands was performed by densitometry using NIH image. The relative expression levels of RANKL protein were normalized by β-actin protein levels. The fold induction was measured by the following: the intensity of relative RANKL protein expression at each time-point/the intensity of relative RANKL protein expression in the day 0 sample. a p < 0.05 vs. control value at same time-point.
Chondrocytes support osteoclastogenesis through RANKL production
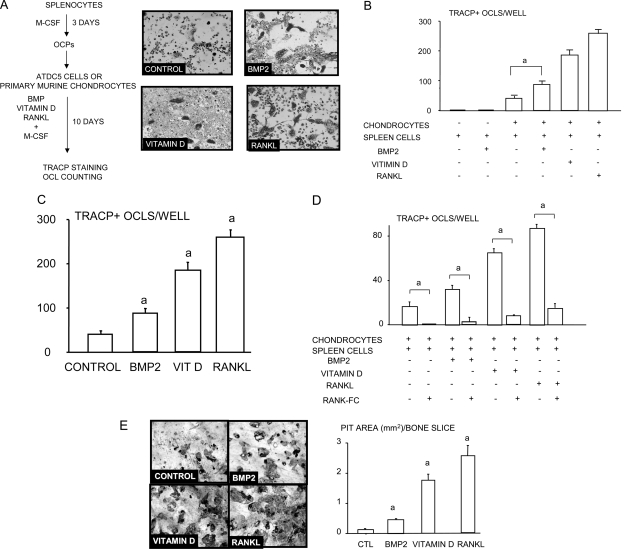
To determine whether chondrocytes can directly promote differentiation of osteoclast precursors to mature osteoclasts, we co-cultured ADTC5 cells or primary mouse sternal chondrocytes with spleen-derived WT murine osteoclast precursors (Fig. 3A). Spleen cells cultured alone or with BMP2 failed to form osteoclasts, as expected. However, osteoclasts formed spontaneously in co-cultures of chondrocytes and spleen cells in the absence of RANKL or BMP2 (Fig. 3A). Addition of BMP2 significantly increased osteoclast numbers in these co-cultures, although the numbers formed were lower than those treated with vitamin D or RANKL, which were used as positive controls in the ADTC5 (Fig. 3B) or primary mouse sternal chondrocyte cultures (Fig. 3C). Furthermore, RANK-Fc significantly inhibited osteoclast formation in all groups including chondrocyte/spleen co-cultures alone and co-cultures with BMP2, vitamin D3, or RANKL (Fig. 3D), indicating that RANKL is essential for chondrocyte-induced osteoclast formation. The osteoclasts formed in these co-cultures also formed resorption pits after 15 days, and BMP-2 treatment significantly increased the area of pits formed as did RANKL and vitamin D3, consistent with their effects on osteoclast numbers (Fig. 3E).
FIG. 3.
BMP2-stimulated chondrocytes induce osteoclast formation in vitro. ATDC5 cells were plated in subconfluent condition and were treated with or without BMP2 for 12 h. Spleen cells were cultured with conditioned medium containing M-CSF (1:20) for 3 days to generate osteoclast precursors. Osteoclast precursors were seeded on top of the chondrocytes. Co-cultures were maintained for an additional 10 days in the presence of BMP2, vitamin D3, or RANKL. Osteoclasts were detected by TRACP staining. Representative pictures from the co-cultures (A). The number of osteoclasts in BMP2-, vitamin D3– or RANKL-treated groups (B) and in RANK:Fc-treated groups (C). Co-cultures of chondrocytes and splenocytes were performed on bone slices for 14 days. After cells were brushed off, dentine slices were stained with 0.1% toluidine blue, and the total area of the pits per slice was measured (D). The same experiments were repeated twice with similar results. a p < 0.05 where indicated in B and C and vs. the control value in D.
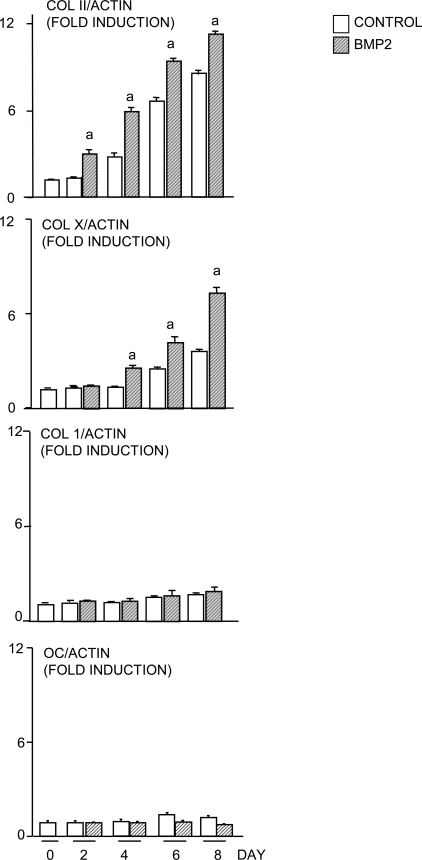
BMP2 is known to stimulate RANKL production by osteoblasts.(23) To eliminate the possibility that ATDC5 cells contain osteoblast precursors, we examined expression of cell lineage markers using cells that have been cultured with BMP2 for 8 days, the same length of time as the co-cultures on plastic. Type II and type X collagens were used as markers for proliferating and hypertrophic chondrocytes, respectively. Type 1 collagen and osteocalcin were used as markers for osteoblasts. We found that the expression of type II collagen increased until day 6 and decreased in the presence and absence of BMP2, whereas type X collagen expression increased later than that of type II and remained elevated at 8 days. However, we found no change in type 1 collagen or osteocalcin expression in response to BMP2, confirming the chondrocytic nature of ATDC5 cells (Fig. 4). We also examined expression of bone sialoprotein (BSP) expression in primary chick upper and lower chondrocytes in response to BMP2. We found no response to BMP2 in the lower sternal chondrocytes, but we did observe a significant increase in BSP expression in response to BMP2 after 2 days of treatment of upper sternal chondrocytes. This effect was not sustained after 4 days when BSP expression was also increased in the control cultures, consistent with published data that hypertrophic chondrocytes express BSP and that BSP is a maturation marker for chondrocytes.(24,25)
FIG. 4.
ATDC5 cells have characteristics of hypertrophic chondrocytes and not osteoblasts. ATDC5 cells were treated with BMP2 (100 ng/ml) for various times. The expression levels of Col II, Col X, Col 1, osteocalcin, and β-actin mRNA were determined by real-time RT-PCR. The relative expression levels of Col II, Col X, Col I, and osteocalcin were normalized by β-actin levels in the same sample. The fold induction level of individual genes was calculated as described in Fig. 1. a p < 0.05 vs. control value at same time-point.
BMP2 signaling–induced RANKL expression is mediated through the Runx2-binding sites in the RANKL promoter
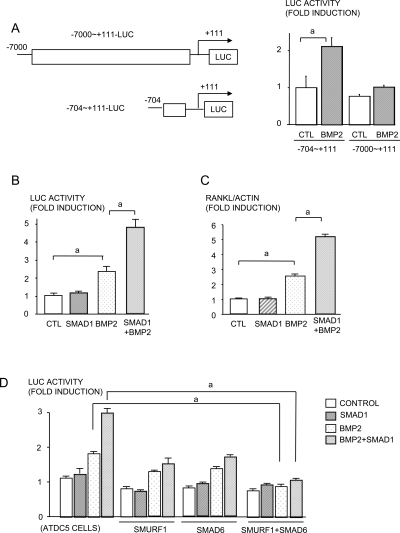
To explore the molecular mechanism by which BMP2 induces RANKL expression, we transiently transfected a RANKL-luciferase reporter construct,(21) which contains the −704 to +111 region of the promoter (−704-luc) into ADTC5 cells and found that BMP2 significantly increased luciferase activity in the cells (Fig. 5A), We used this −704-luc reporter construct in subsequent studies.
FIG. 5.
RANKL activation is regulated by BMP signaling. A schematic presentation of RANKL promoter reporters. The top panel is the long promoter reporter (−7000 to +111 bp) and the bottom panel is the short promoter reporter (−704 to +111 bp) (A). ATDC5 cells were transfected with RANKL promoter plasmids (−704 to +111 and −7000 to +111) and treated with BMP2 (100 ng/ml) for 48 h. A luciferase assay was conducted (B). ATDC5 cells were co-transfected with short-form RANKL promoter plasmids (−704 to +111) and a Smad1 expression vector in the presence or absence of BMP2 (100 ng/ml) for 48 h. A luciferase assay was performed (C), and RANKL mRNA expression was determined by real-time RT-PCR (D). ATDC5 cells were co-transfected with RANKL-Luc, Smurf1, Smad6, and Smad1 and were treated with BMP2. A luciferase assay was performed (E). a p < 0.05.
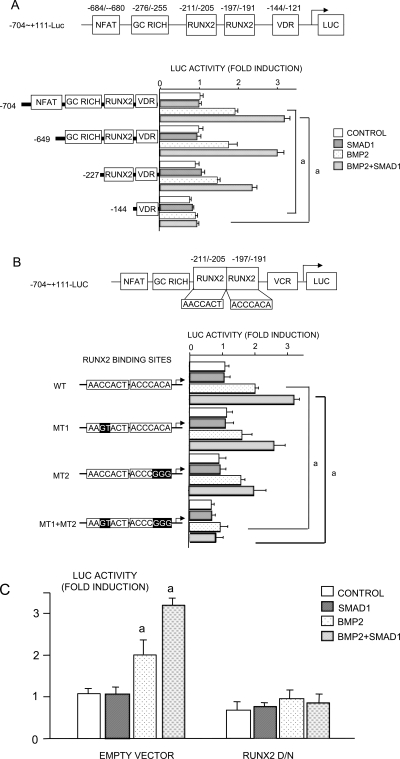
Smad1 is the major transcription factor that is positively regulated by BMP signaling,(26) Overexpression of Smad1 alone had no effect on luciferase activity of the −740-luc construct, but Smad1 plus BMP2 enhanced BMP2-induced activation of this reporter synergistically (Fig. 5B), which was confirmed at the mRNA level (Fig. 5C). Smad1 is phosphorylated after it is recruited to the type I BMP receptor and translocates to the nucleus where it interacts with other transcription factors, particularly Runx2, to regulate transcription of target genes.(27) Smurf1, an E3 ubiquitin ligase, negatively regulates BMP signaling by promoting degradation of Smad1 and Runx2 proteins and Smurf1-mediated Runx2 degradation can be enhanced by Smad6 overexpression.(20) Overexpression of Smurf1 or Smad6 alone significantly reduced the effect of BMP2 and of BMP2/Smad1 on −704-luc activation (Fig. 5D). However, Smurf1 plus Smad6 overexpression completely abolished RANKL activation by BMP2 or BMP2/Smad1 (Fig. 5D). There are two adjacent Runx2 binding sequences within the −704 to +110 region of the RANKL promoter. However, overexpression of Runx2 alone does not affect the basal level of RANKL expression in osteoblast/stromal cells, suggesting that other factors must interact with Runx2 to affect RANKL gene transcription.(21) Smad1 enhances BMP2-induced RANKL expression and may be one of these factors. To test this, we assessed putative binding sites for transcription factors within the −704 to +110 region of the RANKL promoter and identified a GC rich region located in the −276 to −255 region, which could be a Smad binding site (Fig. 6A). We also confirmed the presence of NFAT, vitamin D receptor, and two Runx2-binding sites, which have been characterized previously.(21) We generated three deletion mutants of these constructs and tested the effect of BMP2/Smad1 on their activation (Fig. 6A). Deletion of the region containing the NFAT binding sequence and the GC-rich region did not change RANKL promoter activity. However, deletion of the region containing the NFAT, GC-rich regions, and the Runx2-binding sites abolished BMP2 or BMP2/Smad1-induced RANKL promoter activity altogether (Fig. 6A), suggesting that the BMP/Smad response is through the Runx2 regions and not through the GC-rich region of the proximal RANKL promoter.
FIG. 6.
Runx2 is required for BMP2 and Smad1-induced RANKL activation. ATDC5 cells were transiently co-transfected with Smad1 expression vector and RANKL reporter deletion mutants (A), RANKL reporter with point mutations in the Runx2-binding sequence (B), or dominant negative Runx2 ± Smad1 expression vector and RANKL reporter (C). Cells were treated with BMP2 (100 ng/ml) for 48 h, and a luciferase assay was performed. a p < 0.05 where indicated in A and B and against control value in C.
To confirm that Runx2 binding sites are critical for BMP signaling–induced RANKL expression, we used three reporter plasmids where the two Runx2 binding sites were mutated alone or together. Mutation of either Runx2-binding site alone did not affect BMP-induced RANKL promoter activity (Fig. 6B), whereas mutations in both Runx2 binding sites inhibited this activity completely (Fig. 6B). Furthermore, overexpression of a dominant negative version of Runx2 completely blocked BMP2 and Smad1-induced RANKL promoter activity (Fig. 6C). Thus, Runx2 is responsible for BMP-induced RANKL activation.
Hypertrophic chondrocytes respond to BMP2 to produce RANKL
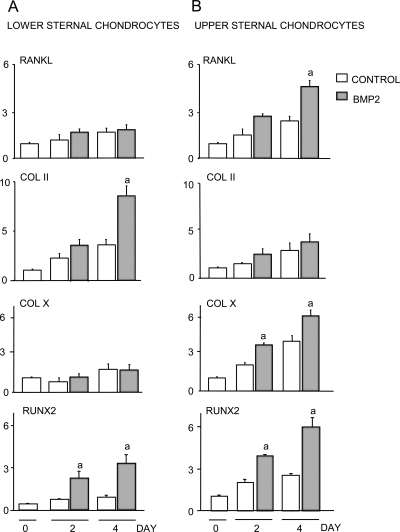
In growth plates, chondrocytes are organized from proliferating chondrocytes to hypertrophic chondrocytes, and expression of RANKL protein is detected predominantly in hypertrophic chondrocytes.(5) Thus, we treated primary chondrocytes from chick sterna with BMP2 because cells isolated from the upper sternum are undergoing hypertrophic differentiation, whereas those in the lower sternum are immature, proliferating chondrocytes.(28) As expected, lower sternal proliferating chondrocytes express high levels of type II collagen and low levels of type X collagen (Fig. 7A), whereas upper sternal hypertrophic chondrocytes have high type X and low type II collagen expression (Fig. 7B). BMP2 or BMP2/Smad1 overexpression stimulated upper sternal chondrocytes to produce RANKL, and this was associated with higher basal and BMP2-stimulated Runx2 expression (Fig. 7B). In contrast, BMP2 did not affect RANKL expression in lower sternal chondrocytes, which have lower Runx2 mRNA (Fig. 7A).
FIG. 7.
BMP2 stimulates chicken upper sternal chondrocytes to express RANKL. Lower sternal chondrocytes (A) and upper chondrocytes (B) from 15-day-old chick embryos were isolated and plated in 6-well plates for 7 days. The cells were treated with BMP2 (100 ng/ml) for various times. The expression levels of RANKL, Col II, Col X, Runx2, and GAPDH mRNA were determined by real-time RT-PCR. The fold induction of levels of individual genes was calculated as described in Fig. 1. a p < 0.05 vs. control value at the same time-point.
DISCUSSION
In this study, we provided the first evidence that chondrocytes express RANKL constitutively in vitro in amounts sufficient for them to support osteoclastogenesis directly from splenic precursors when both cell types are cultured together in the presence of M-CSF but no other cytokines. BMP2 increased this osteoclastogenic action of chondrocytes by significantly increasing RANKL expression by the chondrocyte cell line, ATDC5, and by primary chondrocytes isolated from neonatal mice. Published immunohistochemical and in situ hybridization data have shown that BMP2 is expressed by proliferating chondrocytes and that RANKL is expressed by hypertrophic chondrocytes in murine growth plates.(29,30) We confirmed this RANKL protein expression by hypertrophic chondrocytes using immunohistochemistry (data not shown). We found that chicken sternal hypertrophic chondrocytes express higher levels of RANKL both in control and BMP2-treated cultures than proliferating chondrocytes, supporting these observations. Thus, our studies suggest a new role for BMP2 to regulate osteoclast formation at the growth plate during endochondral ossification.
Because bone growth during embryonic development and in the first 2 wk after birth occurs at a very high rate, it is essential that osteoclasts are induced to form at the growth plate and rapidly remove calcified matrix to maintain the development of a medullary cavity and prevent osteopetrosis. Hypertrophic chondrocytes are the cells in embryonic bones and in growth plates that are closest to the blood vessels that bring osteoclast precursors from the circulation into developing bones. They secrete vascular endothelial growth factor (VEGF) and other factors that direct vascular ingrowth. In addition to promoting osteoclast formation, chondrocytes could also promote osteoclast precursor egression from these newly formed blood vessels through RANKL expression. For example, RANKL has been shown to have a chemo-attractant effect on peripheral blood monocytes.(31,32) Thus, local release of RANKL by chondrocytes and induction of its expression by BMP2 could serve this essential function. Endothelial cells also express RANKL,(33) and these are present close by hypertrophic chondrocytes and osteoclasts at the growth plate. Thus, they too could be closely involved in the recruitment of osteoclasts to this site during endochondral ossification. However, further studies will be required to determine whether the endothelial cells next to the growth plate express higher levels of RANKL than those distant from this site within the marrow cavity to account for the accumulation of osteoclasts close by hypertrophic chondrocytes as we propose here.
Osteoblastic cells in bone marrow also express RANKL. Thus, one potential concern about our findings is that our chondrocyte cultures could be contaminated by osteoblastic cells to account for the effects we observed. We found no change in osteocalcin expression in ATDC5 cells in response to BMP during the first 8 days of culture when type X collagen expression was already increased, suggesting that these cells are not contaminated by osteoblastic cells. However, we and others have reported that some chondrocytes can increase their expression of osteocalcin as they mature.(34,35) We also examined expression of BSP, another putative osteoblast marker by primary chick chondrocytes, and found that BMP2 transiently increased its expression in upper sternal chondrocytes after 2 days and before it had any effect on type X collagen expression. However, BSP expression also increased in the control and BMP2-treated cultures of these cells 2 days later to similar degrees. Interestingly, although BSP has been used as a marker of osteoblastic differentiation by many investigators, as proliferating chondrocytes differentiate to hypertrophic chondrocytes, their BSP levels also go up, and in some studies, BSP has been used as a maturation marker for chondrocytes.(24,25) Thus, although it could be concluded that the changes we observed in expression levels of osteocalcin or BSP do not entirely exclude the possibility of some osteoblastic cells being in our cultures, overall, our findings support our contention that hypertrophic chondrocytes increase their expression of RANKL in response to BMP2 and that the cells we studied are chondrocytic rather than osteoblastic.
We found that induction of RANKL by BMP-2 is mediated by Runx2, and this effect was augmented synergistically in chondrocytes by overexpression of Smad1. We used the −704 to +111 region of the RANKL promoter to show BMP2 induction of RANKL. It has a number of binding domains, including two Runx2-binding sites, a vitamin D receptor binding site, and a single NFAT and putative Smad binding domains. Using deletion constructs of this promoter, we showed that the Runx2-binding domains, but none of the other domains, were required for BMP2 induction of RANKL. Interestingly, overexpression of a WT or a dominant negative form of Runx2 had no effect on basal promoter activity, suggesting that Runx2 must work with other transcription factors to activate the RANKL promoter. We also found that, like Runx2, Smad1 overexpression alone does not affect RANKL promoter activity, but Smad1 and Runx2 together significantly increase RANKL activation to a degree similar to that of BMP2 (Fig. 5C). Furthermore, Smad1 acted synergistically with BMP2 to increase RANKL expression. These findings suggest that Runx2 interacts with Smad1 and that Smad1 does not need to interact directly with DNA to induce RANKL expression.
Runx2 and Smad1 have been found to act cooperatively to mediate the effect of BMP2 on promoters of target genes through different mechanisms.(36–38) For example, elimination of either Runx2 or Smad1 binding sites from the type X collagen promoter abolishes responsiveness to BMP2,(36) whereas only the Runx2 binding site is needed in the Smad6 promoter.(37) Our findings suggest that BMP2-regulated RANKL promoter activity may work similarly to this latter mechanism. Expression of Runx2 is required for the invasion of hypertrophic chondrocytes by blood vessels and osteoclasts in developing bones during endochondral ossification because this process fails to initiate in Runx2−/− mice(39–41) Consequently Runx2−/− mice do not form bone or a marrow cavity. Thus, an essential role for Runx2 signaling in chondrocytes seems to be induction of cartilage invasion by endothelial cells. Our findings suggest that it also promotes resorption of calcified hypertrophic cartilage by osteoclasts.
Although osteoblast/stromal cells are considered to be the major regulators of osteoclast formation during bone development through their expression of M-CSF and RANKL, a role for chondrocytes to directly induce osteoclast formation through vitamin D3 induction of RANKL expression has been reported recently.(6) We also found that vitamin D3 increases RANKL expression in ATDC5 clonal chondrocytes and in primary sternal chondrocytes and that BMP2 increased this RANKL induction and osteoclast formation (Fig. 5B). However, unlike us, these investigators did not find that chondrocytes supported osteoclast formation constitutively. This may represent differences in the experimental conditions. Smads have been shown to interact with the vitamin D receptor.(42,43) Thus, BMP 2 and vitamin D could act cooperatively by this mechanism to induce RANKL expression during endochondral ossification.
BMP2 has been reported to directly enhance osteoclast activity(44) and to stimulate formation of osteoclasts derived from bone marrow macrophages by enhancing the effect of RANKL.(14,15) However, there are no published data that BMP2 induces osteoclast formation directly. We confirmed that BMP2 does not induce osteoclast formation directly using spleen cells. We also found that BMP2 does not induce M-CSF expression in ATDC5 cells, although it did upregulate its expression in pre-osteoblastic C2C12 cells. The role of BMPs in endochondral ossification has been studied extensively using a variety of genetic manipulations, and these have shown a variety of functions for BMP signaling.(12) For example, Smad6 transgenic and Smurf1/Smad6 double transgenic mice generated recently using the type II collagen promoter to inhibit BMP signaling in chondrocytes have delayed chondrocyte hypertrophy and mineralization and dwarfism.(45) They also have osteopenia 3 wk after birth, associated with reduced bone formation rates and increased osteoclast numbers in their metaphyses, suggesting that reduced BMP signaling in chondrocytes might stimulate rather than reduce osteoclast formation. However, osteoclast formation from bone marrow cells from these mice was normal, and this study did not report osteoclast numbers at the growth plates of the mice during embryonic development when chondrocyte hypertrophy and mineralization were delayed. Thus, the mechanism whereby inhibition of BMP signaling in chondrocytes increased osteoclast numbers in metaphyseal bone and reduced bone formation in these Smad6 transgenic mice remains unexplained.
In conclusion, our findings showed that BMP2 induces RANKL expression in hypertrophic chondrocytes and regulates osteoclast formation and bone remodeling during long bone growth. This novel finding further supports the integration of signals that control chondrocyte maturation and bone remodeling in the developing limb.
ACKNOWLEDGMENTS
The authors thank Dr C O'Brien (University of Arkansas for Medical Sciences) for providing RANKL-luciferase reporter constructs; Dr Hiromu Ito (Kyoto University) for providing ATDC5 cells; Dr Sunao Takeshita (National Institute for Longevity Sciences, Japan) for an M-CSF–producing cell line; and Yan Lu and Xiaoyun Zhang for technical assistance with the histology. This study was funded by National Institutes of Health Grants PHS AR43510 and AR49305 to BFB and AR 48697 to LX and the Research Fellowships of the Japan Society for the Promotion of Science for Young Scientists (to MU).
Footnotes
The authors state that they have no conflicts of interest.
REFERENCES
- 1.van der Eerden BC, Karperien M, Wit JM. Systemic and local regulation of the growth plate. Endocr Rev. 2003;24:782–801. doi: 10.1210/er.2002-0033. [DOI] [PubMed] [Google Scholar]
- 2.Boyle WJ, Simonet WS, Lacey DL. Osteoclast differentiation and activation. Nature. 2003;423:337–342. doi: 10.1038/nature01658. [DOI] [PubMed] [Google Scholar]
- 3.Yasuda H, Shima N, Nakagawa N, Yamaguchi K, Kinosaki M, Mochizuki S, Tomoyasu A, Yano K, Goto M, Murakami A, Tsuda E, Morinaga T, Higashio K, Udagawa N, Takahashi N, Suda T. Osteoclast differentiation factor is a ligand for osteoprotegerin/osteoclastogenesis-inhibitory factor and is identical to TRANCE/RANKL. Proc Natl Acad Sci USA. 1998;95:3597–3602. doi: 10.1073/pnas.95.7.3597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Simonet WS, Lacey DL, Dunstan CR, Kelley M, Chang MS, Lüthy R, Nguyen HQ, Wooden S, Bennett L, Boone T, Shimamoto G, DeRose M, Elliott R, Colombero A, Tan HL, Trail G, Sullivan J, Davy E, Bucay N, Renshaw-Gegg L, Hughes TM, Hill D, Pattison W, Campbell P, Sander S, Van G, Tarpley J, Derby P, Lee P, Boyle WJ. Osteoprotegerin: A novel secreted protein involved in the regulation of bone density. Cell. 1997;89:309–319. doi: 10.1016/s0092-8674(00)80209-3. [DOI] [PubMed] [Google Scholar]
- 5.Kishimoto K, Kitazawa R, Kurosaka M, Maeda S, Kitazawa S. Expression profile of genes related to osteoclastogenesis in mouse growth plate and articular cartilage. Histochem Cell Biol. 2006;125:593–602. doi: 10.1007/s00418-005-0103-z. [DOI] [PubMed] [Google Scholar]
- 6.Masuyama R, Stockmans I, Torrekens S, Van Looveren R, Maes C, Carmeliet P, Bouillon R, Carmeliet G. Vitamin D receptor in chondrocytes promotes osteoclastogenesis and regulates FGF23 production in osteoblasts. J Clin Invest. 2006;116:3150–3159. doi: 10.1172/JCI29463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kong YY, Yoshida H, Sarosi I, Tan HL, Timms E, Capparelli C, Morony S, Oliveira-dos-Santos AJ, Van G, Itie A, Khoo W, Wakeham A, Dunstan CR, Lacey DL, Mak TW, Boyle WJ, Penninger JM. OPGL is a key regulator of osteoclastogenesis, lymphocyte development and lymph-node organogenesis. Nature. 1999;397:315–323. doi: 10.1038/16852. [DOI] [PubMed] [Google Scholar]
- 8.Franzoso G, Carlson L, Xing L, Poljak L, Shores EW, Brown KD, Leonardi A, Tran T, Boyce BF, Siebenlist U. Requirement for NF-kappaB in osteoclast and B-cell development. Genes Dev. 1997;11:3482–3496. doi: 10.1101/gad.11.24.3482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Roodman GD, Windle JJ. Paget disease of bone. J Clin Invest. 2005;115:200–208. doi: 10.1172/JCI24281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Takuma S. Electron microscopy of cartilage resorption by chondroclasts. J Dent Res. 1962;41:883–889. doi: 10.1177/00220345620410042101. [DOI] [PubMed] [Google Scholar]
- 11.Bettex-Galland M, Boillat C, Bettex M. Chondroclasts in osteoneogenesis. Tissue Cell. 1990;22:93–100. doi: 10.1016/0040-8166(90)90093-o. [DOI] [PubMed] [Google Scholar]
- 12.Tsumaki N, Yoshikawa H. The role of bone morphogenetic proteins in endochondral bone formation. Cytokine Growth Factor Rev. 2005;16:279–285. doi: 10.1016/j.cytogfr.2005.04.001. [DOI] [PubMed] [Google Scholar]
- 13.Chen D, Zhao M, Mundy GR. Bone morphogenetic proteins. Growth Factors. 2004;22:233–241. doi: 10.1080/08977190412331279890. [DOI] [PubMed] [Google Scholar]
- 14.Kaneko H, Arakawa T, Mano H, Kaneda T, Ogasawara A, Nakagawa M, Toyama Y, Yabe Y, Kumegawa M, Hakeda Y. Direct stimulation of osteoclastic bone resorption by bone morphogenetic protein (BMP)-2 and expression of BMP receptors in mature osteoclasts. Bone. 2000;27:479–486. doi: 10.1016/s8756-3282(00)00358-6. [DOI] [PubMed] [Google Scholar]
- 15.Itoh K, Udagawa N, Katagiri T, Iemura S, Ueno N, Yasuda H, Higashio K, Quinn JMW, Gillespie MT, Martin TJ, Suda T, Takahashi N. Bone morphogenetic protein 2 stimulates osteoclast differentiation and survival supported by receptor activator of nuclear factor-kappaB ligand. Endocrinology. 2001;142:3656–3662. doi: 10.1210/endo.142.8.8300. [DOI] [PubMed] [Google Scholar]
- 16.Shukunami C, Shigeno C, Atsumi T, Ishizeki K, Suzuki F, Hiraki Y. Chondrogenic differentiation of clonal mouse embryonic cell line ATDC5 in vitro: Differentiation-dependent gene expression of parathyroid hormone (PTH)/PTH-related peptide receptor. J Cell Biol. 1996;133:457–468. doi: 10.1083/jcb.133.2.457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Grimsrud CD, Romano PR, D'Souza M, Puzas JE, Reynolds PR, Rosier RN, O'Keefe RJ. BMP-6 is an autocrine stimulator of chondrocyte differentiation. J Bone Miner Res. 1999;14:475–482. doi: 10.1359/jbmr.1999.14.4.475. [DOI] [PubMed] [Google Scholar]
- 18.Kaneki H, Guo R, Di Chen D, Yao Z, Schwarz EM, Zhang YE, Boyce BF, Xing L. Tumor necrosis factor promotes Runx2 degradation through up-regulation of Smurf1 and Smurf2 in osteoblasts. J Biol Chem. 2006;281:4326–4333. doi: 10.1074/jbc.M509430200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Xing L, Bushnell TP, Carlson L, Tai Z, Tondravi M, Siebenlist U, Young F, Boyce BF. NF-kappaB p50 and p52 expression is not required for RANK-expressing osteoclast progenitor formation but is essential for RANK- and cytokine-mediated osteoclastogenesis. J Bone Miner Res. 2002;17:1200–1210. doi: 10.1359/jbmr.2002.17.7.1200. [DOI] [PubMed] [Google Scholar]
- 20.Shen R, Chen M, Wang YJ, Kaneki H, Xing L, O'Keefe RJ, Chen D. Smad6 interacts with Runx2 and mediates Smad ubiquitin regulatory factor 1-induced Runx2 degradation. J Biol Chem. 2006;281:3569–3576. doi: 10.1074/jbc.M506761200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.O'Brien CA, Kern B, Gubrij I, Karsenty G, Manolagas SC. Cbfa1 does not regulate RANKL gene activity in stromal/osteoblastic cells. Bone. 2002;30:453–462. doi: 10.1016/s8756-3282(01)00692-5. [DOI] [PubMed] [Google Scholar]
- 22.Horwood NJ, Elliott J, Martin TJ, Gillespie MT. Osteotropic agents regulate the expression of osteoclast differentiation factor and osteoprotegerin in osteoblastic stromal cells. Endocrinology. 1998;139:4743–4746. doi: 10.1210/endo.139.11.6433. [DOI] [PubMed] [Google Scholar]
- 23.Otsuka E, Notoya M, Hagiwara H. Treatment of myoblastic C2C12 cells with BMP-2 stimulates vitamin D-induced formation of osteoclasts. Calcif Tissue Int. 2003;73:72–77. doi: 10.1007/s00223-002-1071-0. [DOI] [PubMed] [Google Scholar]
- 24.Barnes GL, Della Torre T, Sommer B, Young MF, Gerstenfeld LC. Transcriptional regulation restricting bone sialoprotein gene expression to both hypertrophic chondrocytes and osteoblasts. J Cell Biochem. 2002;87:458–469. doi: 10.1002/jcb.10351. [DOI] [PubMed] [Google Scholar]
- 25.Stanton L-A, Sabari S, Sampaio AV, Underhill TM, Beier F. p38 MAP kinase signalling is required for hypertrophic chondrocyte differentiation. Biochem J. 2004;378:53–62. doi: 10.1042/BJ20030874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kawabata M, Imamura T, Miyazono K. Signal transduction by bone morphogenetic proteins. Cytokine Growth Factor Rev. 1998;9:49–61. doi: 10.1016/s1359-6101(97)00036-1. [DOI] [PubMed] [Google Scholar]
- 27.Lee KS, Kim HJ, Li QL, Chi XZ, Ueta C, Komori T, Wozney JM, Kim EG, Choi JY, Ryoo HM, Bae SC. Runx2 is a common target of transforming growth factor beta1 and bone morphogenetic protein 2, and cooperation between Runx2 and Smad5 induces osteoblast-specific gene expression in the pluripotent mesenchymal precursor cell line C2C12. Mol Cell Biol. 2000;20:8783–8792. doi: 10.1128/mcb.20.23.8783-8792.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pacifici M, Golden EB, Oshima O, Shapiro IM, Leboy PS, Adams SL. Hypertrophic chondrocytes. The terminal stage of differentiation in the chondrogenic cell lineage. Ann NY Acad Sci. 1990;599:45–57. doi: 10.1111/j.1749-6632.1990.tb42363.x. [DOI] [PubMed] [Google Scholar]
- 29.Kartsogiannis V, Zhou H, Horwood NJ, Thomas RJ, Hards DK, Quinn JMW, Niforas P, Ng KW, Martin TJ, Gillespie MT. Localization of RANKL (receptor activator of NF kappa B ligand) mRNA and protein in skeletal and extraskeletal tissues. Bone. 1999;25:525–534. doi: 10.1016/s8756-3282(99)00214-8. [DOI] [PubMed] [Google Scholar]
- 30.Silvestrini G, Ballanti P, Patacchioli F, Leopizzi M, Gualtieri N, Monnazzi P, Tremante E, Sardella D, Bonucci E. Detection of osteoprotegerin (OPG) and its ligand (RANKL) mRNA and protein in femur and tibia of the rat. J Molec Histol. 2005;36:59–67. doi: 10.1007/s10735-004-3839-1. [DOI] [PubMed] [Google Scholar]
- 31.Breuil V, Schmid-Antomarchi H, Schmid-Alliana A, Rezzonico R, Euller-Ziegler L, Rossi B. The receptor activator of nuclear factor (NF)-kappaB ligand (RANKL) is a new chemotactic factor for human monocytes. FASEB J. 2003;17:1751–1753. doi: 10.1096/fj.02-1188fje. [DOI] [PubMed] [Google Scholar]
- 32.Mosheimer BA, Kaneider NC, Feistritzer C, Sturn DH, Wiedermann CJ. Expression and function of RANK in human monocyte chemotaxis. Arthritis Rheum. 2004;50:2309–2316. doi: 10.1002/art.20352. [DOI] [PubMed] [Google Scholar]
- 33.Collin-Osdoby P, Rothe L, Anderson F, Nelson M, Maloney W, Osdoby P. RANKL and OPG expression by human microvascular endothelial cells, regulation by inflammatory cytokines, and role in human osteoclastogenesis. J Biol Chem. 2001;276:20659–20672. doi: 10.1074/jbc.M010153200. [DOI] [PubMed] [Google Scholar]
- 34.Zhang X, Ziran N, Goater JJ, Schwarz EM, Puzas JE, Rosier RN, Zuscik M, Drissi H, O'Keefe RJ. Primary murine limb bud mesenchymal cells in long-term culture complete chondrocyte differentiation: TGF-beta delays hypertrophy and PGE2 inhibits terminal differentiation. Bone. 2004;34:809–817. doi: 10.1016/j.bone.2003.12.026. [DOI] [PubMed] [Google Scholar]
- 35.Gartland A, Mechler J, Mason-Savas A, MacKay CA, Mailhot G, Marks SC, Jr, Odgren PR. In vitro chondrocyte differentiation using costochondral chondrocytes as a source of primary rat chondrocyte cultures: An improved isolation and cryopreservation method. Bone. 2005;37:530–544. doi: 10.1016/j.bone.2005.04.034. [DOI] [PubMed] [Google Scholar]
- 36.Drissi MH, Li X, Sheu TJ, Zuscik MJ, Schwarz EM, Puzas JE, Rosier RN, O'Keefe RJ. Runx2/Cbfa1 stimulation by retinoic acid is potentiated by BMP2 signaling through interaction with Smad1 on the collagen X promoter in chondrocytes. J Cell Biochem. 2003;90:1287–1298. doi: 10.1002/jcb.10677. [DOI] [PubMed] [Google Scholar]
- 37.Wang Q, Wei X, Zhu T, Zhang M, Shen R, Xing L, O'Keefe RJ, Chen D. BMP-2 activates Smad6 gene transcription through bone-specific transcription factor Runx2. J Biol Chem. 2007;282:10742–10748. doi: 10.1074/jbc.M610997200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yanagi Y, Suzawa M, Kawabata M, Miyazono K, Yanagisawa J, Kato S. Positive and negative modulation of vitamin D receptor function by transforming growth factor-beta signaling through smad proteins. J Biol Chem. 1999;274:12971–12974. doi: 10.1074/jbc.274.19.12971. [DOI] [PubMed] [Google Scholar]
- 39.Komori Y, Yagi H, Nomura S, Yamaguchi A, Sasaki K, Deguchi K, Shimizu Y, Bronson RT, Gao YH, Inada M, Sato M, Okamoto R, Kitamura Y, Yoshiki S, Kishimito T. Targeted disruption of Cbfa 1 results in a complete lack of bone formation owing to maturational arrest of osteoblasts. Cell. 1997;89:755–764. doi: 10.1016/s0092-8674(00)80258-5. [DOI] [PubMed] [Google Scholar]
- 40.Ducy P, Zhang R, Geoffroy V, Ridall AL, Karsenty G. Osf2/Cbfa 1: A transcriptional activator of osteoblast differentiation. Cell. 1997;89:747–754. doi: 10.1016/s0092-8674(00)80257-3. [DOI] [PubMed] [Google Scholar]
- 41.Choi JY, Pratap J, Javed A, Zaidi SK, Xing L, Balint E, Dalamangas S, Boyce BF, van Wijnen AJ, Lian JB, Stein JL, Jones SN, Stein GS. Subnuclear targeting of Runx/Cbfa/AML factors is essential for tissue-specific differentiation during embryonic development. Proc Natl Acad Sci USA. 2001;98:8650–8655. doi: 10.1073/pnas.151236498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.MacDonald PN, Baudino TA, Tokumaru H, Dowd DR, Zhang C. Vitamin D receptor and nuclear receptor coactivators: Crucial interactions in vitamin D-mediated transcription. Steroids. 2001;66:171–176. doi: 10.1016/s0039-128x(00)00200-2. [DOI] [PubMed] [Google Scholar]
- 43.Yanagisawa J, Yanagi Y, Masuhiro Y, Suzawa M, Watanabe M, Kashiwagi K, Toriyabe T, Kawabata M, Miyazono K, Kato S. Convergence of transforming growth factor-beta and vitamin D signaling pathways on SMAD transcriptional coactivators. Science. 1999;283:1317–1321. doi: 10.1126/science.283.5406.1317. [DOI] [PubMed] [Google Scholar]
- 44.Kanatani M, Sugimoto T, Kaji H, Kobayashi T, Nishiyama K, Fukase M, Kumegawa M, Chihara K. Stimulatory effect of bone morphogenetic protein-2 on osteoclast-like cell formation and bone-resorbing activity. J Bone Miner Res. 1995;10:1681–1690. doi: 10.1002/jbmr.5650101110. [DOI] [PubMed] [Google Scholar]
- 45.Horiki M, Imamura T, Okamoto M, Hayashi M, Murai J, Akira Myoui A, Ochi T, Miyazono K, Yoshikawa H, Tsumaki N. Smad6/Smurf1 overexpression in cartilage delays chondrocyte hypertrophy and causes dwarfism with osteopenia. J Cell Biol. 2004;165:433–445. doi: 10.1083/jcb.200311015. [DOI] [PMC free article] [PubMed] [Google Scholar]