Abstract
Ectopic expression of Smurf2 in chondrocytes and perichondrial cells accelerated endochondral ossification by stimulating chondrocyte maturation and osteoblast development through upregulation of β-catenin in Col2a1-Smurf2 embryos. The mechanism underlying Smurf2-mediated morphological changes during embryonic development may provide new mechanistic insights and potential targets for prevention and treatment of human osteoarthritis.
Introduction
Our recent finding that adult Col2a1-Smurf2 mice have an osteoarthritis-like phenotype in knee joints prompted us to examine the role of Smurf2 in the regulation of chondrocyte maturation and osteoblast differentiation during embryonic endochondral ossification.
Materials and Methods
We analyzed gene expression and morphological changes in developing limbs by immunofluorescence, immunohistochemistry, Western blot, skeletal preparation, and histology. A series of markers for chondrocyte maturation and osteoblast differentiation in developing limbs were examined by in situ hybridization.
Results
Ectopic overexpression of Smurf2 driven by the Col2a1 promoter was detected in chondrocytes and in the perichondrium/periosteum of 16.5 dpc transgenic limbs. Ectopic Smurf2 expression in cells of the chondrogenic lineage inhibited chondrocyte differentiation and stimulated maturation; ectopic Smurf2 in cells of the osteoblastic lineage stimulated osteoblast differentiation. Mechanistically, this could be caused by a dramatic increase in the expression of β-catenin protein levels in the chondrocytes and perichondrial/periosteal cells of the Col2a1-Smurf2 limbs.
Conclusions
Ectopic expression of Smurf2 driven by the Col2a1 promoter accelerated the process of endochondral ossification including chondrocyte maturation and osteoblast differentiation through upregulation of β-catenin, suggesting a possible mechanism for development of osteoarthritis seen in these mice.
Key words: Smurf2, β-catenin, chondrocyte, osteoblast, endochondral ossification
INTRODUCTION
The process of endochondral ossification is a complex one that consists of multiple stages. First, the mesenchymal cells aggregate to form condensations, which subsequently differentiate into two types of cells: chondrocytes that form cartilage elements and osteoblast lineage cells that form perichondrium surrounding the cartilage rudiment. At 13.5 days postcoitum (dpc) of mouse development, the condensed mesenchymal cells in the center of the condensation undergo differentiation into chondrocytes, which produce cartilage-specific matrix proteins including type 2 collagen (Col2a1) and aggrecan. Chondrocytes undergo proliferation and become hypertrophic chondrocytes around 14.5 dpc, and the primary ossification center is formed by 14.5–16.5 dpc. Furthermore, this process of chondrocyte maturation is coupled to osteoblast recruitment and differentiation. In parallel to the process of chondrocyte maturation and matrix ossification, mesenchymal cells in the periphery of the cartilage differentiate and mature into the osteoblast lineage to form perichondrium or periosteum flanking chondrocytes and the primary spongiosa. Overall, this process of endochondral ossification happens in the majority of the skeletal elements, except the flat bones of the skull.
The process of endochondral ossification is carefully regulated by an array of growth factors and transcription factors. Sox9, a high-mobility-group (HMG-box) transcription factor, is required for chondrogenic entry of mesenchyme and chondrocyte differentiation and maturation during embryonic development.(1–5) For example, inactivation of Sox9 before chondrogenic mesenchymal condensation results in an early chondrocyte differentiation arrest; removal of Sox9 after chondrogenetic condensation leads to lack of proliferating chondrocytes and a severe reduction of cartilage matrix production in the growth plate.(1,2) In addition, Sox9 also plays a role in negatively regulating the transition of chondrocytes into hypertrophic chondrocytes in the growth plate, leading to an enlarged hypertrophic zone and accelerated immature ossification in the heterozygous Sox9 mutant mice.(2,6) Consistent with the in vivo studies that Sox9 is essential for chondrogenesis, in vitro studies have shown that Sox9 binds to and activates chondrocyte-specific enhancer elements in Col2a1, aggrecan, and CD-AP genes.(7–10) Indian Hedgehog (Ihh) is indispensable for endochondral ossification. Ihh expressed by prehypertrophic chondrocytes coordinates with PTH-like peptide (PthrP;Pthlh) expressed by periarticular chondrocytes to form a negative feedback loop regulating the growth and differentiation of chondrocytes.(11) In addition, Ihh directly signals to both chondrocytes and osteoblast lineage cells in perichondrium and regulates chondrocyte proliferation and osteoblast development.(12,13) Runx2, a transcription factor with Runt domain, is required for osteoblast cell fate determination and sufficient to induce osteoblast-specific marker genes including collagen type 1 (Col1a1), bone sialoprotein (BSP), and osteocalcin (OC).(14–16) Osterix (Osx1), an osteoblast-specific transcription factor, acts downstream of Runx2 to ensure full differentiation of Runx2-expressing osteoprogenitor cells along the osteoblast lineage.(17)
Canonical Wnt signaling has been implicated in multiple steps of the endochondral bone formation cascade. The canonical Wnt signaling pathway signals through β-catenin. At the chondrogenic mesenchymal condensation stage (E11–E13), β-catenin is normally expressed in perichondrium surrounding the cartilage rudiments, with relatively lower levels detected in the condensed chondrogenic mesenchymal cells, suggesting a potential role of β-catenin in differentiation of mesenchymal precursors toward the osteoblast lineage.(4,5) More direct evidence for the involvement of β-catenin in osteoblastogenesis is that removal of β-catenin from either mesenchymal progenitors for both chondrocytes and osteoblast (β-cateninflox/flox;Dermo-Cre) or chondrogenic progenitors (β-cateninflox/flox;Col2a1-Cre) results in a complete loss of mature osteoblasts and bone formation.(5,13) Interestingly, in both types of mutant embryos, along with other two similar studies from β-cateninflox/flox;Prx1-Cre and β-cateninflox/flox;Osx1-Cre mouse mutants,(4,18) ectopic chondrocyte differentiation instead of osteoblast commitment occurred in the area where bone is normally formed, suggesting a fundamental role for β-catenin in repression of chondrogenic potential in osteochondroprogenitors located within the periosteum. Similar to loss-of-function studies, stabilization of β-catenin in either mesenchymal cells (β-catenin Δex3Prx/+) or chondrogenic precursors (β-catenin+/floxp(ex3);Col2a1-Cre) also results in defects in cartilage and bone formation.(3,4) However, ectopic overexpression of Wnt14 driven by the Col2a1 promoter promotes osteoblast differentiation and chondrocyte maturation, leading to accelerated endochondral ossification in the Col2a1-Wnt14 mutant long bones.(5) Thus, β-catenin level in the mesenchymal precursors or osteochondroprecursors is critical for controlling cell fate determination, cell differentiation, and maturation through regulation of Sox9 and Runx2 expression.(3–5) In general, β-catenin levels are upregulated by Wnt ligands. β-catenin levels are normally kept low through continuous proteasome-mediated degradation of phosphorylated β-catenin, which is catalyzed by the enzymes glycogen synthase kinase 3β (GSK-3β) in a “destruction complex.”(19,20) On Wnt ligand binding to its receptors, the activity of GSK-3β is suppressed, and nonphosphorylated β-catenin accumulates in the cytoplasm and translocates into the nucleus as a co-activator to regulate expression of Wnt target genes.(21) Thus, any mechanism by which normal β-catenin regulation is uncoupled from Wnt signaling control could result in dysregulation of osteoblast/chondrocyte function in the human, leading to abnormal bone mass or degenerative joints.(22–24) However, our understanding of the abnormal regulation of β-catenin in these diseases and others remains incomplete.
Smurf2, an E3 ubiquitin ligase, induces multiple targets for ubiquitination and degradation and modulates various signaling pathways in vitro. Smurf2 directly interacts with Smad1 and Smad2 and indirectly interacts with the type I receptor of TGFβ and β-catenin through an adaptor Smad7 for ubiquitination and proteasome-dependent degradation.(25–28) Our previous study that overexpression of Smurf2 in articular chondrocytes activated cell maturation indicated an important role of Smurf2 in development of osteoarthritis (OA).(29) Because activation of articular chondrocyte maturation and cartilage degeneration during OA recapitulates many of the events that occur during embryonic development, Smurf2 may also regulate the process of embryonic development. In this study, we showed that overexpression of Smurf2 under the control of Col2a1 promoter accelerates endochondral ossification through upregulation of β-catenin during embryonic development.
MATERIALS AND METHODS
Skeletal preparation
Embryos at 13.5, 14.5, 16.5, and 18.5 dpc and newborns were harvested in cold PBS, fixed in 95% ethanol for 2–5 days, and stained with 0.3% Alcian blue 8GS and 0.1% alizarin red S for 3–6 days. The embryos were cleared in 1% KOH in a series concentration of glycerol solution.
Histology and in situ hybridization
Embryonic limbs at 16.5 and18.5 dpc, as well as adult knee joints, were dissected in cold PBS, fixed in 4% paraformaldehyde at 4°C for 1–3 days, and embedded in paraffin for histology and in situ hybridization. Immunofluorescence staining was performed according to a modified protocol.(30) Briefly, antigen was unmasked by incubating the sections in sodium citrate buffer at 95°C for 15 min followed by cooling to room temperature. The primary monoclonal anti-flag M2 antibody (Sigma) was diluted at 1:600, and polyclonal anti-β-catenin antibody (Cell Signaling) was diluted at 1:120 in PBS/Triton. The secondary antibodies used were FITC-conjugated donkey anti-mouse or -rabbit (H&L; Rockland) diluted at 1:2000. The coverslip slides and Vectashied mounting medium were from Vector Laboratories. Immunohistochemistry was performed as described previously.(13) Briefly, sections were incubated with a monoclonal anti-GSK-3β (Cell Signaling) diluted at 1:100 after antigen retrieval. Signal was detected using horseradish peroxidase (HRP) streptavidin and romulin AEC chromagen (Biocare Medical). Alizarin red, safranin O, and hematoxylin staining were performed using standard histological methods. We assessed chondrocyte number per unit area by counting cell number from three randomly chosen regions of each section. We used six sections at a similar level from six independent samples. For in situ hybridization, after fixation, the samples were decalcified with diethylpyrocarbonate (DEPC)-treated 10% EDTA and embedded in paraffin, and 6-μm sections were cut. A riboprobe was labeled with [35S]UTP (Amersham), and in situ hybridization was performed using a technique that we have described previously.(31)
Western blot
Protein was extracted from limbs as described previously.(32) Briefly, the 16.5 dpc limbs were pulverized in liquid nitrogen using a mini-pestle (Clontech) before protein extraction with RIPA buffer. Protein concentration of the disrupted tissues was determined using the Bradford method. Fifty-microgram aliquots of protein extract were separated by SDS-PAGE and transferred to a PVDF membrane (Schleider and Schuell). The blots were probed overnight at 4°C with anti-β-catenin pAb and anti-GSK-3β mAb (Cell Signaling) at a 1:1000 dilution. Blots were further incubated for 1 h at room temperature in the presence of HRP-conjugated secondary antibodies against rabbit or mouse (BioRad) at a dilution of 1:2000. The immune complexes were detected using ECL-Femto (Pierce) and visualized through exposure of X-OMAT AR film (Kodak).
RESULTS
Acceleration of endochondral ossification in Col2a1-Smurf2 embryos
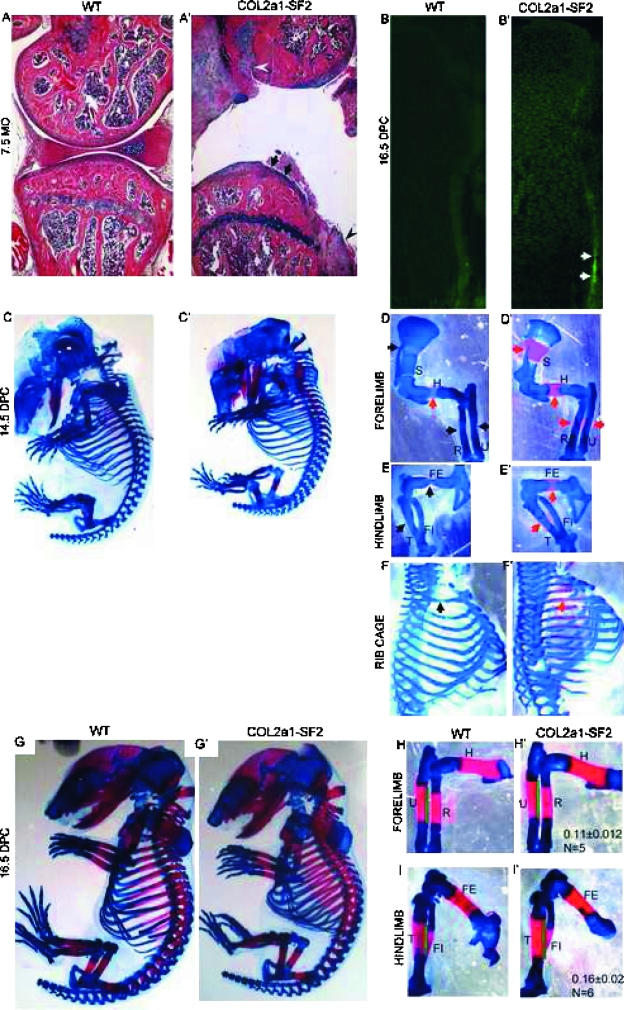
The human flag-Smurf2 cDNA was cloned downstream of Col2a1 promoter, and Col2a1-Smurf2 transgenic mouse lines were generated. The adult transgenic mice showed OA phenotype. Hematoxylin and Alcian blue staining indicated that normal articular cartilage surface is smooth in 7.5-mo-old wildtype mice (Fig. 1A), whereas cartilage degeneration and osteophytes were detected in the transgenic littermates (Fig. 1A′, arrows and arrowheads, respectively). The Smurf2 transgene was mainly expressed by the articular chondrocytes and much less by the growth plate chondrocytes in the adult mice (data not shown); however, it was highly expressed in all chondrocytes in the16.5 dpc transgenic limbs compared with that in the wildtype littermates (Figs. 1B and 1B′). In addition, consistent with previous findings about the Col2a1 promoter function,(5,33) the expression of Smurf2 was highly detected in the osteoprecursors in the perichondrium/periosteum where osteoblasts arise (Fig. 1B′, arrows). This suggests that Col2a1 promoter not only works in all chondrocytes but also in some osteoprogenitors, which have the potential to differentiate into the chondrogenic linage.(4,5,18) Therefore, we took the advantage of the Col2a1 expression profile and studied the impact of Smurf2 on chondrocytes and osteoprogenitors during embryonic development. During embryogenesis, no apparent difference was observed between wildtype and transgenic embryos at 13.5 dpc by whole mount preparation stained with Alcian blue, which specifically stains cartilage matrix blue (data not shown). This suggests that mesenchymal condensation and differentiation into the skeletal anlagen is not affected. However, from 14.5 dpc onward, we found that the ossification in all skeletal elements derived by endochondral bone formation was accelerated in the transgenic embryos, and the size of transgenic embryos was shortened over the wildtype littermates, which is shown by skeletal preparation stained with Alcian blue and Alizarin red, which specifically stains mineralized matrix in a red color (Figs. 1C–1F′). In general, at 14.5 dpc, chondrocyte hypertrophic differentiation occurred in the middle of most long bones (Figs. 1D–1F, black arrows), or ossification had just started in some proximal long bones such as the humerus (Fig. 1D, red arrow). In contrast, in the 14.5 dpc transgenic mouse embryos, the ossification domain was apparent and expanded in proximal long bones such as scapula, humerus of the forelimb, and femurs of the hindlimb, as well as ribs (Figs. 1D′–1F′, red arrows). The ossification in the transgenic distal long bones such as radius and ulna of the forelimb, and tibia and fibula of the hindlimb was less accelerated than that in the transgenic proximal long bones (Fig. 1, compare E′ with D′), which is caused by the dynamic mesenchyme migration from the lateral plate mesoderm, leading to delayed development of distal versus the proximal elements (Fig. 1, compare E with D).(4,19) By 16.5 dpc, the difference in overall appearance between the wildtype and transgenic embryos was less than that at 14.5 dpc (Figs. 1G and 1G′), but the continuous acceleration of endochondral ossification in the transgenic long bones still could be observed. Specifically, the length of the ossification domain in the 16.5 dpc transgenic forelimbs and hindlimbs is increased 11% and 16%, respectively, compared with wildtype littermates (Fig. 1, compare H′ with H and I′ with I), although the full length of the transgenic limbs was shorter (Figs. 1H′, 1H, 1I′, and 1I; data not shown). Despite this, no gross phenotypic difference was observed between the wildtype and transgenic littermates after birth (data not shown). These data suggest that ectopic overexpression of Smurf2 under the control of Col2a1 promoter enhances the process of endochondral ossification during the early stage of skeletal development.
FIG. 1.
Ectopic overexpression of Smurf2 under Col2a1 promoter promotes endochondral ossification. (A and A′) Phenotype of knee joints from 7.5-mo-old wildtype (A) and Col2a1-Smurf2 (A′) mice. Arrows and arrowheads indicate cartilage degeneration and osteophytes, respectively. (B and B′) Immunofluorescence of wildtype (B) and Col2a1-Smurf2 (B′) tibias at 16.5 dpc. Arrows indicate overexpression of Smurf2 in the perichondrial cells. (C–I′) Skeletal preparation of mouse embryos. Wildtype (C and G) and Col2a1-Smurf2 (C′ and G′) embryos were stained with Alcian blue and alizarin red at 14.5 (C and C′) and 16.5 dpc (G and G′). Forelimbs (D, D′, H, and H′), hindlimbs (E, E′, I, and I′), and rib cages (F and F′) were separated from C, C′, G, and G′ with the aid of a dissecting microscope. Black arrows indicate the hypertrophic region; red arrows indicate the ossification region. S, scapula; H, humerus; R, radius; U, ulna; FE, femur; T, tibia; FI, fibula. Green line indicates the length of ossification domain.
Smurf2 promotes osteoblast development
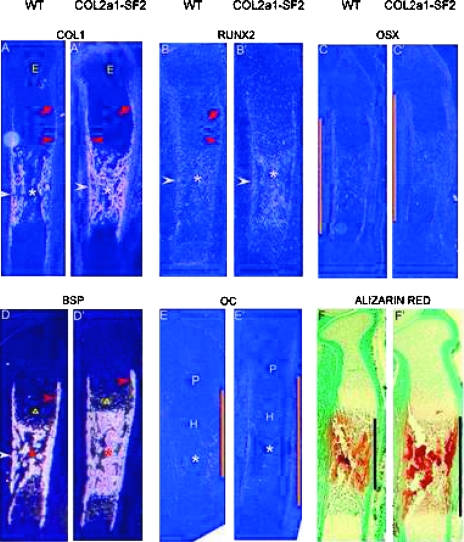
Accelerated endochondral ossification could have been caused by advanced osteoblast differentiation or/and maturation in the transgenic mice. We tested this hypothesis by examining a panel of markers activated at various stages of osteoblast development. Col1a1, an early marker for the osteoblast lineage,(34) was expressed at a lower level in early osteoblast progenitors in the perichondrium toward the epiphysis (Fig. 2A, red arrow) and was subsequently upregulated in more mature osteoblast lineage cells in the perichondrium surrounding the prehypertrophic and hypertrophic chondrocytes (Fig. 2A, red arrowhead) in the bone collar flanking the marrow cavity (Fig. 2A, white arrowhead), as well as in the primary spongiosa (Fig. 2A, asterisk), consistent with the previous studies.(13,32) In the 16.5 dpc transgenic littermates, however, the expression levels of Col1a1 were dramatically increased in the perichondrium, which is from the immature area toward the epiphysis (Fig. 2A′, red arrow) to the mature area flanking the hypertrophic chondrocytes (Fig. 2A′, red arrowhead), in the bone collar (Fig. 2A′, white arrowhead), and the primary spongiosa (Fig. 2A′, asterisk). Like Col1a1, Runx2, another marker of early stage of osteoblast lineage, was normally expressed in a steady low level in the perichondrium toward the epiphysis and flanking the prehypertrophic and hypertrophic chondrocytes (Fig. 2B, red arrow and arrowhead, respectively), in the bone collar, and the primary spongiosa at 16.5 dpc (Fig. 2B, white arrowhead and asterisk, respectively). Similar to the expression pattern of Col1a1 in the 16.5 dpc wildtype and its transgenic siblings, the Runx2 levels were also significantly increased in the bone collar and the primary spongiosa in the transgenic limbs (Fig. 2B′, white arrowhead and asterisk, respectively) compared with that in the wildtype counterparts. Osx, a specific marker of osteoblast lineage and functioning downstream of Runx2,(17) was normally expressed in the perichondrium where Runx2 was expressed (Fig. 2C). By comparing the expression pattern of Osx in the 16.5 dpc wildtype with that in the transgenic siblings, we found that the expression domain in the transgenic limbs was enlarged (Fig. 2, orange line, compare C′ with C). BSP, a marker gene for late stage of the osteoblast lineage, was expressed in the perichondrium surrounding the prehypertrophic and hypertrophic chondrocytes, in the bone collar, and primary spongiosa (Fig. 2D, red-, white-arrowhead, and asterisk, respectively). In addition, BSP was also expressed in hypertrophic chondrocytes (Fig. 2D, triangle). Consistent with the overexpression pattern of Col1a1, and Runx2 in the 16.5 dpc transgenic limbs, BSP expression levels were significantly increased in the primary spongiosa and slightly increased in the perichondrium in the transgenic limbs (Fig. 2D′, asterisk and red arrowhead, respectively). In addition, BSP expression was also increased in the transgenic hypertrophic chondrocytes over the wildtype counterparts (Fig. 2, triangle, D′ and D), which was further confirmed by the histological evidence that more hypertrophic cells were in the hypertrophic zone of the transgenic limbs (Figs. 2C and 2D). Oc, an osteoblast terminal differentiation marker, was detected in the perichondrium flanking hypertrophic chondrocytes and in the bone collar and primary spongiosa in the 16.5 dpc wildtype limbs (Fig. 2E). Although the expression levels of Oc in the transgenic limbs were low, the expression domain was enlarged over the wildtype littermates (Fig. 2, orange line, E′ and E). Notably, we found the similar expression pattern of these marker genes in the 18.5 dpc wildtype and their transgenic littermates (data not shown). All these data suggest that ectopic overexpression of Smurf2 under the control of Col2a1 promoter accelerated osteoblast differentiation and stimulated immature osteoblast maturation. Consistent with this, Alizarin red staining showed that the mineralization domain is significantly expanded in the 16.5 dpc transgenic limbs versus the wildtype littermates (Fig. 2, black line, F′, and F), further confirming that Smurf2 accelerated endochondral ossification by promoting osteoblast development.
FIG. 2.
Accelerated osteoblast differentiation and bone matrix mineralization inCol2a1-Smurf2 embryos. (A–E′) Analyses of osteoblast differentiation by in situ hybridization. In situ hybridization was performed on longitudinal sections through the tibia from wildtype (A–E) and Col2a1-Smurf2 (A′–E–) embryos. Red arrow and red arrowhead indicates the perichondrium toward the epiphysis and that surrounding the prehypertrophic and hypertrophic chondrocytes, respectively. Triangles indicate the prehypertrophic zone. White arrowheads and asterisks indicate the perichondrium/periosteum surrounding the primary spongiosa and the primary spongiosa, respectively. E, epiphysis; P, proliferation zone; H, hypertrophic zone. The orange line indicates the length of the domain of gene expression. (F and F′) Alizarin red staining was performed on the sections from wildtype (F) and Col2a1-Smurf2 (F′) embryos. Black line indicates ossification domain.
Smurf2 accelerates chondrocyte maturation
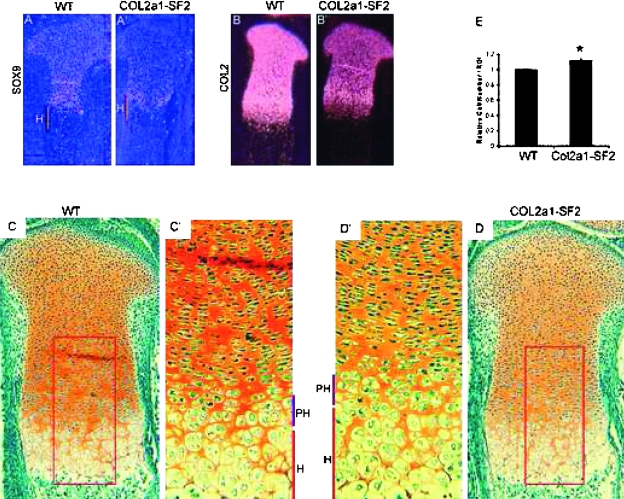
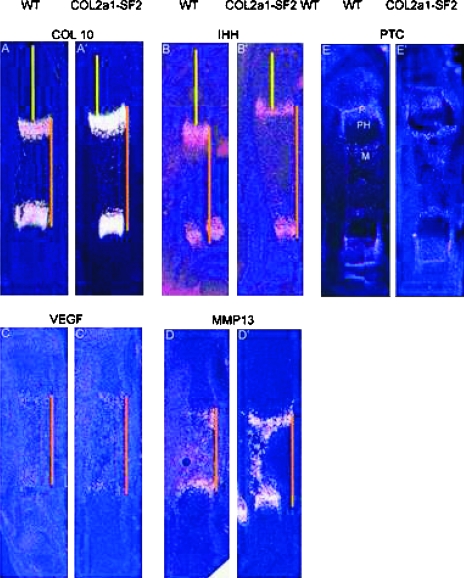
The process of endochondral ossification consists of development and maturation of both chondrocytes and osteoblasts; therefore, an increase in chondrocyte maturation rate could be another reason for the acceleration of endochondral ossification. To test this hypothesis, we examined a panel of stage-specific chondrocyte maturation markers by in situ hybridization and performed histological analysis. In the 16.5 dpc transgenic limbs, we found that the hypertrophic zone is longer than that of the wildtype littermates (Figs. 4D, 4D′, 4C, and 4C′). Furthermore, we observed that the number of prehypertrophic and hypertrophic chondrocytes in a defined area was increased in the transgenic growth plates versus that in the wildtype counterparts (Figs. 4D′, 4C′, and 4E). These morphological changes suggest that chondrocytes are undergoing immature hypertrophic differentiation, similar to the phenotype in the heterozygous Sox9 mutant mice.(6) In situ hybridization showed that the domain of type 10 collagen (Col10a1), a marker gene for hypertrophic chondrocytes, was stronger and wider in the 16.5 dpc transgenic limbs than that in the wildtype counterparts (Figs. 3A′ and 3A), consistent with the histological evidence that the transgenic embryo possessed a longer hypertrophic zone with more hypertrophic chondrocytes in the hypertrophic zone of the growth plates (Figs. 4C and 4D). Ihh, a marker for prehypertrophic chondrocytes, was expressed at normal levels in the 16.5 dpc transgenic limbs, compared with that in the wildtype littermates (Figs. 3B′ and 3B). However, the two domains of either Col10a1 or Ihh were further from each other in the transgenic limbs than that in the wildtype littermates, although the entire length of the transgenic limb was shorter (Fig. 3, orange line, compare A′ with A and B′ with B). Furthermore, the distance from the domain of either Col10a1 or Ihh to the end of the long bones was reduced in size in the transgenic limbs over the wildtype counterparts (Fig. 3, yellow line, compare A′ with A and B′ with B). Like Col10a1 and Ihh, the region between the two domains of VEGF and MMP13, markers for the terminally differentiated hypertrophic chondrocytes, was expanded in the 16.5 dpc transgenic limbs versus that in the wildtype littermates, and the distance between the VEGF/MMP13 domain and the articular surface was shortened in the 18.5 dpc transgenic limbs versus that in the wildtype littermates (Fig. 3, orange line, compare C′ with C and D′ with D; data not shown). All these results indicated that overexpression of Smurf2 in chondrogenic lineage stimulated chondrocyte maturation during endochondral ossification.
FIG. 4.
Inhibition of chondrocyte differentiation in Col2a1-Smurf2 limbs. (A–B′) Expression of chondrocyte differentiation markers Sox9 (A and A′) and Col2 (B and B′) was analyzed on sections through the tibia from wildtype (A and B) and Col2a1-Smurf2 (A′ and B′) embryos by in situ hybridization. H, hypertrophic zone (orange line). (C–D′) Safranin O staining of tibias from wildtype (C) and Col2a1-Smurf2 (D) embryos. C′ and D′ are higher-magnification images of boxed areas in C and D. PH, prehypertrophic zone (purple line); H, hypertrophic zone (red line). (E) Relative cell number per unit area of hypertrophic zone. Data are expressed as mean ± SD (n = 6). Statistical significance is assessed by an unpaired Student's t-test (p < 0.01).
FIG. 3.
Advanced chondrocyte maturation in Col2a1-Smurf2 embryos. (A–D′) Chondrocyte maturation markers were analyzed on longitudinal sections through the tibia from wildtype (A–D) and Col2a1-Smurf2 (A′–D′) embryos by in situ hybridization. The orange line indicates the distance between the separated domains of Col10 (A and A′), Ihh (B and B′), VEGF (C and C′), and MMP13 (D and D′), and the yellow line indicates that from the domain to the articular surface. (E and E′) Ptc expression was analyzed on sections from wildtype (E) and Col2a1-Smurf2 (E′) by in situ hybridization. P, proliferation zone; PH, prehypertrophic zone; M, marrow cavity.
Chondrocyte differentiation is inhibited in Col2a1-Smurf2 embryos
To understand further whether chondrocyte differentiation and proliferation was affected by overexpression of Smurf2 under the control of Col2a1 promoter, we analyzed chondrocyte differentiation markers and performed BrdU in vivo pulse and proliferating cell nuclear antigen staining. Sox9, a marker gene for early chondrocyte differentiation and essential for activation of cartilage matrix expression, was highly detected in all chondrocytes including the resting chondrocytes in the articular region and growth plate chondrocytes from the proliferative zone and the prehypertrophic zone. However, Sox9 was absent from the hypertrophic chondrocytes at 16.5 dpc (Fig. 4A). In the 16.5 dpc transgenic littermates, the Sox9 mRNA expression levels were significantly decreased in these chondrocytes (Figs. 4A′ and 4A). Col2a1 was normally expressed coincidentally with the expression of Sox9. Similar to the expression pattern of Sox9 mRNA in both wildtype and transgenic littermates at 16.5 dpc, Col2a1 mRNA was highly expressed in all chondrocytes except hypertrophic chondrocytes in the wildtype limbs, and a significant decrease was detected in all chondrocytes in the transgenic littermates (Figs. 4B′ and 4B). Safranin O staining (stains aggrecan protein in a red color) showed that the protein levels of aggrecan, a specific marker for chondrocytes, was decreased in the 16.5 dpc transgenic limbs compared with that in the wildtype littermates (Figs. 4D, 4D′, 4C, and 4C′). The similar expression pattern of these genes was also found in the 18.5 dpc transgenic embryos and wildtype littermates (data not shown). These results suggest that chondrocyte differentiation is inhibited in the Col2a1-Smurf2 transgenic limbs. We also examined whether chondrocyte proliferation was affected in the transgenic embryos. BrdU and proliferating cell nuclear antigen staining showed that chondrocyte proliferation was not significantly affected in the transgenic limbs compared with that in the wildtype littermates (data not shown).
β-catenin protein levels are upregulated in Col2a1-Smurf2 limbs
To study the molecular mechanism underlying Smurf2-mediated acceleration of endochondral ossification, we analyzed several signaling pathways. Ihh, expressed by prehypertrophic chondrocytes, regulates the growth and differentiation of chondrocytes directly or through the control of other factors such as Pthrp;Pthlh. In addition, Ihh also acts directly on perichondrium to initiate an osteogenic program in osteoblast progenitors. Ihh signaling is initiated by binding to Patched (ptc), which is a receptor and transcriptional target, and normally expressed in the proliferating chondrocytes, the primary spongiosa within the marrow cavity, and the perichondrium surrounding the Ihh domain (Fig. 3E).(11,13) We examined the mRNA levels of Ihh, Ptc, PthrP;PthIh, and PthrP receptor (PP-R) by in situ hybridization; however, no significant difference could be found between the transgenic limbs and the wildtype littermates at 16.5 dpc (Figs. 3B′, B, E, and E′; data not shown), suggesting that Ihh and Ihh-Pthrp signaling pathways were not responsible for the phenotypic changes in osteoblasts and chondrocytes in the Col2a1-Smurf2 transgenic embryos.
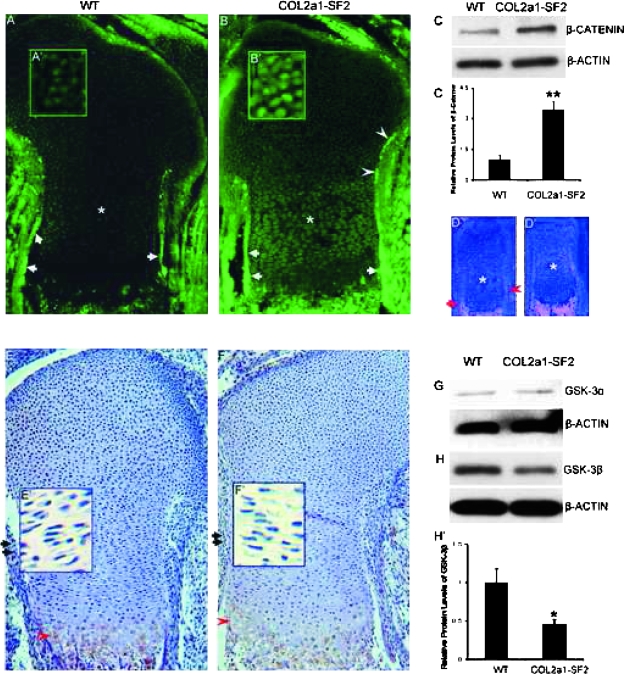
The phenotypic similarities between Col2a1-Wnt14 and Col2a1-Smurf2 mouse embryos prompted us to examine β-catenin in the Col2a1-Smuuf2 embryos. Immunofluorescence staining showed that β-catenin protein was highly expressed in the perichondrium/periosteum flanking the prehypertrophic and hypertrophic chondrocytes, as well as the primary spongiosa (Fig. 5A, arrows, data not shown), whereas its expression was relatively low in the chondrocytes in the 16.5 dpc wildtype limbs (Fig. 5A, asterisk). In the 16.5 dpc transgenic littermates, however, the protein levels of β-catenin in the perichondrium/periosteum adjacent the prehypertrophic and hypertrophic chondrocytes was dramatically increased (Fig. 5B, arrows). Notably, β-catenin was also expressed with relatively higher levels in the perichondrium flanking the immature chondrocytes toward the epiphysis in the transgenic limbs (Fig. 5B, arrowheads) than that in the wildtype counterparts. Upregulation of β-catenin was detected not only in the perichondrium/periosteum but also in the chondrocytes including resting, proliferating and prehypertrophic chondrocytes of the growth plate in the Col2a1-Smurf2 limbs (Figs. 5B, 5B′, 5A, and 5A′). In addition, high magnification of the immunofluorescence staining showed that β-catenin nuclear translocation was increased in the perichondrial cells and the chondrocytes of the growth plate in the transgenic limbs (Figs. 5B′ and 5A′; data not shown). To confirm the observed upregulation of β-catenin in the transgenic limbs shown by immunofluorescence, we extracted protein from limbs containing cartilage and perichondrium/periosteum and performed Western blot analysis. As expected, the transgenic limbs showed a 3-fold increase in the protein levels of β-catenin compared with the wildtype littermates (Figs. 5C and 5C′).
FIG. 5.
Upregulation of β-catenin protein levels inCol2a1-Smurf2 limbs. (A–C′) β-catenin protein levels were analyzed by immunofluorescence (A–B′) and Western blot (C and C′) in wildtype and Col2a1-Smurf2 limbs as indicated. A′ and B′ are higher magnification of resting chondrocytes. (D and D′) Analysis of β-catenin expression by in situ hybridization in wildtype (D) and Col2a1-Smurf2 (D′) limbs. Arrows and arrowheads indicate the perichondrium surrounding the hypertrophic and the prehypertrophic chondrocytes, respectively. Asterisks indicate the proliferating chondrocytes. (E–H′) Analysis of GSK-3β (E–F′ and H and H′) and GSK-3α (G) by immunohistochemistry (E–F′) and Western blot (G and H) in wildtype and Col2a1-Smurf2 limbs as indicated. Arrows and arrowheads indicate perichondrium and maturing chondrocytes, respectively. E′ and F′ are higher magnification of proliferating chondrocytes. The protein levels of β-catenin and GSK-3 were quantitated by scanning densitometry (normalized β-actin). Data are expressed as mean ± SD of three independent experiments. Statistical significance is assessed by an unpaired Student's t-test (**p < 0.01; *p < 0.05).
Upregulation of β-catenin by Smurf2 occurs at protein levels
To understand whether the increase in β-catenin protein levels in the perichondrium/periosteum and chondrocytes was caused by an increase in transcription of the gene, we examined the mRNA levels of β-catenin in the limbs by in situ hybridization. Consistent with the expression pattern of protein in the 16.5 dpc wildtype limbs, the mRNA levels of β-catenin were very high in the perichondrium/periosteum surrounding the hypertrophic chondrocytes, and the marrow cavity (Fig. 5D, arrow; data not shown); the mRNA levels were relatively low in the perichondrium flanking the prehypertrophic chondrocytes (Fig. 5D, arrowhead) and very low in the chondrocytes (Fig. 5D, asterisk) in the 16.5 dpc wildtype limbs. In contrast to the dramatic increase in the protein levels in the transgenic limbs, no significant increase in the mRNA levels of β-catenin could be detected in the transgenic limbs including the perichondrium/periosteum, the primary spongiosa, and the chondrocytes (Figs. 5D′ and 5D), indicating that the increase in β-catenin protein in the transgenic limbs is not caused by an increase in its transcription.
Because β-catenin protein levels are regulated through continuous phosphorylation and degradation of β-catenin by GSK-3β,(19) we examined the protein levels of GSK-3β in 16.5 dpc limbs. Immunohistochemistry showed that GSK-3β is detected in the perichondrium and the proliferating chondrocytes (Figs. 5E, arrow, and 5E′; data not shown) and is strongly detected in the maturing chondrocytes (Fig. 5E, arrowhead) in the 16.5 dpc wildtype limbs. However, the protein levels of GSK-3β were significantly decreased in either perichondrium or chondrocytes (Figs. 5F, 5F′, 5E, and 5E′; data not shown) in the transgenic littermates. Western blot analysis showed that the protein levels of GSK-3β in the 16.5 dpc transgenic limbs were 2.2-fold lower than that in the wildtype littermates (Figs. 5H and 5H′), consistent with the result by immunohistochemistry. No significant difference in the protein levels of GSK-3α could be detected between the transgenic limbs and the wildtype littermates (Fig. 5G). These data suggested that upregulation of β-catenin in Col2a1-Smurf2 transgenic limbs is caused by a decrease in GSK-3β by Smurf2.
DISCUSSION
Stimulation of osteoblast differentiation and chondrocyte maturation in Col2a1-Smurf2 limbs
Perichondrial cells that surround the developing growth plate differentiate into osteoblasts during endochondral ossification. Recently, several lines of evidence have shown that Col2a1, a marker gene for chondrocyte differentiation, is also expressed in the perichondrium where osteoblasts arise.(5,13) The Col2a1-expressing osteochondroprecursors in the perichondrium normally differentiate into osteoblasts, but will indeed differentiate into chondrocytes after removal of β-catenin from the cells.(4,5,13,18) Therefore, β-catenin signaling is needed to repress the chondrogenic potential of osteochondroprogenitors and to ensure full differentiation along osteoblast lineage by upregulation of Runx2 and Osx in the perichondrium. Indeed, we found that the transgene Smurf2 driven by the Col2a1 promoter was expressed not only in chondrocytes but also in the perichondrium surrounding the prehypertrophic and the hypertrophic chondrocytes at 16.5 dpc (Fig. 1). Thus, the Col2a1-Smurf2 mouse model has allowed us to study the impact of Smurf2 on both chondrocytes and osteochondroprogenitors in a developing long bone. A panel of osteoblast lineage markers is expressed by the perichondrial cells while these cells are undergoing osteoblast differentiation. The expression levels of Col1a1, Runx2, and BSP, markers for early and late stages of osteoblast differentiation, were dramatically increased in the transgenic perichondrium/periosteum and the primary spongiosa (Fig. 2), suggesting that overexpression of Smurf2 in the perichondrial cells stimulates osteoblast differentiation. The expression levels of Oc, a marker gene for the terminally differentiated osteoblasts located in the bone collar and the primary spongiosa, was not increased in the transgenic bone collar or the primary spongiosa at 16.5 dpc, but its expression domain was extended from the perichondrium/periosteum adjacent to the marrow cavity and hypertrophic chondrocytes to that around the proliferating chondrocytes (Figs. 2E and 2E′). Therefore, ectopic expression of Smurf2 in the perichondrium/periosteum appears to stimulate immature osteoblast maturation through upregulation of Runx2/Osx signaling (Fig. 2).(5) In addition, progression of chondrocyte maturation was also accelerated in the Col2a1-Smurf2 long bones. Specifically, the expression domains of Col10 and Ihh, markers for the hypertrophic and prehypertrophic chondrocytes, respectively, were separated faster so that the distance between the Col10 or Ihh domain and the long bone end was shorter than that in the wildtype littermates. VEGF and MMP13, markers for terminally differentiated chondrocytes, show a similar expression pattern in the transgenic and wildtype littermates to that of Col10 and Ihh, suggesting that chondrocyte maturation was accelerated (Fig. 3). Consistent with accelerated osteoblast development and chondrocyte maturation in the Col2a1-Smurf2 embryos, endochondral ossification was advanced accordingly (Figs. 1 and 2F′).
Inhibition of chondrocyte differentiation through downregulation of Sox9 in Col2a1-Smurf2 chondrocytes
Sox9, a transcription factor essential for chondrocyte differentiation and cartilage formation, marks the earliest stage of the chondrocyte lineage.(1) Col2a1, a specific marker for the chondrocyte lineage, is normally co-expressed with Sox9. The transcription levels of both Sox9 and Col2a1 were significantly decreased in the transgenic limbs (Figs. 4A′ and 4B′), suggesting an inhibition of chondrocyte differentiation in the transgenic embryos. Like Col2a1, aggrecan, another major component of the cartilage matrix, is also decreased in the transgenic limbs (Fig. 4D). It is likely that the decrease in the expression levels of Col2a1 and aggrecan is caused by the decrease in Sox9 signaling in the transgenic chondrocytes, because Sox9 is responsible for activation of the promoter and enhancer of these cartilage matrix genes.(7,8,10) Consistent with the phenotypic changes of an enlarged hypertrophic zone and immature endochondral ossification observed in the Sox9 heterozygous mutant mice,(6) we found similar histological evidence in the Col2a1-Smurf2 transgenic embryos, namely, a longer hypertrophic zone with more hypertrophic chondrocytes and advanced endochondral ossification (Figs. 4D, 4C, 4E, and 1C–1I′). Thus, it seems that overexpression of Smurf2 under the Col2a1 promoter inhibits chondrocyte differentiation and stimulates chondrocyte maturation through downregulation of Sox9 in the chondrocytes.
Upregulation of β-catenin at the protein level in Col2a1-Smurf2 embryos
A recent study by Day et al.(5) showed that ectopic overexpression of Wnt14 under the Col2a1 promoter stimulated osteoblast differentiation, chondrocyte maturation, and endochondral ossification through β-catenin signaling in a developing long bone, which is largely similar to the phenotype we observed in the developing long bones of Col2a1-Smurf2 embryos (Figs. 1–3). The phenotypic similarities between Col2a1-Wnt14 and Col2a1-Smurf2 prompted us to examine β-catenin mRNA and protein levels in the limbs. Immunofluorescence staining showed that the protein levels of β-catenin were dramatically increased in all stages of the osteoblast lineage in the perichondrium/periosteum in the Col2a1-Smurf2 transgenic limbs, which may be responsible for the observed immature osteoblast maturation (Fig. 2) and consistent with the previous study by Day et al.(5) Most importantly, upregulation of β-catenin occurred in all chondrocytes of the transgenic growth plates (Fig. 5B). However, its mRNA level was not increased accordingly in the transgenic limbs shown by in situ hybridization (Fig. 5D′), suggesting a mechanism underlying upregulation of β-catenin protein in the transgenic limbs occurred at the protein levels. Supporting this hypothesis, the protein levels of GSK-3β in the transgenic limbs were significantly decreased versus that in the wildtype counterparts (Figs. 5E, 5F, 5H, and 5H′). Given Smurf2 is an E3 ubiquitin ligase, Smurf2 may decrease GSK-3β through induction of GSK-3β ubiquitination and proteasomal degradation, leading to an increase in β-catenin.
ACKNOWLEDGMENTS
This research was supported by NIH RO1 AR045700 (RNR), NIH/IAMS P50 AR054041 (RNR), Aircast Foundation (QW), and NIH/NIAMS R01 AR054465 (DC). The authors thank Dr Fanxin Long for data analysis and discussion.
Footnotes
The authors state that they have no conflicts of interest.
REFERENCES
- 1.Bi W, Deng JM, Zhang Z, Behringer RR, de Crombrugghe B. Sox9 is required for cartilage formation. Nat Genet. 1999;22:85–89. doi: 10.1038/8792. [DOI] [PubMed] [Google Scholar]
- 2.Akiyama H, Chaboissier MC, Martin JF, Schedl A, de Crombrugghe B. The transcription factor Sox9 has essential roles in successive steps of the chondrocyte differentiation pathway and is required for expression of Sox5 and Sox6. Genes Dev. 2002;16:2813–2828. doi: 10.1101/gad.1017802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Akiyama H, Lyons JP, Mori-Akiyama Y, Yang X, Zhang R, Zhang Z, Deng JM, Taketo MM, Nakamura T, Behringer RR, McCrea PD, de Crombrugghe B. Interactions between Sox9 and beta-catenin control chondrocyte differentiation. Genes Dev. 2004;18:1072–1087. doi: 10.1101/gad.1171104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hill TP, Später D, Taketo MM, Birchmeier W, Hartmann H. Canonical Wnt/beta-catenin signaling prevents osteoblasts from differentiating into chondrocytes. Dev Cell. 2005;8:727–738. doi: 10.1016/j.devcel.2005.02.013. [DOI] [PubMed] [Google Scholar]
- 5.Day TF, Guo X, Garrett-Beal L, Yang Y. Wnt/beta-catenin signaling in mesenchymal progenitors controls osteoblast and chondrocyte differentiation during vertebrate skeletogenesis. Dev Cell. 2005;8:739–750. doi: 10.1016/j.devcel.2005.03.016. [DOI] [PubMed] [Google Scholar]
- 6.Bi W, Huang W, Whitworth DJ, Deng JM, Zhang Z, Behringer RR, de Crombrugghe B. Haploinsufficiency of Sox9 results in defective cartilage primordia and premature skeletal mineralization. Proc Natl Acad Sci USA. 2001;98:6698–6703. doi: 10.1073/pnas.111092198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lefebvre V, Huang W, Harley VR, Goodfellow PN, de Crombrugghe B. SOX9 is a potent activator of the chondrocyte-specific enhancer of the pro alpha1(II) collagen gene. Mol Cell Biol. 1997;17:2336–2346. doi: 10.1128/mcb.17.4.2336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bridgewater LC, Lefebvre V, de Crombrugghe B. Chondrocyte-specific enhancer elements in the Col11a2 gene resemble the Col2a1 tissue-specific enhancer. J Biol Chem. 1998;273:14998–15006. doi: 10.1074/jbc.273.24.14998. [DOI] [PubMed] [Google Scholar]
- 9.Xie WF, Zhang X, Sakano S, Lefebvre V, Sandell LJ. Trans-activation of the mouse cartilage-derived retinoic acid-sensitive protein gene by Sox9. J Bone Miner Res. 1999;14:757–763. doi: 10.1359/jbmr.1999.14.5.757. [DOI] [PubMed] [Google Scholar]
- 10.Sekiya I, Tsuji K, Koopman P, Watanabe H, Yamada Y, Shinomiya K, Nifuji A, Noda M. SOX9 enhances aggrecan gene promoter/enhancer activity and is up-regulated by retinoic acid in a cartilage-derived cell line, TC6. Biol Chem. 2000;275:10738–10744. doi: 10.1074/jbc.275.15.10738. [DOI] [PubMed] [Google Scholar]
- 11.Vortkamp A, Lee K, Lanske B, Segre GV, Kronenberg HM, Tabin CJ. Regulation of rate of cartilage differentiation by Indian hedgehog and PTH-related protein. Science. 1996;273:613–622. doi: 10.1126/science.273.5275.613. [DOI] [PubMed] [Google Scholar]
- 12.St-Jacques B, Hammerschmidt M, McMahon AP. Indian hedgehog signaling regulates proliferation and differentiation of chondrocytes and is essential for bone formation. Genes Dev. 1999;13:2072–2086. doi: 10.1101/gad.13.16.2072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hu H, Hilton MJ, Tu X, Yu K, Ornitz DM, Long F. Sequential roles of Hedgehog and Wnt signaling in osteoblast development. Development. 2005;132:49–60. doi: 10.1242/dev.01564. [DOI] [PubMed] [Google Scholar]
- 14.Ducy P, Zhang R, Geoffroy V, Ridall AL, Karsenty G. Osf2/Cbfa1: A transcriptional activator of osteoblast differentiation. Cell. 1997;89:747–754. doi: 10.1016/s0092-8674(00)80257-3. [DOI] [PubMed] [Google Scholar]
- 15.Komori T, Yagi H, Nomura S, Yamaguchi A, Sasaki K, Deguchi K, Shimizu Y, Bronson RT, Gao YH, Inada M, Sato M, Okamoto R, Kitamura Y, Yoshiki S, Kishimoto T. Targeted disruption of Cbfa1 results in a complete lack of bone formation owing to maturational arrest of osteoblasts. Cell. 1997;89:755–764. doi: 10.1016/s0092-8674(00)80258-5. [DOI] [PubMed] [Google Scholar]
- 16.Otto F, Thornell AP, Crompton T, Denzel A, Gilmour KC, Rosewell IR, Stamp GW, Beddington RS, Mundlos S, Olsen BR, Selby PB, Owen MJ. Cbfa1, a candidate gene for cleidocranial dysplasia syndrome, is essential for osteoblast differentiation and bone development. Cell. 1997;89:765–771. doi: 10.1016/s0092-8674(00)80259-7. [DOI] [PubMed] [Google Scholar]
- 17.Nakashima K, Zhou X, Kunkel G, Zhang Z, Deng JM, Behringer RR, de Crombrugghe B. The novel zinc finger-containing transcription factor osterix is required for osteoblast differentiation and bone formation. Cell. 2002;108:17–29. doi: 10.1016/s0092-8674(01)00622-5. [DOI] [PubMed] [Google Scholar]
- 18.Rodda SJ, McMahon AP. Distinct roles for Hedgehog and canonical Wnt signaling in specification, differentiation and maintenance of osteoblast progenitors. Development. 2006;133:3231–3244. doi: 10.1242/dev.02480. [DOI] [PubMed] [Google Scholar]
- 19.Logan CY, Nusse R. The Wnt signaling pathway in development and disease. Annu Rev Cell Dev Biol. 2004;20:781–810. doi: 10.1146/annurev.cellbio.20.010403.113126. [DOI] [PubMed] [Google Scholar]
- 20.Clevers H. Wnt/beta-catenin signaling in development and disease. Cell. 2006;127:469–480. doi: 10.1016/j.cell.2006.10.018. [DOI] [PubMed] [Google Scholar]
- 21.Price MA. CKI, there's more than one: Casein kinase I family members in Wnt and Hedgehog signaling. Genes Dev. 2006;20:399–410. doi: 10.1101/gad.1394306. [DOI] [PubMed] [Google Scholar]
- 22.Boyden LM, Mao J, Belsky J, Mitzner L, Farhi A, Mitnick MA, Wu D, Insogna K, Lifton RP. High bone density due to a mutation in LDL-receptor-related protein 5. N Engl J Med. 2002;346:1513–1521. doi: 10.1056/NEJMoa013444. [DOI] [PubMed] [Google Scholar]
- 23.Little RD, Carulli JP, Del Mastro RG, Dupuis J, Osborne M, Folz C, Manning SP, Swain PM, Zhao SC, Eustace B, Lappe MM, Spitzer L, Zweier S, Braunschweiger K, Benchekroun Y, Hu X, Adair R, Chee L, FitzGerald MG, Tulig C, Caruso A, Tzellas N, Bawa A, Franklin B, McGuire S, Nogues X, Gong G, Allen KM, Anisowicz A, Morales AJ, Lomedico PT, Recker SM, Van Eerdewegh P, Recker RR, Johnson ML. A mutation in the LDL receptor-related protein 5 gene results in the autosomal dominant high-bone-mass trait. Am J Hum Genet. 2002;70:11–19. doi: 10.1086/338450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Loughlin J, Dowling B, Chapman K, Marcelline L, Mustafa Z, Southam L, Ferreira A, Ciesielski C, Carson DA, Corr M. Functional variants within the secreted frizzled-related protein 3 gene are associated with hip osteoarthritis in females. Proc Natl Acad Sci USA. 2004;101:9757–9762. doi: 10.1073/pnas.0403456101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kavsak P, Rasmussen RK, Causing CG, Bonni S, Zhu H, Thomsen GH, Wrana JL. Smad7 binds to Smurf2 to form an E3 ubiquitin ligase that targets the TGF beta receptor for degradation. Mol Cell. 2000;6:1365–1375. doi: 10.1016/s1097-2765(00)00134-9. [DOI] [PubMed] [Google Scholar]
- 26.Lin X, Liang M, Feng XH. Smurf2 is a ubiquitin E3 ligase mediating proteasome-dependent degradation of Smad2 in transforming growth factor-beta signaling. J Biol Chem. 2000;275:36818–36822. doi: 10.1074/jbc.C000580200. [DOI] [PubMed] [Google Scholar]
- 27.Zhang Y, Chang C, Gehling DJ, Hemmati-Brivanlou A, Derynck R. Regulation of Smad degradation and activity by Smurf2, an E3 ubiquitin ligase. Proc Natl Acad Sci USA. 2001;98:974–979. doi: 10.1073/pnas.98.3.974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Han G, Li AG, Liang YY, Owens P, He W, Lu S, Yoshimatsu Y, Wang D, Ten Dijke P, Lin X, Wang XJ. Smad7-induced beta-catenin degradation alters epidermal appendage development. Dev Cell. 2006;11:301–312. doi: 10.1016/j.devcel.2006.06.014. [DOI] [PubMed] [Google Scholar]
- 29.Zuscik MJ, Baden JF, Wu Q, Sheu TJ, Schwarz E, Drissi H, O'Keefe RJ, Puzas JE, Rosier RN. 5-azacytidine alters TGF-beta and BMP signaling and induces maturation in articular chondrocytes. J Cell Biochem. 2004;92:316–331. doi: 10.1002/jcb.20050. [DOI] [PubMed] [Google Scholar]
- 30.Wu Q, Chen Q. Mechanoregulation of chondrocyte proliferation, maturation, and hypertrophy: Ion-channel dependent transduction of matrix deformation signals. Exp Cell Res. 2000;256:383–391. doi: 10.1006/excr.2000.4847. [DOI] [PubMed] [Google Scholar]
- 31.Zhang X, Schwarz EM, Young DA, Puzas JE, Rosier RN, O'Keefe RJ. Cyclooxygenase-2 regulates mesenchymal cell differentiation into the osteoblast lineage and is critically involved in bone repair. J Clin Invest. 2002;109:1405–1415. doi: 10.1172/JCI15681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hilton MJ, Tu X, Cook J, Hu H, Long F. Ihh controls cartilage development by antagonizing Gli3, but requires additional effectors to regulate osteoblast and vascular development. Development. 2005;132:4339–4351. doi: 10.1242/dev.02025. [DOI] [PubMed] [Google Scholar]
- 33.Long F, Chung UI, Ohba S, McMahon J, Kronenberg HM, McMahon AP. Ihh signaling is directly required for the osteoblast lineage in the endochondral skeleton. Development. 2004;131:1309–1318. doi: 10.1242/dev.01006. [DOI] [PubMed] [Google Scholar]
- 34.Aubin JE. Advances in the osteoblast lineage. Biochem Cell Biol. 1998;76:899–910. [PubMed] [Google Scholar]