Abstract
The cannabinoid receptor one (CB1) is prevalent in the brains of many species. Receptor binding, in situ hybridization and immunohistochemical surveys have described the distribution of this receptor in a limited number of species. The current study used in situ hybridization to examine the expression of CB1 mRNA in the chick brain, a non-mammalian vertebrate. The results were compared to the observed patterns of expression for CB1 mRNA, protein, and agonist binding that have been reported for other avian species and mammals. Importantly, since CB1 receptors are typically located on neuronal terminals, comparison of the somatic mRNA expression with previously reported descriptions of the location of functional receptors, allows speculation about the circuits that make use of these receptors. The expression pattern for CB1 mRNA appears to be highly conserved across species in key areas such as the cerebellum and portions of the forebrain. For example, high levels of expression were observed in the avian amygdala and hippocampus, areas which express high levels of CB1 in mammals. The avian substantia nigra and ventral tegmental area, however, showed specific labeling. This finding is in stark contrast to the high levels of receptor binding or CB1 protein, but not CB1 mRNA in these areas of the mammalian brain. Moderate labeling was also seen throughout the hyperpallium and mesopallium. Throughout the brain, a number of regions that are known to be involved in visual processing displayed high levels of expression. For example, the tectum also had strong mRNA expression within layers 9-11 of the stratum griseum et fibrosum superficale and stratum album centrale.
Keywords: avian, comparative anatomy, tectum, striatum, sensory systems
1. Introduction
Cannabinoid (CB) signaling is rapidly gaining recognition as a widespread and influential form of signal modulation in the brain. At least two subtypes of receptors, CB1 (Matsuda et al., 1990) and CB2 (Munro et al., 1993), have been identified. CB2 is predominately found in cells of the immune system, and it is only present in microglia within the brain (Nunez et al., 2004). CB1, however, is highly abundant in the brain and is involved in synaptic signaling. Analysis of sequence conservation has revealed high sequence identity of orthologs with human CB1 (97% rat, 91% zebra finch, and 83% newt salamander) (McPartland and Glass, 2003; Soderstrom and Johnson, 2000). The genome of the puffer fish, Fugu rubripes, also contains a CB1 ortholog (Yamaguchi et al., 1996), albeit with much lower sequence identity to human CB1, 59% (McPartland and Glass, 2003). These findings suggest that CB1 orthologs are present in a wide variety of vertebrate species, and that sequence conservation is high.
While the sequence of the CB1 receptor appears to be highly conserved across vertebrates, it is unclear if the localization of cells producing this receptor in the brain is likewise conserved. The pattern of mammalian CB1 localization has been well characterized utilizing receptor binding (Devane et al., 1988; Herkenham et al., 1991), in situ hybridization (ISH) (Mailleux et al., 1992; Matsuda et al., 1993), and immunohistochemistry (IHC) (Egertova and Elphick, 2000; Pettit et al., 1998; Tsou et al., 1998). However, relatively little is known of CB1 localization in non-mammalian species. The goal of this investigation was to determine the localization of CB1 mRNA in the chick brain, Gallus domesticus, a non-mammalian vertebrate. The publication of the chick genome (Consortium, 2004) makes this animal a potentially useful model for the study of gene expression in non-mammalian vertebrates. The localization of CB1 was examined throughout the chick brain using ISH and regions displaying distinct, above-background labeling were objectively analyzed by densitometry.
CB1 mRNA and protein have been observed in regions of the zebra finch telencephalon, and this receptor appears to be important for vocal production (Soderstrom and Johnson, 2000; Soderstrom et al., 2000; Soderstrom and Tian, 2006). Localization of CB1 receptor has also been described through binding assays in the budgerigar brain (Alonso-Ferrero et al., 2006). The present study of mRNA expression in the chick brain allows for two important comparisons with these previous studies. The first important comparison comes from the fact that the previous studies have focused on receptor localization in avian species capable of vocal learning. The present analyses of the chick, a non-vocal learning species, will aid in understanding which aspects of CB1 expression are general across avian species and which aspects may be specialized for vocal learning. The second important comparison comes from the fact that CB1 receptors are typically inserted at axon terminals, whereas mRNA is typically localized in cell bodies. Consequently, if CB1 is contained in projection neurons, then mRNA expression and protein expression patterns will not be identical. Thus, the comparison of the present mRNA results with prior results on the localization of functional receptors in birds (Alonso-Ferrero et al., 2006) not only provides information about which areas of the brain produce CB1 receptors, but also allows speculation into the functional circuitries that may utilize these receptors.
2. Results
2.1 Probe Specificity
Probe specificity was confirmed in three ways. First, Northern analysis using our CB1 probe revealed a single band on lanes containing RNA from cerebellum and brain stem, but lanes containing heart and liver RNA had no labeling. Since CB1 is believed to be selectively expressed in the nervous system, the selective labeling of RNA from the nervous system tissue suggests that the probe was specifically labeling CB1. Hybridization of the 28S rRNA probe showed the presence of 28S rRNA in all lanes. This indicates that the lack of CB1 labeling in heart and liver was not because of insufficient RNA. Relative positions of the bands revealed that the CB1 mRNA was of greater length than 28S rRNA (4.7kb). This is consistent with previous reports of CB1 mRNA in zebra finch (∼5.5kb) (Soderstrom and Johnson, 2000).
Quantitative RT-PCR was carried out to further establish the specificity and validity of our new chick CB1 mRNA probe. The CB1 primers with cerebellar cDNA showed detectable signal at 17.7 cycles. Both liver and cerebellum cDNA with RP27 primers (household gene) displayed a signal first at 20.6 and 20.8 cycles respectively. The CB1 primers with liver cDNA did not produce a signal until 26.4 cycles. Negative control wells showed no significant signal. PCR product was run on an agarose gel and a single band of the appropriate length was confirmed. Furthermore, the PCR product identity was confirmed through sequencing (FSU DNA Sequencing Laboratory).
Finally, the specificity of the ISH was confirmed using a riboprobe version of the CB1 probe. The anti-sense CB1 riboprobe produced a pattern of labeling that was identical to that obtained using the cDNA CB1 probe. A sense-control riboprobe, however, resulted in only diffuse, non-specific labeling at background levels.
2.2 Qualitative in situ Hybridization results
The distribution of CB1 throughout the brain is widespread and heterogeneous. Labeling can be seen in every section but high levels of labeling were observed only in specific subregions of the brain. Anatomical schematics were created using both autoradiographs and nissl stain images from the same tissue sections. Chick stereotaxic atlases (Kuenzel and Masson, 1988; Puelles et al., 2007) were used to identify neural structures, and the revised avian nomenclature (Reiner et al., 2004) was used in reference to these regions. Qualitative impressions of all autoradiographs (ARs) were made and objective measurements were performed on selected regions.
ARs from post-hatch day 20 chicks confirmed that the pattern of CB1 mRNA labeling in post-hatch day 10 chicks was consistent at this older age. No quantitative measurements were made on films from 20 day old chicks. The labeling pattern did appear more distinct with a lower background. This improved signal to noise ratio, however, could be due to any number of processing variables.
Hindbrain
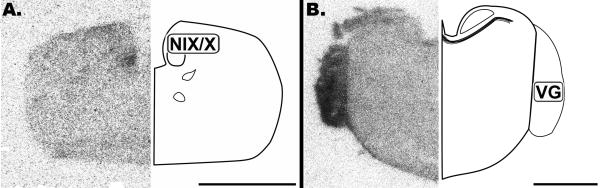
The brainstem overall contained little CB1 mRNA (Figure 1 and 2). However, the glossopharyngeal/vagal nucleus (NIX/X) and vestibular ganglion (VG) were found to strongly express CB1 (Figure 1). Some slight labeling could only be discerned upon close examination of the cochlear nuclei, nucleus magnocellularis (NM) and angularis (NL) (Figure 2).
Figure 1.
Coronal 20μm autoradiographs showing CB1 mRNA expression (left) and corresponding anatomical schematic (right). Low levels of labeling are observed in the brain stem except for cranial nerve nuclei. NIX/X: Glossopharyngeal/Vagal Nucleus, VG: Vestibular Ganglion. Scale bar = 2mm.
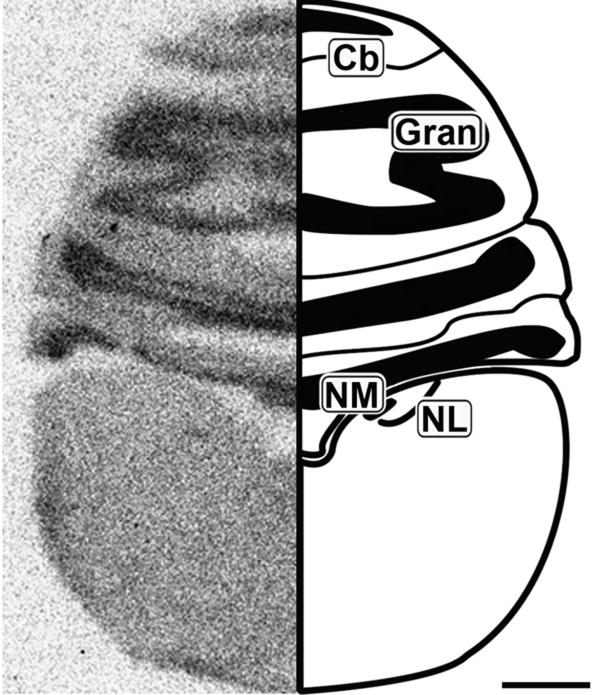
Figure 2.
Caudal coronal 20μm autoradiograph showing CB1 mRNA expression (left) and corresponding anatomical schematic (right). High levels of expression are observed in the granule cell region of the cerebellum. Cb: Cerebellum, Gran: Granule Cell Layer, NM: Nucleus Magnocellularis, NL: Nucleus Laminaris. Scale bar = 1mm.
Cerebellum
The coronal section in Figure 2 shows robust layer-specific labeling of the cerebellum (Cb). The granule cell layer (Gran) of the Cb contains dense CB1 expression, but the molecular layer also contains light to moderate labeling.
Midbrain
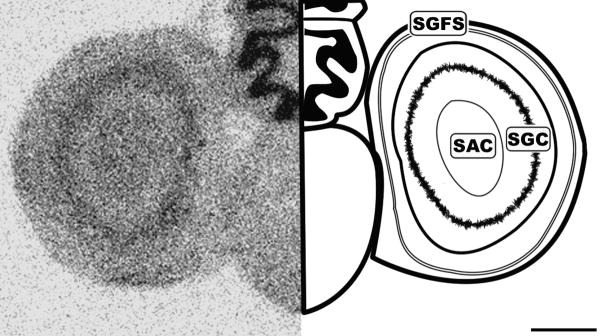
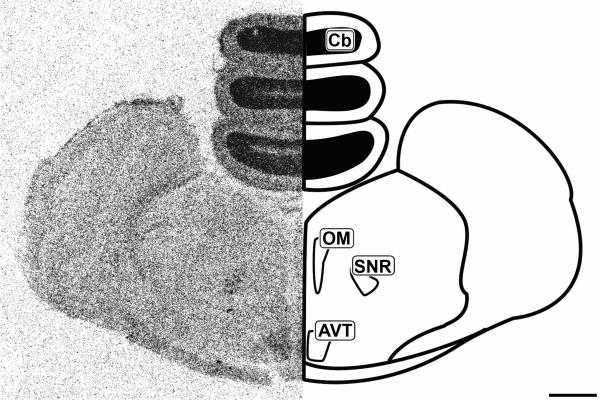
Histologically, the tectum (TeO) is divided into layers arranged in a semi-concentric fashion. In line with the morphological organization, CB1 expression was restricted to very specific regions of the tectum. The most external layers labeled are of the stratum griseum et fibrosum superficale layers 9-11 (SGFS). The SGFS labeling was light, but very distinct in many sections (Figures 3 and 4). More central and darker tectal labeling near the tectal ventricle was seen in the stratum album centrale (SAC) and the stratum griseum central (SGC) (Figures 3 and 4). Figure 5 clearly shows the area ventralis tegmentalis (AVT) and substantia nigra, pars reticulata (SNR) are labeled.
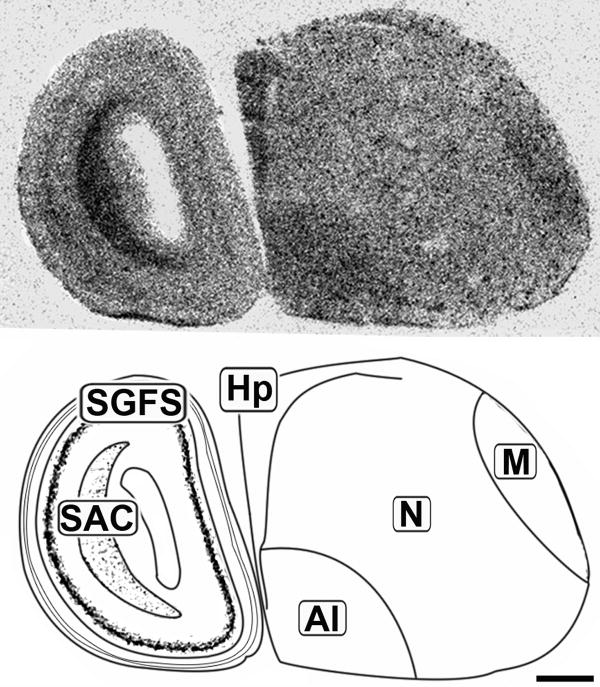
Figure 3.
Lateral sagittal 20μm autoradiograph showing CB1 mRNA expression (left) and corresponding anatomical schematic (right). Areas showing high levels of expression are highlighted on the schematic. Hp: Hippocampus, M: Mesopallium, N: Nidopallium, AI: Arcopallium Intermedium, SGFS: Stratum Griseum et Fibrosum, SAC: Stratum Album Centrale. Scale bar = 1mm.
Figure 4.
Caudal coronal 20μm autoradiograph showing CB1 mRNA expression (left) and corresponding anatomical schematic (right). Note the specific labeling within the tectum. SGFS: Stratum Griseum et Fibrosum, SAC: Stratum Album Centrale, SGC: Stratum Griseum Centrale. Scale bar = 1mm.
Figure 5.
Mid-coronal 20μm autoradiograph showing CB1 mRNA expression (left) and corresponding anatomical schematic (right). Areas showing high levels of expression are highlighted on the schematic. Cb: Cerebellum, OM: Occulomotor complex, SNR: Substantia Nigra pars reticulata, AVT: Area Ventralis Tegmenti. Scale bar = 1mm.
Forebrain
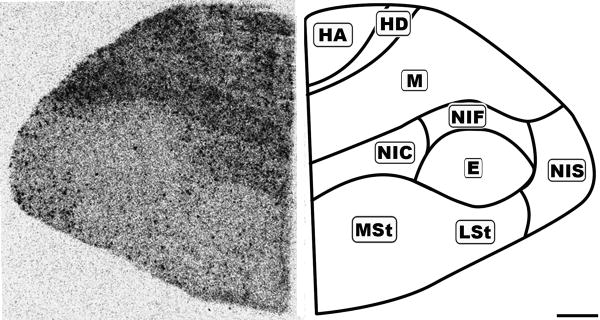
While the forebrain labeling was not as uniformly dense as in the granule cell layer of the Cb, it displayed diffuse, moderate labeling with an overlay of dark punctate labeling evenly distributed throughout (Figures 6-8). The resolution of the autoradiographs did not permit cellular localization and therefore it could not be determined if the punctate labeling was associated with any specific neuronal subtype. The nidopallium (N) contained light labeling. The only region lacking the underlying diffuse labeling was the entopallium (E), which still contained the distributed punctate labeling.
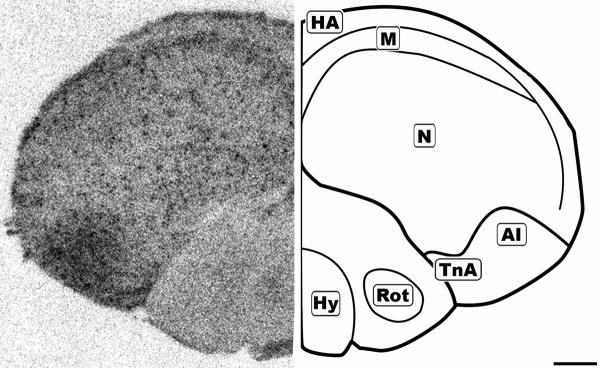
Figure 6.
Mid-coronal 20μm autoradiograph showing CB1 mRNA expression (left) and corresponding anatomical schematic (right). Areas showing high levels of expression or notable absense of expression are highlighted on the schematic. HA: Hyperpallium Apicale, M: Mesopallium, N: Nidopallium, AI: Arcopallium Intermedium, TnA: Nucleus Taeniae Amygdalae, Hy: Hypothalamus, Rot: Nucleus Rotundus. Scale bar = 1mm.
Figure 8.
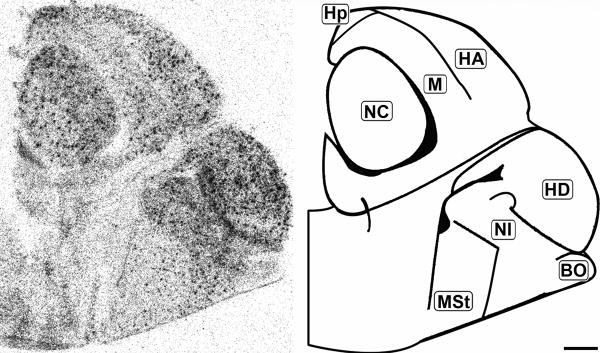
Rostral coronal 20μm autoradiograph showing CB1 mRNA expression (left) and corresponding anatomical schematic (right). Note the regional distribution of labeling. HA: Hyperpallium Apicale, HD: Hyperpallium Densocellulare, M: Mesopallium, NC: Nidopallium Intermedium Centrale, NF: Nidopallium Intermedium Frontale, NI: Nidopallium Intermedium Superficale, StM: Striatum Mediale, StL: Striatum Laterale. Scale bar = 1mm.
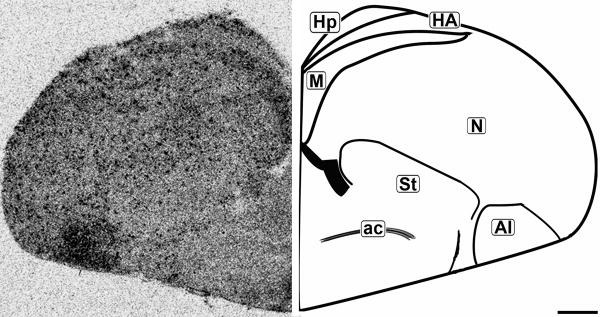
The concentration of CB1 mRNA was relatively high in the apical part of the hyperpallium (HA), mesopallium (M), and hippocampus (Hp) (Figures 3, 6-8). Within the Hp the superficial neurons were more darkly labeled than in the rest of this structure (Figure 3, 7 and 9). Figure 6 and 7 display coronal sections of the caudal forebrain. The dark ventromedial labeling was found in the contiguous structures arcopallium intermedium (AI) and nucleus taeniae amygdalae (TnA) which comprise the majority of the avian amygdala. Figures 8 and 9 show several, more distinct neuroanatomical subdivisions within the chick forebrain.
Figure 7.
Coronal 20μm autoradiograph showing CB1 mRNA expression (left) and corresponding anatomical schematic (right). Note the punctate pattern of labeling throughout the telecephelon, with darker labeling in specific subregions. Hp: Hippocampus, HA: Hyperpallium Apicale, M: Mesopallium, N: Nidopallium, St: Striatum, AI: Arcopallium Intermedium, ac: Anterior Commissure. Scale bar = 1mm.
Figure 9.
Mid-sagittal 20μm autoradiograph showing CB1 mRNA expression (left) and corresponding anatomical schematic (right). Hp: Hippocampus, HA: Hyperpallium Apicale, NC: Nidopallium Centrale, HD: Hyperpallium Densocellulare, NI: Nidopallium Intermedium, StM: Striatum Mediale, BO: Olfactory Bulb. Scale bar is 1mm.
2.3 Quantitative Results
To provide an objective measure of relative labeling intensity, the mean gray scale density of labeling was measured for selected “labeled” nuclei. Mean gray scale values were measured with NIH ImageJ. At a minimum, two measurements of a given region of interest (ROI) were taken per tissue section. The lightest tissue gray scale value was determined and that background value was subtracted from mean gray scale value over the ROIs on the same section. When regions could be sampled on multiple sections in the set of slides, the gray scale values were averaged across sections. The gray scale measurements were normalized to the gray scale density measured over the granule cell layer of the cerebellum (the area of most consistent and highest labeling) to account for processing variations, and the ratios of selected areas are presented in Figure 10. The objective data were consistent with the qualitative impressions.
Figure 10.
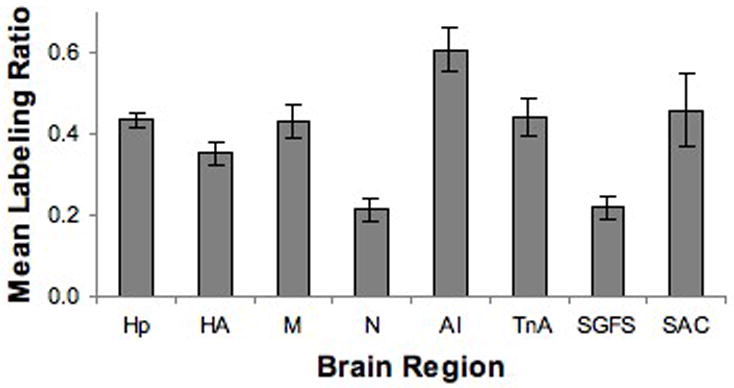
Mean density of selected brain regions relative to the labeling density of the cerebellum in the same brain. The objective analyses of labeling density corresponded with the visual impressions. Hp: Hippocampus, HA: Hyperpallium, M: Mesopallium, N: Nidopallium, AI: Arcopallium Intermedium, TnA: Nucleus Taeniae Amygdalae, SGFS: Stratum Griseum et Fibrosum, SAC: Stratum Album Centrale. Error bars = SEM.
3. Discussion
Expression of CB1 mRNA occurs throughout the chick brain, but distinct labeling is present within a variety of inter-region circuits involved in sensory processing, motor control, and sensorimotor integration. The present analysis was restricted to regions that showed clear labeling in all samples. This does not rule out the possibility that a more thorough analysis will reveal additional areas of CB1 expression. Nevertheless, the patterns of expression observed provide some general trends. While it cannot be known for certain that the CB1 localization observed in the 10-to-20-day-old chick is the same as would be observed in the adult, developmental analyses of a variety of neurotransmitter systems suggest that adult-like patterns are typically established by late embryonic or early posthatch ages. This includes studies of GABA (Burger et al., 2005; Fan et al., 1997; Rodriguez Gil et al., 2005), NMDA (Tang and Carr, 2004), bombesin, (Maekawa et al., 2004), acetylcholine (Kaneko et al., 1998), dopamine (Soares et al., 2000) and serotonin (Wallace, 1985) receptors. With this caveat in mind, the following discussion will summarize the labeling patterns observed in the chick and compare these findings to those reported for other species.
Discrepancies are expected between various studies of CB1 localization. As noted previously, ISH will only reveal the location of cell bodies producing CB1 mRNA, whereas analyses of the location of the functional receptor will show the location of the cells' terminals. Autoradiography (AR) from CB1 receptor agonists will have a labeling pattern more consistent with those seen when using IHC. However, the epitope of the different antibodies used also affects the results produced by IHC labeling. (Ashton et al., 2004; Egertova and Elphick, 2000; Pettit et al., 1998).
3.1 Sensory Processing
CB1 appears to be present in areas that perform the earliest stages of sensory processing. Previous studies utilizing ISH, IHC, or receptor binding have found the presence of this receptor in the nucleus of the solitary tract (Herkenham et al., 1991), olfactory bulb (Pettit et al., 1998; Tsou et al., 1998), sensory ganglia (Hohmann and Herkenham, 1999; Salio et al., 2002; Sanudo-Pena et al., 1999; Straiker et al., 1999), and retina (Straiker et al., 1999) in a number of species. In addition, the spinal cord (Salio et al., 2002), dorsal root ganglion (Bridges et al., 2003), and trigeminal ganglion (Price et al., 2003) have been found to produce high levels of CB1 mRNA or to contain CB1 protein in mammalian species. IHC localization of CB1 has also been observed in the vestibular and auditory brain stem nuclei (Ashton et al., 2004). Our finding of robust CB1 mRNA in the vestibular ganglion is consistent with these data and suggests that the CB1 IHC labeling in these brain stem nuclei may be on the terminals of the VIIIth nerve. Together, the localization data suggest that CB signaling may allow a form of modulation important in the earliest stages of sensory processing.
3.2 Visual system
The present study revealed a trend for CB1 to be highly expressed in areas of the chick brain that process visual information. This is consistent with previous findings of CB influences in visual processing. A detailed localization study found CB1 in the retina across several species including monkey, mouse, chick, tiger salamander, and goldfish (Straiker et al., 1999).
In the present study, the TeO was found to contain CB1 mRNA in distinct subregions. The TeO of vertebrates receives retinal input and is organized into layers (Cajal, 1909, 1910). The avian TeO is distinguished from the mammalian by both a greater relative size and increased laminar organization. 15 layers of the avian TeO are numbered external to internal as described by the chick brain atlas (Kuenzel and Masson, 1988). Within these layers exists a highly organized set of connections receiving input from the retina (Layers 2-5, 7) which relay this information through both ascending and descending pathways.
Specific labeling was observed in layers 9-11 of the SGFS. The majority of cells within these layers relay information to brain regions outside the TeO. This is consistent with the relatively low CB1 receptor binding within the SGFS proper of the budgerigar (Alonso-Ferrero et al., 2006).
A band of cells between layers 9 and 10 provides the sole input for the nucleus isthmo-opticus (ION) (Woodson et al., 1991). The ION represents an important nucleus in the closed feedback loop between the brain and retina, modulating early sensory processing (Cowan and Clarke, 1976; Gardino et al., 2004).
Layers 10 and 11 exclusively output to the ventral part of the lateral geniculate nucleus (GLv) (Hu et al., 2004b). Ascending information is sent through the thalamofugal pathway, which projects from the TeO to the GLv and onto the area pretectalis (Hu et al., 2004a), and is ultimately important for color vision (Maturana and Varela, 1982) and oculomotor functions (Gioanni et al., 1991; Pateromichelakis, 1979).
Neurons involved in a parallel ascending pathway from the TeO may also contain CB1 mRNA. Some of the cells found throughout layers 8-12 innervate Layer 13 cells which in turn project to nucleus triangularis (Hellmann and Gunturkun, 2001). There has been little functional characterization of the nucleus triangularis, but it does project to the E and may modulate tectofugal processing. Nucleus rotundus (Rot) is the main relay point in the tectofugal pathway and a region of discrepancy between budgerigar and chick CB1 data. While the budgerigar displayed both high receptor and binding stimulation values (Alonso-Ferrero et al., 2006), no significant mRNA signal was observed in the chick Rot. Furthermore, we observed no signal in nucleus subpretectalis (SP) which projects to Rot. These findings leave open the question as to whether the cell bodies with CB1 terminals in Rot are from a source other than SP or if this is a point of divergence between chick and budgerigar.
CB1 signaling may also be functional in synapses between the SAC and portions of M (Bradley et al., 1985). We observed strong labeling in the SAC. While there is little CB1 immunoreactivity reported in M and only moderate receptor AR, the agonist stimulated rates of binding suggest strong CB1 activity within M (Alonso-Ferrero et al., 2006). The IMM (Intermediate Medial Mesopallium), a subregion of M, is implicated in imprinting (Bateson, 1966; Bolhuis, 1991).
3.3 Motor Control
Brain motor regions such as the cerebellum also contain very high concentrations of CB1 mRNA. Our finding of uniformly strong labeling of the granule cell layer of the chick Cb is consistent with mammalian studies (Herkenham et al., 1991; Herkenham et al., 1990; Mailleux et al., 1992; Matsuda et al., 1993). The chick Cb had greater CB1 mRNA labeling than any other measured region. A contrasting pattern of CB1 mRNA and protein labeling is seen in the rat. CB1 mRNA is located in cells of the granular layer (Mailleux et al., 1992; Mailleux and Vanderhaeghen, 1992), whereas the protein is located in the molecular layer (Egertova and Elphick, 2000). Receptor binding in the budgerigar and IHC in the zebra finch also show high levels of CB1 in the molecular layer (Alonso-Ferrero et al., 2006; Soderstrom and Tian, 2006). This pattern of results suggests that CB1 is localized to the terminals of parallel fibers, which synapse with Purkinje cells. CB1 activation is able to cause a cessation of nearly all presynaptic input to the Purkinje cells (Takahashi and Linden, 2000). This is not mediated through a mechanism which affects the firing of granule cells or fiber kinetics, but rather, through suppression of neurotransmitter release. The consistency of the CB1 mRNA labeling across species would suggest protein localization would be similar between the avian and mammalian cerebellum and, importantly, a conserved functional role.
The mammalian Cb has strong CB1 labeling, but the basal ganglia is home to specific subregions having the greatest CB1 mRNA density in the brain (Herkenham et al., 1991; Herkenham et al., 1990; Mailleux et al., 1992). However, disparity does exist between the labeling seen with ISH and receptor binding/IHC (Moldrich and Wenger, 2000; Westlake et al., 1994), suggesting translocation of protein down axons to the terminals where the CB1 is inserted in the membrane. Excitotoxic lesion studies have shown that striatal and subthalamic nucleus cells produce the majority of CB1 in the basal ganglia (Herkenham et al., 1991). Double-label ISH has shown that the majority of CB1 mRNA in the striatum is found in striatal efferent projection neurons (Hohmann and Herkenham, 2000). The axons of these neurons terminate in the substantia nigra and pallidal region.
CB1 expression is observed in both the SN and AVT of the chick. Localization of CB1 mRNA in cells of the chick SN contrasts with the presence of CB1 only in terminals innervating the mammalian SN. Although the dorsal telencephalon, containing associative regions, has high CB1 expression, the ventral motor regions have relatively weak labeling. Neither the VTA (homolog of AVT) nor the SN is labeled in mammalian brains (Westlake et al., 1994). The mammalian SN is labeled with receptor AR (Herkenham et al., 1991), suggesting that the cells of the SN do not produce CB1, but receive input from terminals containing the receptor.
Consistent with the mammalian pattern (Matsuda et al., 1993), CB1 mRNA was not observed in the globus pallidus (GP). In contrast to the mammalian pattern, however, there was little labeling in areas of the medial or lateral striatum that are believed to project to the GP (Reiner et al., 1984). It remains possible that a more detailed examination of these motor regions in the chick may reveal that some small nuclei were overlooked, but no pervasive labeling was observed in these areas. The contrast between avian and mammalian brains is intriguing in that it suggests that cellular signaling may differ greatly even where neural circuitry is conserved.
3.4 Integrative Pathways
The forebrain has the greatest total concentration of CB1 mRNA labeling, both in mammalian and avian species. Combining knowledge of afferent and efferent projections within the forebrain with CB1 mRNA localization allows some speculation regarding the functional significance of this receptor. Significant expression in the telencephalon was expected not only due to the previous reports in mammals, but specifically in light of extensive investigations into the localization of CB1 within the zebra finch telencephalon and the effect of CBs on vocal learning (Soderstrom and Johnson, 2001; Soderstrom and Johnson, 2003; Soderstrom and Tian, 2004). The distinct spots seen with ISH are not reflected in receptor binding data (Alonso-Ferrero et al., 2006). This suggests that projections from these sub-areas may be spreading out diffusely across the telencephalon.
The avian hippocampal formation, while morphologically dissimilar to that of mammals, shares many functional characteristics (Szekely, 1999). Like the mammalian hippocampus, high levels of CB1 receptor binding are observed in the avian brain (Alonso-Ferrero et al., 2006). The present study revealed that the chick Hp contains moderate to heavy CB1 mRNA labeling. This suggests that the CB1 receptors in the hippocampus may arise from cells within the Hp, rather than from projection neurons. The localization of CB1 mRNA to the hippocampus is not surprising since the effects of CBs on memory have been observed numerous times (Diana et al., 2003; Molina-Holgado et al., 1995).
While the mammalian amygdala displays moderate CB activity (Herkenham et al., 1990), the chick amygdala contains high levels of CB1 mRNA (present data) and agonist binding (Alonso-Ferrero et al., 2006). The avian amygdala complex contains portions of the region previously known as the archistriatum, AI and TnA (Consortium, 2004). The labeling in the AI was intense and greater than the labeling observed in TnA. The pattern of labeling in the chick is to an extent similar to that observed in the zebra finch with the exception of the robust nucleus of the arcopallium (RA), which is only found in songbirds (Soderstrom and Johnson, 2000). The AI has reciprocal connections to Hp, HA, M, and N as well as sending additional afferents to APH, HD, and SAC, notably AI directly projects to the ventral striatum (Csillag, 1999; Davies et al., 1997).
TnA receives input from the hippocampus and sends major projections to the hypothalamus (Cheng et al., 1999). In light of the ability of cannabis use to induce paranoia in humans and studies demonstrating that CBs affect fear conditioning in rats and mice (Chhatwal et al., 2005; Marsicano et al., 2002) labeling of these limbic regions are not surprising. Receptor binding in the budgerigar displayed quite a bit more binding in TnA versus AI, but agonist stimulated GTPγS binding values were much more on par with each other (Alonso-Ferrero et al., 2006).
The M constitutes a major area of integration for the chick brain, receiving a large number of afferent projections (Brauth et al., 2001; Krutzfeldt and Wild, 2004; Krutzfeldt and Wild, 2005; Montagnese et al., 2003) and sending out many efferents. The M was marked by moderate labeling, higher than that in the HA, but lower than that observed in TnA and AI The M has been implicated in a variety of functions which may differ between parrots, songbirds, and non-songbirds.
The majority of afferent input to M comes ipsilaterally, however there is significant contralateral input as well. In addition to a role in imprinting, the IMM is also involved in passive avoidance learning (Csillag et al., 1993; Patel et al., 1988; Patel and Stewart, 1988; Rose and Csillag, 1985). CB1 antagonists disrupt passive avoidance, further supporting the role of cannabinoid signaling in memory consolidation (Adam et al., 2008). Efferent projections are sent to parts of the chick limbic system.
In addition, the arcopallium, TnA, and Hp relay information from the IMM, a region believed to play a role in imprinting, to the medial and lateral striatum, formerly Parolfactory lobe (Bradley et al., 1981; Horn, 1981; Horn et al., 1985). While IHC and AR labeling in M is unremarkable the agonist stimulated GTPγS binding has a very high ratio, suggesting strong cannabinoid activity despite a relatively modest concentration of CB1 (Alonso-Ferrero et al., 2006; Soderstrom and Tian, 2006). Therefore, cannabinoids could potentially affect the process of imprinting.
The avian Wulst (W) is comparable to some areas of the mammalian neocortex. Higher sensory processing and integration occurs here including auditory, visual, and somatosensory (Funke, 1989; Wild, 1987; Wild, 1993). The W is composed of three pseudolayers, the HA, intercalculated part of the H (HI), and the densocellular part of the H (HD), located within the medial ridge of the avian telencephalon. Labeling appeared throughout this region, but was especially dense and distinct within the HA. The HA is especially linked with visual processing, but does contain some overlap with somatosensory related neurons (Deng and Wang, 1992). Therefore, CB1 activation in this area might be expected to result in effects on more complex visual tasks such as visual discrimination.
Lastly, while there are many subregions of the N could, these could not be easily determined through most coronal sections. However, sagittal sections revealed that the frontal, intermediate, and caudal portions of the N did differ slightly in their low CB1 mRNA concentration, which was approximately half to a quarter of the intensity observed in the M.
The N is a large region of the telencephalon involved in higher sensory processing, especially auditory, and used in associative learning (Braun et al., 1999). The N also contains auditory and vocal control nuclei involved in both the learning and production of song in some avian species (Nottebohm et al., 1976). The majority of connections between N and other telencephalic regions are reciprocal. These regions include the W, E, AI, and IMM (Nixdorf and Bischof, 1982). As the presence of CB1 mRNA would suggest, CB exposure does affect vocal learning in songbirds (Soderstrom and Johnson, 2003; Soderstrom and Tian, 2004). The labeling within the N differs greatly between chick and zebra finch. A potential small species differences may be seen in the N of the chick, which has a fairly regular pattern, unlike the zebra finch N which contains relatively low CB1 message except in the song learning region, HVC (proper name) (Soderstrom and Johnson, 2000).
A major difference in the chick labeling pattern may be in the relative lack of labeling in the areas analogous to the caudate-putamen or GP in the chick, compared to the robust labeling reported in the mammalian brain. In addition, specific labeling of CB1 mRNA was observed in the AVT and SN of the chick. This finding is in contrast to mammalian SN cells, which do not produce CB1 mRNA, but rather receive input from cells that do express CB1.
A second region that seems to show species-related differences is the TeO. High levels of CB1 expression were particularly localized to visual regions of the chick's TeO. In contrast, little message for CB1 is observed in the superior colliculus of the rat, and it appears that there is no obvious expression in either the nucleus parabigeminalis or the lateral tegmentum (Mailleux et al., 1992; Mailleux and Vanderhaeghen, 1992; Matsuda et al., 1993), two areas that have been suggested to be homologous to areas of the avian TeO (Wang et al., 2004).
Finally, robust expression was found in VG. This is consistent with CB1 IHC labeling in the mammalian vestibular and auditory brain stem nuclei (Ashton et al., 2004), suggesting that the source of this labeling is the VIIIth cranial nerve. This is consistent with the trend for CB1 localization at early stages of sensory processing, such as in the retina. Together these findings suggest that in addition to the acknowledged roles of CB signaling in motor control and higher functions such as learning and memory, CB signaling may also play a significant role in the earliest stages of sensory processing across a variety of sensory systems.
4. Experimental Procedures
4.1 In situ hybridization
Chickens (Ross X Ross) were hatched and reared to post-hatch day 10 at the Florida State University. Subjects were anesthetized with halothane, and sacrificed by decapitation. Whole brains were quickly extracted and rapidly frozen in 2-methylbutane cooled on dry ice. Brains were stored at -80°C until cryostat sectioning. The brains were allowed to equilibrate to -20°C in the cryostat at least 45 minutes before sectioning at 20 μm. A full series of sections was collected from six subjects. Three brains were cut in the sagittal plane. Sections were taken approximately every 150 μm. Three brains were cut coronally and sections were taken every 250 μm. Tissue sections were directly mounted on Fisher Superfrost Plus slides (Fisher Scientific, Pittsburg, PA), desiccated overnight, and stored at -80°C until use. Partial series of sections from an additional 3 subjects were qualitatively analyzed to confirm localization at selected brain regions. Another 3 subjects, post-hatch day 20, were later examined (2 coronal, 1 sagittal) in order to confirm consistency of CB1 mRNA pattern through latter stages of development.
A 702 base pair fragment of CB1 cDNA was isolated from a chicken brain cDNA library (Accession #AF80088) and sub-cloned into the pCR 2.1 vector (Invitrogen, Carlsbad, CA). The fragment was removed from the vector with restriction enzymes and isolated. The fragment was then used as a template for the Prime-It II Random Primer Labeling Kit (Stratagene, La Jolla, CA) to produce a cDNA probe. The probe was labeled with 33P dCTP (Perkin Elmer Life Sciences Inc., Boston, MA) and run through Micro Bio-spin purification columns (Bio-Rad, Hercules, CA) to remove any unincorporated dNTPs. The 33P isotope was chosen due to its balance of resolution, signal strength, and relative safety.
Specificity of the probe was determined through northern blot analysis. Total cellular RNA was isolated from the cerebellum, brain stem, heart, and liver of a post-hatch day 10 chick using Trizol reagent (Invitrogen, Carlsbad, CA). RNA from each region (25 μg) was subjected to agarose gel (1% agarose with 0.66 M formaldehyde) electrophoresis and transferred to a nylon membrane (GeneScreen, NEN/Dupont, Boston, MA) by capillary blotting. RNA transfer was confirmed by visualization of eithidium bromide-stained RNA under UV light. Blots were UV cross-linked and stored at 4°C until hybridization overnight at 55°C. Northern blot was hybridized to a 32 P-labeled cDNA probe based on the 702bp chick CB1 fragment (Prime-It II Random Primer Labeling Kit, Stratagene, La Jolla, CA) purified with Micro Bio-spin purification columns (Bio-Rad, Hercules, CA). The blot was exposed to Kodak MR film for 2-6 hrs. A 28S rRNA probe was created in the same manner as the CB1 probe and hybridized to the same blot following decay of the radioactivity in the CB1 probe.
A sense control was performed using a riboprobe version of the chick CB1 probe (data not shown) using a protocol adapted from previously published work (Krause et al., 2006; McCullumsmith et al., 2003). Alternating tissue sections were hybridized with either sense or anti-sense 35S riboprobe and exposed to film.
qRT-PCR provides additional support for the specificity of the CB1 probe. Primers were designed based on the genebank sequence for RP27 and our 702bp fragment of chick CB1 cDNA. Total RNA was extracted from both chick liver and cerebellum using Trizol (Invitrogen), quantified with a spectrophotometer, and examined on a agarose gel. RNA was used in conjunction with the iScript kit (Bio-Rad) to produce cDNA libraries. RT-PCR was carried out on an iCycler using iQ Supermix with sybr-green (Bio-Rad). RP27 and CB1 primers were used separately in wells containing liver cDNA, cerebellar cDNA, or no cDNA.
An entire set of slides from a given subject was processed using the same labeled probe. A set consisted of 7-12 slides with multiple sections on each slide. They were removed from the -80°C freezer and placed in 4% (weight/volume) paraformaldehyde at room temperature for 60 minutes. The slides were briefly dipped in 2× 300 mM NaCl/30 mM sodium citrate, pH 7.2 (Standard Sodium Citrate, SSC) to rinse off excess fixative and then washed in 2× SSC for 10 minutes. Sections were dehydrated with 30-second washes in graded ethanols (50%, 70%, 85%, 90%, 90% 100%, and 100%). Slides were allowed to air dry for at least 30 minutes before hybridization.
Purified probe was boiled with salmon sperm DNA for 2 minutes and added to 75% formamide buffer (75% formamide, 10% dextran sulfate, 3× SSC, 50 mM Na2HPO4, 10 mM dithiothreitol, 1× Denhardt's solution, 100 μg/ml yeast tRNA) warmed with a 68°C water bath. The probe/formamide mixture was kept warm at 68°C for 60 minutes before the hybridization procedure to provide time for the probe to become evenly distributed.
Each slide was placed face down on a coverslip containing 110 μl of 2.5 million counts/minute cDNA probe and formamide buffer. After aligning the coverslip and removing any air bubbles, the slides were placed in a covered plastic hybridization trays containing filter paper soaked in 75% formamide. Incubation at 51.6°C continued overnight. The coverslips were then removed and the slides underwent the following washes: 1) 2 minutes, 2× SSC, room temperature with rotation 2) 10 minutes, 2× SSC, room temperature, 3) 10 minutes 1× SSC, 4) 20 minutes, 0.5× SSC, 48°C, 5) 10 minutes, 0.5× SSC, room temperature. Washed slides were then dehydrated through graded ethanols. Slides were allowed to air-dry, placed in X-ray cassettes, and apposed to Kodak Biomax MR film for 3 days. After the film was developed, sections were stained for Nissl using thionin.
4.2 Image and data analyses
Film was developed using an automated film processor. Autoradiographs were imaged on a uniform illumination light box using a black and white Sony CCD camera connected to a G4 Macintosh computer through a framegrabber card (Scion Corporation, Frederick, MD). Images of the autoradiographs were captured and aligned with the matching Nissl stained section. The capturing and alignment of the images was done using the ImageJ program, a Java version of NIH Image. Once aligned, a region on the stained section was circled and the average gray scale value of the corresponding area on the autoradiograph was measured.
Average gray scale values (GSV) were measured for the selected regions of interest (ROI). ROIs were chosen by examining other reports of CB1 mRNA expression, visual inspection of labeling, and the interests of our laboratory. Tissue backgrounds were measured from white matter areas and these values were subtracted from ROI measurements. Consequently all areas with positive GSVs are, by definition, above background. When possible, both sides of a bilateral structure were measured and averaged. A minimum of two sections were measured and averaged for each region per subject. Large regions such as the mesopallium were sampled up to 10 times per subject. Nearly all ROIs appeared in several adjacent sections. In such cases the GSVs were measured for the ROI on two darkest, adjacent tissue sections and then averaged.
Normalization was done by considering each ROI GSV as a proportion of the labeling in the granular layer of each subject's cerebellum [(GSVROI)/(GSVcerebellum)]. For tissue sections containing cerebellum, the GSV of the cerebellum was used for that section's ROI measurements. On tissue sections containing no cerebellum, the subject's average GSV for the granule cell layer of the cerebellum was used in the calculation. The granule cell layer of the cerebellum was chosen due to the consistent, strong labeling and the presence of this region in many of the sections.
Table 1.
Abbreviations of avian brain regions.
| AI | Arcopallium Intermedium | NIS | Nidopallium Intermedium Superficiale |
| AVT | Area Ventralis Tegmenti | NIX/X | Glossopharyngeal/Vagal Nucleus |
| Cb | Cerebellum | NL | Nucleus Laminaris |
| E | Entopallium | NM | Nucleus Magnocellularis |
| GLv | Lateral Geniculate Nucleus | OM | Occulomotor complex |
| GP | Globus Pallidus | Rot | Nucleus Rotundus |
| Gran | Granule Cell Layer of the Cerebellum | SAC | Stratum Album Centrale |
| HA | Hyperpallium Apicale | SGC | Stratum Griseum Centrale |
| HD | Hyperpallium Densocellulare | SGFS | Stratum Griseum et Fibrosum |
| HI | Hyperpallium Intercalatum | SNR | Substantia Nigra, pars reticulata |
| Hp | Hippocampus | SP | Nucleus Subpretectalis |
| StL | Striatum Laterale | St | Striatum |
| StM | Striatum Mediale | TeO | Optic Tectum |
| N | Nidopallium | TnA | Nucleus Taeniae Amygdalae |
| NC | Nidopallium Centrale | VG | Vestibular Ganglion |
| NIC | Nidopallium Intermediium Centrale | W | Wulst |
| NIF | Nidopallium Intermedium Frontale |
Acknowledgments
We thank Dr. Ken Soderstrom PhD from East Carolina University for his isolation and subcloning of the chick CB1 cDNA fragment (Accession# AF80088), Rani Dhanarajan from the Florida State University Molecular Cloning Laboratory for subcloning of CB1 fragment, and Dr. Cathy Levenson PhD from Florida State University for assistance with northern blot analysis of the CB1 probe. This work was supported by NIDCD grant DC 000858 and NIH Jointly Sponsored Training Grant T32 NS07437
Abbreviations
- CB
Cannabinoid
- CB1
Cannabinoid Receptor 1
- IHC
immunohistochemistry
- ISH
in situ hybridization
- ROI
region of intererst
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Adam AS, Wenger T, Csillag A. The cannabinoid CB1 receptor antagonist rimonabant dose-dependently inhibits memory recall in the passive avoidance task in domestic chicks (Gallus domesticus) Brain Res Bull. 2008;76:272–4. doi: 10.1016/j.brainresbull.2008.02.004. [DOI] [PubMed] [Google Scholar]
- Alonso-Ferrero ME, Paniagua MA, Mostany R, Pilar-Cuellar F, Diez-Alarcia R, Pazos A, Fernandez-Lopez A. Cannabinoid system in the budgerigar brain. Brain Res. 2006;1087:105–13. doi: 10.1016/j.brainres.2006.02.119. [DOI] [PubMed] [Google Scholar]
- Ashton JC, Zheng Y, Liu P, Darlington CL, Smith PF. Immunohistochemical characterisation and localisation of cannabinoid CB1 receptor protein in the rat vestibular nucleus complex and the effects of unilateral vestibular deafferentation. Brain Res. 2004;1021:264–71. doi: 10.1016/j.brainres.2004.06.065. [DOI] [PubMed] [Google Scholar]
- Bateson PP. The characteristics and context of imprinting. Biol Rev Camb Philos Soc. 1966;41:177–211. doi: 10.1111/j.1469-185x.1966.tb01489.x. [DOI] [PubMed] [Google Scholar]
- Bolhuis JJ. Mechanisms of avian imprinting: a review. Biol Rev Camb Philos Soc. 1991;66:303–45. doi: 10.1111/j.1469-185x.1991.tb01145.x. [DOI] [PubMed] [Google Scholar]
- Bradley P, Davies DC, Horn G. Connections of the hyperstriatum ventrale of the domestic chick (Gallus domesticus) J Anat. 1985;140(Pt 4):577–89. [PMC free article] [PubMed] [Google Scholar]
- Bradley P, Horn G, Bateson P. Imprinting. An electron microscopic study of chick hyperstriatum ventrale. Exp Brain Res. 1981;41:115–20. doi: 10.1007/BF00236600. [DOI] [PubMed] [Google Scholar]
- Braun K, Bock J, Metzger M, Jiang S, Schnabel R. The dorsocaudal neostriatum of the domestic chick: a structure serving higher associative functions. Behav Brain Res. 1999;98:211–8. doi: 10.1016/s0166-4328(98)00086-2. [DOI] [PubMed] [Google Scholar]
- Brauth SE, Liang W, Roberts TF. Projections of the oval nucleus of the hyperstriatum ventrale in the budgerigar: relationships with the auditory system. J Comp Neurol. 2001;432:481–511. [PubMed] [Google Scholar]
- Bridges D, Rice AS, Egertova M, Elphick MR, Winter J, Michael GJ. Localisation of cannabinoid receptor 1 in rat dorsal root ganglion using in situ hybridisation and immunohistochemistry. Neuroscience. 2003;119:803–12. doi: 10.1016/s0306-4522(03)00200-8. [DOI] [PubMed] [Google Scholar]
- Burger RM, Pfeiffer JD, Westrum LE, Bernard A, Rubel EW. Expression of GABA(B) receptor in the avian auditory brainstem: ontogeny, afferent deprivation, and ultrastructure. J Comp Neurol. 2005;489:11–22. doi: 10.1002/cne.20607. [DOI] [PubMed] [Google Scholar]
- Cajal SR. Histologie du systeme nerveux de l'homme et des vertebres, vol. Maloine; Paris: 1909 1910. [Google Scholar]
- Cheng M, Chaiken M, Zuo M, Miller H. Nucleus taenia of the amygdala of birds: anatomical and functional studies in ring doves (Streptopelia risoria) and European starlings (Sturnus vulgaris) Brain Behav Evol. 1999;53:243–70. doi: 10.1159/000006597. [DOI] [PubMed] [Google Scholar]
- Chhatwal JP, Myers KM, Ressler KJ, Davis M. Regulation of gephyrin and GABAA receptor binding within the amygdala after fear acquisition and extinction. J Neurosci. 2005;25:502–6. doi: 10.1523/JNEUROSCI.3301-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Consortium ICGS. Sequence and comparative analysis of the chicken geome provide unique perspectives on vertebrate evolution. Nature. 2004;432:695–716. doi: 10.1038/nature03154. [DOI] [PubMed] [Google Scholar]
- Cowan WM, Clarke PG. The development of the isthmo-optic nucleus. Brain Behav Evol. 1976;13:345–75. doi: 10.1159/000123821. [DOI] [PubMed] [Google Scholar]
- Csillag A. Striato-telencephalic and striato-tegmental circuits: relevance to learning in domestic chicks. Behav Brain Res. 1999;98:227–36. doi: 10.1016/s0166-4328(98)00088-6. [DOI] [PubMed] [Google Scholar]
- Csillag A, Stewart MG, Szekely AD, Magloczky Z, Bourne RC, Steele RJ. Quantitative autoradiographic demonstration of changes in binding to delta opioid, but not mu or kappa receptors, in chick forebrain 30 minutes after passive avoidance training. Brain Res. 1993;613:96–105. doi: 10.1016/0006-8993(93)90459-z. [DOI] [PubMed] [Google Scholar]
- Davies DC, Csillag A, Szekely AD, Kabai P. Efferent connections of the domestic chick archistriatum: a phaseolus lectin anterograde tracing study. J Comp Neurol. 1997;389:679–93. doi: 10.1002/(sici)1096-9861(19971229)389:4<679::aid-cne10>3.0.co;2-7. [DOI] [PubMed] [Google Scholar]
- Deng C, Wang B. Overlap of somatic and visual response areas in the Wulst of pigeon. Brain Res. 1992;582:320–2. doi: 10.1016/0006-8993(92)90149-4. [DOI] [PubMed] [Google Scholar]
- Devane WA, Dysarz FA, 3rd, Johnson MR, Melvin LS, Howlett AC. Determination and characterization of a cannabinoid receptor in rat brain. Mol Pharmacol. 1988;34:605–13. [PubMed] [Google Scholar]
- Diana G, Malloni M, Pieri M. Effects of the synthetic cannabinoid nabilone on spatial learning and hippocampal neurotransmission. Pharmacol Biochem Behav. 2003;75:585–91. doi: 10.1016/s0091-3057(03)00127-8. [DOI] [PubMed] [Google Scholar]
- Egertova M, Elphick MR. Localisation of cannabinoid receptors in the rat brain using antibodies to the intracellular C-terminal tail of CB. J Comp Neurol. 2000;422:159–71. doi: 10.1002/(sici)1096-9861(20000626)422:2<159::aid-cne1>3.0.co;2-1. [DOI] [PubMed] [Google Scholar]
- Fan SS, Chang NC, Chang A, Yin HS. Differential expression of the GABAA receptor alpha 1 subunit in developing chicken brain. Neuroreport. 1997;8:2399–404. doi: 10.1097/00001756-199707070-00059. [DOI] [PubMed] [Google Scholar]
- Funke K. Somatosensory areas in the telencephalon of the pigeon. I. Response characteristics. Exp Brain Res. 1989;76:603–19. doi: 10.1007/BF00248917. [DOI] [PubMed] [Google Scholar]
- Gardino PF, Schmal AR, Calaza Kda C. Identification of neurons with acetylcholinesterase and NADPH-diaphorase activities in the centrifugal visual system of the chick. J Chem Neuroanat. 2004;27:267–73. doi: 10.1016/j.jchemneu.2004.04.004. [DOI] [PubMed] [Google Scholar]
- Gioanni H, Palacios A, Sansonetti A, Varela F. Role of the nucleus geniculatus lateralis ventralis (GLv) in the optokinetic reflex: a lesion study in the pigeon. Exp Brain Res. 1991;86:601–7. doi: 10.1007/BF00230533. [DOI] [PubMed] [Google Scholar]
- Hellmann B, Gunturkun O. Structural organization of parallel information processing within the tectofugal visual system of the pigeon. J Comp Neurol. 2001;429:94–112. doi: 10.1002/1096-9861(20000101)429:1<94::aid-cne8>3.0.co;2-5. [DOI] [PubMed] [Google Scholar]
- Herkenham M, Lynn AB, de Costa BR, Richfield EK. Neuronal localization of cannabinoid receptors in the basal ganglia of the rat. Brain Res. 1991;547:267–74. doi: 10.1016/0006-8993(91)90970-7. [DOI] [PubMed] [Google Scholar]
- Herkenham M, Lynn AB, Little MD, Johnson MR, Melvin LS, de Costa BR, Rice KC. Cannabinoid receptor localization in brain. Proc Natl Acad Sci U S A. 1990;87:1932–6. doi: 10.1073/pnas.87.5.1932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hohmann AG, Herkenham M. Localization of central cannabinoid CB1 receptor messenger RNA in neuronal subpopulations of rat dorsal root ganglia: a double-label in situ hybridization study. Neuroscience. 1999;90:923–31. doi: 10.1016/s0306-4522(98)00524-7. [DOI] [PubMed] [Google Scholar]
- Hohmann AG, Herkenham M. Localization of cannabinoid CB(1) receptor mRNA in neuronal subpopulations of rat striatum: a double-label in situ hybridization study. Synapse. 2000;37:71–80. doi: 10.1002/(SICI)1098-2396(200007)37:1<71::AID-SYN8>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- Horn G. Neural mechanisms of learning: an analysis of imprinting in the domestic chick. Proc R Soc Lond B Biol Sci. 1981;213:101–37. doi: 10.1098/rspb.1981.0057. [DOI] [PubMed] [Google Scholar]
- Horn G, Bradley P, McCabe BJ. Changes in the structure of synapses associated with learning. J Neurosci. 1985;5:3161–8. doi: 10.1523/JNEUROSCI.05-12-03161.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu M, Naito J, Chen Y, Ohmori Y, Fukuta K. Afferent and efferent connections of the nucleus geniculatus lateralis ventralis demonstrated by WGA-HRP in the chick. Anat Histol Embryol. 2004a;33:192–5. doi: 10.1111/j.1439-0264.2004.00534.x. [DOI] [PubMed] [Google Scholar]
- Hu M, Naito J, Chen Y, Ohmori Y, Fukuta K. Morphological characteristics of tectal neurons of layer I projecting to the nucleus geniculatus lateralis ventralis in chick. J Vet Med Sci. 2004b;66:1015–6. doi: 10.1292/jvms.66.1015. [DOI] [PubMed] [Google Scholar]
- Kaneko WM, Britto LRG, Lindstrom JM, Karten HJ. Distribution of the alpha7 nicotinic acetylcholine receptor subunit in the developing chick cerebellum. Brain Res Dev Brain Res. 1998;105:141–5. [PubMed] [Google Scholar]
- Krause EG, Curtis KS, Stincic TL, Markle JP, Contreras RJ. Oestrogen and weight loss decrease isoproterenol-induced Fos immunoreactivity and angiotensin type 1 mRNA in the subfornical organ of female rats. J Physiol. 2006;573:251–62. doi: 10.1113/jphysiol.2006.106740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krutzfeldt NO, Wild JM. Definition and connections of the entopallium in the zebra finch (Taeniopygia guttata) J Comp Neurol. 2004;468:452–65. doi: 10.1002/cne.10972. [DOI] [PubMed] [Google Scholar]
- Krutzfeldt NO, Wild JM. Definition and novel connections of the entopallium in the pigeon (Columba livia) J Comp Neurol. 2005;790:40–56. doi: 10.1002/cne.20627. [DOI] [PubMed] [Google Scholar]
- Kuenzel W, Masson M. A Stereotaxic Atlas of the Brain of the Chick (Gallus domesticus), vol 1988 [Google Scholar]
- Maekawa F, Tsukahara S, Tanaka K, Ohki-Hamazaki H. Distributions of two chicken bombesin receptors, bombesin receptor subtype-3.5 (chBRS-3.5) and gastrin-releasing peptide receptor (chGRP-R) mRNAS in the chicken telencephalon. Neuroscience. 2004;125:569–82. doi: 10.1016/j.neuroscience.2004.01.057. [DOI] [PubMed] [Google Scholar]
- Mailleux P, Parmentier M, Vanderhaeghen JJ. Distribution of cannabinoid receptor messenger RNA in the human brain: an in situ hybridization histochemistry with oligonucleotides. Neurosci Lett. 1992;143:200–4. doi: 10.1016/0304-3940(92)90265-9. [DOI] [PubMed] [Google Scholar]
- Mailleux P, Vanderhaeghen JJ. Distribution of neuronal cannabinoid receptor in the adult rat brain: a comparative receptor binding radioautography and in situ hybridization histochemistry. Neuroscience. 1992;48:655–68. doi: 10.1016/0306-4522(92)90409-u. [DOI] [PubMed] [Google Scholar]
- Marsicano G, Wotjak CT, Azad SC, Bisogno T, Rammes G, Cascio MG, Hermann H, Tang J, Hofmann C, Zieglgansberger W, Di Marzo V, Lutz B. The endogenous cannabinoid system controls extinction of aversive memories. Nature. 2002;418:530–4. doi: 10.1038/nature00839. [DOI] [PubMed] [Google Scholar]
- Matsuda LA, Bonner TI, Lolait SJ. Localization of cannabinoid receptor mRNA in rat brain. J Comp Neurol. 1993;327:535–50. doi: 10.1002/cne.903270406. [DOI] [PubMed] [Google Scholar]
- Matsuda LA, Lolait SJ, Brownstein MJ, Young AC, Bonner TI. Structure of a cannabinoid receptor and functional expression of the cloned cDNA. Nature. 1990;346:561–4. doi: 10.1038/346561a0. [DOI] [PubMed] [Google Scholar]
- Maturana HR, Varela FJ. Color-opponent responses in the avian lateral geniculate: a study in the quail (Coturnix coturnix japonica) Brain Res. 1982;247:227–41. doi: 10.1016/0006-8993(82)91248-3. [DOI] [PubMed] [Google Scholar]
- McCullumsmith RE, Stincic TL, Agrawal SM, Meador-Woodruff JH. Differential effects of antipsychotics on haloperidol-induced vacuous chewing movements and subcortical gene expression in the rat. Eur J Pharmacol. 2003;477:101–12. doi: 10.1016/j.ejphar.2003.08.018. [DOI] [PubMed] [Google Scholar]
- McPartland JM, Glass M. Functional mapping of cannabinoid receptor homologs in mammals, other vertebrates, and invertebrates. Gene. 2003;312:297–303. doi: 10.1016/s0378-1119(03)00638-3. [DOI] [PubMed] [Google Scholar]
- Moldrich G, Wenger T. Localization of the CB1 cannabinoid receptor in the rat brain. An immunohistochemical study. Peptides. 2000;21:1735–42. doi: 10.1016/s0196-9781(00)00324-7. [DOI] [PubMed] [Google Scholar]
- Molina-Holgado F, Gonzalez MI, Leret ML. Effect of delta 9-tetrahydrocannabinol on short-term memory in the rat. Physiol Behav. 1995;57:177–9. doi: 10.1016/0031-9384(94)00201-f. [DOI] [PubMed] [Google Scholar]
- Montagnese CM, Mezey SE, Csillag A. Efferent connections of the dorsomedial thalamic nuclei of the domestic chick (Gallus domesticus) J Comp Neurol. 2003;459:301–26. doi: 10.1002/cne.10612. [DOI] [PubMed] [Google Scholar]
- Munro S, Thomas KL, Abu-Shaar M. Molecular characterization of a peripheral receptor for cannabinoids. Nature. 1993;365:61–5. doi: 10.1038/365061a0. [DOI] [PubMed] [Google Scholar]
- Nixdorf BE, Bischof HJ. Afferent connections of the ectostriatum and visual wulst in the zebra finch (Taeniopygia guttata castanotis Gould)--an HRP study. Brain Res. 1982;248:9–17. doi: 10.1016/0006-8993(82)91142-8. [DOI] [PubMed] [Google Scholar]
- Nottebohm F, Stokes TM, Leonard CM. Central control of song in the canary, Serinus canarius. J Comp Neurol. 1976;165:457–86. doi: 10.1002/cne.901650405. [DOI] [PubMed] [Google Scholar]
- Nunez E, Benito C, Pazos MR, Barbachano A, Fajardo O, Gonzalez S, Tolon RM, Romero J. Cannabinoid CB2 receptors are expressed by perivascular microglial cells in the human brain: an immunohistochemical study. Synapse. 2004;53:208–13. doi: 10.1002/syn.20050. [DOI] [PubMed] [Google Scholar]
- Patel SN, Rose SP, Stewart MG. Training induced dendritic spine density changes are specifically related to memory formation processes in the chick, Gallus domesticus. Brain Res. 1988;463:168–73. doi: 10.1016/0006-8993(88)90542-2. [DOI] [PubMed] [Google Scholar]
- Patel SN, Stewart MG. Changes in the number and structure of dendritic spines 25 hours after passive avoidance training in the domestic chick, Gallus domesticus. Brain Res. 1988;449:34–46. doi: 10.1016/0006-8993(88)91021-9. [DOI] [PubMed] [Google Scholar]
- Pateromichelakis S. Response properties of units in the lateral geniculate nucleus of the domestic chick (Gallus domesticus) Brain Res. 1979;167:281–96. doi: 10.1016/0006-8993(79)90823-0. [DOI] [PubMed] [Google Scholar]
- Pettit DA, Harrison MP, Olson JM, Spencer RF, Cabral GA. Immunohistochemical localization of the neural cannabinoid receptor in rat brain. J Neurosci Res. 1998;51:391–402. doi: 10.1002/(SICI)1097-4547(19980201)51:3<391::AID-JNR12>3.0.CO;2-A. [DOI] [PubMed] [Google Scholar]
- Price TJ, Helesic G, Parghi D, Hargreaves KM, Flores CM. The neuronal distribution of cannabinoid receptor type 1 in the trigeminal ganglion of the rat. Neuroscience. 2003;120:155–62. doi: 10.1016/S0306-4522(03)00333-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puelles L, Martinez-de-la-Torre M, Paxinos G, Watson C, Martinez S. The Chick Brain in Stereotaxic Coordinates: An Atlas featuring Neuromeric Subdivisions and Mammalian Homologies, vol. Academic Press; 2007. [Google Scholar]
- Reiner A, Davis BM, Brecha NC, Karten HJ. The distribution of enkephalinlike immunoreactivity in the telencephalon of the adult and developing domestic chicken. J Comp Neurol. 1984;228:245–62. doi: 10.1002/cne.902280210. [DOI] [PubMed] [Google Scholar]
- Reiner A, Perkel DJ, Bruce LL, Butler AB, Csillag A, Kuenzel W, Medina L, Paxinos G, Shimizu T, Striedter G, Wild M, Ball GF, Durand S, Gunturkun O, Lee DW, Mello CV, Powers A, White SA, Hough G, Kubikova L, Smulders TV, Wada K, Dugas-Ford J, Husband S, Yamamoto K, Yu J, Siang C, Jarvis ED. Revised nomenclature for avian telencephalon and some related brainstem nuclei. J Comp Neurol. 2004;473:377–414. doi: 10.1002/cne.20118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez Gil DJ, Vacotto M, Rapacioli M, Scicolone G, Flores V, Fiszer de Plazas S. Development and localisation of GABA(A) receptor alpha1, alpha2, beta2 and gamma2 subunit mRNA in the chick optic tectum. J Neurosci Res. 2005;81:469–80. doi: 10.1002/jnr.20579. [DOI] [PubMed] [Google Scholar]
- Rose SP, Csillag A. Passive avoidance training results in lasting changes in deoxyglucose metabolism in left hemisphere regions of chick brain. Behav Neural Biol. 1985;44:315–24. doi: 10.1016/s0163-1047(85)90324-3. [DOI] [PubMed] [Google Scholar]
- Salio C, Fischer J, Franzoni MF, Conrath M. Pre- and postsynaptic localizations of the CB1 cannabinoid receptor in the dorsal horn of the rat spinal cord. Neuroscience. 2002;110:755–64. doi: 10.1016/s0306-4522(01)00584-x. [DOI] [PubMed] [Google Scholar]
- Sanudo-Pena MC, Strangman NM, Mackie K, Walker JM, Tsou K. CB1 receptor localization in rat spinal cord and roots, dorsal root ganglion, and peripheral nerve. Zhongguo Yao Li Xue Bao. 1999;20:1115–20. [PubMed] [Google Scholar]
- Soares HC, de Melo Reis RA, De Mello FG, Ventura AL, Kurtenbach E. Differential expression of D(1A) and D(1B) dopamine receptor mRNAs in the developing avian retina. J Neurochem. 2000;75:1071–5. doi: 10.1046/j.1471-4159.2000.0751071.x. [DOI] [PubMed] [Google Scholar]
- Soderstrom K, Johnson F. CB1 cannabinoid receptor expression in brain regions associated with zebra finch song control. Brain Res. 2000;857:151–7. doi: 10.1016/s0006-8993(99)02393-8. [DOI] [PubMed] [Google Scholar]
- Soderstrom K, Johnson F. Zebra finch CB1 cannabinoid receptor: pharmacology and in vivo and in vitro effects of activation. J Pharmacol Exp Ther. 2001;297:189–97. [PubMed] [Google Scholar]
- Soderstrom K, Johnson F. Cannabinoid exposure alters learning of zebra finch vocal patterns. Brain Res Dev Brain Res. 2003;142:215–7. doi: 10.1016/s0165-3806(03)00061-0. [DOI] [PubMed] [Google Scholar]
- Soderstrom K, Leid M, Moore FL, Murray TF. Behavioral, pharmacological, and molecular characterization of an amphibian cannabinoid receptor. J Neurochem. 2000;75:413–23. doi: 10.1046/j.1471-4159.2000.0750413.x. [DOI] [PubMed] [Google Scholar]
- Soderstrom K, Tian Q. Distinct periods of cannabinoid sensitivity during zebra finch vocal development. Brain Res Dev Brain Res. 2004;153:225–32. doi: 10.1016/j.devbrainres.2004.09.002. [DOI] [PubMed] [Google Scholar]
- Soderstrom K, Tian Q. Developmental pattern of CB1 cannabinoid receptor immunoreactivity in brain regions important to zebra finch (Taeniopygia guttata) song learning and control. J Comp Neurol. 2006;496:739–58. doi: 10.1002/cne.20963. [DOI] [PubMed] [Google Scholar]
- Straiker A, Stella N, Piomelli D, Mackie K, Karten HJ, Maguire G. Cannabinoid CB1 receptors and ligands in vertebrate retina: localization and function of an endogenous signaling system. Proc Natl Acad Sci U S A. 1999;96:14565–70. doi: 10.1073/pnas.96.25.14565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szekely AD. The avian hippocampal formation: subdivisions and connectivity. Behav Brain Res. 1999;98:219–25. doi: 10.1016/s0166-4328(98)00087-4. [DOI] [PubMed] [Google Scholar]
- Takahashi KA, Linden DJ. Cannabinoid receptor modulation of synapses received by cerebellar Purkinje cells. J Neurophysiol. 2000;83:1167–80. doi: 10.1152/jn.2000.83.3.1167. [DOI] [PubMed] [Google Scholar]
- Tang YZ, Carr CE. Development of NMDA R1 expression in chicken auditory brainstem. Hear Res. 2004;191:79–89. doi: 10.1016/j.heares.2004.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsou K, Brown S, Sanudo-Pena MC, Mackie K, Walker JM. Immunohistochemical distribution of cannabinoid CB1 receptors in the rat central nervous system. Neuroscience. 1998;83:393–411. doi: 10.1016/s0306-4522(97)00436-3. [DOI] [PubMed] [Google Scholar]
- Wallace JA. An immunocytochemical study of the development of central serotoninergic neurons in the chick embryo. J Comp Neurol. 1985;236:443–53. doi: 10.1002/cne.902360403. [DOI] [PubMed] [Google Scholar]
- Wang Y, Major DE, Karten HJ. Morphology and connections of nucleus isthmi pars magnocellularis in chicks (Gallus gallus) J Comp Neurol. 2004;469:275–97. doi: 10.1002/cne.11007. [DOI] [PubMed] [Google Scholar]
- Westlake TM, Howlett AC, Bonner TI, Matsuda LA, Herkenham M. Cannabinoid receptor binding and messenger RNA expression in human brain: an in vitro receptor autoradiography and in situ hybridization histochemistry study of normal aged and Alzheimer's brains. Neuroscience. 1994;63:637–52. doi: 10.1016/0306-4522(94)90511-8. [DOI] [PubMed] [Google Scholar]
- Wild JM. The avian somatosensory system: connections of regions of body representation in the forebrain of the pigeon. Brain Res. 1987;412:205–23. doi: 10.1016/0006-8993(87)91127-9. [DOI] [PubMed] [Google Scholar]
- Wild JM. Descending projections of the songbird nucleus robustus archistriatalis. J Comp Neurol. 1993;338:225–41. doi: 10.1002/cne.903380207. [DOI] [PubMed] [Google Scholar]
- Woodson W, Reiner A, Anderson K, Karten HJ. Distribution, laminar location, and morphology of tectal neurons projecting to the isthmo-optic nucleus and the nucleus isthmi, pars parvocellularis in the pigeon (Columba livia) and chick (Gallus domesticus): a retrograde labelling study. J Comp Neurol. 1991;305:470–88. doi: 10.1002/cne.903050310. [DOI] [PubMed] [Google Scholar]
- Yamaguchi F, Macrae AD, Brenner S. Molecular cloning of two cannabinoid type 1-like receptor genes from the puffer fish Fugu rubripes. Genomics. 1996;35:603–5. doi: 10.1006/geno.1996.0406. [DOI] [PubMed] [Google Scholar]