Abstract
During Drosophila embryogenesis, the transcription factor Prospero is critical for neuronal differentiation and axonal outgrowth. The prospero pre-mRNA undergoes alternative splicing, but is unique in that it harbors a rare twintron whereby one intron lies embedded within another. The innermost intron is excised by the major U2-type spliceosome and the outermost is excised by the minor U12-type spliceosome. Previously, an intronic purine-rich element (PRE) was identified as an enhancer of both U2- and U12-type splicing, with a greater effect on the U2-type pathway. We find that the PRE binds Drosophila homologs of heterogeneous nuclear ribonucleoprotein (hnRNP) A1, Hrp38 and Hrp36. RNAi-mediated knockdown of these proteins in S2 cells specifically decreases U2-type splicing of the twintron, which is surprising because hnRNPs usually are repressive. Conversely, tethering Hrp38 to the twintron increases U2-type splicing. Thus, developmentally regulated alternative splicing of the prospero twintron can be explained by documented changes in the abundance of these hnRNP A1-like proteins during embryogenesis.
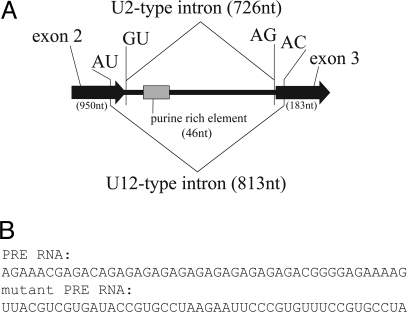
Twintrons are arrangements of introns in which one intron lies embedded within another. They are prevalent in chloroplast pre-mRNAs, as self-splicing group II introns (1). A rare instance of a twintron in a nuclear genome is found in Drosophila prospero, which encodes a transcription factor expressed in many immature neuronal cells (Fig. 1) (2–8). Prospero represses neuroblast-specific and cell-cycle genes, while activating genes important for differentiation into mature neurons. It is also critical for guiding outreaching axons of motor and sensory neurons in the central nervous system (5). The second intron of the prospero pre-mRNA can be excised by either the major U2-type or minor U12-type spliceosome, altering the homeodomain of the protein. The decision to excise either the U2-type intron or the U12-type intron is mutually exclusive. Splicing of the U2-type intron precludes splicing of the U12-type intron (8): pre-mRNAs from which the U2-type intron has been removed, despite containing an 87-nt intron with the U12-type 5′ splice site, 3′ splice site, and branch site consensus sequences, are not further processed by the U12-type spliceosome for reasons that are not fully understood. During embryogenesis, splicing of the prospero twintron is temporally regulated, with U2-type splicing predominating early and U12-type splicing occurring later (9).
Fig. 1.
The prospero pre-mRNA is alternatively spliced. (A) Diagram (not to scale) of the twintron, which constitutes the second prospero intron. U2- and U12-type splice sites and the PRE are indicated. (B) Sequences of the PRE RNA (Upper) and a mutant PRE RNA (Lower) used for the competition experiments shown in Fig. 2A.
Two major classes of splicing regulatory proteins are the heterogeneous nuclear ribonucleoproteins (hnRNPs) and the serine/arginine-rich (SR) proteins (10–15). Classically, hnRNPs have been associated with repression of splicing and SR proteins have been associated with activation of splicing, although many counterexamples exist (10, 16, 17). In Drosophila, the most abundant hnRNPs are Hrp36, Hrp40, and Hrp48 (18–22), which are similar to the hnRNPA/B family of proteins in vertebrates. A fourth major hnRNP in Drosophila, hrp38, was apparently generated by gene duplication and is ≈80% identical to hrp36. Although hrp36 can be deleted with no obvious growth defects, a double hrp36/hrp38 knockout is inviable (23), suggesting that the two proteins have redundant functions. Whereas each of the Drosophila hnRNPs binds to and influences splicing of a specific set of pre-mRNAs, the targets of Hrp36 and Hrp38 overlap (D. Rio, personal communication). Of all of the Drosophila hnRNPs, Hrp36 and Hrp38 are the closest homologs of vertebrate hnRNP A1 (20, 21, 23).
Previously, a 46-nt purine-rich element (PRE), located 29 nt downstream of the U2-type 5′ splice site within the prospero twintron, was identified as important for both U2- and U12-type splicing (Fig. 1B) (9). Mutation of this sequence severely reduced U2-type splicing (to 10%) and moderately reduced U12-type splicing (to 45% of wild-type levels) in vitro. Addition of exogenous PRE RNA to in vitro splicing reactions inhibited both splicing reactions, with a more pronounced effect on the U2-type pathway. This finding suggested that a transacting factor(s) binding to the PRE mediates its stimulatory effects, which are more significant for the U2-type than for U12-type spliceosome. Here, we present data that implicate Hrp38 and Hrp36 as proteins that bind to the PRE and enhance the ratio of U2- versus U12-type splicing of the prospero twintron.
Results
A 40-kDa Protein That Binds Specifically to the PRE in Vitro Is Hrp38.
To identify factors that bind the PRE of the prospero twintron (9), we began with cross-linking experiments. In vitro-transcribed, radiolabeled PRE RNA (46 nt) was added to Drosophila S2 cell nuclear extract and exposed to UV light. Fig. 2A reveals that proteins with molecular masses of ≈40 and 47 kDa became cross-linked, but binding of only the 40-kDa band was blocked by the addition of unlabeled PRE RNA compared with mutant PRE RNA (Fig. 1B), indicating a specific interaction. Similar results were obtained in Kc nuclear extract (data not shown). The mutant RNA sequence was the same as that used in previous studies of the PRE (9) and is an unrelated sequence of the same length as the PRE, but with significantly lower purine content (91.3% purine in the wild-type sequence versus 37.0% purine in the mutant sequence).
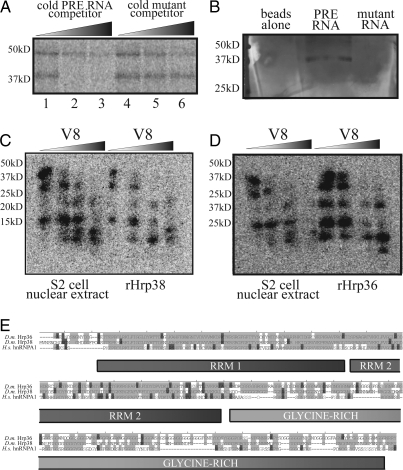
Fig. 2.
Hrp38 and Hrp36 interact with the PRE. (A) A 40-kDa protein in Drosophila Kc cell nuclear extract specifically cross-links to radiolabeled PRE RNA upon UV irradiation. Radiolabeled PRE (30 fmol) was cross-linked to proteins in nuclear extract after incubation with 300 fmol (10×; lanes 1 and 4), 3 pmol (100×; lanes 2 and 5), or 30 pmol (1,000×; lanes 3 and 6) competitor RNA. (B) A ≈40-kDa band was the most abundant selected from Drosophila S2 cell nuclear extract using immobilized wild-type PRE RNA. Mass spectrometric analysis revealed the major protein in this band to be Hrp38 and a minor protein to be Hrp36. (C) Partial proteolysis pattern of the PRE-cross-linked 40-kDa protein from extract and recombinant Hrp38. (D) Partial proteolysis pattern of recombinant Hrp36 is highly similar to that of the 40-kDa protein and Hrp38. In A–D the positions of marker proteins are indicated on the left. (E) Sequence alignment of Drosophila (D.m.) Hrp36 and Hrp38, which are ≈80% identical, and human (H.s.) hnRNP A1. Light gray indicates identical amino acids and dark gray shows similar amino acids.
In complementary experiments, PRE RNA was immobilized on agarose beads via its 3′ end. Affinity purification from S2 nuclear extract again showed a protein of ≈40 kDa retained with PRE RNA, but not by beads alone or beads coated with the mutant RNA sequence (Fig. 2B). Mass spectrometry identified the major protein in this SDS/PAGE band as Hrp38, the Drosophila homolog of hnRNP A1 (20). The highly related Hrp36 protein was present at lower levels.
To confirm that Hrp38 is in fact the protein in nuclear extract that cross-links to the PRE, partial proteolysis mapping was performed (24). Nuclear extract and recombinant Hrp38 protein were individually UV-cross-linked to radiolabeled PRE RNA and purified by SDS/PAGE. After autoradiography, the 2 cross-linked bands were excised, divided into smaller slices, and transferred into the wells of a second SDS gel for in-gel partial proteolysis with the site-specific protease V8 (25, 26). Fig. 2C shows a digestion pattern for the 40-kDa extract protein identical to that of recombinant Hrp38. The digestion pattern of recombinant Hrp36 was likewise very similar (Fig. 2D). This result was expected because Hrp36 and 38 are ≈80% identical, especially in the N-terminal RNA-recognition motif (RRM) domains (Fig. 2E) where most of the V8 protease cleavage sites reside. For comparison, the partial protease digestion pattern for recombinant Transformer-2 protein (Tra-2), another 40-kDa Drosophila protein known to be a splicing regulator that binds purine-rich sequences with high affinity (27, 28), was produced; its pattern was significantly different (Fig. S1). We conclude that Hrp38 and/or Hrp36 are ≈40-kDa proteins that bind the PRE in vitro. Because they are suggested to be functionally redundant, both proteins may interact with the PRE in vivo (29, 30).
RNAi-Mediated Knockdown of Hrp38 and/or Hrp36 Selectively Inhibits U2-Type Splicing of the Prospero Twintron Reporter in Vivo.
To test whether the interactions of Hrp38 and Hrp36 with the PRE have functional consequences for splicing regulation in vivo, a splicing reporter containing the prospero twintron was constructed. Truncated exons 2 and 3 of prospero mRNA were cloned downstream of the constitutively active Drosophila actin 5c promoter (Fig. 3A), separated by a truncated prospero twintron, which harbors both U2- and U12-type splice sites, and the PRE. An antisense, radiolabeled riboprobe was designed to distinguish unspliced precursor from U2- and U12-type spliced products by the RNase protection assay (RPA).
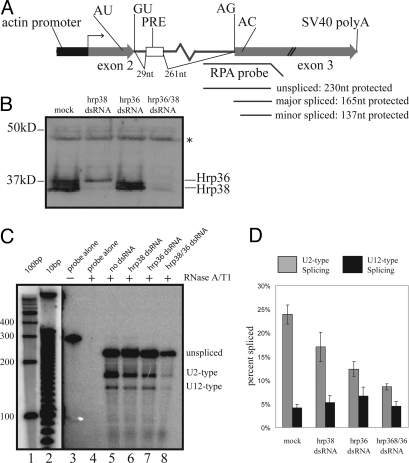
Fig. 3.
dsRNA knockdown of Hrp36 and/or Hrp38 results in lower U2-type splicing efficiency, but does not dramatically influence U12-type splicing efficiency. (A) In vivo splicing substrate used to assess twintron splicing. 5′ and 3′ splice sites of U2-type (GU/AG) and U12-type (AU/AC) introns are indicated, with regions of the antisense probe protected by unspliced precursor, major-spliced product, or minor-spliced product shown. (B) Western blot analysis of lysate from mock- or dsRNA-treated cells indicates reduction in protein expression. The antibody was rabbit polyclonal anti-Hrp38 (see Materials and Methods), which cross-reacts with Hrp36. Asterisk indicates a lysate protein that nonspecifically cross-reacts with the antiserum and therefore serves as a loading control. Hrp36 migrates slower than Hrp38 not only because of intrinsic differences in molecular mass (Hrp36 is ≈39.5kDa, and the 2 major Hrp38 isoforms are 38 and 39 kDa) but perhaps posttranslational modifications of Hrp36 as well (52). (C) RPA shows that the levels of U2-type splicing decrease upon dsRNA-mediated knockdown of Hrp38 and Hrp36 individually (lanes 6 and 7, respectively) or together (lane 8). The yield of total RNA harvested from S2 cells treated with both anti-hrp36 and anti-hrp38 dsRNA was considerably lower than that obtained from untreated or singly treated cells, resulting in lower signals of unspliced, U2-type spliced, and U12-type spliced products in the ensuing RPA (compare lanes 5–7 with lane 8). (D) Quantitation of U2- versus U12-type spliced products. Values are the average of 5 independent experiments, with SDs shown.
Using dsRNA treatment, we attempted knockdown of Hrp36 and Hrp38 in Drosophila S2 cells (Fig. 3B). We also tested simultaneous knockdown of both proteins, because it seemed likely that either might compensate for the other owing to functional redundancy (29, 30). Monitoring with antibodies raised against Hrp38, which cross-react with Hrp36 on Western blots, showed that knockdown of the hrp38 mRNA did not result in diminution of Hrp36 protein or vice versa. However, treatment with dsRNAs targeting both hrp38 and hrp36 mRNAs was effective in simultaneously reducing the levels of the two proteins. Although the cellular concentration of Hrp36 is much higher than that of Hrp38 (18, 19), it does not appear so, probably because the rabbit antibody raised against recombinant Hrp38 reacts with Hrp38 much more efficiently than with Hrp36. dsRNA knockdown specificity was confirmed at the mRNA level (Fig. S2).
The levels of U2- and U12-type spliced products of the prospero twintron reporter were then assessed in S2 cells after Hrp36/38 knockdown. A representative RPA is shown in Fig. 3C. Normalizing for the difference in the number of α-32P uridylate residues present in the protected riboprobe when hybridized to unspliced, U2- or U12-type spliced products revealed a marked decrease in the fraction of U2-type spliced product upon knockdown of either Hrp38 or Hrp36, but particularly upon knockdown of both Hrp38 and Hrp36 (Fig. 3D). The fraction of U12-type spliced product was less affected. The levels of unspliced precursor also increased upon knockdown, increasing from 72% (±2%) of the total in mock-treated samples to 81% (± 3%) and 78% (±4%) in anti-hrp36- and anti-hrp38-treated samples, respectively, and to 87% (±2%) in anti-hrp36/38-treated samples.
Tethering Hrp38 to the Prospero Twintron Specifically Increases U2-Type Splicing in Vivo.
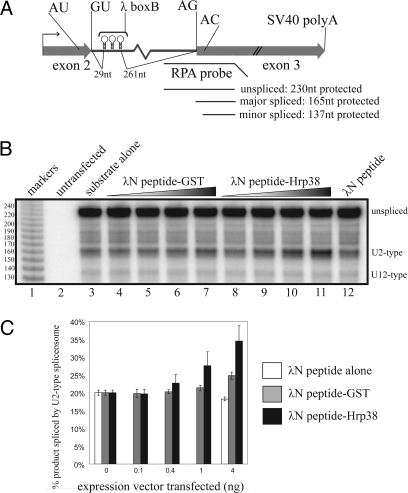
To ask whether Hrp36/38 binding to the prospero twintron is not only necessary but also sufficient to enhance U2-type splicing, we replaced the PRE sequence of the twintron splicing reporter with 3 copies of the boxB hairpin from bacteriophage λ (Fig. 4A). This reporter was cotransfected into S2 cells with vectors encoding: (i) λN peptide alone, (ii) λN peptide fused to GST, or (iii) λN peptide fused to Hrp38. As seen in Fig. 4 B and C, coexpression of λN peptide-Hrp38 specifically increased U2-type splicing in vivo. U12-type splicing levels were relatively unaffected (≈5% under all conditions tested). Two additional lambda peptide-tagged RNA-binding proteins, Ago2 and GW182, were similarly tethered to the PRE site, but neither was able to enhance U2-type splicing (Fig. S3). Furthermore, when the 3 bBox hairpins were substituted with 3 copies of the MS2 hairpin sequence, which binds proteins tagged with the MS2 coat protein, expression of lambda peptide-tagged Hrp36 did not enhance U2-type splicing in vivo (Fig. S4). These results demonstrate that tethering of the hnRNP to the PRE site, and not merely its overexpression, is specifically required to observe enhanced U2-type splicing. Western blotting analysis using anti-lambda-peptide antiserum confirmed that the tagged proteins were expressed at approximately equivalent levels (data not shown). These results confirm that Hrp38 stimulates U2-type splicing in vivo.
Fig. 4.
Tethering of Hrp38 to the twintron enhances U2-type splicing. (A) Splicing reporter containing 3 copies of the boxB hairpin in place of the PRE. (B) RPA of boxB twintron substrate after cotransfection with no λN fusion protein vector (lane 3) or 0, 0.1, 0.4, 1, or 4 ng of vector-encoding λN-GST fusion protein (lanes 4–7), λN-Hrp38 fusion protein (lanes 8–11) or 4 ng of λN peptide alone (lane 12). (C) Quantitation of U2- and U12-type splicing from 3 independent experiments.
Discussion
Hrp38 and Hrp36 Regulate Alternative Splicing of the Prospero Twintron.
Here, we have identified the Drosophila homologs of hnRNP A1 as transacting factors that recognize an intronic enhancer (the PRE) to stimulate U2-type relative to U12-type splicing of the prospero twintron. Hrp38 and the highly related Hrp36 were identified both by UV cross-linking and affinity selection as binding to the PRE. RNAi-mediated knockdown of either Hrp38 or Hrp36, but particularly both, preferentially inhibited U2-type splicing of a twintron splicing reporter in vivo. Finally, tethering Hrp38 to the twintron stimulated production of the U2-type relative to the U12-type splice, arguing that it is not only necessary but also sufficient to regulate alternative splicing. Although there are several reports that hnRNPs bind intronic splicing enhancers (31, 32), our work specifically implicates an hnRNP A1 family member.
These results are in agreement with previous observations. First, in in vitro assays, mutation of the PRE exhibited a greater effect on U2- than U12-type splicing, and competitor PRE more effectively inhibited U2-type splicing (9). Second, hrp38 mRNA levels were highest during early embryogenesis (20, 21) when U2-type splicing of the prospero twintron predominates relative to the U12 type (9). Third, human hnRNP A1 binds purine-rich sequences with nanomolar affinity (33, 34), and purine-rich exonic splicing silencers have been shown to bind hnRNP A1, most notably in the SMN2 pre-mRNA (35). Fourth, mRNA expression of Hrp38 and Hrp36 is strongly enriched in the nerve cord and brain during Drosophila embryogenesis. Fifth, whereas flies that are homozygous mutant for both proteins do not survive to adulthood, some mutant embryos hatch into larvae. These larvae show poor mobility with intermittent twitching, a phenotype potentially consistent with nervous system defects (S. Haynes, personal communication).
Alternative splicing plays an important role in the regulation of the Prospero transcription factor (8, 9, 36). Among the 3 annotated prospero mRNAs in Flybase (http://flybase.bio.indiana.edu), pros-RC and pros-RD differ in that pros-RC, the U2-type spliced product, retains 87 nt that are spliced out of pros-RD, the U12-type spliced product (Fig. 1A); thereby, 29 aa upstream of and 5 aa within the N terminus of the homeodomain differ. The specific effects of alternative splicing on DNA binding affinity and sequence specificity of the Prospero transcription factor are still unclear, but the homeodomain is known to be responsible for both DNA binding and nuclear localization of the protein (37, 38). Indeed, the 3′ splice site of the U12-type intron lies close to the nuclear export signal found within the homeodomain (37). In addition to the pros-RC mRNA predominating early and the pros-RD predominating later in embryogenesis, indirect evidence suggests that pros-RC predominates in the central nervous system whereas pros-RD predominates in the peripheral nervous system (36). Overall, these results suggest that spatial and temporal regulation of alternative splicing of the twintron modifies the functional properties of Prospero in the animal.
hnRNP Activation of Splicing.
Several Drosophila hnRNPs have been shown to regulate alternative splicing. Hrp36 and Hrp38 have been individually overexpressed in flies, resulting in similar phenotypes, e.g., splicing repression in the Dopa decarboxylase pre-mRNA, which is expressed in central nervous system and hypodermal tissues (29, 39). Recently, Blanchette and coworkers knocked down 4 major splicing regulators in Drosophila (two SR proteins, dASF/SF2 and B52/SRp55, and 2 hnRNP-like proteins, P-element somatic inhibitor protein (PSI) and Hrp48) and examined the effects on all known splice junctions by microarray analysis (ref. 17 and M. Blanchette and D. C. Rio, personal communication). The results challenged the preexisting notions that hnRNPs act only as splicing repressors and that hnRNPs nonspecifically affect splicing. Only ≈90 Drosophila splice junctions were strongly regulated by Hrp48, and utilization of approximately half of these was lowered by Hrp48 knockdown, suggesting that they are normally activated by Hrp48. We can now add Hrp38 and Hrp36 to the list of hnRNPs that act as specific splicing activators.
Several reports indicate that hnRNP A1 can act to tether small nuclear ribonucleoproteins (snRNPs) to pre-mRNAs in nonproductive ways, one mechanism by which hnRNPs may regulate alternative splicing. In the instance of exon 2 of the HIV tat pre-mRNA, hnRNP A1 is hypothesized to bind and sequester snRNPs in inactive complexes on the pre-mRNA; formation of nonproductive prespliceosomal complexes has been proposed to inhibit proper splicing elsewhere on the pre-mRNA (40, 41). Similarly, the Drosophila hnRNP A1-like protein Hrp48 and PSI recruit U1 snRNP to “pseudo 5′ splice sites” of the third intron of the P element transposon pre-mRNA, inhibiting spliceosome assembly elsewhere (10, 30, 42).
It is unlikely that Hrp36 and Hrp38 activate U2-type splicing by repressing U12-type splicing because knockdown of these proteins failed to activate U12-type splicing and tethering of Hrp38 did not repress U12-type splicing (Figs. 3 and 4). Rather, they appear to enhance splicing of the U2-type intron, perhaps by recruiting U2 snRNP or some other component specific to the U2-type spliceosome. This idea is consistent with reports that hnRNP A1 cross-links to U2 snRNA, and with evidence that hnRNP molecules recruit snRNPs to specific sites on a pre-mRNA (30, 42, 43). hnRNP A1 might then anneal the U2 snRNA onto the U2-type intron branch point sequence (30, 42, 44, 45). Such selective recruitment might explain the specific inhibition of U2-type splicing relative to U12-type splicing upon knockdown of Hrp38 and Hrp36 proteins, the selective inhibition of U2-type splicing documented in mutational and competition experiments (9), and the enhancement of U2-type splicing seen in the tethering assay (Fig. 4). The work of Chabot and coworkers (46) has revealed that one mechanism by which hnRNP A1 activates splicing of some introns is by binding near the ends of these introns and looping out the intervening sequence. It is possible that the Drosophila Hrp36 and Hrp38 proteins bind not only to the PRE sequence, but also to a more distal sequence, thus looping out the intervening U2-type intron. However, we note that depleting both Hrp36 and Hrp38 by RNAi knockdown leads to an ≈90% reduction in U2-type splicing efficiency, the same level of reduction reported upon mutation of the first 5 residues of the PRE and deletion of the remainder of the element (9). The similar magnitudes in reduction suggest that these hnRNPs are working through the PRE alone and not through other sequences within the intron. However, if hnRNPs require both binding sites to enhance splicing, then a similar reduction would be seen upon deletion of just one site. Although no other PRE-like sequences that might bind a second Hrp36 or Hrp38 protein are readily apparent within the prospero U2-type intron, we nonetheless cannot rule out a looping mechanism as the mode of action. Indeed, other proteins might bind to the PRE and/or other sequences within the twintron to help regulate splicing and could account for the relatively inefficient splicing in in vitro assays compared with splicing levels in vivo during Drosophila embryogenesis (9).
Overall, our results point to an unexpected role for hnRNPs as splicing activators acting via a purine-rich intronic splicing enhancer and important regulators in the alternative splicing of the prospero pre-mRNA during embryogenesis.
Materials and Methods
Plasmid Construction and Oligonucleotide Sequences.
Details on plasmid construction and sequences of oligonucleotides used for cloning and in vitro transcriptions are given in SI Text.
Antibody Production.
Antibodies against Hrb98DE and Hrb87F were generated by expressing GST-Hrb98DE or GST-Hrb87F fusion proteins in Escherichia coli BL21 cells from the pGEX6p vector. Briefly, 200 mL of cell culture was grown to an OD595 of 0.6 and induced with 0.5 mM IPTG for 5 h at 22 °C. Each gram of cell pellet was resuspended in 15 mL of chilled PBS supplemented with 5 mM EDTA, 5 mM β-mercaptoethanol, and protease inhibitor mixture (Calbiochem). Cell suspension was sonicated on ice and then clarified by centrifugation at 11,500 × g for 15 min at 4 °C. Supernatant was nutated with 500 μL of bed volume of glutathione-Sepharose beads preequilibrated with cleavage buffer [50 mM Tris·HCl (pH 7.0), 150 mM NaCl, 1 mM EDTA, 1 mM DTT] for 4 h at 4 °C. Beads were washed with 200 mL of cleavage buffer, and full-length protein was freed from beads by incubating it with 40 units of PreScission Protease (GE) in 1 mL of cleavage buffer for 4 h at 4 °C. Full-length protein was further purified on a 10% SDS glycine gel before raising antibodies in rabbit (Cocalico Biologicals).
Cross-Linking, Affinity Purification of Proteins from Extract, and Partial Protease Mapping.
For in vitro cross-linking studies, 300,000 cpm of RNA containing the 32P-labeled PRE or a mutant sequence (Fig. 1B) was incubated with 9 μL of Kc or S2 cell nuclear extract in a 30-μL reaction containing 2 mM Hepes (pH 7.9), 2 mM creatine phosphate, 1 mM ATP, 2 μg/μL of E. coli tRNA, and 3 μg/μL of yeast total carrier RNA. Reactions were incubated on ice for 30 min, irradiated twice with 860 mJ/cm2, and then treated with 0.6 μg of RNase V1, 18.75 units of RNase T1, and 4.5 units of RNase One at 30 °C for 30 min. Samples were resolved by 10% SDS/PAGE.
Affinity purification of proteins from extract used wild-type PRE RNA or mutant RNA (both 46 nt; Fig. 1B), which had been in vitro-transcribed and gel-purified by standard methods. The 3′ ends of gel-purified RNAs were oxidized and immobilized onto adipic acid dihydrazide beads as described (40, 47). For purification of the 40-kDa protein, 300 μL of nuclear extract was first precleared with 200 μL of adipic acid dihydrazide beads by nutation at 4 °C for 1 h. Then, RNA-coated beads were nutated for 4 h at 4 °C with precleared nuclear extract supplemented with 3 μg/mL of carrier RNA, 2 mg/mL of E. coli tRNA, 1 mM ATP, 2 mM creatine phosphate, and 2 mM Hepes, pH 7.9. Beads were washed and proteins were eluted from the resin by micrococcal nuclease treatment at 37 °C for 1 h and supplemented with 2 mM CaCl2. Eluted proteins were resolved by 10% SDS PAGE and visualized by silver staining.
Partial proteolysis mapping was performed as described (24).
Cell Lines, Transfections, dsRNA Knockdown, and Preparation of Nuclear Extract.
Drosophila S2 cells were grown in Schneider's medium (Invitrogen) supplemented with l-glutamine and penicillin/streptomycin. For transient transfections, Effectene reagent (Qiagen) was used per the manufacturer's instructions.
For dsRNA knockdowns, forward and reverse strands were transcribed by T7 or SP6 RNA polymerase in vitro. After phenol extraction, precipitation, and quantification of yields, the strands were annealed and dosed onto cells as described (48, 49).
Nuclear extract was prepared as described from Kc cells (9, 50) or S2 cells (51).
RNase Protection Assay.
For RPA, total RNA was harvested from transfected S2 cells by using TRIzol reagent (Invitrogen) according to the manufacturer's instructions. Harvested RNA was further treated with RQ1 DNase (Promega) for 30 min at 37 °C, phenol-extracted, and precipitated before use. Fifteen micrograms of harvested RNA was coprecipitated with 20,000 cpm of antisense probe, and then resuspended in 30 μL of hybridization buffer [80% formamide, 400 mM NaCl, 40 mM Pipes (pH 6.5), 1 mM EDTA]. Samples were heated at 95 °C for 10 min, followed by overnight incubation at 45 °C. Then, 300 μL of RNase buffer [300 mM NaCl, 10 mM Tris·HCl (pH 7.5), 5 mM EDTA] was added per sample, along with 1.5 μg of RNase A (Sigma) and 40 units of RNase T1 (Calbiochem). Samples were incubated at 30 °C for 30 min, before the addition of 100 μL of protease K solution [2 mg/mL of protease K, 2% SDS, 1 μL of GlycoBlue (Ambion), 50 μg/mL of yeast carrier RNA]. Samples were treated with protease K for 15 min at 37 °C, followed by phenol extraction and precipitation. Samples were then resolved on a 6% acrylamide/8 M urea TBE gel.
Supplementary Material
Acknowledgments.
We thank Kristen Lynch (University of Texas Southwestern, Dallas) for recombinant Tra-2; Hervé Agaisse (Yale University) for the Drosophila actin 5c promoter; Elisa Izaurralde (Max Planck Institute, Tübingen, Germany) for lambda peptide-tagged Ago2 and GW182; Matthias Hentze (European Molecular Biology Laboratory, Heidelberg) for anti-lambda peptide antiserum; Brenton Graveley, Donald Rio, and Susan Haynes for advice; and Kazimierz Tycowski and J. Kyle Friend for critical reading of the manuscript. This work was supported by National Institutes of Health Grant GM26154 and an Anna Fuller predoctoral fellowship (to S.B.). J.A.S. is an Investigator of the Howard Hughes Medical Institute.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0812826106/DCSupplemental.
References
- 1.Copertino DW, Hallick RB. Group II twintron: An intron within an intron in a chloroplast cytochrome b-559 gene. EMBO J. 1991;10:433–442. doi: 10.1002/j.1460-2075.1991.tb07965.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Desplan C, Theis J, O'Farrell PH. The sequence specificity of homeodomain–DNA interaction. Cell. 1988;54:1081–1090. doi: 10.1016/0092-8674(88)90123-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chu-Lagraff Q, Wright DM, McNeil LK, Doe CQ. The prospero gene encodes a divergent homeodomain protein that controls neuronal identity in Drosophila. Development. 1991;2(Suppl):79–85. [PubMed] [Google Scholar]
- 4.Doe CQ, Chu-LaGraff Q, Wright DM, Scott MP. The prospero gene specifies cell fates in the Drosophila central nervous system. Cell. 1991;65:451–464. doi: 10.1016/0092-8674(91)90463-9. [DOI] [PubMed] [Google Scholar]
- 5.Vaessin H, et al. prospero is expressed in neuronal precursors and encodes a nuclear protein that is involved in the control of axonal outgrowth in Drosophila. Cell. 1991;67:941–953. doi: 10.1016/0092-8674(91)90367-8. [DOI] [PubMed] [Google Scholar]
- 6.Matsuzaki F, Koizumi K, Hama C, Yoshioka T, Nabeshima Y. Cloning of the Drosophila prospero gene and its expression in ganglion mother cells. Biochem Biophys Res Commun. 1992;182:1326–1332. doi: 10.1016/0006-291x(92)91878-t. [DOI] [PubMed] [Google Scholar]
- 7.Li L, Vaessin H. Pan-neural Prospero terminates cell proliferation during Drosophila neurogenesis. Genes Dev. 2000;14:147–151. [PMC free article] [PubMed] [Google Scholar]
- 8.Otake LR, Scamborova P, Hashimoto C, Steitz JA. The divergent U12-type spliceosome is required for pre-mRNA splicing and is essential for development in Drosophila. Mol Cell. 2002;9:439–446. doi: 10.1016/s1097-2765(02)00441-0. [DOI] [PubMed] [Google Scholar]
- 9.Scamborova P, Wong A, Steitz JA. An intronic enhancer regulates splicing of the twintron of Drosophila melanogaster prospero pre-mRNA by two different spliceosomes. Mol Cell Biol. 2004;24:1855–1869. doi: 10.1128/MCB.24.5.1855-1869.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Black DL. Mechanisms of alternative pre-messenger RNA splicing. Annu Rev Biochem. 2003;72:291–336. doi: 10.1146/annurev.biochem.72.121801.161720. [DOI] [PubMed] [Google Scholar]
- 11.Huang Y, Steitz JA. SRprises along a messenger's journey. Mol Cell. 2005;17:613–615. doi: 10.1016/j.molcel.2005.02.020. [DOI] [PubMed] [Google Scholar]
- 12.Graveley BR. Sorting out the complexity of SR protein functions. RNA. 2000;6:1197–1211. doi: 10.1017/s1355838200000960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dreyfuss G, Matunis MJ, Pinol-Roma S, Burd CG. hnRNP proteins and the biogenesis of mRNA. Annu Rev Biochem. 1993;62:289–321. doi: 10.1146/annurev.bi.62.070193.001445. [DOI] [PubMed] [Google Scholar]
- 14.Tenenbaum SA, Aguirre-Ghiso J. Dephosphorylation shows SR proteins the way out. Mol Cell. 2005;20:499–501. doi: 10.1016/j.molcel.2005.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hertel KJ, Graveley BR. RS domains contact the pre-mRNA throughout spliceosome assembly. Trends Biochem Sci. 2005;30:115–118. doi: 10.1016/j.tibs.2005.01.002. [DOI] [PubMed] [Google Scholar]
- 16.Mayeda A, Krainer AR. Regulation of alternative pre-mRNA splicing by hnRNP A1 and splicing factor SF2. Cell. 1992;68:365–375. doi: 10.1016/0092-8674(92)90477-t. [DOI] [PubMed] [Google Scholar]
- 17.Blanchette M, Green RE, Brenner SE, Rio DC. Global analysis of positive and negative pre-mRNA splicing regulators in Drosophila. Genes Dev. 2005;19:1306–1314. doi: 10.1101/gad.1314205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Matunis EL, Matunis MJ, Dreyfuss G. Characterization of the major hnRNP proteins from Drosophila melanogaster. J Cell Biol. 1992;116:257–269. doi: 10.1083/jcb.116.2.257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Matunis MJ, Matunis EL, Dreyfuss G. Isolation of hnRNP complexes from Drosophila melanogaster. J Cell Biol. 1992;116:245–255. doi: 10.1083/jcb.116.2.245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Haynes SR, Raychaudhuri G, Beyer AL. The Drosophila Hrb98DE locus encodes four protein isoforms homologous to the A1 protein of mammalian heterogeneous nuclear ribonucleoprotein complexes. Mol Cell Biol. 1990;10:316–323. doi: 10.1128/mcb.10.1.316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Haynes SR, Johnson D, Raychaudhuri G, Beyer AL. The Drosophila Hrb87F gene encodes a new member of the A and B hnRNP protein group. Nucleic Acids Res. 1991;19:25–31. doi: 10.1093/nar/19.1.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Haynes SR, Raychaudhuri G, Johnson D, Amero S, Beyer AL. The Drosophila Hrb loci: A family of hnRNA binding proteins. Mol Biol Rep. 1990;14:93–94. doi: 10.1007/BF00360430. [DOI] [PubMed] [Google Scholar]
- 23.Zu K, Sikes ML, Beyer AL. Separable roles in vivo for the two RNA binding domains of Drosophila A1-hnRNP homolog. RNA. 1998;4:1585–1598. doi: 10.1017/s135583829898102x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cleveland DW, Fischer SG, Kirschner MW, Laemmli UK. Peptide mapping by limited proteolysis in sodium dodecyl sulfate and analysis by gel electrophoresis. J Biol Chem. 1977;252:1102–1106. [PubMed] [Google Scholar]
- 25.Houmard J, Drapeau GR. Staphylococcal protease: A proteolytic enzyme specific for glutamoyl bonds. Proc Natl Acad Sci USA. 1972;69:3506–3509. doi: 10.1073/pnas.69.12.3506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Drapeau GR, Boily Y, Houmard J. Purification and properties of an extracellular protease of Staphylococcus aureus. J Biol Chem. 1972;247:6720–6726. [PubMed] [Google Scholar]
- 27.Tacke R, Tohyama M, Ogawa S, Manley JL. Human Tra2 proteins are sequence-specific activators of pre-mRNA splicing. Cell. 1998;93:139–148. doi: 10.1016/s0092-8674(00)81153-8. [DOI] [PubMed] [Google Scholar]
- 28.Lynch KW, Maniatis T. Synergistic interactions between two distinct elements of a regulated splicing enhancer. Genes Dev. 1995;9:284–293. doi: 10.1101/gad.9.3.284. [DOI] [PubMed] [Google Scholar]
- 29.Zu K, Sikes ML, Haynes SR, Beyer AL. Altered levels of the Drosophila HRB87F/hrp36 hnRNP protein have limited effects on alternative splicing in vivo. Mol Biol Cell. 1996;7:1059–1073. doi: 10.1091/mbc.7.7.1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hammond LE, Rudner DZ, Kanaar R, Rio DC. Mutations in the hrp48 gene, which encodes a Drosophila heterogeneous nuclear ribonucleoprotein particle protein, cause lethality and developmental defects and affect P-element third-intron splicing in vivo. Mol Cell Biol. 1997;17:7260–7267. doi: 10.1128/mcb.17.12.7260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hovhannisyan RH, Carstens RP. Heterogeneous ribonucleoprotein M (hnRNP M) is a splicing regulatory protein that can enhance or silence splicing of alternatively spliced exons. J Biol Chem. 2007;50:36265–362740. doi: 10.1074/jbc.M704188200. [DOI] [PubMed] [Google Scholar]
- 32.Wang E, Dimova N, Cambi F. PLP/DM20 ratio is regulated by hnRNPH and F and a novel G-rich enhancer in oligodendrocytes. Nucleic Acids Res. 2007;35:4164–4178. doi: 10.1093/nar/gkm387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Burd CG, Dreyfuss G. RNA binding specificity of hnRNP A1: Significance of hnRNP A1 high-affinity binding sites in pre-mRNA splicing. EMBO J. 1994;13:1197–1204. doi: 10.1002/j.1460-2075.1994.tb06369.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Abdul-Manan N, Williams KR. hnRNP A1 binds promiscuously to oligoribonucleotides: Utilization of random and homooligonucleotides to discriminate sequence from base-specific binding. Nucleic Acids Res. 1996;24:4063–4070. doi: 10.1093/nar/24.20.4063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kashima T, Manley JL. A negative element in SMN2 exon 7 inhibits splicing in spinal muscular atrophy. Nat Genet. 2003;34:460–463. doi: 10.1038/ng1207. [DOI] [PubMed] [Google Scholar]
- 36.Guenin L, et al. Spatio-temporal expression of Prospero is finely tuned to allow the correct development and function of the nervous system in Drosophila melanogaster. Dev Biol. 2007;304:62–74. doi: 10.1016/j.ydbio.2006.12.016. [DOI] [PubMed] [Google Scholar]
- 37.Ryter JM, Doe CQ, Matthews BW. Structure of the DNA binding region of prospero reveals a novel homeo-prospero domain. Structure. 2002;10:1541–1549. doi: 10.1016/s0969-2126(02)00883-3. [DOI] [PubMed] [Google Scholar]
- 38.Yousef MS, Matthews BW. Structural basis of Prospero–DNA interaction: Implications for transcription regulation in developing cells. Structure. 2005;13:601–607. doi: 10.1016/j.str.2005.01.023. [DOI] [PubMed] [Google Scholar]
- 39.Shen J, Zu K, Cass CL, Beyer AL, Hirsh J. Exon skipping by overexpression of a Drosophila heterogeneous nuclear ribonucleoprotein in vivo. Proc Natl Acad Sci USA. 1995;92:1822–1825. doi: 10.1073/pnas.92.6.1822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Caputi M, Mayeda A, Krainer AR, Zahler AM. hnRNP A/B proteins are required for inhibition of HIV-1 pre-mRNA splicing. EMBO J. 1999;18:4060–4067. doi: 10.1093/emboj/18.14.4060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dyhr-Mikkelsen H, Kjems J. Inefficient spliceosome assembly and abnormal branch site selection in splicing of an HIV-1 transcript in vitro. J Biol Chem. 1995;270:24060–24066. doi: 10.1074/jbc.270.41.24060. [DOI] [PubMed] [Google Scholar]
- 42.Siebel CW, Kanaar R, Rio DC. Regulation of tissue-specific P-element pre-mRNA splicing requires the RNA-binding protein PSI. Genes Dev. 1994;8:1713–1725. doi: 10.1101/gad.8.14.1713. [DOI] [PubMed] [Google Scholar]
- 43.Buvoli M, Cobianchi F, Riva S. Interaction of hnRNP A1 with snRNPs and pre-mRNAs: Evidence for a possible role of A1 RNA annealing activity in the first steps of spliceosome assembly. Nucleic Acids Res. 1992;20:5017–5025. doi: 10.1093/nar/20.19.5017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kumar A, Wilson SH. Studies of the strand-annealing activity of mammalian hnRNP complex protein A1. Biochemistry. 1990;29:10717–10722. doi: 10.1021/bi00500a001. [DOI] [PubMed] [Google Scholar]
- 45.Pontius BW, Berg P. Renaturation of complementary DNA strands mediated by purified mammalian heterogeneous nuclear ribonucleoprotein A1 protein: Implications for a mechanism for rapid molecular assembly. Proc Natl Acad Sci USA. 1990;87:8403–8407. doi: 10.1073/pnas.87.21.8403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Martinez-Contreras R, et al. Intronic binding sites for hnRNP A/B and hnRNP F/H proteins stimulate pre-mRNA splicing. PLoS Biol. 2006;4:e21. doi: 10.1371/journal.pbio.0040021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Langland JO, Pettiford SM, Jacobs BL. Nucleic acid affinity chromatography: Preparation and characterization of double-stranded RNA agarose. Protein Expression Purif. 1995;6:25–32. doi: 10.1006/prep.1995.1004. [DOI] [PubMed] [Google Scholar]
- 48.Clemens JC, et al. Use of double-stranded RNA interference in Drosophila cell lines to dissect signal transduction pathways. Proc Natl Acad Sci USA. 2000;97:6499–6503. doi: 10.1073/pnas.110149597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Park JW, Parisky K, Celotto AM, Reenan RA, Graveley BR. Identification of alternative splicing regulators by RNA interference in Drosophila. Proc Natl Acad Sci USA. 2004;101:15974–15979. doi: 10.1073/pnas.0407004101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rio DC. Accurate and efficient pre-mRNA splicing in Drosophila cell-free extracts. Proc Natl Acad Sci USA. 1988;85:2904–2908. doi: 10.1073/pnas.85.9.2904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Dignam JD, Lebovitz RM, Roeder RG. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 1983;11:1475–1489. doi: 10.1093/nar/11.5.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Raychaudhuri G, Haynes SR, Beyer AL. Heterogeneous nuclear ribonucleoprotein complexes and proteins in Drosophila melanogaster. Mol Cell Biol. 1992;12:847–855. doi: 10.1128/mcb.12.2.847. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.